Abstract
Transforming growth factor beta-induced (TGFBI/βIG-H3), also known as βig-H3, is a protein inducible by TGFβ1 and secreted by many cell types. It binds to collagen, forms part of the extracellular matrix and interacts with integrins on the cell surface. Recombinant TGFBI and transgenic TGFBI overexpression can promote both islet survival and function. In this study, we generated TGFBI KO mice and further assessed TGFBI function and signaling pathways in islets. Islets from KO mice were of normal size and quantity, and these animals were normoglycemic. However, KO islet survival and function was compromised in vitro. In vivo, KO donor islets became inferior to wild-type donor islets in achieving normoglycemia when transplanted into KO diabetic recipients. TGFBI KO mice were more prone to straptozotocin-induced diabetes than the wild-type counterpart. Phosphoprotein array analysis established that AKT1S1, a molecule linking the AKT and mTORC1 signaling pathways, was modulated by TGFBI in islets. Phosphorylation of four molecules in the AKT and mTORC1 signaling pathway, i.e. AKT, AKT1S1, RPS6 and EIF4EBP1, was upregulated in islets upon TGFBI stimulation. Suppression of AKT activity by a chemical inhibitor, or knockdown of AKT1S1, RPS6 and EIF4EBP1 expression by small interfering RNA, modulated islet survival, proving the relevance of these molecules in TGFBI-triggered signaling. Human genetic studies revealed that in the TGFBI gene and its vicinity, three single-nucleotide polymorphisms were significantly associated with type 1 diabetes risks, and one with type 2 diabetes risks. Our study suggests that TGFBI is a potential risk gene for human diabetes.
INTRODUCTION
Transforming growth factor beta-induced (TGFBI), also known as βig-H3, is a gene cloned from TGFβ-stimulated A549 lung adenocarcinoma cells (1). TGFBI protein has an N-terminal secretory signal (amino acids 1–23), four FAS1 homologous internal domains, and a cell attachment RGD domain at its C terminus (2). A secreted protein, TGFBI, is also found in the cell cytoplasm and nucleus (3). It is produced by several cell types, such as smooth muscle cells, keratinocytes, fibroblasts (3), proximal tubular epithelial epithelial cells (4), upon TGFβ or high-glucose stimulation (5). In addition, in situ hybridization shows that TGFBI is expressed in the heart, blood vessels, intestine, eyes and other tissues of mesenchymal origin (6), indicating that the gene plays a general role in various organs during ontogeny.
Although the biological significance of cytoplasmic and nuclear TGFBI has not yet been elucidated, we have a basic understanding of the function of secreted TGFBI. After secretion, TGFBI binds to the extracellular matrix (ECM) through interaction of the YH motif in its FAS1 domains with ECM collagens I, II, IV and VI (7,8). Its FAS1 domains can interact with α3β1, αVβ3 and αVβ5 integrins on the cell surface (4,9,10). Its RGD domain at the C-terminal binds to integrins, although its interacting integrins have not been pin-pointed (11,12). TGFBI is also known to bind α1β1, α6β4 and α7β1(13,14), but the domain(s) involved in such associations have not been identified. Thus, secreted TGFBI acts as a bi-functional molecule, enhancing ECM and cell interactions, a lack of which culminates in dysfunction of many cell types.
Indeed, when TGFBI is coated on solid phase, it induces adhesion, reduces proliferation and inhibits the differentiation of keratinocytes (10). Solid-phase TGFBI enhances osteoblast adhesion, suppresses their differentiation (9) and blocks corneal epithelial cell adhesion and spreading (15). These biological effects of TGFBI could be explained by triggering the cell surface integrin signaling pathway with solid-phase TGFBI. On the other hand, a soluble fragment containing the TGFBI C-terminal RGD domain induces Chinese hamster ovary cell apoptosis (2,11). In this condition, it is likely that the soluble fragment interferes with normal cell–cell interactions between integrins and fibronectin, as the RGD domain could bind to integrins and dampen their association with fibronectins. This suggests that TGFBI is not only necessary for ECM–cell interaction, but might also regulate cell–cell contact, both of which are essential for cellular function and survival. There is much controversy regarding the effect of TGFBI on tumorigenesis. Enhanced as well as reduced TGFBI expression has been found to be associated with increased malignancy in clinical settings and animal models (3,16). Opposite effects in integrin-triggering by the intact TGFBI molecule and integrin-blocking by its fragments might be one of the possible reasons for these seemingly controversial outcomes. Point mutations of the TGFBI gene in humans lead to several types of corneal dystrophy (3,17,18), although the underlying mechanisms are not understood.
Our earlier study demonstrated that recombinant TGFβi could preserve the integrity and enhance the function of cultured islets (19). Islets from TGFBI transgenic (Tg) mice with actin promoter-driven TGFBI overexpression showed better integrity and insulin release after in vitro culture. In vivo, β-cell proliferation was detectable in Tg but not wild-type (WT) pancreata. At the age above 12 months, Tg pancreata contained giant islets. Tg mice displayed better glucose tolerance than the controls. Tg islets were more potent in lowering blood glucose when transplanted into syngeneic mice with streptozotocin-induced diabetes, and these transplanted islets also underwent regeneration. Our results indicate that TGFβi is a vital trophic factor promoting islet survival, function and regeneration.
In the current study, we generated TGFBI gene knockout (KO) mice. In vitro, KO islet survival and function were compromised. In vivo, KO mice were more susceptible to low-dose streptozotocin (STZ)-induced diabetes. KO donor islets were inferior to WT donor islets in achieving normoglycemia when transplanted into KO diabetic recipients. At the molecular level, TGFBI activated the FAK/AKT/ATK1S1/RPS6/EIF4EBP1 signaling pathway in the islets. Genetic studies in humans indicate that TGFBI was a risk factor for both type 1 and type 2 diabetes (T1D and T2D).
RESULTS
Generation and general features of TGFBI KO mice
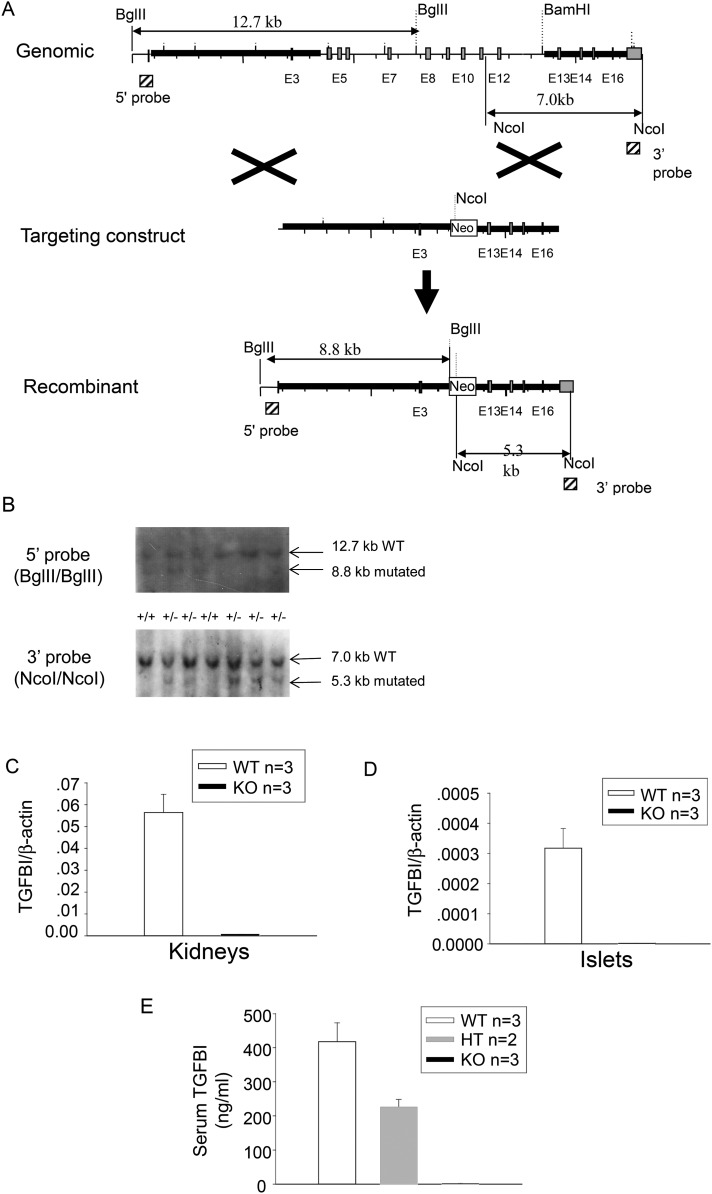
To definitively understand TGFBI's function in islet biology, we generated TGFBI KO mice. The targeting strategy is illustrated in Figure 1A. We deleted exon 4–11 of the TGFBI gene, hence the deletion of amino acid sequence from 101 to 559 of the 683 amino acid TGFBI protein. All the functional domains of the protein were eliminated. Germ-line transmission of the targeted Tgfbi allele was confirmed by Southern blotting of tail DNA (Fig. 1B). The targeted and WT alleles had an 8.8-kb and a 12.7-kb BglII/BglII band, respectively, using the 5′ probe; and a 7.0-kb and a 5.3-kb NcoI/NcoI band, using the 3′ probe.
Figure 1.
Generation of TGFBI KO mice. (A) Targeting strategy generating TGFBI KO mice. The light gray vertical bars are exons. The hatched squares on 5′ and 3′ sides of the mouse TGFBI WT genome represent the sequences serving as probes for genotyping by Southern blotting. The sizes of WT and mutated alleles cut by BglII and NcoI are indicated. (B) Genotyping of TGFBI mutant mice. Tail DNA was digested with BglII or NcoI, and analyzed by Southern blotting, with the 5′ and 3′ probes, respectively. The locations of the probes are indicated in (A). A 7.0-kb band representing the WT allele and a 5.3-kb band representing the recombinant allele are indicated by arrows. (C and D) Absence of mRNA expression in TGFBI KO tissues. mRNA levels from WT and KO kidneys (C) and islets (D) were analyzed by RT–qPCR for TGFBI mRNA expression. The results are expressed as ratios of TGFBI versus β-actin signals with means ± SD from three pairs of KO and WT mice. (E) Absence of TGFBI protein in sera of Tgfbi KO mice. Sera from WT, heterozygous and KO mice were assayed by TGFBI ELISA. Samples were assessed in duplicate by ELISA and mouse numbers were indicated. Means ± SD for each group are reported.
KO mice were backcrossed to the C57BL/6 background for two generations when islet transplantation experiments were carried out. For the rest of the experiments, KO and WT mice in the C57BL/6 background (backcrossed to C57BL/6 for 8–10 generations) were studied.
TGFBI deletion in KO mice at the mRNA level in the kidneys and islets was verified by reverse transcription–quantitative polymerase reaction (RT–qPCR), as illustrated in Figure 1C and D, respectively. Deletion of KO mice TGFBI protein was demonstrated by serum TGFBI levels (Fig. 1D). TGFBI concentration in the sera of heterozygous mice was ∼50% that of WT mice, while it was not detectable in KO mice. The TGFBI KO mice were fertile and had no visible anomalies. Although a previous report showed that TGFBI KO mice have reduced body size which is caused by smaller bones (20), we did not notice obvious differences in body sizes between our KO and WT mice.
TGFBI KO islet survival and function are compromised in vitro
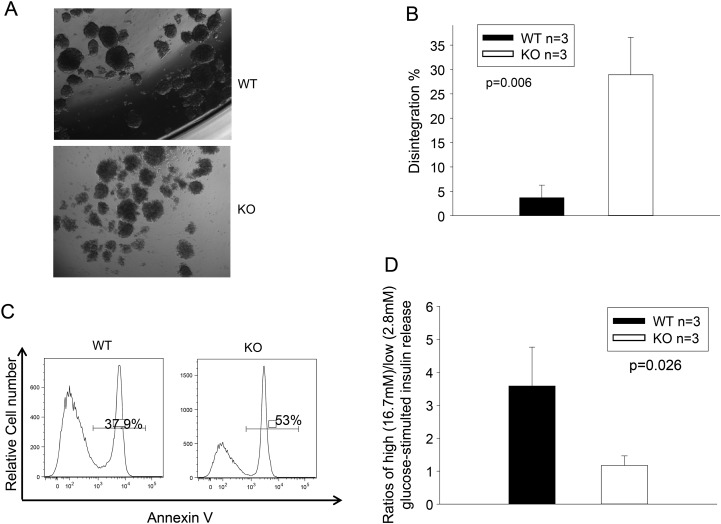
Our previous study showed that islets can produce TGFBI by themselves, and TGFBI can promote islet survival and function (13,14,19). We questioned whether, conversely, TGFBI deletion in islets was detrimental to their survival and function. WT and KO islets were cultured in serum-free medium for 24 h, and their morphology, apoptosis and insulin release were then assessed. As seen in Figure 2A, KO islets quickly lost their integrity after 24-h culture in serum-free medium, unlike WT islets. Quantitative assessment of islet integrity is illustrated in Figure 2B. Fewer than 5% of WT islets disintegrated after 24 h, compared with >25% of KO islets. At this time point, much higher percentages of KO islet cells than WT cells underwent apoptosis (53 versus 38%; Fig. 2C). KO islets presented a significantly lower ratio of high glucose- versus low glucose-stimulated insulin release than WT islets (Fig. 2D), corroborating the apoptosis results, and strongly suggesting that apoptotic islet cells were β-cells.
Figure 2.
TGFBI KO islet survival and function is compromised in vitro. (A and B) TGFBI KO islets quickly lose their integrity in culture. Freshly isolated WT and TGFBI KO islets were cultured in F-12K serum-free medium containing 2% BSA and 1X MEM non-essential amino acids for 24 h. Their morphology is illustrated in micrographs (A). About 150–200 islets from each mouse were assessed visually, and the percentages of those that lost integrity (defined as having lost 50% of capsule surrounding the islets) were registered. Means ± SD of the percentage of each group (based on three independent experiments) are illustrated (B), and statistical significance of the difference is indicated (P = 0.006; Student's t test). (C) TGFBI KO islets are prone to apoptosis in culture. WT and KO islets were cultured in serum-free medium for 24 h, as described above, dispersed and stained with annexin V, followed by flow cytometry analysis. Percentages of annexin V-positive cells are indicated. The experiment was performed 3 times, and histograms of a representative experiment are shown, in which the WT and KO islet cells showed 37.9% and 53% apoptosis, respectively. (D) TGFBI KO islet function is compromised after culture. WT and KO islets were cultured in serum-free medium for 24 h. Ten islets were picked and stimulated sequentially with low- and high-glucose concentrations (2.8 and 16.7 mm, respectively). Insulin released into medium after stimulation was assessed by ELISA in duplicate samples. Ratios of released insulin upon low- and high-glucose stimulation were calculated. Means ± SD of the ratios of each group (from three independent experiments) are reported, and statistically significant differences are indicated (P = 0.026; Student's t test).
TGFBI KO islets function is compromised in vivo
We conducted a series of in vivo experiments to assess KO islet function. Islets in KO pancreata were of similar size and number as those in WT pancreata according to histology (Supplementary Material, Fig. S1A) and islet count after isolation (Supplementary Material, Fig. S1B), blood glucose was normal in KO mice (Supplementary Material, Fig. S1C). Their glucose tolerance was similar to that of WT mice (Supplementary Material, Fig. S1D). These data indicate that a lack of TGFBI does not affect islet development and function under normal circumstances in vivo.
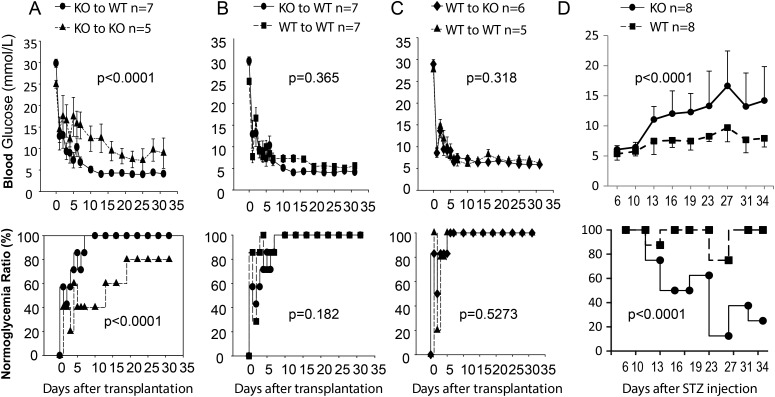
However, the necessity of TGFBI for islet function was revealed when islets were stressed. We transplanted KO islets into diabetic KO and WT recipients. Transplanted KO islets were significantly less potent in reversing the diabetic condition of KO recipients than that of WT recipients. The former group presented higher glucose levels (Fig. 3A, upper panel) and a lower percentage of recipients returning to normoglycemia (Fig. 3A, lower panel). Interestingly, such a detrimental effect due to a lack of TGFBI was obvious only when KO islets were transplanted to KO recipients. When KO islets were transplanted to diabetic WT recipients, they were no worse than WT islets in terms of lowering blood glucose (Fig. 3B, upper panel) and rendering the recipients euglycemic (Fig. 3B, lower panel). The lack of TGFBI alone in the recipients was not sufficient to reveal the effect of this protein on islet protection, because WT islets functioned well in diabetic KO recipients as they did in diabetic WT recipients, again in terms of lowering blood glucose (Fig. 3C, upper panel) and rendering the recipients euglycemic (Fig. 3C, lower panel).
Figure 3.
KO islet function is compromised after transplantation into KO diabetic recipients. Numbers of mice (n) per group are indicated. Blood glucose levels are shown in the upper row and % of mice with normoglycemia is shown at the bottom row. Data from different groups were assessed by ANOVA. For percentages of normoglycemic mice in different groups, ANOVA was performed after they were square-rooted, followed by arcsin transformation. P values are reported. (A) KO islets perform poorly after transplantation into KO recipients. A dose-limiting number of KO islets (350 islets/recipient) were transplanted under the renal capsules of syngeneic WT and KO recipients, which were rendered diabetic by prior STZ treatment. Blood glucose levels of the recipients were monitored for 31 days. Blood glucose levels of the different groups and the percentages of mice that reached normoglycemia (<12 mm) are reported. (B) KO and WT donor islets have comparable function in WT recipients. Islets from KO and WT donors were transplanted into WT diabetic recipients; their blood glucose levels and percentages of mice becoming normoglycemic were monitored for 31 days, as described in (A). (C) WT donor islets have comparable function in WT and KO recipients. Islets from WT donors were transplanted into diabetic WT and KO recipients; their blood glucose levels and percentages of mice becoming normoglycemic were monitored for 31 days, as described in A. (D) KO mice are prone to multiple low-dose STZ-induced diabetes. KO and WT male mice were treated i.p. with low dose STZ (40 mg/kg, q.d. for 5 days). Their blood glucose levels and percentages of mice with normoglycemic were monitored once every 3 days from Day 6 until Day 34.
Collectively, these data indicate that TGFBI in the WT milieu surrounding transplanted KO islets is sufficient for donor KO islets to function normally, and TGFBI produced by WT islets is also sufficient for them to function normally in the recipient KO milieu. The detrimental effect of TGFBI deletion is revealed only when islets are deprived of TGFBI produced both by themselves and the milieu around them under stress conditions, such as islet transplantation.
In an additional experiment, we assess the consequence of a lack of TGFBI in both the islets and their environment under a simulated T1D condition, i.e. multiple low-dose STZ-induced diabetes. Such manipulation causes an chronic inflammatory condition in the pancreata (10,21,22). The KO and WT mice were treated with suboptimal multiple low doses of STZ. This caused a slow increase of blood glucose over a period of 20–25 days in WT mice, although we titrated the STZ dose so that most of the WT mice did not reach the 12 mmol/l threshold of diabetes (Fig. 3D). However, there was significantly higher blood glucose levels and diabetes incidence in the KO mice. This suggests that individuals with TGFBI loss-of-function mutations might have increased T1D risks.
TGFβ1 stimulates TGFBI secretion by islets and by tissues surrounding transplanted islets
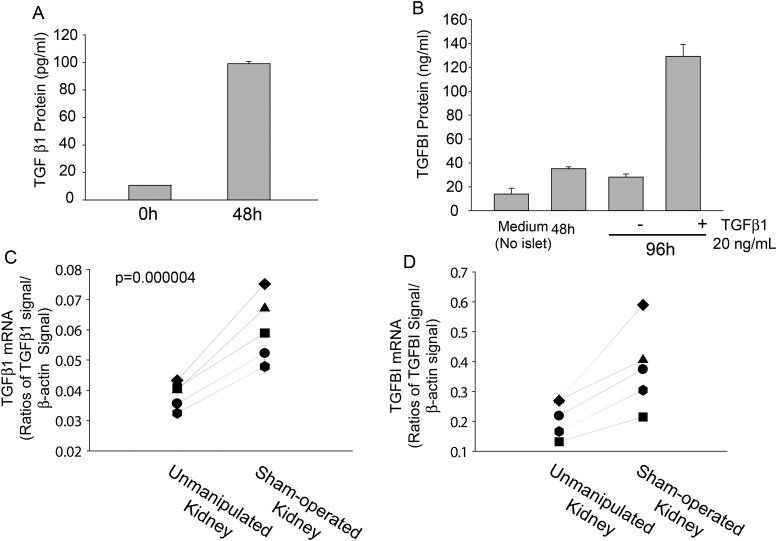
TGFBI was first identified as a molecule produced by TGFβ1-stimulated lung carcinoma cells (1,9). TGFβ1 production is upregulated at trauma sites as part of wound-healing processes (15,23). It is logical to expect that isolated islets, which have experienced mechanical and enzymatic insults, will upregulate TGFβ1 secretion. We have demonstrated that this was indeed the case: isolated WT islets produced significantly high TGFβ1 levels within 48 h of isolation in the serum-free culture condition (Fig. 4A). We further showed that under the serum-free condition, employed to exclude the possible influence of TGFβ1 from bovine serum, TGFBI secretion was upregulated by WT islets within 48 h after isolation, and such secretion could be drastically boosted by a near-physiological concentration of exogenous TGFβ1 (20 ng/nl) within another 48-h culture (Fig. 4B). Taken together, it proved that TGFβ1 could upregulate TGFBI secretion by islets and that upregulated TGFBI secretion shortly after islet isolation is, at least in part, caused by endogenously upregulated TGFβ1 stimulation.
Figure 4.
TGFβ1 and TGFBI production by islets and renal parenchyma. (A) TGFβ1 production by isolated islets. WT islets were cultured in F-12K serum-free medium (200 islets/0.2 ml/well in 48-well plates) for the durations indicated. TGFβ1 in supernatants was measured by ELISA of duplicate samples. Means ± SD of TGFβ1 levels are reported. (B) TGFBI production by WT islets upon exogenous TGFβ1 stimulation. WT islets were cultured as described in (A), and TGFBI in supernatants was measured at different time points by ELISA of duplicate samples. In some culture, recombinant mouse TGFβ1 (20 ng/ml) was added at 48 h after the initiation of culture. Means ± SD of TGFBI levels are reported. In (A and B), experiments were performed more than twice, and data from representative experiments are reported. (C and D) TGFβ1 and TGFBI production by renal parenchyma after trauma. The left kidneys of WT mice were sham-operated to mimic islet transplantation procedures, After 48 h, the renal parenchyma near the wound was pared, and its RNA extracted to measure TGFβ 1 (C) and TGFBI (D) mRNA levels by RT–qPCR in duplicate. The results are expressed as ratios of TGFβ1 or TGFBI versus β-actin signals. Unmanipulated right kidneys served as controls, and lines link the ratios of two kidneys (operated versus unmanipulated) from the same mouse. P values (paired Student's t test) are reported.
We wondered whether WT tissues surrounding transplanted islets, in this case the renal parenchyma, could also produce TGFBI. Due to technical difficulties, mainly the interference of high serum TGFBI levels, it is not feasible to accurately measure the TGFβ1 and TGFBI proteins in tissue fluid exodus under the renal capsule where the islets were transplanted. We quantified the mRNA levels of these two molecules in tissues (i.e. renal parenchyma) adjacent to where the islets were implanted. As shown in Figure 4C, renal parenchyma near the capsule of sham-operated kidneys had significantly upregulated TGFβ1 mRNA levels 24 h after the operation, compared with unmanipulated kidneys. They were accompanied by upregulated TGFBI mRNA levels in this tissue (Fig. 4D).
Phospho-protein array analysis of TGFBI-triggered signaling pathways
TGFBI is known to bind integrins. In our previous study, we demonstrated that FAK, which is positioned upstream in the integrin signaling pathway, is activated in islets upon TGFBI stimulation (19). We interrogated the Full Moon BioSystem phospho-protein array, which contains Abs against 402 phosphorylated kinases, adaptor proteins, and transcription factors as well as Abs against the total proteins of these molecules, looking for additional molecules involved in the TGFBI signaling pathway. Islets from transgenic mice with actin-promoter-driven TGFBI overexpression were used as starting material as they receive excessive TGFBI stimulation; islets from WT mice were used as controls. Ratios of phospho-protein signals versus total protein signals were first determined, and fold changes of Tg versus WT islet ratios were calculated. Molecules with fold changes >1.5 or <0.67 are listed in Table 1. Considering the relevance of these molecules to islet function based on the existing literature, we selected six molecules, i.e. DAXX, p27Kip1, AKT1S1, MAPKAPK2, SMAD1 and calmodulin, for validation by western blotting. Only AKT1S1 and p27Kip1 phosphorylation showed discernible and reproducible differences between Tg and WT islets (Fig. 5 and data not shown). The former was further investigated in the following section.
Table 1.
Phospho-protein array analysis of TGFBI Tg versus WT islets
| Protein (Phosphorylation sites) | Phospho-protein versus total protein ratio (P/T ratio) |
P/T ratio changes | |
|---|---|---|---|
| TGFB1 Tg | WT | Tg/WT | |
| BRCA1 (Phospho-Ser1457) | 0.97 | 0.54 | 1.80 |
| DAXX (Phospho-Ser668) | 0.08 | 0.05 | 1.76 |
| VASP (Phospho-Ser157) | 0.47 | 0.27 | 1.75 |
| p27Kip1 (Phospho-Ser10) | 1.66 | 1.02 | 1.62 |
| Histone H3.1 (Phospho-Ser10) | 1.03 | 0.65 | 1.58 |
| MEF2A (Phospho-Ser408) | 2.27 | 1.48 | 1.54 |
| AKT1S1 (Phospho-Thr246) | 0.26 | 0.18 | 1.49 |
| Paxillin (Phospho-Tyr31) | 1.41 | 0.95 | 1.49 |
| BLNK (Phospho-Tyr96) | 0.22 | 0.35 | 0.63 |
| Cyclin B1 (Phospho-Ser147) | 0.49 | 0.85 | 0.58 |
| MAPKAPK2 (Phospho-Thr334) | 0.05 | 0.11 | 0.41 |
| Smad1 (Phospho-Ser465) | 0.31 | 1.08 | 0.28 |
| Calmodulin (Phospho-Thr79/Ser81) | 0.14 | 0.74 | 0.18 |
Islets from Tg and WT mice were isolated, and cultured for 4 h in F-12K serum-free medium. Islet lysates were analyzed by Full Moon BioSystem (Sunnyvale, CA, USA) phospho-protein array. Samples were run in triplicate. The names of proteins and their phosphorylation sites assessed are indicated in column 1. The ratios of mean signals of phospho-protein versus total protein (P/T ratio) are listed in columns 2 (Tg islets) and 3 (WT islets). Fold differences between the Tg ratio over the WT ratio of each protein (P/T ratio changes) were calculated and listed in the last column. Only proteins with fold differences between Tg and WT islets above 1.5 or less than 0.67 are included in the table.
Figure 5.
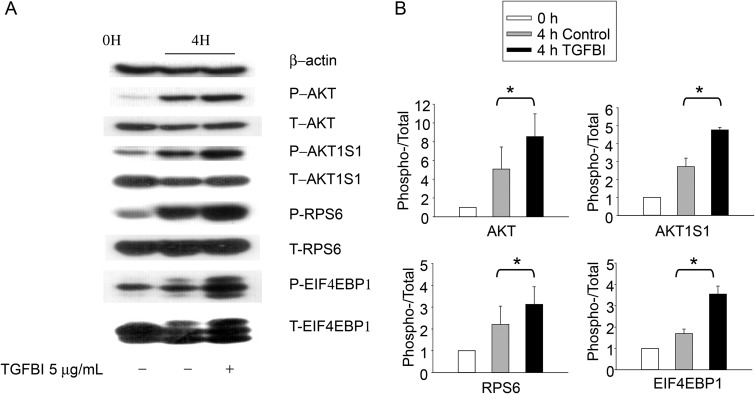
Phosphorylation of molecules putatively involved in TGFBI signaling according to immunoblotting. WT islets were either lysed directly or cultured for 4 h in presence of TGFBI (5 µg/ml) in F-12K serum-free medium and then lysed, as indicated. Lysates were analyzed for phospho-AKT (P-AKT; Ser473), total AKT (T-AKT), phospho-AKT1S1 (P-AKT1S1; Thr246), total AKT1S1 (T-AKT1S1), phospho-RPS6 (P-RPS6; Ser235/236), total RPS6 (T-S6), phospho-EIF4EBP1(P-EIF4EBP1; Thr37/46), and total EIF4EBP1 (T-EIF4EBP1). Beta-actin was included as an additional loading control. The experiments were conducted three times in total, and data from a representative experiment are reported. All blotting in (A) were derived from 1 SDS–PAGE run and one membrane. The data from all three experiments were analyzed by densitometry, and the ratios of signal strength of phosphoproteins versus that of total proteins were illustrated in bar graphs in (B). The differences between TGFBI-treated verses the controls are significant (*P < 0.05, paired Student's t test).
TGFBI activates the FAK/AKT/AKT1S1/mTOR pathway
We reported previously that TGFBI retains higher FAK activity in islets after their isolation, and FAK knockdown compromises islet function (19), establishing an upstream event in the TGFBI signaling pathway. We elected to further study the role of AKT1S1 in TGFBI signaling, because it is a substrate of AKT which is downstream of FAK, and it is also part of the mTOR signaling pathway which is critical in cell survival and proliferation (24). As seen in Figure 5, we confirmed that AKT1S1 phosphorylation in islets was increased within 4 h of TGFBI stimulation. Such upregulation is accompanied by higher AKT phosphorylation, reflecting enhanced AKT activity which is responsible for AKT1S1 phosphorylation. AKT1S1 phosphorylation allows it to dissociate from mTOR complex 1 (mTORC1) and renders mTOR active (24). Indeed, we demonstrated that two downstream molecules of mTOR, i.e. RPS6 and EIF4EBP1, underwent increased phosphorylation in TGFBI-treated islets, indicating mTOR signaling pathway activation. Taken together, these data suggest that TGFBI could trigger signaling along the FAK/AKT/AKT1S1/mTOR pathway.
It needs to be mentioned that within 4 h of islet isolation, AKT1S1 and RPS6 phosphorylation was increased, as was, to a small extent, AKT phosphorylation, even in the absence of exogenous TGFBI (Fig. 5, second column). It is likely that the islet isolation process and/or the new culture environment were capable of activating these molecules through so-far unknown mechanisms. Also, TGFBI secreted by islets themselves during this period could act as paracrine to stimulate the islets. Nevertheless, the exogenous TGFBI could further enhance the augmented phosphorylation of these molecules (Fig. 5, third column).
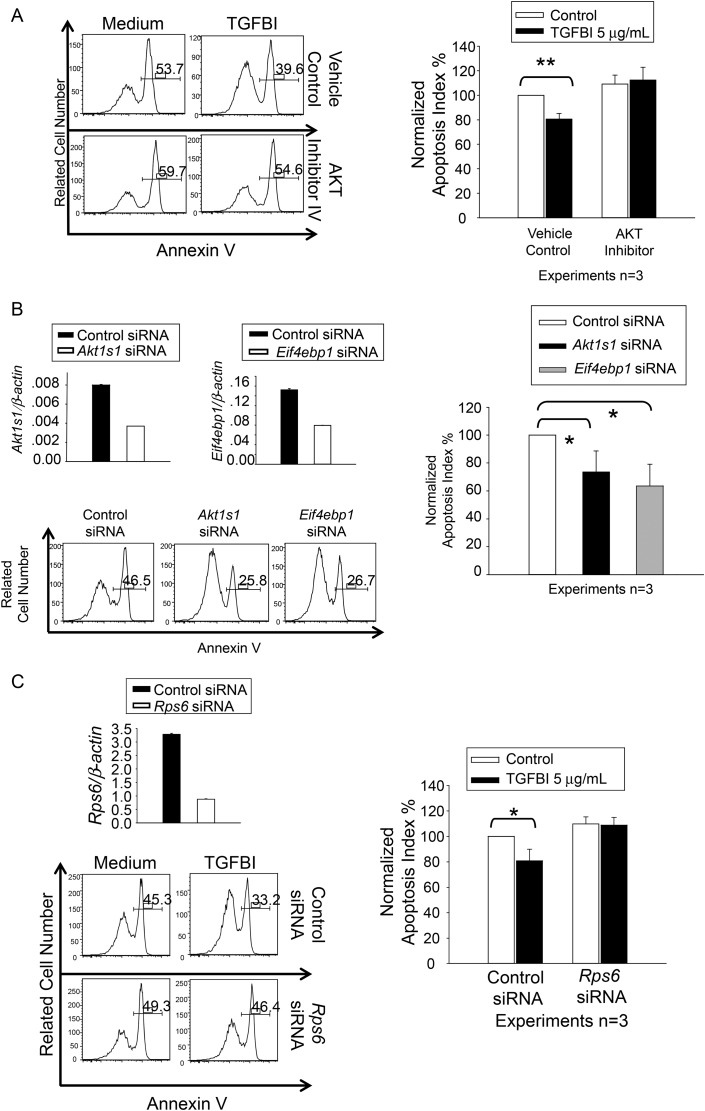
To prove that FAK/AKT/AKT1S1/mTOR pathway activation was relevant to the observed role of TGFBI in islet survival, we used kinase inhibitors and small interfering RNA (siRNA) knockdown in islets to assess whether these molecules could blunt TGFBI's beneficial effect on islets. We have already demonstrated that FAK knockdown by siRNA compromises islet function (19), proving its importance in islet function. The next molecule in the pathway, AKT, underwent augmented phosphorylation in islets upon TGFBI stimulation. The relevance of AKT function to islet survival was ascertained with an inhibitor (AKT inhibitor IV). Islets were cultured in serum-free medium to avoid possible influence of exogenous TGFBI from bovine serum. In the absence of TGFBI, AKT inhibitor IV at 1 µM had no significant effect on islet cell survival (Fig. 6A: left column of histograms and empty bars of right panel), indicating that the inhibitor, by itself at this concentration, did not constitutively impact islet survival. TGFBI significantly improved islet survival (Fig. 6A: top row of histograms and left group in bar graph on the right), but the AKT inhibitor revoked its beneficial effect, and islet cell apoptosis returned to a level similar to that without TGFBI treatment (Fig. 6A: lower row of histograms and left group in bars of the bar graph), suggesting that AKT is in the TGFBI signaling pathway with regard to TFGBI's function in islet survival.
Figure 6.
Islet survival upon modulation of AKT, AKT1S1, RPS6 and EIF4EBP1 activity and expression. Three independent experiments were conducted in (A), (B) and (C), and histograms of a representative experiment are presented, with percentages of annexin V-positive cells. (A) AKT inhibitor IV revokes the beneficial effect of TGFBI on islet survival. WT islets were cultured in F-12K serum-free medium for 72 h in presence and absence of human recombinant TGFBI (5 µg/ml) and AKT inhibitor IV (1 µM), as indicated. Islet cell apoptosis was measured according to annexin V staining, followed by flow cytometry. (B) AKT1S1 and EIF4EBP1 siRNA transfection reduces islet apoptosis. WT islets were transfected with siRNAs targeting AKT1S1 or EIF4EBP1, or with control siRNA. They were cultured in F-12K serum-free medium for 72 h. AKT1S1 and EIF4EBP1 mRNA knockdown was confirmed by RT–qPCR (upper row), with expression as ratios (means ± SD) of targeting gene signals versus β-actin signals. Islet cell apoptosis was measured by annexin V staining, followed by flow cytometry. (C) RPS6 siRNA transfection revokes the beneficial effect of TGFBI on islet survival. WT islets were transfected with siRNAs targeting RPS6, or with control siRNA. They were cultured in F-12K serum-free medium for 72 h in absence or presence of TGFBI (5 µg/ml), as indicated. Rps6 mRNA knockdown was confirmed by RT–qPCR (upper row), with expression as ratios (means ± SD) of targeting gene signals versus β-actin signals. Islet cell apoptosis was measured by annexin V staining, followed by flow cytometry. The experiments in (A), (B), and (C) were conducted a total of three times, and the data are summarized in bar graphs at the right of each panel. Differences with statistical significance (paired Student's t tests) are indicated with asterisks (*P < 0.05; **P < 0.01).
AKT1S1 (PRAS40) is a known substrate of AKT. It likely connects the AKT signaling pathway to the mTOR pathway. AKT1S1 binds mTORC1 via Raptor and can inhibit mTORC1 autophosphorylation as well as its kinase activity (25). AKT1S1 phosphorylation at Thr246 by AKT leads to its dissociation from mTORC1 (26), and releases its inhibitory effect on mTORC1 (27). We demonstrated that TGFBI-induced AKT1S1 phosphorylation at Thr246 (Fig. 5). Reduced unphosphorylated AKT1S1 levels in cells, in theory, would equate with AKT1S1 dephosphorylation, and release the mTORC1 inhibition. Would such a decrease of unphosphorylated AKT1S1 be beneficial to islets? We conducted siRNA knockdown of AKT1S1 to answer this question. AKT1S1 knockdown at the mRNA level was ascertained by RT–qPCR (Fig. 6B: upper left panel). WT islets cultured in serum-free medium for 48 h resulted in a high rate of apoptosis, but AKT1S1 knockdown significantly reduced such apoptosis, suggesting that the TGFBI-induced decline of unphosphorylated AKT1S1 level would exert a similar protective influence on islet apoptosis.
mTORC1 has two kinase substrates: EIF4EBP1 and p70S6K, whose phosphorylation and activation will lead to RPS6 phosphorylation. EIF4EBP1 is a repressor of protein translation (28). It has seven identified phosphorylation sites, i.e. Thr37, Thr46, Ser65, Thr70, Ser83, Ser101 and Ser112, phosphorylation of the first two being a priming event (29,30). Hypophosphorylated EIF4EBP1 interacts with EIF4E and prevents recruitment of the translation machinery to mRNA, inhibiting translation. EIFE4BP1 phosphorylation results in the disassociation of this protein with EIF4E, releasing its translation inhibitory effect, which has a broad, positive influence on cell biology, such as proliferation, survival and function. While TGFBI KO islets were prone to apoptosis (Fig. 2), WT islets upon TGFBI stimulation presented improved survival (19), accompanied by upregulated EIF4EBP1 phosphorylation (Fig. 6). Such phosphorylation should reduce the amount of hypophosphorylated EIF4EBP1 in a given cell and result in release of the inhibitory effect of EIFEBP1 on translation. Is this a relevant event for better islet survival? We undertook siRNA knockdown of EIF4EBP1 in islets. mRNA knockdown was confirmed by RT–qPCR (Fig. 6B: left panel of first row). Islets transfected with Eif4ebp1 siRNA showed significantly reduced apoptosis (Fig. 6B: histograms in second row and bar graph on the right), indicating a beneficial effect of reduced total EIF4EBP1 levels including unphosphorylated one, similar to that caused by EIF4EBP1 phosphorylation.
The other mTORC1 downstream signaling molecule RPS6 is a component of the ribosome complex machinery for protein synthesis. It has been proposed that ribosomes with the highest proportion of phosphorylated RPS6 become active to serve as part of polysomes (31). RPS6 phosphorylation sites have been mapped to Ser235, Ser236, Ser240, Ser244 and Ser247. Knock-in mice with these residues mutated to alanines suffer from small pancreatic β-cells, diminished levels of pancreatic insulin, hypoinsulinemia and impaired glucose tolerance (32), revealing essential roles of RPS6 phosphorylation in islet biology. We demonstrated that TGFBI stimulation upregulated RPS6 phosphorylation (Fig. 5). To answer the question whether such upregulation is relevant to the observed beneficial effect of TGFBI on islet survival, we transfected WT islets with RPS6 siRNA. RPS6 mRNA knockdown was confirmed by RT–qPCR (Fig. 6C: upper left panel). TGFBI increased islet survival (Fig. 6C: upper right panel of histograms and bar graph on the left). WT islets with RPS6 knockdown by siRNA manifested no significant difference in apoptosis from control siRNA-transfected islets (Fig. 6C: left column of histograms and empty bars of the bar graph on the right), indicating that at this level of knockdown, the survival of islets was not directly affected. However, such RPS6 knockdown revoked the beneficial effect of TGFBI on islet survival (Fig. 6C: right column of histograms and solid bars in the bar graph on the left), indicating that RPS6 protein levels and, consequently, phosphorylated RPS6 protein levels are critical for TGFBI's role in islet survival.
TGFBI as a diabetes-risk gene in T1D and T2D patients
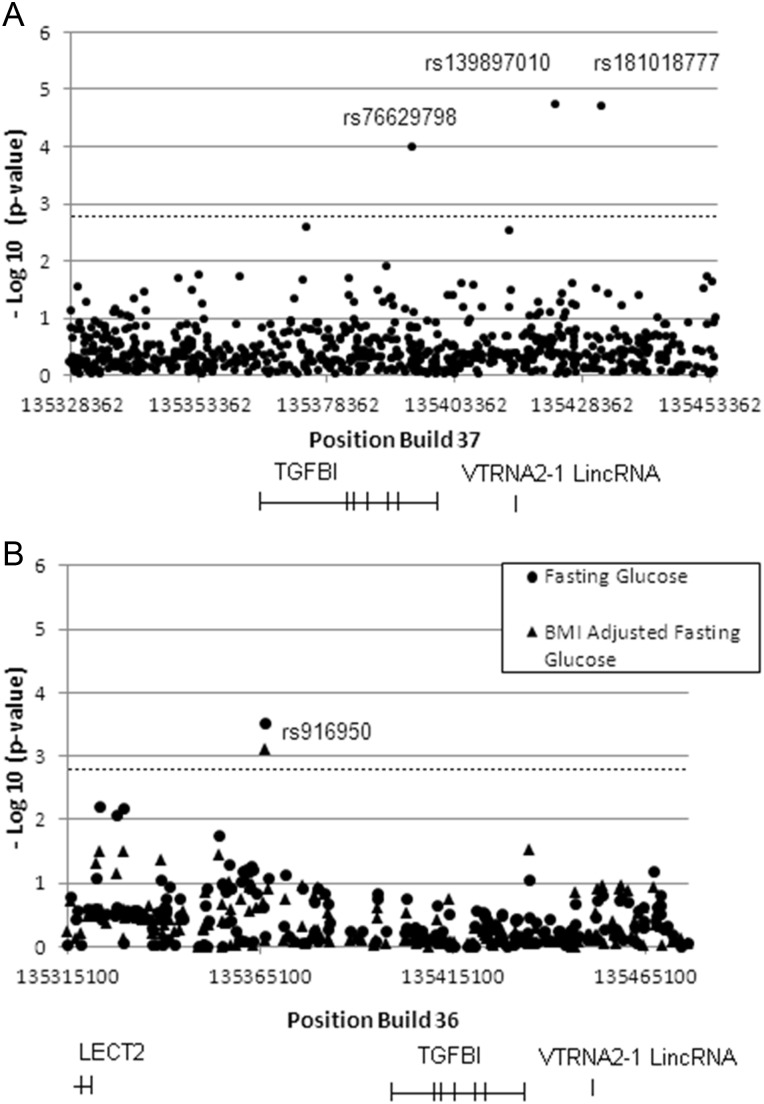
Single-nucleotide polymorphisms (SNPs) that were observed to be significantly associated with diabetes phenotypes are presented in Table 2. The locations of the observed significant SNPs within the queried region are shown in Figure 7A and B.
Table 2.
TGFBI SNPs significantly associated with T1D and T2D phenotypes
| rs Number | Phenotype/study | Position (Build 36) | Position (Build 37) | P-value | MAF | Description |
|---|---|---|---|---|---|---|
| rs76629798 | T1D/WTCCC | 135 423 309 | 135 395 410 | 0.00010619 | 0.0666 | Intronic |
| rs139897010 | T1D/WTCCC | 135 451 470 | 135 423 571 | 0.0000195 | 0.0014 | 24.064 kb 3′ of TGFBI |
| rs181018777 | T1D/WTCCC | 135 460 208 | 135 432 309 | 0.0000209 | 0.0014 | 32.802 kb 3′ of TGFBI in very high LD with rs139897010 |
| rs916950 | Fasting Glucose/MAGIC | 135 366 498 | 135 338 599 | 0.0003104 | 0.2621 | 25.985 kb 5′ of TGFBI |
| rs916950 | Fasting Glucose Adjusted by BMI/MAGIC | 135 366 498 | 135 338 599 | 0.0007926 | 0.2621 | 25.985 kb 5′ of TGFBI |
Five SNPs located within a region from 50 kb upstream to 50 kb downstream of the TGFBI gene showed significant association with T1D or fasting blood glucose (a T2D-related phenotype). There are 42 independent LD blocks within this region, and the calculated Bonferroni-corrected P-value is 0.00119 (0.05/42 = 0.00119). MAF of these SNPs is indicated.
Figure 7.
Association of SNPs in the TGFBI gene and its vicinity with T1D and T2D-related phenotype in WTCCC and MAGIC GWAS studies. (A) Significant association of 3 SNPs in the TGFBI gene and its 3′ untranslated region with T1D. Positions and –log (P-values) of SNPs from 50 kb upstream to 50 kb downstream of the TGFBI gene are indicated. The dotted line represents the Bonferroni-corrected P-value at 1.19 × 10−3. The names of 3 SNPs with P-values below the corrected P-value are indicated. (B) Significant association of a SNP in the 5′ untranslated region of the TGFBI gene with fasting glucose levels. Positions and –log (P-values) of SNPs from 50 kb upstream to 50 kb downstream of the TGFBI gene are indicated. The dotted line represents the Bonferroni-corrected P-value at 1.19 × 10−3 for one phenotype test. The name of SNP with P-value below the corrected P-value without (diamond) and with BMI-adjustment (dot) is shown.
Three SNPs, rs139897010, rs181018777 and rs76629798, were found to be associated with T1D in the Wellcome Trust Case Control Consortium (WTCCC) genome-wide association study (GWAS) dataset (33) at P-values of 0.0000193, 0.0000209 and 0.00010619, respectively, well below the Bonferroni-corrected P-value at 0.00119 (dotted line in Fig. 7A). The odds ratios for these three SNPs were 2.45, 2.44 and 1.98, respectively. rs139897010 is located 24.064 kb 3′ of TGFBI; rs181018777 is located 32.802 kb 3′ of TGFBI and appears to be in high linkage disequilibrium (LD) with rs139897010; rs76629798 is intronic within TGFBI. SNPs rs139897010 and rs181018777 are rare variants recently discovered in the 1000 genomes sequencing project (http://www.1000genomes.org) (34).
Fasting glucose levels in nondiabetic adults can reveal pre-T2D conditions and impaired glucose tolerance. GWAS have been conducted for the association of SNPs with fasting glucose levels with a view to identify T2D-risk genes, and indeed some of these are identical to known T2D-risk genes (35,36). One SNP, rs916950, was associated with both fasting glucose and body mass index (BMI)-adjusted fasting glucose among nondiabetics in the Meta-Analysis of Glucose- and Insulin-related traits Consortium (MAGIC) meta-analysis data (22) at P-values 3.104 × 10−4 and 7.926 × 10−4, respectively. If we correct for one phenotype test, this SNP is significant (dotted line in Fig. 7B represents the corrected P-value using one phenotype test) for both fasting glucose and BMI-adjusted fasting glucose. If we correct for two phenotype tests which is highly conservative for these correlated phenotypes, rs916950 is significant only for fasting glucose. rs916950 is located 25.985 kb 5′ of TGFBI gene.
The locations of the SNPs in relation to the known H3K4Me1 marks (mono-methylation of lysine 4 of the H3 histone protein) H3K4Me2 marks (tri-methylation of lysine 4 of the H3 histone protein), DNase I hypersensitive clusters and transcription factor-binding sites in the TGFBI gene and its 5′ and 3′ adjacent regions are shown in Supplementary Material, Figure S2. rs76629798, rs139897010, and to a lesser extent, rs916950, are in the H3K4Me1 mark-rich, DNase I-sensitive clusters and transcription factor-binding regions, according to the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/) (37).
DISCUSSION
In this study, we employed genetically manipulated mice and analyzed human GWAS datasets to determine the role of TGFBI in diabetes pathogenesis.
We compared islet integrity, survival and function of KO and WT islets in vitro. All these parameters were compromised in KO islets, but such compromise was evident only when they were cultured in serum-free medium. A very likely reason is that fetal calf serum (FCS) contains bovine TGFBI, which might be cross-reactive to murine TGFBI. It is conceivable that exogenous bovine TGFBI in serum could minimize the difference between KO and WT islets with regard to TGFBI availability, and make KO islets as healthy and functional as WT islets. KO islets became inferior to WT islets only in serum-free medium, because no exogenous or endogenous TGFBI was available. Of course, islets cultured in serum-free medium were not as healthy as those in medium with 10% FCS, and up to 40–50% of the islets became apoptotic in 2 days (Figs 2 and 7), compared with ∼10% of those cultured in medium with 10% FCS (19). On the other hand, such a high rate of apoptosis was a suitable condition to test the survival-promoting effect of TGFBI.
TGFBI generated by isolated islets seems to be part of a self-protecting mechanism for them to cope with the damage they experience. TGFBI production is known to be stimulated by TGFβ1 (1). We demonstrated that islets which experienced trauma during isolation significantly upregulated their TGFβ1 production (∼10-fold) within 48 h (Fig. 5A). Subsequent TGFBI production by isolated islets in vitro could be a consequence of the initial TGFβ1 upregulation, although we cannot exclude the possibility that there are TGFβ1-independent TGFBI upregulation mechanisms in islets. Previously, we reported that islets after isolation presented 4–5-fold upregulation of TGFBI mRNA (19) within 16 h. Despite the increase of TGFβ1 and TGFBI mRNA upregulation, we observed only a moderate increment of TGFBI protein levels after 48-h culture. Possible explanations of such discrepancies are: (i) in our current experiments, we employed serum-free medium that was not optimal for cellular function, including TGFBI protein synthesis, unlike our previous experiments on medium with 10% FCS; (ii) TGFβ1 secretion was too diluted by the culture medium (only about 100 pg/ml) to drastically stimulate islet TGFBI production. When 20 ng/ml TGFβ1, which was still within the range of physiological and pathophysiological concentrations (10–20 ng/ml, as seen in normal and diabetic patients (38), was added to culture, a significant, additional 4-fold increase of TGFBI secretion was achieved within another 48 h (Fig. 4B). In islet transplantation, it is conceivable that the local TGFβ1 produced by islets and accumulated in their vicinity could well reach physiological and pathophysiological concentrations.
TGFBI generated by the islets alone was not sufficient to confer apparent in vivo benefit, as KO and WT islets transplanted into WT recipients showed no significant difference in terms of lowering blood glucose (Fig. 3C). KO donor islets became only inferior when the recipients were also KO (Fig. 3A). This suggests that TGFBI at transplantation sites in WT recipients is sufficient to support KO donor islet function. One source of TGFBI was serum, which contained ∼400 ng/ml TGFBI in WT mice (Fig. 1E). Another source of TGFBI in vivo might be the tissues surrounding transplanted islets in the islet transplantation setting. In our animal model, we transplanted islets under the renal capsule. As with islets, trauma during islet transplantation at transplantation sites could trigger local TGFβ1 release which, in turn, could stimulate TGFBI secretion by islets or tissues surrounding the islets. Such local TGFBI, plus that from serum in WT mice, confers beneficial and protective effects on transplanted WT and KO islets. This explains why the detrimental effect of missing TGFBI on KO islets was only revealed in KO and not in WT recipients (Fig. 3), because, in the former, no TGFBI was available from the milieu surrounding transplanted islets. In a clinical situation, islets were transplanted into the liver through the portal vein. In this case, trauma to the liver tissue is limited, and, consequently, there would not be trauma-induced TGFβ1 secreted by the liver parenchyma surrounding transplanted islets. However, recipient patients all have underlying diabetes, and diabetics tend to manifest elevated serum TGFβ1 levels, especially if renal damage is involved (39,40). The increased TGFβ1 level in such patients might trigger TGFBI secretion by various tissues, including those surrounding transplanted islets and, paradoxically, provide some protection to islets.
AKT and mTORC1 are both critical kinases, to each of which multiple upstream signaling events converge, and of which many downstream cellular events, such as cell proliferation, differentiation, survival and function, are affected (41). AKT1S1, a substrate of AKT, could be considered as a link between the two signaling pathways involving AKT and mTORC1, respectively, as its phosphorylation by AKT at Thr246 makes it dissociate from mTORC1 and release its inhibition of mTORC1 activity. We identified several signaling molecules in the TGFBI signaling pathway and showed that TGFBI-triggered signals starting from FAK were propagated to AKT, and were then linked to the mTORC1 pathway, in which RPS6 and EIF4BP1 were phosphorylated. Our finding that the beneficial effect of TGFBI on islet survival is due in part to its signaling through the mTORC1 pathway is compatible with a previous report that upregulation of mTORC1 activity improves β-cell function and increases islet mass (42). We are aware that signaling pathways are highly connected and interactive, with each molecule in the pathway networking with multiple substrates/associating proteins. For example, AKT1S1 knockdown in HeLa cells could protect them from apoptosis, similarly to what we see in islets, but such protection is independent of its inhibitory effect on mTORC1 (25), suggesting that AKT1S1 interacts with other signaling molecules in addition to mTORC1. Therefore, we cannot rule out the possibility that AKT1S1 might affect events in addition to the mTORC1 pathway, and such interaction confers beneficial effects to islets upon TGFBI stimulation.
Furthermore, the consequence of activation of a given molecule depends on cell type, differentiation status and other signals that cells receive. In myocytes, AKT1S1 knockdown results in proliferation blockage at the G1 phase accompanied by cell size increment (43). Since β-cells in islets are not actively proliferating cells, we did not see any changes in cell division and size after AKT1S1 knockdown (data not shown). We previously demonstrated that rapamycin, an inhibitor of the mTORC1, reduces inflammatory cytokine-triggered apoptosis of insulinoma NIT-1 cells (44). On the contrary, we showed in this study that AKT1S1 knockdown and, consequently, mTORC1 pathway activation benefited islet survival; others also reported that mTORC1 pathway activation by transgenic overexpression of Rheb increases islet function and mass (42). Such discrepant results of mTORC1 signaling pathway activation could be due to: (i) different cell types used (insulinoma cells versus islet cells); (ii) different stimulation events from the milieu (inflammatory cytokines in (44) versus serum-free medium in this study), (iii) different points of intervention [AKT1S1, RPS6 and EIF4EBP1 knockdown, and AKT inhibition in this study; mTORC1 inhibition in (44); and more upstream Rheb activation in (42)].
In T1D, the death of islet β cells is the major cause of the disease. In T2D, insulin resistant is the initial mechanism of pathogenesis, but at the later stage, islets also undergo apoptosis. We demonstrated in vitro and in vivo in animal models that a lack of TGFBI compromised islet survival. This prompted us to assess whether TGFBI mutation is a risk factor for both T1D and T2D in humans.
We queried the WTCCC dataset and found that three SNPs (rs76629798, rs139897010 and rs181018777) in the TGFBI gene and its vicinity were found to be highly significantly associated with T1D risks. rs139897010 and rs181018777 are rare variants with minor allele frequency (MAF) of 0.0014, meaning that only 0.14% of the tested population carry this mutation. T1D is a polygenic disease. It is expected that many such rare polymorphisms might contribute to diabetes risk for different small subpopulations of patients.
rs76629798 is intronic, and rs139897010 is in the 3′ untranslated region ∼25 kb downstream of the TGFBI gene. Both these SNPs are located in sites rich in H2K4Me1 marks, DNase I hypersensitive clusters and transcription factor-binding regions, which are features indicative of regulatory elements (enhancer/promoter) for gene expression (45–47).
rs76629798 is intronic, and rs139897010 is in the 3′ untranslated region ∼25 kb downstream of the TGFBI gene. Both these SNPs are located in sites rich in H2K4Me1 marks, DNase I hypersensitive clusters and transcription factor-binding regions, which are features indicative of regulatory elements (enhancer/promoter) for gene expression (45–47).
In the MAGIC study, we found that rs916950, located ∼25 kb upstream of the TGFBI gene, is significantly associated with fasting glucose levels, which is a parameter with predictive values for T2D. The SNP has MAF of 0.26, indicating a sizable subset (26%) of the study population has this polymorphism. This SNP is again located in a site rich in H3K4Me1, DNase I hypersensitive clusters and transcription factor-binding regions. Therefore, the strategic locations of these SNPs with regard to TGFBI expression regulation raises the possibility that such mutations could reduce TGFBI expression levels in a sub-group of the studied populations and contribute to pathogenesis of T2D.
In summary, using transgenic TGFBI overexpressing mouse model in our previous study (19), and a series experiments using TGFBI gene KO mouse model in this study, we demonstrated that TGFBI produced by islets and their surrounding milieu favored islet survival and function. Further investigation, based on protein array findings, showed the involvement of AKT and mTORC1 pathways in TGFBI signaling in islets. Genetics studies in two human cohorts revealed that four SNPs in the TGFBI gene and its vicinity were significantly associated with T1D and T2D risks. These multiple indirect evidences from animal and human studies suggest that TGFBI is likely a diabetes-risk gene. Functional studies of the SNPs with regard to TGFBI expression regulation and T1D and T2D incidences and severity will be required to fully establish TGFBI as a bona fide diabetes risk gene.
MATERIALS AND METHODS
Generation of TGFBI KO mice
A PCR fragment amplified from the TGFBI (NM_009369.4) cDNA sequence was used as probe to isolate genomic BAC DNA clone 5N13 from the 129/sv mouse BAC genomic library RPCI-22. The targeting vector was constructed by recombination and routine cloning methods with a 21-kb TGFBI (NM_009369.4) genomic fragment from clone 5N13 as starting material. A 9.67-kb (6 bp before exon 4 to the BamHI site in intron 12) genomic fragment containing exons 4–11 was replaced by a 1.1-kb Neo cassette from pMC1Neo-Poly A, as illustrated in Figure 1A. The final targeting fragment was excised from its cloning vector backbone by NotI digestion and electroporated into R1 ES cells for G418 selection (48). The targeted ES cell clones were injected into C57BL/6 blastocysts. Chimeric male mice were mated with C57BL/6 females to establish mutated TGFBI (NM_009369.4) allele germline transmission.
As illustrated in Figure 1A, Southern blotting with probes corresponding to the 5′ and 3′ sequence (hatched rectangles) outside the targeting region were used to screen for gene-targeted ES cells and, eventually, confirmed gene deletion in mouse tail DNA. The targeted and WT alleles had an 8.8- and a 12.7-kb BglII/BglII band, respectively, using the 5′ probe; and a 7.0-kb and a 5.3-kb NcoI/NcoI band, respectively, using the 3′ probe.
PCR was used for routine genotyping of the targeted allele(s). The following PCR conditions were applied: 4 min at 94°C, followed by 30 cycles of 15 s at 94°C, 20 s at 56°C, and 20 s at 72°C. The forward primer 5′-CTGCGTGTTCGAATTCGCCAATGA-3′ and reverse primer 5′-GCCTGGAATGTTTCCACCCAATGT-3′ detected a 144-bp fragment from the targeted allele. The forward primer 5′-CACAAAGCACACAAGTGGCAGTGA-3′ and reverse primer 5′-AACATTCCCTTGCAGACCCAGAGA-3′ detected a 187-bp fragment from the WT allele.
All mice were housed under specific pathogen-free conditions and studied according to protocols approved by the Institutional Animal Protection Committee of the CRCHUM.
Reverse transcription–quantitative PCR
Total RNA from the islets or kidneys was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA). RNA was reverse-transcribed into cDNA with iScript cDNA Synthesis kits (Bio-Rad Laboratories, Hercules, CA, USA). iQ SYBR Green Supermix PCR kits (Bio-Rad Laboratories) were employed for real-time PCR amplification of cDNA templates. The PCR amplification program was as follows: 95°C, 3 min, 1 cycle; 95°C, 10 s, 59°C, 20 s, 72°C, 30 s, 45 cycles. Primers for TGFBI (NM_009369.4) TGFβ1 (NM_011577.1), RPS6 (ribosomal protein S6, NM_009096.3) and EIF4EBP1 (eukaryotic translation initiation factor 4E binging protein 1, NM_007918.3) mRNA quantification are listed in Supplementary Material, Table S1. All samples were tested in duplicate. β-Actin mRNA levels were measured as internal controls. In addition to melting curve analysis, qPCR products were verified by agarose gel electrophoresis for expected band sizes. The data are expressed as signal ratios of mRNA of the tested genes/β-actin mRNA.
Enzyme-linked immunosorbent assay
Mouse TGFBI (NM_009369.4) and TGFβ1 (NM_011577.1) ELISA kits were purchased from R&D Systems (Minneapolis, MN, USA) and eBioscience (San Diego, CA, USA), respectively. Assays were performed according to the manufacturers' instructions. All samples were assayed in duplicate. Sensitivity was 8 pg/ml with TGFBI (NM_009369.4) enzyme-linked immunosorbent assay (ELISA) and 5 pg/ml with TGFβ1 (NM_011577.1) ELISA.
Islet isolation, culture, flow cytometry and insulin-release assays
These experimental methods are detailed in our earlier publications (19). KO and WT islets were cultured in serum-free F-12K medium containing 2% BSA and 1× MEM non-essential amino acids before flow cytometry analysis and insulin release assays. In some experiments, islets were cultured in the presence of AKT inhibitor IV ((49), Calbiochem, Darmstadt, Germany) or vehicle.
Syngeneic islet transplantation
KO and WT male mice in the second generation of 129/sv to C57BL/6 backcrossing (10–12 weeks old) were treated i.p. 3 times with 90 mg/kg STZ every other day to chemically induce diabetes. Two weeks after STZ injection, blood glucose was monitored daily with an Ascensia Contour glucose meter (Toronto, Ontario, Canada) until it was >20 mm for 2 consecutive days, and these mice were then grouped as islet transplantation recipients. Three hundred and fifty KO and WT islets of the same background as that of the recipients in 20 µl phosphate-buffered saline (PBS) were transplanted underneath the left renal capsule of the recipients. The surgical procedure is described in our publication (6,19). Recipient blood glucose was monitored daily for 7 days and then once every 3 days until Day 31.
Multiple-low dose STZ-induced diabetes in KO mice
KO and WT male mice in the C57BL/6 background (10–12 weeks old) were treated i.p. with low-dose STZ (40 mg/kg, q.d. for 5 days) to chemically induce diabetes. Blood glucose was monitored once every 3 days from Day 6 until Day 34.
TGFβ1 secretion and TGFβ1-induced TGFBI secretion by islets
Freshly isolated WT islets were starved in F-12K serum-free medium (containing 2% BSA and 1× MEM non-essential amino acids) for 48 h. Parts of the culture supernatants were harvested at 0 and 48 h for TGFβ1 (NM_011577.1) measurements by ELISA. Exogenous mouse recombinant TGFβ1 (NM_011577.1) at 20 ng/ml (final concentration) was added at 48 h, and the islets were cultured for another 48 h. TGFBI (NM_009369.4) levels in supernatants were measured at different time points by ELISA.
TGFβ1 and TGFBI expression in tissues adjacent to islet transplantation sites
An incision of 5–8 mm in length was made on the renal capsule of the left kidney of mice, and the capsule of 15–25 mm2 in surface size was freed from its underneath renal parenchyma near the incision, to mimic the lacerated condition of islet transplantation. Both the left (lacerated) and right (control, unmanipulated) kidneys were harvested 48 h later. Thin tissue slices were pared from the left kidney in the original lacerated area and from the corresponding area of the control right kidney. They were homogenized and their total RNA was extracted for the measurement of TGFβ1 (NM_011577.1) and TGFBI (NM_009369.4) mRNA levels.
Phospho-protein array analysis of 402 kinases, transcription factors and adaptor proteins in transgenic (Tg) versus WT islets
Islets from Tg and WT mice were isolated and cultured for 4 h. Islets from individual mice were pooled (two mice per pool; three pools islets from Tg mice and three pools of islets from WT mice were used), lysed and lysate proteins were analyzed by Full Moon BioSystem (Sunnyvale, CA, USA) phospho-protein array which contains antibodies (Abs) against 402 phosphorylated kinases, adaptor proteins and transcription factors as well as Abs against total proteins of the said molecules. The list of all Abs used can be found in http://www.fullmoonbiosystems.com/DataSheets/AntibodyArrays/PEX100_AbList.xls
Immunoblotting
WT islets were cultured in F-12K serum-free medium in the presence of recombinant human TGFBI (NM_009369.4) (5 µg/ml) for 4 h and then lysed. Fifty micrograms of lysate protein per lane were loaded for 12% SDS–PAGE. Proteins in the gels were transferred to polyvinylidene fluoride membranes after electrophoresis. The membranes were hybridized with rabbit Abs against total or phosphorylated mouse AKT (NM_001165894, S473), AKT1S1 (NM_001253920.1, Thr246), RPS6 (NM_009096.3, S235/S236) and EIF4EBP1 (NM_007918.3, Thr37/Thr46). All Abs were from Cell Signaling Technology (Danvers, MA, USA) and used at 1:1000 dilution. The hybridization procedure was performed according to the manufacturer's instructions. Signals were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
siRNA transfection
Mouse AKT1S1 (NM_001253920.1) and EIF4EBP1 (NM_007918.3) siRNAs were synthesized by Integrated DNA Technologies (Coralville, IA, USA). RPS6 (NM_009096.3) siRNA was from Thermo Scientific (Pittsburgh, PA, USA). The sequences of siRNAs specific to AKT1S1 (NM_001253920.1), RPS6 (NM_009096.3) and EIF4EBP1 (NM_007918.3) and control siRNA are listed in Supplementary Material, Table S2. siRNAs were transfected into islets with X-tremeGENE transfection reagent (Roche Diagnostics GmbH, Mannheim, Germany) at a final concentration of 80 nm siRNA for 200 islets at 0.2 ml/well in 48-well plates. mRNA knockdown was confirmed by RT–qPCR 48 h after transfection.
Analysis of SNPs in the TGFBI gene for their association with T1D and T2D
A GWAS and a meta-analysis dataset were queried for the significance of association for all SNPs located within the genomic region containing the TGFBI (NM_009369.4) gene and adjacent regions 50 kb upstream and downstream of the gene (chr5:135,315,100–135,482,840, Build 36; chr5;135,287,201–135,454,941, Build37) with either T1D or T2D phenotypes. The GWAS was conducted by the WTCCC, comprising 2000 T1D patients and 3000 control individuals, who are Great Britain Caucasians (33). The meta-analysis was performed by the MAGIC, and includes 96 496 non-diabetic individuals of European ancestry from 52 GWAS, using fasting blood glucose levels without or with correction for BMI as a phenotype (34). Such a glycemic phenotype might be associated with T2D risks. The number of tag SNPs representing independent LD blocks within the test region was calculated using the Pairwise Tagger Program (50,51) on the HapMap website (http://hapmap.ncbi.nlm.nih.gov) with the r2 parameter set at 0.8 and no minimum allele frequency. This analysis determined that there were 42 Tag SNPs within the region using these parameter settings. These Tag SNPs can proxy for the total of 795 SNPs queried (231 for T2D, 737 for T1D and 173 common to both T1D and T2D) and give a statistically conservative estimate (using r2 of 0.8) of the number of independent tests. This estimate of independent LD blocks was used to determine the Bonferroni-corrected P-value at P = 0.05/42 = 1.19 × 10−3. Only one phenotype was examined for the WTCCC GWAS and two correlated phenotypes were queried for the MAGIC meta-analysis. Correcting for two phenotypes in the latter analysis gives a corrected P-value of 5.85 × 10−4, which is conservative given the correlation between fasting glucose and BMI-corrected fasting glucose.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the Canadian Institutes of Health Research (MOP57697 and MOP123389), the Heart and Stroke Foundation of Quebec, the Natural Sciences and Engineering Research Council of Canada (203906-2012), Juvenile Diabetes Research Foundation (17-2013-440) and the Jean-Louis Levesque Foundation to J.W.
Supplementary Material
REFERENCES
- 1.Skonier J., Neubauer M., Madisen L., Bennett K., Plowman G.D., Purchio A.F. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol. 1992;11:511–522. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- 2.Skonier J., Bennett K., Rothwell V., Kosowski S., Plowman G., Wallace P., Edelhoff S., Disteche C., Neubauer M., Marquardt H. Beta ig-h3: a transforming growth factor-beta-responsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA Cell Biol. 1994;13:571–584. doi: 10.1089/dna.1994.13.571. [DOI] [PubMed] [Google Scholar]
- 3.Billings P.C., Herrick D.J., Kucich U., Engelsberg B.N., Abrams W.R., Macarak E.J., Rosenbloom J., Howard P.S. Extracellular matrix and nuclear localization of beta ig-h3 in human bladder smooth muscle and fibroblast cells. J. Cell. Biochem. 2000;79:261–273. doi: 10.1002/1097-4644(20001101)79:2<261::aid-jcb90>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Park S.-W., Bae J.-S., Kim K.-S., Park S.-H., Lee B.-H., Choi J.-Y., Park J.-Y., Ha S.-W., Kim Y.-L., Kwon T.-H., et al. Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin. Exp. Mol. Med. 2004;36:211–219. doi: 10.1038/emm.2004.29. [DOI] [PubMed] [Google Scholar]
- 5.Lee S.-H., Bae J.-S., Park S.-H., Lee B.-H., Park R.-W., Choi J.-Y., Park J.-Y., Ha S.-W., Kim Y.-L., Kwon T.-H., et al. Expression of TGF-beta-induced matrix protein betaig-h3 is up-regulated in the diabetic rat kidney and human proximal tubular epithelial cells treated with high glucose. Kidney Int. 2003;64:1012–1021. doi: 10.1046/j.1523-1755.2003.00158.x. [DOI] [PubMed] [Google Scholar]
- 6.Schorderet D.F., Menasche M., Morand S., Bonnel S., Büchillier V., Marchant D., Auderset K., Bonny C., Abitbol M., Munier F.L. Genomic characterization and embryonic expression of the mouse Bigh3 (Tgfbi) gene. Biochem. Biophys. Res. Commun. 2000;274:267–274. doi: 10.1006/bbrc.2000.3116. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto K., Noshiro M., Ohno S., Kawamoto T., Satakeda H., Akagawa Y., Nakashima K., Okimura A., Ishida H., Okamoto T., et al. Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochim. Biophys. Acta. 1997;1355:303–314. doi: 10.1016/s0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- 8.Hanssen E., Reinboth B., Gibson M.A. Covalent and non-covalent interactions of betaig-h3 with collagen VI. Beta ig-h3 is covalently attached to the amino-terminal region of collagen VI in tissue microfibrils. J. Biol. Chem. 2003;278:24334–24341. doi: 10.1074/jbc.M303455200. [DOI] [PubMed] [Google Scholar]
- 9.Thapa N., Kang K.-B., Kim I.-S. Beta ig-h3 mediates osteoblast adhesion and inhibits differentiation. Bone. 2005;36:232–242. doi: 10.1016/j.bone.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Oh J.-E., Kook J.-K., Min B.-M. Beta ig-h3 induces keratinocyte differentiation via modulation of involucrin and transglutaminase expression through the integrin alpha3beta1 and the phosphatidylinositol 3-kinase/Akt signaling pathway. J. Biol. Chem. 2005;280:21629–21637. doi: 10.1074/jbc.M412293200. [DOI] [PubMed] [Google Scholar]
- 11.Kim J.-E., Kim S.-J., Jeong H.-W., Lee B.-H., Choi J.-Y., Park R.-W., Park J.-Y., Kim I.-S. RGD peptides released from beta ig-h3, a TGF-beta-induced cell-adhesive molecule, mediate apoptosis. Oncogene. 2003;22:2045–2053. doi: 10.1038/sj.onc.1206269. [DOI] [PubMed] [Google Scholar]
- 12.Ruoslahti E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 13.Ohno S., Noshiro M., Makihira S., Kawamoto T., Shen M., Yan W., Kawashima-Ohya Y., Fujimoto K., Tanne K., Kato Y. RGD-CAP ((beta)ig-h3) enhances the spreading of chondrocytes and fibroblasts via integrin alpha(1)beta(1) Biochim. Biophys. Acta. 1999;1451:196–205. doi: 10.1016/s0167-4889(99)00093-2. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson J.W., Thoma B.S., Mikesh M.F., Kramer R.H., Bennett K.L., Purchio A., Bellard B.J., LeBaron R.G. The extracellular matrix protein betaIG-H3 is expressed at myotendinous junctions and supports muscle cell adhesion. Cell Tissue Res. 2003;313:93–105. doi: 10.1007/s00441-003-0743-z. [DOI] [PubMed] [Google Scholar]
- 15.Kim J.E., Kim S.J., Lee B.H., Park R.W., Kim K.S., Kim I.S. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J. Biol. Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- 16.Ween M.P., Oehler M.K., Ricciardelli C. Transforming Growth Factor-Beta-Induced Protein (TGFBI)/(βig-H3): A Matrix Protein with Dual Functions in Ovarian Cancer. Int. J. Mol. Sci. 2012;13:10461–10477. doi: 10.3390/ijms130810461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt-Bernard C.F., Guittard C., Arnaud B., Demaille J., Argiles A., Claustres M., Tuffery-Giraud S. BIGH3 exon 14 mutations lead to intermediate type I/IIIA of lattice corneal dystrophies. Invest. Ophthalmol. Vis. Sci. 2000;41:1302–1308. [PubMed] [Google Scholar]
- 18.Warren J.F., Abbott R.L., Yoon M.K., Crawford J.B., Spencer W.H., Margolis T.P. A new mutation (Leu569Arg) within exon 13 of the TGFBI (BIGH3) gene causes lattice corneal dystrophy type I. Am. J. Ophthalmol. 2003;136:872–878. doi: 10.1016/s0002-9394(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 19.Han B., Qi S., Hu B., Luo H., Wu J. TGF-beta i promotes islet beta-cell function and regeneration. J. Immunol. 2011;186:5833–5844. doi: 10.4049/jimmunol.1002303. [DOI] [PubMed] [Google Scholar]
- 20.Yu H., Wergedal J.E., Zhao Y., Mohan S. Targeted disruption of TGFBI in mice reveals its role in regulating bone mass and bone size through periosteal bone formation. Calcif. Tissue Int. 2012;91:81–87. doi: 10.1007/s00223-012-9613-6. [DOI] [PubMed] [Google Scholar]
- 21.Liadis N., Murakami K., Eweida M., Elford A.R., Sheu L., Gaisano H.Y., Hakem R., Ohashi P.S., Woo M. Caspase-3-dependent beta-cell apoptosis in the initiation of autoimmune diabetes mellitus. Mol. Cell. Biol. 2005;25:3620–3629. doi: 10.1128/MCB.25.9.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pighin D., Karabatas L., Pastorale C., Dascal E., Carbone C., Chicco A., Lombardo Y.B., Basabe J.C. Role of lipids in the early developmental stages of experimental immune diabetes induced by multiple low-dose streptozotocin. J. Appl. Physiol. 2005;98:1064–1069. doi: 10.1152/japplphysiol.00559.2004. [DOI] [PubMed] [Google Scholar]
- 23.Werner S., Grose R. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 24.Wiza C., Nascimento E.B.M., Ouwens D.M. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am. J. Physiol. Endocrinol. Metab. 2012;302:E1453–E1460. doi: 10.1152/ajpendo.00660.2011. [DOI] [PubMed] [Google Scholar]
- 25.Thedieck K., Polak P., Kim M.L., Molle K.D., Cohen A., Jenö P., Arrieumerlou C., Hall M.N. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nascimento E.B.M., Snel M., Guigas B., van der Zon G.C.M., Kriek J., Maassen J.A., Jazet I.M., Diamant M., Ouwens D.M. Phosphorylation of PRAS40 on Thr246 by PKB/AKT facilitates efficient phosphorylation of Ser183 by mTORC1. Cell. Signal. 2010;22:961–967. doi: 10.1016/j.cellsig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez Canedo C., Demeulder B., Ginion A., Bayascas J.R., Balligand J.-L., Alessi D.R., Vanoverschelde J.-L., Beauloye C., Hue L., Bertrand L. Activation of the cardiac mTOR/p70(S6K) pathway by leucine requires PDK1 and correlates with PRAS40 phosphorylation. Am. J. Physiol. Endocrinol. Metab. 2010;298:E761–E769. doi: 10.1152/ajpendo.00421.2009. [DOI] [PubMed] [Google Scholar]
- 28.Gingras A.C., Kennedy S.G., O'Leary M.A., Sonenberg N., Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt (PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingras A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee V.H.Y., Healy T., Fonseca B.D., Hayashi A., Proud C.G. Analysis of the regulatory motifs in eukaryotic initiation factor 4E-binding protein 1. FEBS J. 2008;275:2185–2199. doi: 10.1111/j.1742-4658.2008.06372.x. [DOI] [PubMed] [Google Scholar]
- 31.Fumagalli S., Thomas G. S6 Phosphorylation and signal transduction. In: Sonenberg N., Hershey J.W.B., Mathews M., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 695–717. [Google Scholar]
- 32.Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J., Ellinghaus D., Franke A., Howie B., Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning A.K., Hivert M.-F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.-T., Bielak L.F., Prokopenko I., et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zorena K., Malinowska E., Raczyńska D., Myśliwiec M., Raczyńska K. Serum concentrations of transforming growth factor-Beta 1 in predicting the occurrence of diabetic retinopathy in juvenile patients with type 1 diabetes mellitus. J. Diabetes. Res. 2013 doi: 10.1155/2013/614908. May 31, doi:10.1155/2013/614908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Border W.A., Noble N.A. Transforming growth factor beta in tissue fibrosis. N. Engl. J. Med. 1994;331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 40.Ketteler M., Noble N.A., Border W.A. Increased expression of transforming growth factor-beta in renal disease. Curr. Opin. Nephrol. Hypertens. 1994;3:446–452. doi: 10.1097/00041552-199407000-00011. [DOI] [PubMed] [Google Scholar]
- 41.Altomare D.A., Khaled A.R. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr. Med. Chem. 2012;19:3748–3762. doi: 10.2174/092986712801661130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamada S., Hara K., Hamada T., Yasuda H., Moriyama H., Nakayama R., Nagata M., Yokono K. Upregulation of the mammalian target of rapamycin complex 1 pathway by Ras homolog enriched in brain in pancreatic beta-cells leads to increased beta-cell mass and prevention of hyperglycemia. Diabetes. 2009;58:1321–1332. doi: 10.2337/db08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazi A.A., Lang C.H. PRAS40 regulates protein synthesis and cell cycle in C2C12 myoblasts. Mol. Med. 2010;16:359–371. doi: 10.2119/molmed.2009.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao J., Luo H., Han B., Bertrand R., Wu J. Drak2 is upstream of p70S6 kinase: its implication in cytokine-induced islet apoptosis, diabetes, and islet transplantation. J. Immunol. 2009;182:4762–4770. doi: 10.4049/jimmunol.0802255. [DOI] [PubMed] [Google Scholar]
- 45.Hon G.C., Hawkins R.D., Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotea V., Visel A., Westlund J.M., Nobrega M.A., Pennacchio L.A., Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kau T.R., Schroeder F., Ramaswamy S., Wojciechowski C.L., Zhao J.J., Roberts T.M., Clardy J., Sellers W.R., Silver P.A. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor cells. Cancer Cell. 2003;4:463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- 50.de Bakker P.I.W., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 51.Tagger Website. http://www.broadinstitute.org/mpg/tagger/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.