Abstract
Throughout the 20th century a body of literature concerning the long lasting effects of early environment was produced. Adverse experiences in early life, or early life stress (ELS), is associated with a higher risk for developing various psychiatric illnesses. The mechanisms driving the complex interplay between ELS and adult phenotype has baffled many investigators for decades. Over the last decade, the new field of neuroepigenetics has emerged as one possible mechanism by which ELS can have far reaching effects on adult phenotype, behavior, and risk for psychiatric illness. Here we review two commonly investigated epigenetic mechanisms, histone modifications and DNA methylation, and the emerging field of neuroepigenetics as they relate to ELS. We discuss the current animal literature demonstrating ELS induced epigenetic modulation of gene expression that results in altered adult phenotypes. We also briefly discuss other areas in which neuroepigenetics has emerged as a potential mechanism underlying environmental and genetic interactions.
Keywords: epigenetics, stressful events, physiological stress reactivity, early life stress, life stress, psychological stress, neuroepigenetics, maternal separation
Introduction
Visionary psychologists of the 20th century produced findings that prompted a movement of research demonstrating that early life experiences help shape an individual’s development into maturation. At the turn of the 20th century, pioneering psychologist Gates (1904) referred to early life experiences mediating brain development as a “process of brain-building”. A few decades later, Hebb (1947) found that cognitive performance in rats was enhanced by different, particularly more enriched, rearing environments. The next decade produced literature from Dennenberg’s lab demonstrating a lasting neuroendocrine effect of early life handling in rodents (Levine et al., 1967). Around the same time, Harlow demonstrated that the quality of maternal care had long lasting detrimental effects on behavior in non-human primates (Harlow et al., 1965). Meaney (1985) elaborated on Dennenberg’s findings by establishing a relationship between early handling and adult hippocampal glucocorticoid receptor expression. Meaney expanded upon this work in a seminal paper (Plotsky and Meaney, 1993) in which a group with extended maternal separation was added. Soon afterwards, it was discovered that maternal care, specifically licking, grooming, and arch back nursing (LG or LG-abn), was driving the differences seen in handling groups (Liu et al., 1997). The majority of rodent paradigms modeling early life stress used today are modified versions of paradigms based on this collection of work.
Exposure to early life stress (ELS) such as traumatic events, abuse, and neglect is highly related to health in adulthood. Multiple variables within the home such as parenting style and the quality of the parent/child relationship can affect a child’s physical and intellectual development (Anda et al., 2006; Bremne & Vermetten, 2001; Lovallo et al., 2012; Schimmenti & Bifulco, 2013). For example, in individuals with a history of ELS, there is an increased risk for the development of physical illnesses such as obesity, diabetes, seizures, and cardiovascular diseases (Gluckman et al., 2008; Gunstad et al., 2006; Huang, 2014). Furthermore, ELS is a risk factor for the development of various psychiatric illnesses such as personality disorders, major depression, post-traumatic stress disorder, addictive disorders, and schizophrenia (De Bellis, 2002; Pechtel & Pizzagalli, 2011; Teicher et al., 2003). Additionally, aggressive and suicidal behaviors are strongly associated with ELS (McEwen, 2003). It is clear that the consequences of ELS are highly detrimental to the individual and his/her family, and affect societies worldwide (De Bellis & Zisk, 2014).
Understanding the neural underpinnings of the relationship between ELS and the mental health of the victims is of great value in the pursuit of possible treatment and intervention. One way to achieve this is via investigation using animal models. In the rat, it is well known that ELS is profoundly impactful because neurological development is plastic during this time. In one example, rat pups undergo a stress hyporesponsive period (SHRP) during the first two weeks of life that is considered necessary for normal neurodevelopment (Levine, 1994). The critical period of the two week SHRP is the common time point in which ELS is induced in rodent research. There may be a similar sensitive period in human infants (Gunnar & Donzella, 2002). Preclinical studies show that stress hormones are abnormally elevated during the SHRP when maternal care is disrupted. Exposing the developing brain to stress and stress hormones during the SHRP period causes a multitude of neurobiological changes. Rats exposed to stress during the SHRP demonstrate alterations in monoaminergic systems, as well as the primary excitatory and inhibitory neurotransmitter systems and neuroendocrine stress responses in adulthood (Meaney, 2001).
The deleterious effects of ELS on neurodevelopment are abundant, yet many questions have gone unanswered for decades. How does an experience early in life effectively change the development of a complex system such as the brain? Why do some but not all who experience ELS develop illnesses in adulthood? Recent evidence has suggested epigenetics as a possible mechanism to answer these and many other questions. One of the leaders of the field, Sweatt (2013), provided a graphical representation of the increase in epigenetics research over the last 15 years in an eloquent discussion of the topic. Within this influx of research is an impressive body of literature demonstrating that DNA methylation and histone modifications in neuronal cells are modulated by experiences, both in early life and adulthood. This new field, referred to as ‘neuroepigenetics’ (Sweatt, 2013) or ‘neurobehavioral epigenetics (Lutz & Turecki, 2013), indicates that a myriad of external factors including nutritional status, education and socioeconomic status (SES), exposure to environmental toxins and drugs of abuse, withdrawal from chronic drug intake, and stress can all influence epigenetic regulators of gene expression in the central nervous system. Gene expression in turn influences behavior, phenotypes, and vulnerability to various psychiatric illnesses. This interactive view of behavior was championed by Kuo (1967) when he criticized the commonly narrow interpretations and grossly over-simplified generalizations of behavior by different schools of psychology. He particularly stressed this issue when evaluating social species, arguing that behavior is a dynamic functional product of a bidirectional relationship between organism and the environment.
In this article we review the recent literature concerning the effects of ELS on the epigenome later in life. The concept that our adult genetic profile is shaped in part by earliest experiences is an innovative paradigm shift from traditional views of neurodevelopment (Sweatt, 2013). It also addresses the long-standing debate of nature versus nurture in the field of psychology; it is no longer nature versus nurture, but now it is undeniably nature and nurture that determine the sum of the complex indivizual.
Epigenetic mechanisms
The word “epigenetics” comes from the Greek root ‘epi’ meaning ‘above’, ‘over’, or ‘in addition to’. This refers to the regulation of gene expression outside of genomic information in the DNA nucleotide sequence (Biliński et al., 2012). In 1942, the developmental biologist Waddington introduced this term, and the ‘epigenetic landscape’, to explain the process by which multicellular organisms develop different phenotypes independent of their identical genome (Waddington, 1942). Through activation and repression of specific genes, cells develop distinct phenotypes such that a liver cell is distinct from a muscle cell and so forth. The epigenetic cellular profile is then inherited by the daughter cells and is maintained through cellular division; thus liver cells remain liver cells and muscle cells remain muscle cells for the lifespan of the organism. This traditional definition of epigenetics included ‘heritability’ as a qualifier. In the most recent decades it has become apparent that post-mitotic cells, such as neurons, go through rapid dynamic processes that modulate gene expression (Narayan & Dragunow, 2010; Renthal & Nestler, 2008; Roth et al., 2012; Tsankova et al., 2007). Yet, these epigenetic markers are not heritable either in a procreative or mitotic fashion. Hence the term ‘epigenetics’ and the debate over its accurate definition spans many disciplines (Ho & Burggren, 2010).
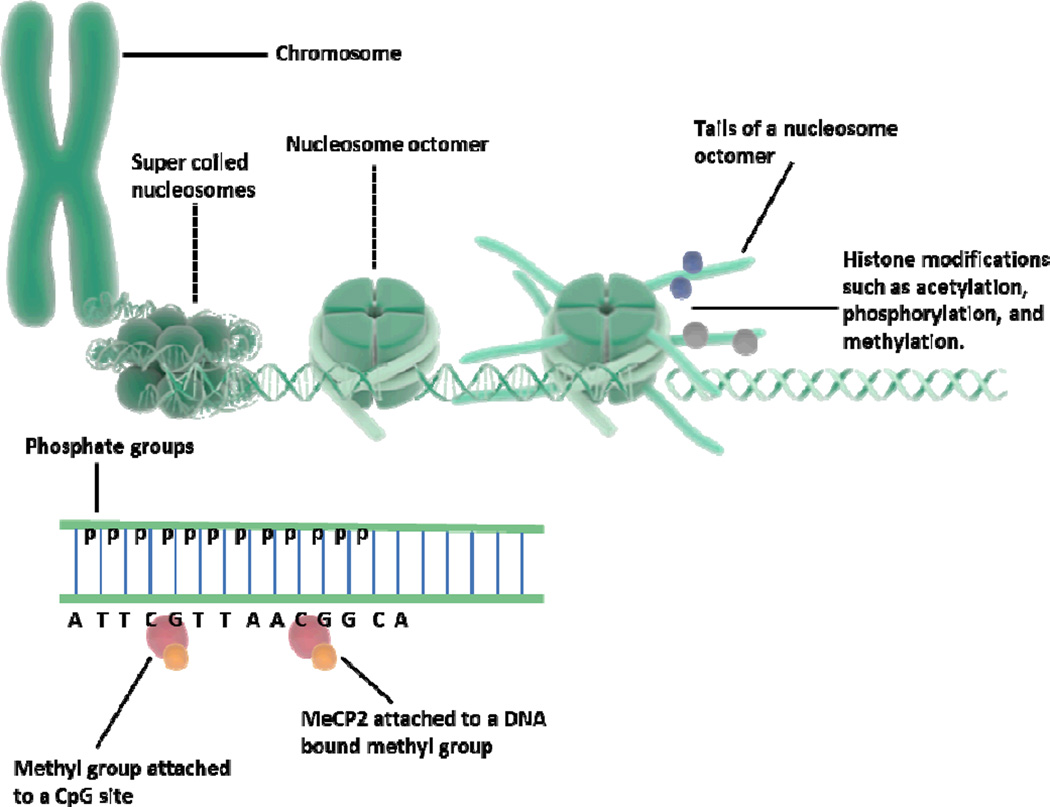
There are numerous epigenetic mechanisms, but the most commonly studied are post-translational modifications to histones and DNA methylation (Figure 1). Other mechanisms such as non-coding RNAs (e.g., microRNAs and others), prion proteins, and histone remodeling, though emerging as relevant processes, are beyond the scope of this review. Briefly described here are the mechanisms underlying post-translational histone modifications and DNA methylation in regulating gene expression. (For readings on other epigenetic mechanisms please see other reviews: Bannister & Kouzarides, 2011; Cohen et al., 2011; Strahl & Allis, 2000; Sweatt, 2013; Zheng & Hayes, 2003).
Figure 1.
Schematic representation of nucleosomes and DNA methylation.
Histone modifications
Eukaryotic DNA is packaged into chromatin that consists of units known as nucleosomes, which are comprised of the DNA double helix wrapping around a protein complex made of eight histones. Chromatin can exist in the form of heterochromatin or euchromatin (Tamaru, 2010). In the heterochromatin state, the DNA is tightly packaged which blocks transcriptional machinery and silences gene expression. In contrast, euchromatin is more loosely packaged, permitting gene transcription. Histone complexes are protein octamers with 2 copies each of histone H3, H4, H2A, and H2B. Histone variants such as H2A.Z and H3.3 have been associated with replacement of the typical histone proteins during transcription and chromatin structure (Henikoff et al., 2004). The double-stranded DNA helix is wrapped around the histone octamer ~1.5 times, equivalent to ~147 base pairs, and histone H1 serves as a connector histone between nucleosomes (Thoma et al., 1979). Histones have protruding N- and C-terminal tails in which specific amino acids undergo covalent post-translational modifications, of which the most commonly studied are methylation, acetylation, and phosphorylation (Bode & Dong, 2005; Kouzarides, 2007; Morales et al., 2001).
Histone modifications regulate chromatin structure in various ways, including the recruitment of remodeling enzymes and altering the overall charge of the histone protein. Histone phosphorylation state is regulated by kinases and phosphatases that add or remove phosphate groups, respectively, significantly altering the overall charge of the histone, which in turn influences chromatin structure. Histone phosphorylation is associated with both transcriptional repression and activation. Histone methylation and acetylation are by far the most studied modifications of these proteins. Histone acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetylation at lysine (K) residues is generally associated with euchromatin and is permissive of gene expression (Bode & Dong, 2005; Kouzarides, 2007; Morales et al., 2001).
Histone methylation was traditionally thought to be a static process, but it is now widely accepted as being dynamic (Bannister & Kouzarides, 2005; Bannister et al., 2002). Histone methyltransferases (HMTs) regulate the complexity of histone methylation; lysine residues can be mono-, di-, or tri-methylated and arginine (R) residues can be mono- and di-methylated (either symmetrically or asymmetrically). Methylation is different from acetylation and phosphorylation since it does not change the overall charge of the nucleosome. Unlike acetylation, methylation is associated with both transcriptional activation and repression, which are dependent on various factors including the specific histone subunit, residue, and methylation state. Furthermore, the overall ‘histone code’ or histone ‘crosstalk’ between modifications and DNA methylation makes for an almost infinite level of possible gene expression regulatory processes (Ben-Porath & Cedar, 2001; Kondo, 2009).
DNA methylation
Most eukaryotic cells contain methylated DNA that is distributed across the genome, referred to as global methylation (Bird, 2002). In mammals, DNA methyltransferases (DNMTs) mostly target a 5’ cytosine (C) adjacent to guanine (G) on the same strand (i.e., not complementary C–G base pairing), which is also referred to as a CpG dinucleotide site, although there are exceptions. Regions of DNA with greater than ~55% CpG site content are referred to as CpG islands and tend to appear around promoter regions, with ~40% of genes having CpG islands in their promoter sequences (Miranda & Jones, 2007). It is estimated that the human genome has ~29,000 CpG islands (Bird, 2002). Methylation of CpG sites is associated with gene silencing in cell fate determination, genomic imprinting, and X chromosome inactivation, although many genes with unmethylated CpG sites can also be silenced. DNA methylation is stable and inherited by daughter cells after division even when histone modifications are not, thus maintaining the same gene expression patterns that lead to cell type determination and perpetuation. DNA methylation represses gene transcription on a multitude of levels: by inhibiting transcription factor access; by attracting methylcytosine-binding proteins (MBDs), one of the most commonly studied being methyl-CpG-binding protein 2 (MeCP2); and by recruiting protein complexes that contain other repressors and chromatin remodelers such as HDACs and HMTs (Jones et al., 1998). DNA methylation may also influence nucleosome occupancy over promoter sites and prevent gene activation (Miranda & Jones, 2007). The notion that DNA methylation patterns in the central nervous system and germ cells are highly dynamic is now widely accepted (Bohacek et al., 2013; Ho & Burggren, 2010; Jiang et al., 2008; Kovalchuk, 2012; Miller & Sweatt, 2007; Roth et al., 2009). Methylation patterns may be “rapid and reversible” by various mechanisms, allowing for previously methylated CpG sites to become de-methylated and vice versa. (For reviews on several proposed mechanisms of active DNA demethylation please see: Klengel et al., 2014; Ooi & Bestor, 2008).
Animal models of ELS
There is abundant literature investigating the lasting neurobiological and behavioral effects of ELS during the SHRP in rodents. Within this body of literature, there are multiple paradigms, including maternal separation, maternal deprivation, naturally occurring low levels of maternal behavior, and brief exposure to an abusive female. In this review, we will discuss the literature derived from three primary rodent models of ELS. First, within the maternal separation/deprivation paradigms, modeled after Plotsky and Meaney’s (1993) original paradigm, there is an unfortunate amount of variability in various parameters, including manipulation of litter composition, cross-fostering, number of days of separation, time and length of separation, separation environment, day of weaning, and housing conditions during adolescence. The appropriate control group to use has also received considerable debate (Matthews et al., 1999, 2001). In general, the adult phenotype of pups exposed to ELS consists of a hyperactive HPA axis, increased anxiety-like and depressive behaviors, increased drug intake, a compromised immune system, and alterations in monoaminergic systems (Ladd et al., 2000; Lippmann et al. 2007; Meaney, 2001). Second, Meany and colleagues went on to postulate that the change in adult offspring phenotype after maternal separation was due to a change in maternal behavior (Liu et al., 1997). This spurred an investigation into the natural variation of maternal care, which led to another paradigm to investigate the effects of early life experiences. In this paradigm, maternal care is coded daily in order to define which litters receive high maternal care in the form of licking, grooming, and arch-back nursing (designated “H-LG” for high licking and grooming) or low maternal care (designated “L-LG” for low licking and grooming) (Liu et al., 1997). Adult offspring receiving L-LG maternal behavior develop similar phenotypes as pups exposed to separation/deprivation procedures. A third, more recently developed ELS paradigm, pioneered by Roth et al. (2009), exposed pups to a stressed female who displayed abusive behaviors (discussed in more detail below). For descriptions of other less frequently used ELS rodent paradigms, please see Lutz and Turecki (2013). A consistent phenomenon across these three paradigms is the disruption of maternal care during the SHRP (see above), followed by the evaluation of effects in adulthood. Since much of this literature focuses on the effects of ELS on the hypothalamic-pituitary-axis (HPA) stress response, we will briefly review the neurobiology of this system.
The HPA response is initiated by either a real or perceived stressful stimulus activating the cells in the paraventricular nucleus (PVN) of the hypothalamus that project to various limbic brain regions. The parvocellular projections of the PVN secrete corticotrophin-releasing factor (CRF, or sometimes CRH for corticotrophin-releasing hormone) and arginine vasopressin (AVP) via the median eminence into the anterior pituitary. CRF and AVP in the anterior pituitary stimulate the synthesis and release of adrenocorticotropic hormone (ACTH) into circulation. ACTH in the blood stream acts on the adrenal cortex to initiate the synthesis and release of glucocorticoids (GC) (primarily cortisol in humans and corticosterone in rodents, collectively referred to as CORT) into the circulation. GCs in the systemic circulation then act upon cytosolic and plasma membrane receptors, most notably glucocorticoid and mineralocorticoid receptors (the latter also known as aldosterone receptors). GCs regulate the termination of the stress response via a negative feedback loop both directly and indirectly in the central nervous system.
We will now review the current literature that has utilized these paradigms to investigate epigenetics as a mechanism by which ELS influences adult phenotype and psychiatric vulnerability (Table 1). Additionally, we will briefly survey recent human and non-human primate studies that complement this work.
ELS and epigenetic effects
Glucocorticoid receptor (Nr3c1)
The most extensively studied effect of ELS and epigenetics is of the GC receptor gene (nuclear receptor subfamily 3, group C, member 1, or Nr3c1). In a ground-breaking study by Weaver et al. (2004) it was shown that variation in maternal care altered methylation of exon 17 of the GC receptor (GR) promoter in hippocampal neurons. More specifically, maternal care was associated with variations in methylation of a specific CpG site in which the immediate early gene nerve-growth factor inducible protein A (NGFI-A) binds DNA as a transcription factor, altering GR expression levels in the hippocampus. A change in GR expression then influences HPA axis function and the negative feedback loop that plays a role in terminating the stress response (see above). This initial research was highly influential because it demonstrated for the first time a relationship between ELS-altering epigenetic markers that in turn alter protein expression that directly affects neurophysiological systems that are implicated in psychiatric disorders. Multiple laboratories have continued research investigating the effects of ELS on epigenetic regulation of the Nr3c1 gene. This avenue of research continues to be promising, although some have failed to replicate Weaver’s initial findings (Daniels et al., 2009; Kember et al., 2012). This could be due to different ELS paradigms, or different rodent species or strains utilized. As others have demonstrated, the effects of ELS on epigenetic regulation of the GR gene are complex and dependent on multiple factors, including the strain genetic background, litter composition, sex, cell type, and brain region of interest (Kember et al., 2012; Kosten et al., 2013; Kundakovic et al., 2013; Liberman et al., 2012; McGowan et al., 2011). Interestingly, acute and chronic stress in adult rats does not mimic the change in methylation patterns of the Nr3c1 gene as observed after ELS, demonstrating the importance of the developmental window in this phenomenon (Witzmann et al., 2012).
The similar adult phenotypes observed in rodents and humans with a history of ELS has long suggested that the underlying mechanisms may be conserved across species. Accordingly, McGowan and colleagues were the first to investigate a relationship between ELS and epigenetic regulation of the GR gene in humans, assessing methylation of Nr3c1 exon 1F, which is the human homolog of rat exon 17 (McGowan et al., 2009). Postmortem hippocampal tissue from suicide completers with a history of abuse was evaluated. Increased DNA methylation was found at specific CpG sites in exon 1F of the Nr3c1 promoter in suicide victims with a history of ELS, compared to controls and suicide victims without a history of abuse. The increased DNA methylation was related to decreased rates of GR expression. Other groups have extended this work by demonstrating a relationship between ELS and methylation status of the Nr3c1 gene, with both increased and decreased methylation states for different GR transcripts (Labonté et al., 2012; Perroud et al., 2011; Steiger et al., 2013; Suderman et al., 2012; Tyrka et al., 2012). The functional impact on the HPA axis and adult behaviors of these epigenetic alterations begs further investigation. While the previous data concerning ELS epigenetically modulating the GR gene is intriguing, McGowan et al. (2011) argued for the importance of broadening the investigative scope of this work, as ELS affects numerous systems and behaviors into adulthood. In this latter study, a ~7 million base pair locus around the GR gene in the rat hippocampus was examined, and findings revealed that maternal care influenced DNA methylation in non-promoter regions in non-random, gene-specific and bi-directional manners.
AVP
AVP is a hormone involved in HPA regulation, and also in depression (London et al., 1997). Murgatroyd et al. (2009) found that mice exposed to ELS had decreased methylation of the Avp enhancer and increased AVP expression in the PVN as adults. It is likely that the decrease in methylation in adulthood was due to reduced MeCP2 occupancy at the Avp enhancer (see above) during ELS. ELS activates HPA circuits that trigger phosphorylation of MeCP2 and its release from methylated DNA. This leads to lifelong hypo-methylation and overexpression of AVP in the PVN. This functional consequence of epigenetic regulation was associated with a hyperactive stress response as it was partially reversed by treatment with an AVP V1b receptor antagonist. Additionally, ELS modulates methylation patterns of the Avp gene in the hippocampus (Kember et al., 2012) and DNA methylation of other HPA regulatory genes. For example, increased mRNA expression of the pituitary pro-opiomelanocortin (Pomc) gene after ELS was associated with a decrease in DNA methylation (Wu et al., 2014). Clearly, the interaction between ELS and epigenetic markers is exceedingly complex, dynamically modulating the HPA axis via multiple avenues in multiple brain regions throughout development.
BDNF
Brain derived neurotrophic factor (BDNF) is a protein belonging to the neurotrophin family of growth factors, and its gene Bdnf is another affected by ELS. Roth et al., (2009) used a novel paradigm targeting the effects of abuse during postnatal days 1 – 7. Rat pups were exposed to a stressed female that displayed multiple abuse behaviors, including rough physical contact and active avoidance. In adulthood, these rats showed increased DNA methylation of the BDNF gene in the prefrontal cortex (PFC) and decreased BDNF expression. The DNA methylation status and expression levels of BDNF in the PFC were rescued by chronic intracerebroventricular treatment with a DNMT inhibitor. BDNF has emerged as having somewhat of a ubiquitous role in brain functioning, and is implicated in various diseases, including depression, stroke, Alzheimer’s, and addictive disorders (Biliński et al., 2012; Nagahara & Tuszynski, 2011). Thus, evidence that ELS can trigger BDNF expression changes in adulthood has immense implications concerning vulnerability towards the development of these disorders.
5-HT (Slc6a4)
The serotonin system is also highly involved with emotion regulation and various psychiatric illnesses such as major depression, autism, and addictive disorders (Uaswani et al., 2003). Variation in the serotonin transporter (5-HTT) gene (solute carrier family 6, subfamily a, member 4, Slc6a4) moderates individual reactions to stress and potential risk for development of psychiatric illness (Caspi et al., 2003; Kendler et al., 2005), and determines clinical responsiveness to certain antidepressants (Kim et al., 2000). ELS influences the development of the serotonergic system, and this regulation may be epigenetically determined (Beach et al., 2010; Vijayendran, Beach et al., 2012). In non-human primates, higher levels of methylation of the Slc6a4 gene has been associated with higher stress reactivity in females exposed to ELS but not controls (Kinnally et al., 2011). Another study conducted in adopted humans demonstrated that DNA methylation levels and the specific 5HTT allele (long or short) interact to influence psychological coping with loss and trauma (IJzendoorn et al., 2010). These data highlight the complex interaction of genetics, epigenetics, and ELS in shaping individual characteristics.
GABA and glutamate
ELS also affects the major excitatory and inhibitory neurotransmitter systems, glutamate and GABA, respectively (Bagot et al., 2009; Meaney, 2001) in part via epigenetic mechanisms. Low maternal care was associated with an increase in DNA methylation of the promoter region of the GAD1 gene in the hippocampus of adult rats (Zhang et al., 2010). The GAD1 gene codes for glutamic acid decarboxylase (67 kDa isoform), the rate limiting enzyme of GABA synthesis. Methylation status of the GAD1 promoter in postmortem tissue samples has been associated with schizophrenia (Zhang et al., 2010), implicating a potential mechanism by which ELS may confer vulnerability to this disease. These findings, along with those showing that ELS induced long-term alterations in GABAA and central benzodiazepine (CBZ) receptor subunit profiles in the hippocampus (Meaney, 2001), paint an intriguing picture as to the wide scope in which ELS can modify brain systems. Furthermore, recent studies from Bagot et al. (2012), demonstrate that ELS epigenetically affects hippocampal long-term-potentiation and depression through regulation of a glutamate receptor gene. High maternal care was associated with a decrease in DNA methylation and increased levels of histone 3 lysine 9 acetylation and histone 3 lysine 4 trimethylation of the gene coding for the type 1 metabotropic glutamate (mGluR1) receptor (Grm1) in the hippocampus of adult offspring. Accordingly, these animals displayed an increase in both mRNA and protein levels of the mGluR1 receptor (Bagot et al., 2012). Collectively, these data implicate epigenetic regulation as a mechanism by which ELS can have long lasting effects on excitatory and inhibitory amino acid neurotransmission. Given the ubiquitous distribution of GABA and glutamate systems across the brain, the ability of ELS to epigenetically modulate their development and function has profound and long reaching implications for resultant adult phenotypes.
Epigenetic machinery
ELS can also alter the expression of epigenetic machinery. For example ELS was shown to decrease mRNA levels for multiple HDACs in the cortex in a strain-dependent manner (Levine et al., 2012). This decrease in HDAC expression was accompanied by increased acetylation of H4, and altered emotional phenotypes and responsivity to an antidepressant. Other labs have also demonstrated a relationship between ELS and HDAC expression (Tesone-Coelho et al., 2013). Additionally, ELS decreased nucleus accumbens DNMT expression rates, which were associated with hyper-methylation of neuronal plasticity genes (Anier et al., 2013). It is thus apparent that ELS modulates the epigenome by numerous mechanisms. These alterations can persist into adulthood and regulate gene expression via bi-directional changes in histone acetylation, DNA methylation, and gene expression in both promoter and non-promoter regions (McGowan et al., 2011).
Thus far, we have discussed the current literature investigating the long term epigenetically mediated consequences of ELS exposure. These early experience-driven epigenetic markers have the capacity to modulate neuronal gene transcription. Consequent changes in gene expression have been demonstrated to functionally alter multiple neuroendocrine and neurotransmitter systems. In turn, these alterations influence and affect behavior, future responses to various environments, and vulnerability towards various psychiatric illnesses. Although this intriguing body of literature supports the notion that epigenetic modifications encode ELS into the epigenome, it is certain that the story is much more complex. The diversity in biologically inherited alleles, post-early life environment, and multiple other factors not yet fully understood, are all integral players in the conglomerate human psyche (Klengel et al., 2014). Below, we will briefly discuss some other areas in which the role of environment and epigenetic interactions are becoming apparent in shaping individual differences in development. This is by no means a comprehensive list: there are many other relevant researcg areas, such as nutrition, aging, environmental toxins, and memory, which are beyond the scope of the current review (for reviews on these topics, see Huidobro et al., 2013; Landry et al., 2013; Wang et al., 2012).
Prenatal stress
An emerging body of literature demonstrates that in utero exposure to stress may influence the epigenome of the fetus. One study investigated the HPA stress response and DNA methylation status of the Nr3c1 gene in human infants whose mother was depressed only during the third trimester, and found methylation levels to be positively correlated with maternal depression and predicted higher HPA stress response at 3 months of age (Oberlander et al., 08). Many other reports have echoed the notion that maternal stress during pregnancy influences the methylation status of the GR gene in offspring (Hompes et al., 2013; Mulligan et al., 2012; Radtke et al., 2011). (For a more in depth summary of the literature relating to this topic please see: Hao & Metz, 2013; Lutz & Turecki, 2013).
Transgenerational epigenetics
The field of ‘transgenerational epigenetic inheritance’ can be misleading due to interchangeable use of the term ‘transgenerational’ for two different empirical questions. The first of these investigates the epigenetic inheritance via early behavioral/social interactions with maternal care that in turn regulate the female offspring’s style of maternal care. Cross-fostering studies demonstrate that these effects are completely independent of the germ line, and thus are not permanent and need to be reinstated through maternal interaction with each subsequent generation (Bohacek et al., 2013). This field is easily observed in animal models and is an established and accepted epigenetic phenomenon. The second field investigating the heritability of epigenetic markers within the parental germ cells, however, has unique challenges. Traditionally, it has been thought that epigenetic markers are reset or erased during gametogenesis, but recent evidence has elucidated multiple mechanisms in which epigenetic markers may be conserved in the embryo (Bohacek et al., 2013). Isolating germ line inheritance is still challenging as it is difficult to parse out the in utero environment from its influences on the epigenome of the fetus. The idea that experiences may change epigenetic markers in germ cells, and thus the epigenetic makeup that is passed on to future generations, is nothing short of ground-breaking. Research in this field is very new and only starting to uncover the possibilities.
While the second form of transgenerational research is still scarce, it is a growing field. An experiment of note from Vassoler et al. (2013) demonstrated that voluntary cocaine administration of male rats increased histone H3 acetylation with Bdnf promoter in their sperm. Offspring of these sires had an increase of cortical BDNF expression and a cocaine-resistant phenotype. Additionally, ELS has also been shown to modulate the promoter methylation status of several genes investigated in the germline of males (Franklin et al., 2010).)For a review of the current literature and further discussion of this area please see: Bohacek et al., 2013; Ho & Burggren, 2010; Kovalchuk, 2012; Saab & Mansuy, 2014).
Adult stress
Although epigenetic influences on neuronal plasticity are highest early in development, stress during adulthood also influences the epigenome and subsequent gene expression (Witzmann et al., 2012). Klengel et al. (2014) provides an impressive summary of research that relates DNA methylation status of target genes with psychiatric disorders. The adaptations of the adult epigenome in response to stress is stressor-, duration-, and tissue-specific. For example, Hunter et al. (2009) found regionally specific alterations in histone H3 methylation following 1, 7, and 21 days of restraint stress in male rats. Additionally, chronic but not acute social defeat stress down regulates HDAC expression in the nucleus accumbens of male rats (Renthal et al., 2007). This work brings to mind the difficulties in predicting the development of psychiatric illnesses at an individual level. The epigenome exists in a state dependent on the history of the organism while in constant flux in response to new experiences. It is therefore no wonder that the depth and mystery of the human brain and psyche have evaded medical understanding for so long!
Central and peripheral cells
Thus far, we have discussed data from animal and human studies in which the epigenetic landscape of neuronal tissue is directly evaluated We have also discussed studies in which the epigenome of peripheral cells was investigated (mostly blood cells). The unknown relationship between tissue- and cell-specific epigenetic markers is a concern that needs further investigation. Although animal studies are invaluable to our scientific understanding of neuroepigenetic processes, the development of non-invasive techniques to study the human epigenome pre-mortem is essential. In order to do so, the relationship between central and peripheral epigenetic landscapes must be more firmly established. Liberman et al. (2012) recently tackled this issue by comparing methylation of the Nr3c1 gene between hippocampal and fecal cells in mice. They found maternal care to correspond with methylation status in hippocampal cells but not fecal cells. Another group investigated ELS in male non-human primates and found that epigenetic markers of cortical cells and T-lymphocytes had a small amount of overlap (Provençal et al., 2012). Furthermore, conserved epigenetic markers across tissue may not have identical functional implications for transcription regulation (Fan & Zhang, 2009). These are just a few of the many immense difficulties surrounding the task of developing techniques and understanding the relationship between the epigenome of central and peripheral cells.
Drugs of abuse
Chronic drug use leads to specific alterations in gene expression across many cell types and brain regions (Robison & Nestler, 2011). These changes in gene expression are thought to drive the changes in brain structure, neuronal function, impaired cognition, and maladaptive behavioral characteristics of addictive disorders. Many laboratories, including those of Nestler and colleagues, have established epigenetic links between drug exposure and subsequent changes in immediate early gene expression (Bibb et al., 2001; Cassel et al., 2006; Kumar et al., 2005; LaPlant & Nestler, 2011; Maze & Nestler, 2011; Renthal & Nestler, 2009; Renthal et al., 2009). Drug-induced epigenetic alterations may be crucial to the process by which a casual intermittent drug use transitions to compulsive habitual drug use in addiction (Nielsen et al., 2012; Schmidt et al., 2013; Taylor et al., 2013). Also, the epigenetic landscape prior to drug exposure may play a role in addiction vulnerability (Deng et al., 2010; Im et al., 2010; Tesone-Coelho et al., 2013). There is thus an interesting and circular relationship, in which the epigenome prior to drug use may predispose the individual to higher addiction vulnerability or resiliency, and the epigenetic response to drugs of abuse further modulates risk and progression towards addiction. Interestingly, despite the established link between ELS and addictive disorders, investigation of ELS-induced epigenetic alterations that predispose an individual to a high risk for addiction is still very much in its infancy (Anier et al., 2013; Lewis et al., 2013; Romano-López et al., 2012; Tesone-Coelho et al., 2013).
Epigenetically based treatments
Pharmacological manipulation of the epigenome has been used in many of the aforementioned studies. Multiple mechanisms for pharmacological modulation exist, including DMT inhibitors, DNMT modulators, HDAC inhibitors, and histone methyltransferases inhibitors (Szyf, 2009). Many of these pharmacological ligands for regulating epigenetic processes are in development for the treatment of cancer: for example, vorinostat and romidepsin are HDAC inhibitors that are used for the treatment of lymphoma. In addition, various drugs currently or previously used to treat psychiatric illnesses are now known to be epigenetic modulators. For example, the mood stabilizer valproic acid, which affects GABAergic transmission as well as voltage-gated sodium and calcium channels, is an HDAC1 inhibitor (Rosenberg, 2007). Certain monoamine oxidase inhibitors (such as tranylcypromine) used as antidepressants inhibit histone demethylases (Lee et al., 2006). The ability to manipulate epigenetic markers pharmacologically opens up the possibility of therapeutic treatment to reverse the adverse effects of ELS. While animal work demonstrates that epigenetic pharmacological intervention has the potential to reverse some of the behavioral effects of ELS (Levine et al. 2012; Weaver et al. 2004, 2005), it is important to note the numerous challenges that exist for use in humans (Narayan & Dragunow, 2010). At the moment, epigenetically-based pharmacological treatment options are limited, as they are not gene-, cell-, or neural-network specific. Although this same non-specificity issue did not prevent the clinical use of traditional psychoactive drugs, the complexity of the epigenome and its alteration by psychopharmacological agents should warrant caution. Clearly, the possible long-term, perhaps transgenerational, effects of altering neuronal gene regulation via psychotherapeutic interventions necessitate further investigation. Moreover, future work may establish epigenetic markers as novel biomarkers for predicting and improving various psychiatric diagnoses (Kolshus et al., 2014; Yehuda et al., 2014). Alternatively, because epigenetic markers are dynamic and respond to experiences, it stands to reason that environmental and behavioral therapies may be developed into epigenetically driven treatment options. For example, both environmental enrichment and chronic mild stress were shown to epigenetically modulate the Crhr1 gene in male rodents genetically predisposed to high or low anxiety (Sotnikov et al., 2014). These data shed light on the possibility of patient-designed therapies in which the genetic predisposition, early life experiences, personality traits, and epigenetic interactions are all considered (LeeRaby & Roisman, 2014). The implications of behavioral intervention epigenetically reprograming neurosystems are nothing short of extraordinary. If ELS epigenetic markers can be reversed through treatment options such as behavioral therapy, mindfulness practices, exercise, or other positive experiences the difficulties of pharmacological intervention may be avoided. Furthermore, this possibility deserves to be noted as a likely paradigm shift in all fields of psychology and psychiatry.
Conclusions
Traditional Waddington epigenetics that was originally developed in the context of developmental biology has now expanded into the new field of neuroepigenetics. It is apparent that the epigenome serves a much broader purpose beyond cell fate determination and propagation. The emerging field of neuroepigenetics departs from the notion that DNA methylation or histone modification are static, and embraces the concept that these phenomena are rapidly dynamic in response to constantly changing external influences. The extraordinary concept of an ever-changing and adaptive epigenome that influences an organism’s behavior and phenotype is now clear. The implications, for various fields from genetics to psychology, of a neuroepigenome that is in constant flux and influenced by events prior to conception, highly pliable during development, constantly changing through the lifespan, and intimately adaptive to a legion of experiences, are nothing short of extraordinary. Lifelong adaptive responses to the environment in the form of a complex interplay between various modes of epigenetic regulation will undoubtedly be a fascinating new frontier in the understanding of human behavior.
Supplementary Material
Footnotes
Conflicts: none declared
Contributor Information
Candace Renee Lewis, Arizona State University, Tempe, AZ, 930 S McAllister Ave, Tempe, AZ 85281, Candace.lewis@asu.edu, Phone: (602) 680 – 8786.
Michael Foster Olive, Arizona State University, Tempe, AZ, 930 S McAllister Ave, Tempe, AZ 85281, Foster.olive@asu.edu, Phone: (480) 727-9557.
References
- Anda RF, Felitti VJ, Bremner JD, Walker JD, Whitfield C, Perry BD, et al. The enduring effects of abuse and related adverse experiences in childhood. A convergence of evidence from neurobiology and epidemiology. Eur Arch Psychiatry Clin Neurosci. 2006;256:174–186. doi: 10.1007/s00406-005-0624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Pruus K, Aonurm-Helm A, Zharkovsky A, Kalda A. Maternal separation is associated with DNA methylation and behavioural changes in adult rats. Eur Neuropsychopharm. 2013;24:459–468. doi: 10.1016/j.euroneuro.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Bagot RC, van Hasselt FN, Champagne DL, Meaney MJ, Krugers HJ, Joëls M. Maternal care determines rapid effects of stress mediators on synaptic plasticity in adult rat hippocampal dentate gyrus. Neurobiol Learn Mem. 2009;92:292–300. doi: 10.1016/j.nlm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Bagot RC, Zhang T, Wen X, Thu T, Nguyen T, Nguyen H, et al. Variations in postnatal maternal care and the epigenetic regulation of metabotropic glutamate receptor 1 expression and hippocampal function in the rat. Proceedings of the National Academy of Sciences. 2012;109:17200–17207. doi: 10.1073/pnas.1204599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Todorov A, Gunter TD, Philibert RA. Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa Adoptee sample. Am J Med Genet B Neuropsychiatr Genet. 2010;153:710–713. doi: 10.1002/ajmg.b.31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Cedar H. Epigenetic crosstalk. Mol Cell. 2001;8:933–935. doi: 10.1016/s1097-2765(01)00399-9. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Biliński P, Wojtyła A, Kapka-Skrzypczak L, Chwedorowicz R, Cyranka M, Studziński T. Epigenetic regulation in drug addiction. Ann Agric Environ Med. 2012;19:491–496. [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Devel. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Inducible covalent posttranslational modification of histone H3. Sci STKE. 2005;281:re4. doi: 10.1126/stke.2812005re4. [DOI] [PubMed] [Google Scholar]
- Bohacek J, Gapp K, Saab BJ, Mansuy IM. Transgenerational epigenetic effects on brain functions. Biol Psychiatry. 2013;73:313–320. doi: 10.1016/j.biopsych.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Bremne JD, Vermetten E. Stress and development: behavioral and biological consequences. Devel Psychopathol. 2001;13:473–489. doi: 10.1017/s0954579401003042. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Cassel S, Carouge D, Gensburger C, Anglard P, Burgun C, Dietrich J, et al. Fluoxetine and cocaine induce the epigenetic factors mecp2 and mbd1 in adult rat brain. Mol Pharmacol. 2006;70:487–492. doi: 10.1124/mol.106.022301. [DOI] [PubMed] [Google Scholar]
- Chocyk A, Majcher-Maślanka I, Dudys D, Przyborowska A, Wędzony K. Impact of early-life stress on the medial prefrontal cortex functions - a search for the pathomechanisms of anxiety and mood disorders. Pharmacol Rep. 2013;65:1462–1470. doi: 10.1016/s1734-1140(13)71506-8. [DOI] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton VS, Kolshus E, McLoughlin DM. Epigenetics and depression: return of the repressed. J Affect Disord. 2014;155:1–12. doi: 10.1016/j.jad.2013.10.028. [DOI] [PubMed] [Google Scholar]
- Daniels WMU, Fairbairn LR, van Tilburg G, McEvoy CRE, Zigmond MJ, Russell VA, et al. Maternal separation alters nerve growth factor and corticosterone levels but not the DNA methylation status of the exon 1(7) glucocorticoid receptor promoter region. Metabol Brain Dis. 2009;24:615–627. doi: 10.1007/s11011-009-9163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bellis MD. Developmental traumatology: a contributory mechanism for alcohol and substance use disorders. Psychoneuroendocrinology. 2002;27:155–170. doi: 10.1016/s0306-4530(01)00042-7. [DOI] [PubMed] [Google Scholar]
- De Bellis MD, Zisk A. The biological effects of childhood trauma. Child and Adolesc Psychiatry Clin of N Amer. 2014;23:185–222. doi: 10.1016/j.chc.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Zhang X. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Comm. 2009;383:421–425. doi: 10.1016/j.bbrc.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. New Eng J Medicine. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnar MR, Donzella B. Social regulation of the cortisol levels in early human development. Psychoneuroendocrinology. 2002;27:199–220. doi: 10.1016/s0306-4530(01)00045-2. [DOI] [PubMed] [Google Scholar]
- Gunstad J, Paul RH, Spitznagel MB, Cohen RA, Williams LM, Kohn M, et al. Exposure to early life trauma is associated with adult obesity. Psychiatry Res. 2006;42:31–37. doi: 10.1016/j.psychres.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Hao Y, Metz GAS. Non-genetic inheritance of behaviour and stress resilience. Non-Genet Inherit. 2013;1:9–16. [Google Scholar]
- Harlow HF, Dodsworth RO, Harlow MK. Total social isolation in monkeys. Proc Natl Acad Sci USA. 54:90–97. doi: 10.1073/pnas.54.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb DO. The effects of early experience on problem solving at maturity. Am Psychol. 1947;2:306–307. [Google Scholar]
- Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Ho DH, Burggren WW. Epigenetics and transgenerational transfer: a physiological perspective. J Exp Biol. 2010;21:3–16. doi: 10.1242/jeb.019752. [DOI] [PubMed] [Google Scholar]
- Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. Jf Psychiatric Res. 2013;47:880–891. doi: 10.1016/j.jpsychires.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Huang LT. Early-life stress impacts the developing hippocampus and primes seizure occurrence: cellular, molecular, and epigenetic mechanisms. Front Mol Neurosci. 2014;7 doi: 10.3389/fnmol.2014.00008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huidobro C, Fernandez AF, Fraga MF. Aging epigenetics: causes and consequences. Mol Aspects Med. 2013;34:765–781. doi: 10.1016/j.mam.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Nat Acad Sci USA. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Langley B, Lubin FD, Renthal W, Wood MA, Yasui DH, et al. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kember RL, Dempster EL, Lee THA, Schalkwyk LC, Mill J, Fernandes C. Maternal separation is associated with strain-specific responses to stress and epigenetic alterations to Nr3c1, Avp, and Nr4a1 in mouse. Brain Behav. 2012;2:455–467. doi: 10.1002/brb3.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kuhn JW, Vittum J, Prescott CA, Riley B. The interaction of stressful life events and a serotonin transporter polymorphism in the prediction of episodes of major depression: a replication. Arch Gen Psychiatry. 2005;62:529–535. doi: 10.1001/archpsyc.62.5.529. [DOI] [PubMed] [Google Scholar]
- Kim DK, Lim SW, Lee S, Sohn SE, Kim S, Hahn CG, et al. Serotonin transporter gene polymorphism and antidepressant response. Neuroreport. 2000;11:215–219. doi: 10.1097/00001756-200001170-00042. [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, Coplan JD, et al. DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun. 2011;25:1548–1553. doi: 10.1016/j.bbi.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharm. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J. 2009;50:455–463. doi: 10.3349/ymj.2009.50.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosten TA, Huang W, Nielsen DA. Sex and litter effects on anxiety and DNA methylation levels of stress and neurotrophin genes in adolescent rats. Dev Psychobiol. 2013;56:392–406. doi: 10.1002/dev.21106. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovalchuk I. Transgenerational epigenetic inheritance in animals. Front Genet. 2012;3:76. doi: 10.3389/fgene.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DEH, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Front Psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo ZY. The dynamics of behavioral development. New York: Plenum Press; 1967. [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd CO, Huot RL, Thrivikraman KV, Nemeroff CB, Meaney MJ, Plotsky PM. Long-term behavioral and neuroendocrine adaptations to adverse early experience. Prog Brain Res. 2000;122:81–103. doi: 10.1016/s0079-6123(08)62132-9. [DOI] [PubMed] [Google Scholar]
- Landry CD, Kandel ER, Rajasethupathy P. New mechanisms in memory storage: piRNAs and epigenetics. Trends Neurosci. 2013;36:535–542. doi: 10.1016/j.tins.2013.05.004. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Nestler EJ. CRACKing the histone code: cocaine’s effects on chromatin structure and function. Hormones Behav. 2011;59:321–330. doi: 10.1016/j.yhbeh.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- LeeRaby K, Roisman MG. Gene-environment interplay and risk and resilience during childhood. Resilience. 2014;17:17. [Google Scholar]
- Levine A, Worrell TR, Zimnisky R, Schmauss C. Early life stress triggers sustained changes in histone deacetylase expression and histone H4 modifications that alter responsiveness to adolescent antidepressant treatment. Neurobiol Dis. 2012;45:488–498. doi: 10.1016/j.nbd.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S. The ontogeny of the hypothalamic-pituitary-adrenal axis. The influence of maternal factors. Annal NY Acad Sci. 1994;746:275–288. doi: 10.1111/j.1749-6632.1994.tb39245.x. [DOI] [PubMed] [Google Scholar]
- Levine S, Haltmeyer GC, Karas GG, Denenberg VH. Physiological and behavioral effects of infantile stimulation. Physiol Behav. 1967;2:55–59. [Google Scholar]
- Lewis CR, Staudinger K, Scheck L, Olive MF. The effects of maternal separation on adult methamphetamine self-administration, extinction, reinstatement, and mecp2 immunoreactivity in the nucleus accumbens. Front Psychiatry. 2013;4:55. doi: 10.3389/fpsyt.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman SA, Mashoodh R, Thompson RC, Dolinoy DC, Champagne FA. Concordance in hippocampal and fecal Nr3c1 methylation is moderated by maternal behavior in the mouse. EcolEvolution. 2012;2:3123–3131. doi: 10.1002/ece3.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann M, Bress A, Nemeroff CB, Plotsky PM, Monteggia LM. Long-term behavioural and molecular alterations associated with maternal separation in rats. Eur J Neurosci. 2007;25:3091–3098. doi: 10.1111/j.1460-9568.2007.05522.x. [DOI] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Sharma, et al. Maternal care, hippocampal glucocorticoid responses to stress receptor (GR) expression in the hippocampus. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Londen LV, Goekoop JG, van Kempen GM, Frankhuijzen-Sierevogel AC, Wiegant VM, van der Velde EA, et al. Plasma levels of arginine vasopressin elevated in patients with major depression. Neuropsychopharmacology. 1997;17:284–292. doi: 10.1016/S0893-133X(97)00054-7. [DOI] [PubMed] [Google Scholar]
- Lovallo WR, Farag NH, Sorocco KH, Acheson A, Cohoon AJ, Vincent AS. Early life adversity contributes to impaired cognition and impulsive behavior: studies from the oklahoma family health patterns project. Alcohol Clin Exp Res. 2012;37:616–623. doi: 10.1111/acer.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz PE, Turecki G. DNA methylation and childhood maltreatment: from animal models to human studies. Neurosci. 264:142–156. doi: 10.1016/j.neuroscience.2013.07.069. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Robbins TW, Everitt BJ, Caine SB. Repeated neonatal maternal separation alters intravenous cocaine self-administration in adult rats. Psychopharmacology. 1999;141:123–134. doi: 10.1007/s002130050816. [DOI] [PubMed] [Google Scholar]
- Matthews K, Dalley JW, Matthews C, Tsai TH, Robbins TW. Periodic maternal separation of neonatal rats produces region- and gender-specific effects on biogenic amine content in postmortem adult brain. Synapse. 2001;40:1–10. doi: 10.1002/1098-2396(200104)40:1<1::AID-SYN1020>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. Early life influences on life-long patterns of behavior and health. Ment Retard Dev Disabil Res Rev. 2003;9:149–154. doi: 10.1002/mrdd.10074. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Suderman M, Sasaki A, Huang TCT, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PloS One. 2011;6:1–11. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Aitken DH, Bodnoff SR, Iny LJ, Sapolsky RM. The effects of postnatal handling on the development of the glucocorticoid receptor systems and stress recovery in the rat. Prog Neuropsychopharmacol Biol Psychiatry. 1985;9:731–734. doi: 10.1016/0278-5846(85)90050-8. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;21:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Morales V, Giamarchi C, Chailleux C, Moro F, Marsaud V, Le Ricousse S, et al. Chromatin structure and dynamics: functional implications. Biochimie. 2001;83:1029–1039. doi: 10.1016/s0300-9084(01)01347-5. [DOI] [PubMed] [Google Scholar]
- Mulligan CJ, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7:853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nagahara AH, Tuszynski MH. Potential therapeutic uses of BDNF in neurological and psychiatric disorders. Nat Rev Drug Discov. 2011;10:209–219. doi: 10.1038/nrd3366. [DOI] [PubMed] [Google Scholar]
- Narayan P, Dragunow M. Pharmacology of epigenetics in brain disorders. Br J Pharmacol. 2010;159:285–303. doi: 10.1111/j.1476-5381.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Utrankar A, Reyes JA, Simons DD, Kosten TR. Epigenetics of drug abuse: predisposition or response. Pharmacogenomics. 2012;13:1149–1160. doi: 10.2217/pgs.12.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Ooi SKT, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Pechtel P, Pizzagalli DA. Effects of early life stress on cognitive and affective function: an integrated review of human literature. Psychopharmacology. 2011;214:55–70. doi: 10.1007/s00213-010-2009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olié E, Salzmann A, Nicastro R, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry. 2011;1:1–9. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotsky PM, Meaney MJ. Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Research. Mol Brain Res. 1993;18:195–200. doi: 10.1016/0169-328x(93)90189-v. [DOI] [PubMed] [Google Scholar]
- Provençal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A, et al. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatry. 2011;1:1–6. doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Histone acetylation in drug addiction. Semin Cell Dev Biol. 2009;20:387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg G. The mechanisms of action of valproate in neuropsychiatric disorders: can we see the forest for the trees? Cell Mol Life Sci. 2007;64:2090–2103. doi: 10.1007/s00018-007-7079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL. Epigenetics of neurobiology and behavior during development and adulthood. Dev Psychobiol. 2012;54:590–597. doi: 10.1002/dev.20550. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab BJ, Mansuy IM. Neurobiological disease etiology and inheritance: an epigenetic perspective. J Exp Biol. 2014;217:94–101. doi: 10.1242/jeb.089995. [DOI] [PubMed] [Google Scholar]
- Schimmenti A, Bifulco A. Linking lack of care in childhood to anxiety disorders in emerging adulthood: the role of attachment styles. Child Adolesc Mental Health. doi: 10.1111/camh.12051. (In Press). [DOI] [PubMed] [Google Scholar]
- Schmidt HD, McGinty JF, West AE, Sadri-Vakili G. Epigenetics and psychostimulant addiction. Cold Spring Harb Perspect Med. 3:1–20. doi: 10.1101/cshperspect.a012047. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotnikov SV, Markt PO, Malik V, Chekmareva NY, Naik RR, Sah a, Landgraf R. Bidirectional rescue of extreme genetic predispositions to anxiety: impact of CRH receptor 1 as epigenetic plasticity gene in the amygdala. Transl Psychiatry. 2014;4:1–8. doi: 10.1038/tp.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger H, Labonté B, Groleau P, Turecki G, Israel M. Methylation of the glucocorticoid receptor gene promoter in bulimic women: associations with borderline personality disorder, suicidality, and exposure to childhood abuse. Int J Eat Disord. 2013;46:246–255. doi: 10.1002/eat.22113. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Suderman M, McGowan PO, Sasaki A, Huang TCT, Hallett MT, Meaney MJ, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci USA. 2012;109:17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Review Pharmacold Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tamaru H. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Devel. 2010;24:1465–1478. doi: 10.1101/gad.1941010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher MH, Andersen SL, Polcari A, Anderson CM, Navalta CP, Kim DM. The neurobiological consequences of early stress and childhood maltreatment. Neurosci & Biobehav Rev. 2003;27:33–44. doi: 10.1016/s0149-7634(03)00007-1. [DOI] [PubMed] [Google Scholar]
- Tesone-Coelho C, Morel LJ, Bhatt J, Estevez L, Naudon L, Giros B, et al. Vulnerability to opiate intake in maternally deprived rats: implication of MeCP2 and of histone acetylation. Addict Biol. 2013:1–12. doi: 10.1111/adb.12084. [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PloS One. 2012;7:1–8. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uaswani M, Linda EK, Ramesh S. Role of selective serotonin reuptake inhibitors in psychiatric disorders: a comprehensive review. Prog Neuropsychopharmacol. Biol. Psychiatry. 2003;27:85–102. doi: 10.1016/s0278-5846(02)00338-x. [DOI] [PubMed] [Google Scholar]
- Van IJzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R. Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry. 2010;68:405–407. doi: 10.1016/j.biopsych.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G,Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16:42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetulani J. Early maternal separation: a rodent model of depression and a prevailing human condition. Pharmacol Rep. 2013;65:1451–1461. doi: 10.1016/s1734-1140(13)71505-6. [DOI] [PubMed] [Google Scholar]
- Vijayendran M, Beach SRH, Plume JM, Brody GH, Philibert RA. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front Psychiatry. 2012;3:55. doi: 10.3389/fpsyt.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Wang J, Wu Z, Li D, Li N, Dindot SV, Satterfield MC, et al. Nutrition, epigenetics, and metabolic syndrome. Antioxidants Redox Signaling. 2012;17:282–301. doi: 10.1089/ars.2011.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Diorio J, Seckl JR, Szyf M, Meaney MJ. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Annal N Y Acad Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- Weaver ICG, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann SR, Turner JD, Mériaux SB, Meijer OC, Muller CP. Epigenetic regulation of the glucocorticoid receptor promoter 1(7) in adult rats. Epigenetics. 2012;7:1290–301. doi: 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, et al. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans suffering from post-traumatic stress disorder. Biol Psychiatry. doi: 10.1016/j.biopsych.2014.02.006. (In Press) [DOI] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Hayes JJ. Structures and interactions of the core histone tail domains. Biopolymers. 2003;68:539–546. doi: 10.1002/bip.10303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.