Abstract
As custom arrays are cheaper than generic GWAS arrays, larger sample size is achievable for gene discovery. Custom arrays can tag more variants through denser genotyping of SNPs at associated loci, but at the cost of losing genome-wide coverage. Balancing this trade-off is important for maximizing experimental designs. We quantified both the gain in captured SNP-heritability at known candidate regions and the loss due to imperfect genome-wide coverage for inflammatory bowel disease using immunochip (iChip) and imputed GWAS data on 61 251 and 38 550 samples, respectively. For Crohn's disease (CD), the iChip and GWAS data explained 19 and 26% of variation in liability, respectively, and SNPs in the densely genotyped iChip regions explained 13% of the SNP-heritability for both the iChip and GWAS data. For ulcerative colitis (UC), the iChip and GWAS data explained 15 and 19% of variation in liability, respectively, and the dense iChip regions explained 10 and 9% of the SNP-heritability in the iChip and the GWAS data. From bivariate analyses, estimates of the genetic correlation in risk between CD and UC were 0.75 (SE 0.017) and 0.62 (SE 0.042) for the iChip and GWAS data, respectively. We also quantified the SNP-heritability of genomic regions that did or did not contain the previous 163 GWAS hits for CD and UC, and SNP-heritability of the overlapping loci between the densely genotyped iChip regions and the 163 GWAS hits. For both diseases, over different genomic partitioning, the densely genotyped regions on the iChip tagged at least as much variation in liability as in the corresponding regions in the GWAS data, however a certain amount of tagged SNP-heritability in the GWAS data was lost using the iChip due to the low coverage at unselected regions. These results imply that custom arrays with a GWAS backbone will facilitate more gene discovery, both at associated and novel loci.
INTRODUCTION
Genome-wide association studies (GWAS) have been successful in discovering thousands of trait-SNP associations, across a wide range of complex traits (1). The initial wave of genome-wide arrays covered 300 000 to 600 000 SNP markers and gave good coverage of common variants in the genome (2,3). Genotype imputation can uncover some additional variation, but, in general, the rarer the unobserved variant to be imputed, the larger the imputation error rate (4,5). The results from initial GWAS and subsequent meta-analyses have shown that effect sizes, as measured at associated SNPs, are typically small, and that there are more variants to be discovered using the same experimental designs. Two possible reasons for the ‘missing’ genetic variation are: (i) the effect sizes for many common SNPs are too small to be detected at genome-wide significant thresholds for the current sample sizes and (ii) there is imperfect linkage disequilibrium (LD) between the unobserved causal variants and the genotyped (or imputed) SNPs (6). In both cases, increasing sample size will result in the discovery of more variants associated with the complex trait of interest, whereas for the latter, genotyping or sequencing lower frequency variants will be beneficial. In the period following the first GWAS and meta-analyses (around 2009–2011), the cost of genome-wide SNP arrays and deep sequencing prohibited substantial increases in sample size. However, a compromise was achievable by designing custom arrays that contained dense coverage of SNPs at selected genomic regions. These custom arrays, such as the metabochip (7,8), the iCOGS array (http://ccge.medschl.cam.ac.uk/research/consortia/icogs/), Psychchip, the Human Core Exome array, and the ImmunoChip (abbreviated here as iChip), are an order of magnitude cheaper than genome-wide arrays and allow very large samples to be genotyped.
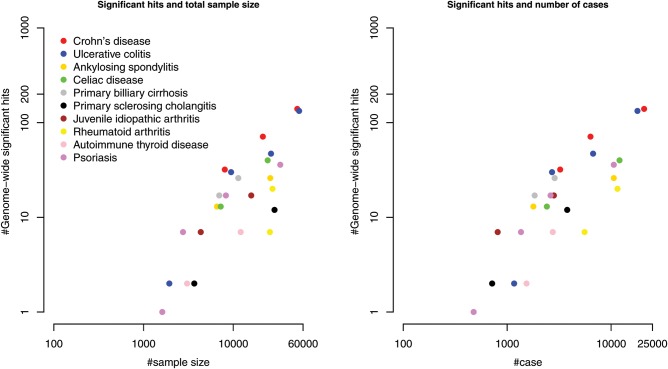
The variants on the iChip (9–11) are distributed across the whole genome, at uneven density. It provides dense coverage of 186 distinct loci in regions flanking SNPs identified to be genome-wide significant for 12 immune-related disorders (10), and includes low coverage for backbone SNPs (∼20 000) which are included as replication for non-immune-related diseases that are part of the WTCCC2 project. The chip in total contains ∼180 000 SNPs and has been successful in gene and variant discovery. For example, 69 additional loci were discovered for Crohn's disease (CD [MIM 266600]) (12), 89 for ulcerative colitis (UC [MIM 191390]) (12), 13 loci for celiac disease (CD [MIM 212750]) (10), 13 ankylosing spondylitis (SPDA1 [MIM 106300]) (11), 3 primary biliary cirrhosis (PBC [MIM 109720]) (13), 9 primary sclerosing cholangitis (PSC [MIM 613806]) (14), 15 psoriasis (PSORS [MIM 142840]) (15), 14 juvenile idiopathic arthritis (MIM 604302) (16), 14 rheumatoid arthritis (RA [MIM 180300]) (17) and 7 auto-immune thyroid disease (AITD [MIM 608175]) (18). As the number of genome-wide significant signals is approximately proportional to the sample size and number of cases on a logarithmic scale (Fig. 1 and Supplementary Material, Table S1), a low-cost genotyping platform such as the iChip enables larger experimental sample sizes and, therefore, greater gene discovery and fine-mapping.
Figure 1.
Increase in the number of GWAS-significant signals for total sample size and the number of cases. Both the axes are on a logarithmic scale. The source of the data is in Supplementary Material, Table S1.
An important question for future studies aiming to discover more variants and explain more heritability is how many of the newly discovered loci reflect increased power as a result of increased sample size versus the deeper coverage at selected loci. We sought to answer this question by estimating how much variation is explained by the iChip and compare and contrast that to the variation explained by past GWAS arrays. Using data on inflammatory bowel disease (IBD) and large sample sizes, we present a range of novel statistical analyses on the heritability of IBD using the variance component method on GWAS data and iChip data, and compared these heritability estimates with those from pooled twin data based on previously published studies (19–23).
Crohn's disease (CD) and UC are diseases with heritabilities of ∼70–80% and 60–70%, respectively, on the scale of liability (Supplementary Material, Note) and gene discovery for these diseases has been particularly productive, with over 163 independent loci identified (12). In addition to the previous GWAS results, where many shared loci were observed between these two diseases, bivariate analysis (24) has shown substantial evidence of widespread pleiotropy. However, the genetic correlation between CD and UC has not previously been quantified. We were able to estimate their genetic correlation with GWAS and iChip data.
RESULTS
SNP array and sample properties
The SNPs featured on the iChip are highly concentrated in a limited number of regions in the genome reflecting the deliberate enrichment of the 186 loci identified in GWAS of 12 auto-immune disorders (Supplementary Material, Fig. S1). We estimated, for the iChip, the effective number of markers, Me (defined in the section Materials and Methods) to be 2822, or ∼2% of the total number of the markers, reflecting the considerable LD between SNPs in the enriched regions. The estimate of the effective number of markers from the variation in pairwise relatedness was 2551 (1.8%) when using a minor allele frequency (MAF) threshold of 0.001. For MAF thresholds of 0.01 and 0.05, the estimates were 2222 (1.7) and 1748 (1.6%), respectively. This implies that the effective number of markers on the iChip is ∼2000–3000. For the CD and UC GWAS data, the effective number of markers are 29 253 and 34 596, respectively, which are lower than those previously reported from an earlier GWAS data (25) possibly because these previous data were not adjusted for population stratification. Stratification increases the variance of measures of relatedness and this reduces the estimate of the effective number of independent SNPs. Alternatively, different criteria for quality control may have contributed to the observed difference in the number of independent SNPs.
For the estimation of relatedness, we generated ∼2 billion pairwise estimates from 61 518 individuals. We have previously used stringent relatedness thresholds of 0.025 or 0.05 to avoid having relatives in the sample (6,26). However, the sampling variance of estimated relatedness, which is 1/Me, is larger for the iChip than for a GWAS array because the smaller effective number of independent SNPs in the iChip (6). Therefore, as the previous thresholds would be too stringent for the iChip, we adopted a threshold of 0.1 (Supplementary Material, Table S6), which is equivalent to thresholds of between 0.025 and 0.05 for GWAS data, which removed 10 055 individuals. The impact of varying MAFs (0.05, 0.01 and 0.001) and relatedness (0.1 and 0.15) was evaluated, but little difference in the estimate of the variance component was observed across these different values for MAF and relatedness (Supplementary Material, Table S7). Unless stated otherwise, results presented below are based on an MAF of 0.001 and a relatedness threshold of 0.1.
Three types of genomic regions were defined for analysis in this study, selected from 4752 genomic bins of 0.6 Mb in size. Classifying Type 1 selected regions (high-density regions) based on genomic bins in which the SNP density in the iChip data were at least 20% larger than the average of the chromosome where a bin was located resulted in 373 selected regions. These 373 regions captured 80% of the total number of iChip SNPs and matched the heterogeneous distribution of the SNPs on the iChip (Supplementary Material, Fig. S1). In the GWAS data, these 373 regions contained approximately 10% of all GWAS SNPs (Table 3). Classifying Type 2 selected regions (GWAS-hit regions) based on genomic regions containing GWAS-significant hits, selected based on the presence of any of the 193 SNPs associated with IBD (12) resulted in 166 genomic regions. The number of markers in these regions for the iChip and GWAS data is summarized in Table 3. Classifying Type 3 genomic regions (HD-GWAS regions) based on regions overlapping between the Type 1 (HD regions) and Type 2 (GWAS-hit regions) selected regions resulted in 116 genomic regions (Table 3).
Table 3.
Estimates of SNP-heritability for three types of selected regions compared with unselected complementary regions for the iChip and the GWAS data
| Sample size |
Type 1a iChip high-density (HD) regions | Type 2b GWAS-hit regions | Type 3c HD-GWAS regions | |||||
|---|---|---|---|---|---|---|---|---|
| Source | Cases | Controls | SNPs | SNPs | SNPs | |||
| iChip CD | ||||||||
| Selected region | 15 230 | 24 106 | 107 006 (76%) | 0.13 (0.0038) | 45 999 (33%) | 0.11 (0.0037) | 44 684 (32%) | 0.09 (0.0034) |
| Complement region | 33 847 (24%) | 0.065 (0.0025) | 94 854 (67%) | 0.09 (0.0037) | 96 169 (68%) | 0.11 (0.0039) | ||
| Sum | 140 853 | 0.195 | 140 853 | 0.20 | 140 853 | 0.20 | ||
| GWAS CD | ||||||||
| Selected region | 5054 | 11 496 | 93 263 (9.3%) | 0.13 (0.006) | 38 858 (3.9%) | 0.11 (0.005) | 28 228 (2.8%) | 0.10 (0.005) |
| Complement region | 914 797 (90.7%) | 0.09 (0.010) | 969 202 (96.1%) | 0.10 (0.010) | 979 832 (97.2%) | 0.12 (0.010) | ||
| Sum | 1 008 060 | 0.23 | 1 008 060 | 0.210 | 1 008 060 | 0.22 | ||
| iChip UC | ||||||||
| Selected region | 11 808 | 24 106 | 107 006 (76%) | 0.10 (0.0036) | 45 999 (33%) | 0.10 (0.0036) | 44 684 (32%) | 0.10 (0.0036) |
| Complement region | 33 847 (24%) | 0.051 (0.0026) | 94 854 (67%) | 0.05 (0.0026) | 96 169 (68%) | 0.05 (0.0026) | ||
| Sum | 140 853 | 0.151 | 140 853 | 0.15 | 140 853 | 0.15 | ||
| GWAS UC | ||||||||
| Selected region | 5799 | 16 201 | 96 598 (9.2%) | 0.09 (0.004) | 40 227 (3.8%) | 0.08 (0.004) | 29 141 (2.8%) | 0.07 (0.004) |
| Complement region | 950 970 (90.8%) | 0.09 (0.008) | 1 007 341 (96.2%) | 0.10 (0.008) | 1 018 427 (97.2%) | 0.10 (0.008) | ||
| Sum | 1 047 568 | 0.18 | 1 047 568 | 0.18 | 1 047 568 | 0.18 | ||
No threshold was applied for the GRM. The heritability in liability was estimated in joint analysis by fitting the GRM for the selected region and the GRM for other regions together.
aThe Type 1 selected bins (iChip high-density regions) were defined as those whose SNP density was 20% higher than the average of the chromosome. 373 bins met this definition.
bThe Type 2 selected bins (GWAS-hit regions) were those containing lead SNPs associated with IBD as previously reported (12). 166 bins met this definition. Of those 166 bins, 110 were overlapped with Type 1 selected regions (iChip HD regions).
cThe Type 3 selected bins (HD-GWAS regions) were those that overlapped between the Type 1 selected bins (iChip HD regions) and Type 2 selected bins (GWAS-hit regions). 110 bins met this definition.
Genetic analyses
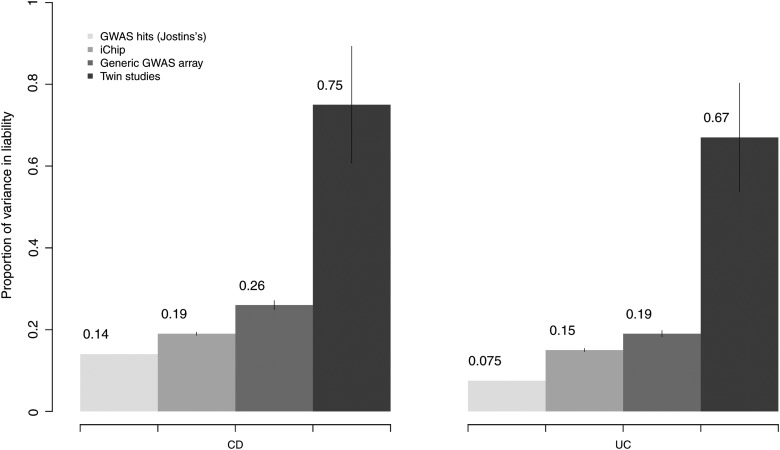
From pooling twin data from four previous studies (19–23,27), we estimated the heritability for liability in CD and UC to be 0.75 and 0.67, respectively (additional data and methods provided in Supplementary Material, Note). In contrast, the estimate of SNP-heritability in liability explained by the iChip SNPs was 0.19 and 0.15 for CD and UC, respectively (Table 1). These estimates indicate that if the total heritability of liability for CD and UC is ∼70% (Supplementary Material, Note), then 0.27 and 0.21 of heritability for CD and UC, respectively, is captured by the iChip SNPs. The corresponding estimates of SNP-heritability for CD and UC based on the imputed GWAS data are 0.37 and 0.27, respectively. All these estimates are significantly different from zero (P << 10−10). The proportion of variation in liability for CD and UC explained by genome-wide significant SNPs is ∼0.13 and 0.08, respectively (12). Comparisons of the proportion of variance in liability explained by GWAS hits (12), iChip, GWAS data (generic GWAS array) and twin studies (Supplementary Material, Note) are illustrated in Figure 2.
Table 1.
Estimated SNP-heritability for the iChip and imputed GWAS data on the observed scale and the scale of liability
| Source | SNPs | Cases | Controls | SNPs | ||
|---|---|---|---|---|---|---|
| Univariate analysis | ||||||
| iChip CD | 140 853 | 15 230 | 24 106 | 140 853 | 0.39 (0.008) | 0.19 (0.004) |
| GWAS CD | 1 008 060 | 5054 | 11 496 | 1 008 060 | 0.46 (0.020) | 0.26 (0.011) |
| iChip UC | 140 853 | 11 808 | 24 106 | 140 853 | 0.33 (0.009) | 0.15 (0.004) |
| GWAS UC | 1 047 568 | 5799 | 16 201 | 1 047 568 | 0.36 (0.016) | 0.19 (0.008) |
| SNPs | Cases CD/UC | Controls CD/UC | CD (SE) | UC (SE) | Correlation (SE) | |
| Bivariate analysis | ||||||
| iChip CD/UCa | 140 853 | 15 230/11 808 | 13 320/10 786 | 0.20 (0.004) | 0.15 (0.004) | 0.75 (0.017) |
| GWAS CD/UCb | 987 572 | 4813/5768 | 6085/11 279 | 0.25 (0.013) | 0.18 (0.009) | 0.62 (0.042) |
The MAF threshold for inclusion of the SNPs was 0.001 and 0.01 for iChip and GWAS, respectively; the GRM threshold was 0.1 and 0.05 for iChip and GWAS, respectively. The prevalence is assumed to be 0.005 and 0.0025 for CD and UC, respectively.
aIn bivariate analysis for iChip data, controls were split in proportion to the number of the CD and UC cases.
bFor GWAS bivariate analysis, the numbers of samples and SNPs are different to those from the univariate analysis because of overlap between the controls samples.
Figure 2.
The comprehensive estimation of the explained variance in liability scale for CD and UC. For each disease, from left to right are the estimates from GWAS hits (12), iChip, generic GWAS array data, and twin studies pooling four published twins cohorts together (Supplementary Material, Note).
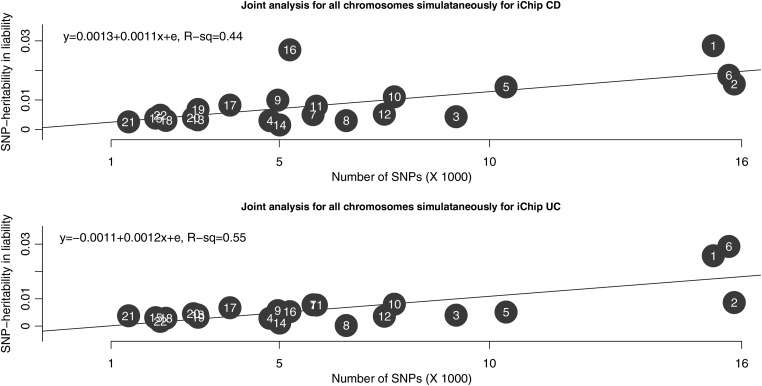
Partitioning the iChip genetic variation into MAF bins indicated that genetic variation is captured across the entire SNP frequency range (Table 2). For both diseases, the variance explained by each chromosome was proportional to the number of the SNPs in the chromosome (Fig. 3) and the linear relationship was highly significant (P = 0.00077 for CD, P = 7.72e−5 for UC; based on an F-statistic with 1 and 20 degrees of freedom for the averaged set). As the iChip data were split into two independent sets of SNPs for autosomal joint analysis, we compared the differences in the variance explained between each subset of SNPs for each chromosome. No statistically significant differences were found at the 0.05 significance level (Supplementary Material, Figs S2 and S3). Chromosomes 6 and 16 were outliers in terms of the SNP-heritability in liability for CD and UC explained by iChip data. For CD, chromosome 16 explained much more variance than would be expected based on its length and on the number of SNPs in this chromosome, a pattern that was also observed in the GWAS data. This could be due to the fact that NOD2, a major causal locus for CD (28,29), is on chromosome 16 and the iChip has a high density of SNPs in the region of this gene (Supplementary Material, Fig. S1). Chromosome 16 had an estimated SNP-heritability of 0.054 (SE 0.0038) for CD and 0.0112 (SE 0.0022) and UC; this difference in the amount of variation explained was statistically significant (P < 10−6). For UC, chromosome 6, which harbors the major histocompatibility complex (MHC), explained more variance than expected. Chromosome 6 had SNP-heritability of 0.037 (SE 0.00328) for CD and 0.063 (SE 0.0049) for UC; this difference in the amount of variation explained was also statistically significant (P = 0.000014). These results imply that the MHC has a much larger effect on UC than CD in European samples.
Table 2.
Partitioning of genetic variation across SNP allele frequencies
| CD |
UC |
|||
|---|---|---|---|---|
| MAF | SNPs (proportion) | SNPs (proportion) | ||
| iChip | ||||
| 0.001–0.1 | 50 993 (36.2%) | 0.034 (0.0044) | 50 993 (36.2%) | 0.028 (0.0042) |
| 0.1–0.2 | 27 656 (19.6%) | 0.027 (0.0040) | 27 656 (19.6%) | 0.021 (0.0040) |
| 0.2–0.3 | 22 441 (15.9%) | 0.043 (0.0045) | 22 441 (15.9%) | 0.035 (0.0045) |
| 0.3–0.4 | 20 080 (14.3%) | 0.047 (0.0047) | 20 080 (14.3%) | 0.025 (0.0044) |
| 0.4–0.5 | 19 683 (14.0%) | 0.043 (0.0048) | 19 683 (14.0%) | 0.034 (0.0046) |
| Sum | 140 853 | 0.194 | 140 853 | 0.143 |
| GWAS | ||||
| <0.1 | 181 442 (18.0%) | 0.014 (0.0074) | 192 492 (18.4%) | 0.021 (0.0061) |
| 0.1–0.2 | 230 847 (22.9%) | 0.037 (0.0088) | 239 861 (22.9%) | 0.039 (0.0068) |
| 0.2–0.3 | 209 141 (20.7%) | 0.070 (0.0095) | 215 573 (20.6%) | 0.038 (0.0069) |
| 0.3–0.4 | 196 930 (19.5%) | 0.076 (0.0095) | 203 732 (19.4%) | 0.047 (0.0070) |
| 0.4–0.5 | 189 700 (18.8%) | 0.047 (0.0082) | 195 910 (18.7%) | 0.038 (0.0062) |
| Sum | 1 008 060 | 0.243 | 1 047 568 | 0.184 |
The iChip sample was split into two equal-sized subsets for computational reasons. The mean of the estimated heritability was used in the table. Individuals with relatedness <0.1, estimated from markers with MAF > 0.001 were used in this analysis. Samples were split into two approximately even subsets. For CD, one sample consisted of 7575 cases and 12 075 controls while the second sample consisted of 7655 cases and 12 031 controls. For UC, one sample consisted of 5894 cases and 12 075 controls, and the second sample consisted of 5914 cases and 12 031 controls.
Figure 3.
Partitioning of genetic variation in liability to CD and UC across chromosomes. The iChip sample was split into two subsets, and the averaged heritability was used in this figure.
The SNP-heritability for each disease estimated from the bivariate analysis was very close to those estimated from univariate analyses, both for the iChip and GWAS data (Table 1). This consistency also demonstrated that the estimated SNP-heritability was robust. The estimated whole-genome genetic correlation between CD and UC from the iChip and GWAS data was 0.75 (SE 0.017) and 0.62 (SE 0.04), respectively. These estimates are significantly different from each other (P = 0.004). Estimations of the genetic correlation between CD and UC based on the iChip Type 1 genomic regions yielded consistent results with those observed using whole-genome estimation (Supplementary Material, Table S8). These results were expected given the high proportion of pleiotropy between CD and UC as previously reported (12).
As the iChip data are enriched for low-frequency genetic variants (MAF < 0.01), we investigated the impact of using three different MAF thresholds (0.001, 0.01 and 0.05) on our results. The influence of different MAF thresholds was minor. The correlations between estimates of genetic relatedness using these three MAF thresholds were 0.994 (MAF 0.001 and 0.01), 0.969 (MAF 0.001 and 0.05), and 0.977 (MAF 0.01 and 0.05). The estimated variance components were similar for MAF thresholds of 0.001, 0.01, and 0.05 (Supplementary Material, Table S7). Hence, low-frequency SNPs appear to contribute little additional information on tagging SNP-heritability in the iChip data.
We quantified the variation explained by the different genomic regions in the three types of genomic partitioning classified above, for both iChip and GWAS data. Based on the Type 1 classification, the HD regions, variation in liability for CD and UC was 0.13 and 0.10, respectively, using iChip data, whereas its complement of the genome (the unselected regions) explained 0.065 (CD) and 0.051 (UC) of liability. In the GWAS data, the corresponding HD regions explained 0.13 and 0.09 of liability for CD and 0.09 and 0.09 for UC (Table 3). Hence the selected high-density SNP regions on the iChip explained at least as much variation as, but not substantially more than, the corresponding regions in the GWAS data, and compared to the GWAS data a substantial amount of information is lost from inadequate tagging of the rest of the genome. This result was also consistent with what was observed in joint bivariate analysis (Supplementary Material, Table S8) for Type 1 HD regions.
For the Type 2 GWAS-hit regions, covering areas of GWAS-significant hits, of the 166 regions in this group, 110 overlapped with Type 1 selected HD regions. For both iChip and GWAS data, the selected regions explained almost the same variation in liability for CD, 0.11 (Table 3). For UC, the selected regions in iChip explained 0.1 of the variation in liability but 0.08 in GWAS data. Heritability estimated from the summation of the GWAS hits for CD and UC were 0.14 and 0.075 (Fig. 2), respectively; in contrast, their estimated SNP-heritability based on GWAS-hit regions was 0.11 and 0.10, respectively (Table 3). For Type 3 HD-GWAS regions, consisting of 110 genomic bins that overlap selected regions of Type 1 HD-regions and Type 2 GWAS-hit regions, the GWAS data explained 0.1 of variation in liability for CD and, using iChip, 0.09. For UC, the variation in liability using iChip was 0.1, which was similar to what was observed for Type 1 and Type 2 genomic regions.
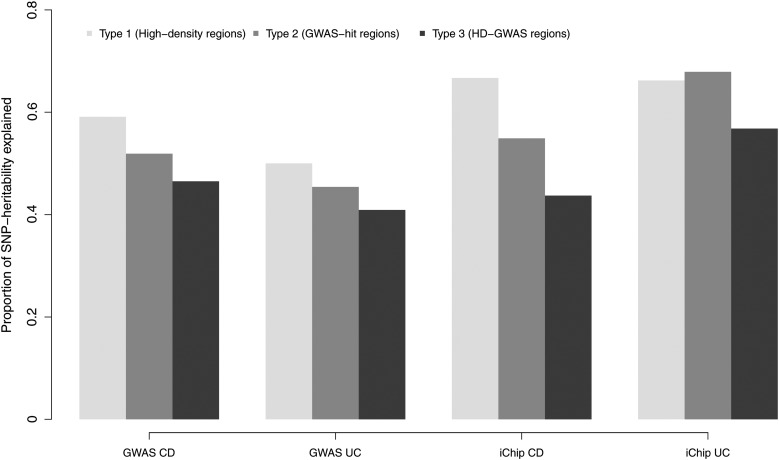
For each of the three types of selected genomic regions, the proportions of variation in liability explained by the selected and the unselected regions are illustrated in Figure 4. In general, the proportion of variance explained was lowest for HD-GWAS regions and highest for HD regions, except for the iChip UC estimate that was slightly higher for the GWAS-hit regions. The largest difference in the proportion of the variation explained was greatest between Type 1 HD regions (67%) and Type 3 HD-GWAS regions (44%), for iChip CD data, while the difference in heritability estimates between these two classifications for the iChip UC data was less (66 and 56%, respectively). For the GWAS CD data, the amount of variation in liability was lower by 15% for the Type 3 selected HD-GWAS-hit regions compared with the Type 1 selected HD regions, and the corresponding difference in estimates for these regions types in the GWAS UC data was 10%.
Figure 4.
Proportion of the genetic variation in liability to CD and UC explained by the selected regions across three types of partitioning of the genome.
DISCUSSION
We have quantified the proportion of liability captured by a custom array that targeted specific regions in the genome known to be associated with auto-immune diseases and have compared and contrasted the estimate with that obtained from genome-wide coverage of marker data. Although densely genotyped iChip regions are expected to tag causal variants better than GWAS arrays at those loci, we found that the estimated SNP-heritability in iChip data does not appear to tag more than the same regions do for imputed GWAS data.
Among the selected bins, allelic heterogeneity and multiple signals may reduce the magnitude of the SNP-heritability. However, we explicitly partitioned the variance into different MAF spectrum and could not find a substantial difference between the sum of estimates from MAF bins and that from all SNPs (Tables 1 and 2). Since the iChip provides dense coverage in the selected genomic regions only, a certain amount of information is lost; we estimated that ∼25% of the SNP-heritability that is tagged in the GWAS data is lost using the iChip. Despite this, the iChip has been highly successful in mapping additional variants for a range of auto-immune diseases, including IBD (12), Celiac disease (10) and ankylosing sponlylitis (11), primary biliary cirrhosis (13), primary sclerosing cholangitis (14), psoriasis (15), juvenile idiopathic arthritis (16) and rheumatoid arthritis (17). This is consistent with expectations since there is increased power from using the iChip as it enables the use of much larger samples sizes. We consequently conclude that the power of custom arrays such as the iChip arises predominantly from enabling larger studies to be carried out. Therefore, we conclude that a custom array that also includes a solid tagging backbone of GWAS hits would enable larger scale association studies to be conducted across the entire genome, and improve capabilities for detecting new loci and explaining more heritability.
In designing a custom array with a fixed number of variants, which is ∼200 000 on custom arrays such as the iChip, the metaboChip, and iCOGS array, determining the optimal strategy for tagging variants across the genome requires careful consideration. For example, should custom arrays aim to tag more rare variants in selected loci or more backbone variants in the rest of the genome? Compared with GWAS data, which tags variants evenly across the MAF spectrum, in the iChip low and rare variants are over-represented in the selected regions, far higher than that of GWAS arrays at these loci. As expected, the iChip SNPs of low MAF, ranging from 0.001 to 0.1, explained proportionally more variance than that of the SNPs of low MAF from the GWAS data. From this study we cannot rule out the existence of rare variants (with large effect sizes) that are not tagged by either the GWAS or iChip arrays. A recently published study investigated the existence of such variants through fine-mapping and indicated that the contribution of rare variants with large effect sizes for auto-immune diseases is negligible at candidate genes (30). These results suggest that tagging an even spread of variants across the entire genome, at the expense of tagging rare SNPs in selected regions, may be more efficient for gene discovery and for capturing genetic variation.
Although in this study we focused on comparing estimates of SNP-heritability between the iChip and GWAS data, a key motivation for designing the iChip was to provide a cost-efficient platform for fine-mapping, particularly at known loci contributing to variation in auto-immune-related diseases. In general, considerations for fine-mapping using iChip are similar to those for optimizing GWAS study designs as discussed by Skol et al. (31). Specifically, as the iChip mainly tags variants that lie within established loci for auto-immune diseases, it will be valuable for fine-mapping provided the selected loci capture a sufficient amount of variation in liability. The iChip was designed by incorporating many candidate loci and GWAS backbone markers, which make up the densely genotyped regions and the low-coverage regions on the iChip. The iChip enables the discovery of more variants through the process of fine-mapping, for example from conditional analysis, in particular in the densely genotyped regions. It also allows the discovery of new variants by enabling much larger experimental sample sizes than GWAS arrays for a fixed budget. If, however, many causal loci are located in the low-coverage regions, then the iChip will be poor at fine-mapping such loci, even if large sample sizes are used. In general, issues of SNP selection and coverage also extend to other custom arrays, such as the metabochip (7), iCOGS chip, Psychchip and the Human Core Exome, some of which have a GWAS backbone. However, those arrays may have different properties from the iChip and therefore more empirical data are needed before extrapolating our results to other custom arrays.
In this study, we conducted comprehensive estimation of the heritability of IBDs using GWAS data and iChip data, and compared these estimates to those from pooled twin data. Twin data gave the highest estimate of heritability and the variance component method using GWAS and iChip data gave lower estimates of heritability. The higher heritability estimate observed from twin data is in line with what is reported for other diseases, such as schizophrenia, bipolar disorder, and autism, and also with other complex traits (1,32). The main likely reason for this discrepancy is that pedigree based heritability captures all additive genetic variation whereas SNP-heritability only captures variation tagged by the SNPs on the array. For other diseases and traits, SNP heritabilities are ∼1/3 to 1/2 of total (pedigree) heritability (1). For UC and CD, the SNP-heritability from GWAS arrays is ∼20–25% whereas pedigree heritability is ∼70% (Fig. 1), and therefore these estimates are in line with what has been reported for other diseases. We cannot rule out biases in the estimates of either the pedigree or the SNP-based heritability. One possible reason for the high estimates of heritability from twin data is that they are obtained under assumptions of no shared environment, no dominance effects, and no epistasis. The contribution of these factors to the estimation of heritability cannot be ruled out, particularly the impact of shared environment, and this may contribute substantially to the higher heritability estimates if the etiology of a disease is driven by environmental factors.
Although clinical diagnosis differs for CD and UC, strong genetic correlation between CD and UC has been observed in previous GWAS that indicate a high proportion (110 of 163 loci) of shared loci between CD and UC (12). In our study, bivariate analysis supports the existence of substantial genetic correlation between the two diseases. The genetic correlation estimated from iChip data was higher than from GWAS data. One possible explanation for these findings is that the iChip is designed to capture variants that affect two or more auto-immune diseases, that is, the iChip positively selects for pleiotropy by design. As demonstrated in previous studies of auto-immune diseases, there is strong evidence for pleiotropy among the different auto-immune conditions (11). Even after partitioning the genome, the genetic correlation between CD and UC remained for low-coverage regions. A possible explanation consistent with these results is that the GWAS and iChip case sets have different proportions of diagnostic misclassification. If the misclassification rate is larger in the iChip set then that will lead to a larger genetic correlation (33). Further research using denser genotyping of low-coverage regions combined with detailed phenotyping may provide more insight into the genetic overlap between these two diseases.
In addition, as the GWAS array and iChip differed from each other with respect to many characteristics, such as SNP density and the effective number of markers, the differences in SNP-heritability observed between them may reflect the sensitivity of the methods to the type of genotyping platform. As recently discussed (34,35), under the current variance component method, estimates of SNP-heritability may be biased due to different underlying genetic architectures of CD and UC. Therefore, adjusting the LD structure for tagged markers may yield more unbiased estimates of the SNP-heritability, as proposed and demonstrated by Gusev et al. (35). In practice, however, unless the underlying genetic architecture is known, it may be difficult to justify the method chosen for estimating heritability. As demonstrated in our recent work (34), the MAF-bin method to partition and estimate SNP-heritability appears robust to different genetic architectures. In our study, the estimated SNP-heritabilities are based on univariate analysis, bivariate analysis, and MAF-bin partitioning analyses were all consistent with another, suggesting that our estimates are robust and reliable.
MATERIALS AND METHODS
SNP array and sample characteristics
Immunochip (iChip) genotype data on case and control samples were provided by the International Inflammatory Bowel Disease Consortium (IIBDGC), release 5 (Nov 2012). Using quality control (QC) guidelines provided by the IIBDGC (12), 174 193 SNPs, and 61 554 individuals were retained, 33 306 cases (15 648 males and 17 658 females) and 28 248 were controls (12 516 males and 15 732 females). To avoid the estimation of spurious genetic variance from case–control data (26,36) we performed additional QC steps. We eliminated an additional 14 784 SNPs with P < 1e-6 in the test of Hardy–Weinberg equilibrium in the remaining data, and 36 individuals whose missing rates were >0.02, leading to a sample size of 61 518 individuals and 159 409 SNPs. Given the sample size used in the iChip data, power calculations (37) indicated that the sample size was sufficient to detect a causal variant explaining 0.1% of SNP-heritability in liability (Supplementary Material, Fig. S4).
For the statistical analyses, we adopted three thresholds for MAF, 0.001, 0.01 and 0.05, and the corresponding numbers of retained SNPs with these MAF thresholds were 140 853, 128 972 and 107 185 SNPs, respectively. Supplementary Material, Table S2 summarizes the QC steps and the number of SNPs and samples that were retained.
For a comparison of results with data from GWAS (38,39), we used HapMap3 imputed data provided by the same consortium. Imputation was undertaken in cohort sets; there were 6 imputation cohorts for CD and 7 for UC. The total number of SNPs was 1 253 071 and 1 253 093 for CD and UC, respectively. After further QC (imputation R2 > 0.6 and MAF > 0.01 for each of the 13 imputation cohorts), the numbers of SNPs that remained were 1 008 060 and 1 047 568 for CD and UC, respectively. The number of GWAS samples was 16 550 (5054 cases and 11 496 controls) and 22 000 (5799 cases and 16 201 controls) for CD and UC, respectively. See Supplementary Material, Table S3 for a summary of the GWAS data. The imputed GWAS datasets for CD and UC had 987 572 SNPs in common, and these were used to estimate relationships across CD and UC case and control samples. One individual per pair was excluded, when the estimated relatedness was >0.05 resulting in sample sizes in the joint CD and UC analyses of 10 898 (4813 cases and 6085 controls) and 17 047 (5768 cases and 11 279 controls) for CD and UC, respectively.
The individuals included for study were all of European descent in both the iChip and GWAS cohorts. The iChip cohort includes many samples from the GWAS cohort (12). We identified samples in common using the genotype data (Supplementary Material, Table S4). In order to investigate the effect of inclusion or exclusion of the individuals in both datasets, we conducted bivariate analysis (24) with and without the sample of overlapping individuals for the iChip data, and the results differed only slightly (Supplementary Material, Table S5). In order to maximize statistical power, we consequently kept all the individuals with iChip data for the study.
SNP array and sample characteristics
The effective number of SNPs for the iChip was estimated in two ways. Firstly, we used a simulation method (25,40) and assigned a phenotype randomly generated from the standard normal distribution for 5000 randomly sampled individuals, performed a GWAS using the actual genotypes and summed the χ2 1-df test statistic for association across all M SNPs If the markers are in linkage equilibrium then and If the SNPs are not in linkage equilibrium then the variance of the test statistics will be higher, i.e., The simulation was replicated 1000 times. The effective number of marker was calculated as .
Secondly, we estimated Me from genetic relatedness between each pair of individuals (6), estimated across the genome as with x the number of reference alleles (0, 1, 2) at an SNP, p the frequency of x and q = 1 − p. For a random sample of conventionally unrelated individuals in the population, the variance of the pairwise relatedness estimates is approximately 1/Me (6,41), and therefore an estimate of the effective number of markers is 1/var(A). These two methods are equivalent and the reciprocal of the sampling variance of the genetic relatedness is the mathematical expectation of Me in the simulation method.
The iChip is a custom-built chip comprising a small core grid of nearly 20 000 backbone SNPs, plus high-density fine-mapping SNPs in distinct loci identified as containing markers reaching genome-wide significance in GWAS of a number of immune-related diseases (including CD and UC). These loci included variants identified in the 1000 Genomes Project low-coverage CEU population, resulting in dense spiking of SNP density in some regions. To study the iChip enriched regions in the GWAS data, we split each chromosome into 0.6 Mb bins, and calculated the iChip SNP density in each bin relative to the average on the chromosome. We then selected the bins with a density of 20% larger than the average density on that chromosome.
Genetic analyses
Estimation of the variance components for CD and UC
Only individuals and SNPs that passed quality control were used in estimating variance components for the two diseases. The genetic relationship between individuals was estimated three ways: (i) using all SNPs (for whole-genome estimation as in Table 1), (ii) using SNPs inside selected and complement regions (for genome partitioning analysis, Tables 3) and (iii) using SNPs on each of the autosome (joint analysis for 22 autosomes, Supplementary Material, Fig. S2 and S3). A linear mixed model was used to estimate the genetic variance associated with SNPs (SNP-heritability). As the estimated SNP-heritability was on the observed case–control risk scale, it was subsequently transformed to the scale of liability, as described previously (24,26). For this transformation we assumed a population prevalence of 0.005 and 0.0025 for CD and UC, respectively, based on the prevalence observed in IIBDGC (Luke Jostins, personal communication). For the iChip data, we estimated variance components fitting the top 10 principal components (PCs) that were calculated and supplied by IIBDGC.
For the imputed GWAS data, PCs were estimated from the data. For the estimation of variance components in the GWAS data, the fixed effects of the overall mean (intercept), sex, cohort and the first 20 ancestry PCs were fitted. In a previous GWAS study (12) that used the same data (but an earlier version), the top 4 PCs were fitted for the iChip data, and 10 and 7 PCs for the imputed GWAS data. In this study, we aimed to be more conservative by adjusting the data with more PCs.
To quantify the variation in liability explained in the GWAS data by the same regions that were densely genotyped in the iChip data, variance components were also estimated for SNPs in the GWAS data that were in the selected bins from the iChip data. Three groups were defined for analysis according to the type of genomic region: type 1, comprised genomic bins that attained a particular level of SNP density in the iChip—at least 20% larger than the average of the chromosome where a bin was located, and we refer to the selected region as a high-density (HD) region; in Type 2, comprised bins that contained GWAS hits reported by Jostins et al. (12), and we refer to the selected region as a GWAS-hit region; and Type 3, comprised bins that overlapped both Type 1 and 2 selected bins, and we refer to the selected region as a HD-GWAS region. The remaining genomic regions that did not meet the above criteria are referred to as complement regions.
Genetic variance partitioned by MAF and by chromosome
We estimated multiple genetic variance components simultaneously by grouping SNPs into MAF bins, as previously described (36). For iChip data, the entire MAF spectrum was partitioned into five intervals for the 140 853 SNPs. MAF bins were defined as 0.001–0.1, 0.1–0.2, 0.2–0.3, 0.3–0.4 and 0.4–0.5. Similar MAF bins were defined for the GWAS data but with the MAF of the first bin ranging from 0.01 to 0.1. The number of SNPs included in each bin was tabulated in Table 2.
Genetic-relatedness matrices (GRMs) were constructed for each autosome and genetic variance for each chromosome was estimated in an analysis in which all chromosomal GRMs were fitted jointly. As joint analysis of all autosomes is computationally intensive, since it requires 22 times the amount of virtual memory, we split the iChip data randomly into two sets: 19 650 and 19 686 individuals for CD analyses, and 17 969 and 17 945 individuals each for UC analyses. For each disease, the estimates of heritability on the liability scale in the two datasets were averaged.
Genetic relationship between CD and UC
We estimated a genetic correlation in risk of CD and UC by fitting a bivariate linear mixed model, as described previously (24,36). For the iChip data, control samples were randomly allocated to CD and UC, but in proportion to the number of cases, and the first 10 PCs were fitted as covariates. This analysis gives a simultaneous estimate of the SNP-heritability of liability to both diseases and an estimate of the SNP-genetic correlation between these liabilities. For the GWAS dataset, 4813 CD cases and 6085 controls and 5768 UC cases and 11 279 controls were used in the bivariate analysis. The same covariates (sex, imputation cohort and 20 PCs) were fitted as for the univariate analyses.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Australian National Health and Medical Research Council (613672 and 1048853 to P.M.V., 1011506 and 1047956 to N.R.W., 1028569 to G.R.S.), the Australian Research Council (DE130100614 to S.H.L.), the Fondation Leducq (FLQ CDA02 to M.J.B.) and the National Institutes of Health (GM099568, GM075091, and MH100141 to P.M.V.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the two anonymous reviewers for their constructive comments which have greatly improved the article.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Wellcome Trust Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Li Y., Browning S.R., Browning B.L., Slater A.J., Kong X., Aponte J.L., Mooser V.E., Chissoe S.L., Whittaker J.C., et al. Performance of genotype imputation for rare variants identified in exons and flanking regions of genes. PLoS One. 2011;6:e24945. doi: 10.1371/journal.pone.0024945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mägi R., Asimit J.L., Day-Williams A.G., Zeggini E., Morris A.P. Genome-wide association analysis of imputed rare variants: application to seven common complex diseases. Genet. Epidemiol. 2012;796:785–796. doi: 10.1002/gepi.21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voight B.F., Kang H.M., Ding J., Palmer C.D., Sidore C., Chines P.S., Burtt N.P., Fuchsberger C., Li Y., Erdmann J., et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong J., Schumacher F., Lim U., Hindorff L.A., Haessler J., Buyske S., Carlson C.S., Rosse S., Bůžková P., Fornage M., et al. Fine Mapping and Identification of BMI Loci in African Americans. Am. J. Hum. Genet. 2013;93:661–671. doi: 10.1016/j.ajhg.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A., Brown M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes A., Hadler J., Pointon J.P., Robinson P.C., Karaderi T., Leo P., Cremin K., Pryce K., Harris J., Lee S., et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jostins L., Ripke S., Weersma R.K., Duerr R.H., Mcgovern D.P.B., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J.Z., Almarri M.A., Gaffney D.J., Mells G.F., Jostins L., Cordell H.J., Ducker S.J., Day D.B., Heneghan M.A., Neuberger J.M., et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J.Z., Hov J.R., Folseraas T., Ellinghaus E., Rushbrook S.M., Doncheva N., Andreassen O.A., Weersma R.K., Weismuller T.J., Eksteen B., et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat. Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., Ding J., Li Y., Tejasvi T., Gudjonsson J.E., et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinks A., Cobb J., Marion M.C., Prahalad S., Sudman M., Bowes J., Martin P., Comeau M.E., Sajuthi S., Andrews R., et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 2013;45:664–669. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre S., Bowes J., Diogo D., Lee A., Barton A., Martin P., Zhernakova A., Stahl E., Viatte S., McAllister K., et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper J.D., Simmonds M.J., Walker N.M., Burren O., Brand O.J., Guo H., Wallace C., Stevens H., Coleman G., Franklyn J.A., et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tysk C., Lindberg E., Jarnerot G., Floderus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halfvarson J., Bodin L., Tysk C., Lindberg E.V.A. Inflammatory Bowel Disease in a Swedish Twin Cohort: A Long-Term Follow-up of Concordance and Clinical Characteristics. Gastroenterology. 2003;124:1767–1773. doi: 10.1016/s0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 21.Halfvarson J. Genetics in twins with Crohn's disease: less pronounced than previously believed? Inflamm. Bowel Dis. 2011;17:6–12. doi: 10.1002/ibd.21295. [DOI] [PubMed] [Google Scholar]
- 22.Thompson N.P., Driscoll R., Pounder R.E., Wakefield A.J. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996;312:95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orholm M., Binder V., Rasmussen L.P., Kyvik K.O., Hospital E. Concordance of inflammatory bowel disease among Danish Twins. Results of a nationwide study. Scand. J. Gastroenterol. 2000;35:1075–1081. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 24.Lee S.H., Yang J., Goddard M.E., Visscher P.M., Wray N.R. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brant S.R. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm. Bowel Dis. 2011;17:1–5. doi: 10.1002/ibd.21385. [DOI] [PubMed] [Google Scholar]
- 28.Duclos B., Dupas J.L., Galmiche J.P., Gendre J.P., Golfain D., Gra C., Malchow H., Lachaux A., Lautraite H., Lenaerts C., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 29.Hugot J., Chamaillard M., Zouali H., Lesage S., Ce J., Macry J. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 30.Hunt K.A., Mistry V., Bockett N.A., Ahmad T., Ban M., Barker J.N., Barrett J.C., Blackburn H., Brand O., Burren O., et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–235. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan P.F., Daly M.J., O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray N.R., Lee S.H., Kendler K.S. Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. Eur. J. Hum. Genet. 2012;20:668–674. doi: 10.1038/ejhg.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.H., Yang J., Chen G.-B., Ripke S., Stahl E.A., Hultman C.M., Sklar P., Visscher P.M., Sullivan P.F., Goddard M.E., et al. Estimation of SNP heritability from dense genotype data. Am. J. Hum. Genet. 2013;93:1151–1155. doi: 10.1016/j.ajhg.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speed D., Hemani G., Johnson M.R., Balding D.J. Response to Lee et al.: SNP-Based Heritability Analysis with Dense Data. Am. J. Hum. Genet. 2013;93:1155–1157. doi: 10.1016/j.ajhg.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S.H., Decandia T.R., Ripke S., Yang J., Sullivan P.F., Goddard M.E., Keller M.C., Visscher P.M., Wray N.R. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S.H., Wray N.R. Novel genetic analysis for case-control genome-wide association studies: quantification of power and genomic prediction accuracy. PLoS One. 2013;8:e71494. doi: 10.1371/journal.pone.0071494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lips E.S., Cornelisse L.N., Toonen R.F., Min J.L., Hultman C.M., Holmans P.A., O'Donovan M.C., Purcell S.M., Smit A.B., Verhage M., et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol. Psychiatry. 2012;17:996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goddard M. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica. 2009;136:245–257. doi: 10.1007/s10709-008-9308-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.