Abstract
Large-scale pharmacogenomic high throughput screening (HTS) studies hold great potential for generating robust genomic predictors of drug response. Two recent large-scale HTS studies have reported results of such screens, revealing several known and novel drug sensitivities and biomarkers. Subsequent evaluation however found only moderate inter-laboratory concordance in the drug response phenotypes, possibly due to differences in the experimental protocols used in the two studies. This highlights the need for community-wide implementation of standardized assays for measuring drug response phenotypes so that the full potential of HTS is realized. We suggest that the path forward is to establish best practices and standardization of the critical steps in these assays through a collective effort to ensure that the data produced from large-scale screens would not only be of high intra-study consistency, so that they could be replicated and compared successfully across multiple laboratories.
Summary
Pharmacogenomic high throughput screening offers tremendous promise in the rational development of targeted therapies, but its full potential will not be realized unless experimental protocols and analysis methods are standardized through a collective effort.
With dropping costs of genetic testing, cutting-edge sequencing technologies are now at the forefront of new cancer drug development. Yet, how to fully incorporate the presence of individual mutations and other specific genomic features to inform personalized therapeutic decisions remains a major challenge. Pharmacogenomics has emerged as a promising strategy to rapidly elucidate the link between genes and drugs by systematically characterizing the effects of pharmacologic agents on whole biological systems, allowing concurrent identification of therapeutic targets and discovery of drug candidates [1].
Advancements in molecular biology and the genomic sciences have had a profound impact on drug discovery, and together with the advent of automated high-throughput screening (HTS) have dramatically changed the drug development endeavor [2]. Pharmacogenomic HTS presents both opportunities and challenges. The goal is to measure the response of hundreds or thousands of cell lines to drugs or other perturbations and to associate the response with the genomic characteristics of each cell line. The extensive data generated from such efforts provide opportunities to search for biomarkers and to explore the mechanism associated with the underlying response. Two recent pharmacogenomic HTS studies, the Cancer Cell Line Encyclopedia (CCLE) [3] and Cancer Genome Project (CGP) [4] evaluated an impressive array of cell lines and drugs (1,036 cell lines and 24 drugs, and 727 cell lines and 138 drugs, respectively), generating gene-expression profiles and drug-sensitivity data for each combination. A subsequent comparative analysis of the CCLE and CGP found that while the gene expression profiles were highly concordant between studies, the measured cell line drug sensitivities were inconsistent [5], which has been further confirmed by an independent research group [6]. While the apparent variability in drug response presents a serious barrier to the ultimate goal of such studies – to develop signatures predictive of response – the high degree of correlation in the gene expression measures provides hope for a potential path forward. Here, we review potential explanations for these findings and extend recommendations for improving the reproducibility of pharmacogenomic HTS studies.
Pharmacogenomics in Cancer Drug Development
With more than 900 new cancer drugs in clinical testing [7] and better technological tools to characterize patient populations and modulation of drug targets, the paradigm of oncology drug development is shifting [8, 9]. A new drug must now demonstrate proof of concept that it can be beneficial as early as possible in clinical development. Pre-clinical pharmacogenomic HTS could identify genomic predictors allowing investigators to enrich early-phase clinical studies for those patients most likely to receive benefit. An early signal of proof of concept activity, such as vemurafenib in BRAF V600 mutant melanoma [10] and crizotinib in ALK fusion non-small cell lung cancer [11], can rapidly reduce the time to clinical testing and subsequent market approval. With a growing repertoire of potential drug combination partners, HTS could help to prioritize genotype-selective drug combinations for clinical testing [12]. Genomic predictors developed from HTS data can support this new paradigm when the number of false positive leads is contained.
HTS has become one of the primary scientific tools used in the pharmaceutical industry to generate new leads [13]. Very large compound libraries are screened for “hits” against a target [14]. Hits are generally target-specific active small molecules, which are further characterized for dose-response effects in secondary screens. Functional characterization and validation in animal models leads to selection of candidates for clinical testing. Pharmacogenomic screening extends the conventional HTS screening by using high-throughput approaches to evaluate the effect of compounds on large panels of cancer cell lines; this approach enables to identify simultaneously druggable targets and biologically active compounds [1, 15]. Effectiveness of a compound is assessed by functional assays that measure changes in such activities as cell proliferation, DNA replication, or cell death. Associating these changes with drug-induced transcriptomic alterations can identify cellular processes that are affected by a given treatment. A key advantage of pharmacogenomic screening is that it can compress the timeline of conventional HTS. By using systems biology methods, at least partial functional characterization and initial validation of the specificity/selectivity of a given compound can be obtained.
Given the complexity of HTS assay, there are many sources of experimental noise that should be carefully controlled to provide sufficient power to detect reliable “hits” [16]. Occasional false positive hits passing through the primary screen will likely be caught in secondary screening steps. However, the biological outcomes measured by pharmacogenomic HTS assays are often associated with increased variability, which reduces our ability to detect weaker target-drug-genotype associations in a single step. Several sources of variation affect these assays and may limit their power to reliably detect a broad range of biological associations, especially where a compound is effective against multiple potential targets in one or more pathways, or when the overall cytotoxic effect reflects crosstalk among interconnected biological pathways.
Lack of Concordance in Large Pharmacogenomic Studies
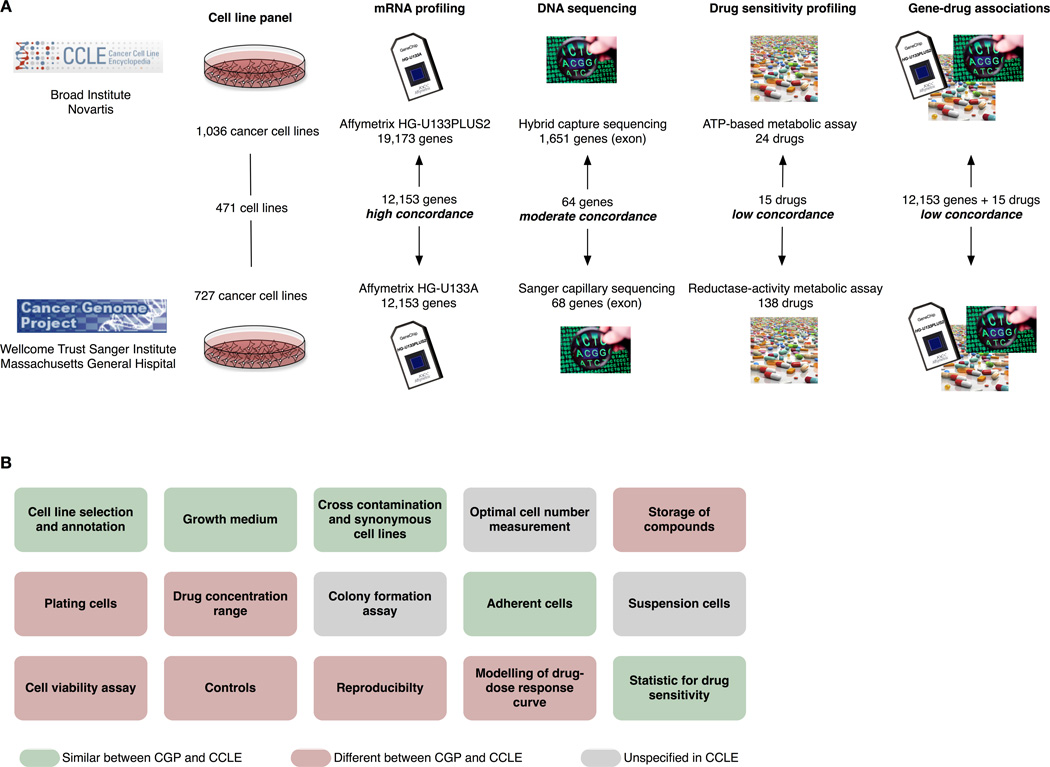
For pharmacogenomics to realize its full potential, the assessment of drug response needs to be robust, reliable, and reproducible. The CCLE and CGP drug screening studies are the largest public pharmacogenomic screens to date. To the credit of the investigators, they shared important details of their experimental protocols and all the pharmacogenomic data generated, allowing a careful reanalysis. Fifteen drugs and 471 cancer cell lines were screened in both studies (Figure 1A). These studies provided a unique opportunity to assess the consistency of both the raw data and the set of significant associations between genomic features, defined by gene expression measurements or mutation calls, and measures of drug response [5].
Figure 1.
(A) Comparison of the Cancer Genome Project [4] and The Cancer Cell Line Encyclopedia [3] as published by Haibe-Kains et al. [5]. Our comparative study revealed a high degree of concordance between the gene- expression datasets, moderate concordance for the mutation data, but low concordance between drug-sensitivity assays; such inconsistency propagates into the gene–drug associations found by the two studies. (B) Main steps in the experimental protocols used in the CCLE and CGP studies; each step is further described in Supplementary Table 1. The choice of experimental procedures vastly differs between the two studies, illustrating the lack of standardization in large pharmacogenomic studies.
The experimental protocols used to profile the transcriptomes at baseline prior to drug treatment were very similar in both studies, with the exception of the different density of the Affymetrix chips used (Figure 1A). Transcriptional profiles from replicate samples within the same study and between identical cell lines across studies were highly correlated, suggesting that the high level of standardization for gene expression profiling and analysis enabled reproducible findings. Only moderate concordance was observed in mutations detected in 64 genes in the 471 common cell lines between CCLE and CGP, which could be due to differences in the platforms used for mutational profiling (Figure 1A), or as a result of genetic divergence of the cell lines during passaging.
CCLE and CGP studies generated large amounts of drug sensitivity data using experimental protocols that differed in many important aspects (Figure 1B; Supplementary Table 1, Supplementary Information), including the specific pharmacological assay used (ATP-based vs. reductase-based metabolic activity for CCLE and CGP, respectively). Although the two studies were not designed specifically to address inter-laboratory reproducibility, the differences reflect a lack of consensus for pharmacogenomic HTS protocols. For 13 out of the 15 drugs tested in common in CCLE and CGP (87%) a high level of inconsistency between drug sensitivity measurements was observed (Spearman’s rank correlation < 0.5), regardless of which characteristic of the drug-dose response curve was used to summarize drug potency (Figure 1A). Intriguingly, when the same protocol was used at two different sites in the CGP study, only a fair inter-site correlation for sensitivity to camptothecin was observed (Spearman’s rank correlation for IC50 of 0.57). These results led us to conclude that the two experimental protocols are not equivalent and that measuring drug response is a complex and poorly reproducible process. While compounds were assayed at least in duplicate for each cell line, unfortunately, the raw data from these technical replicates were not publicly released by either study, preventing a direct assessment of assay reproducibility.
To confirm our findings, we compared the data for two drugs, lapatinib and paclitaxel, against a third large pharmacogenomic dataset generated by GlaxoSmithKline (GSK) that used the same ATP-based assay as in the CCLE study [17]. As expected, the correlation was higher between CCLE and GSK compared with CGP and GSK, suggesting that the choice of assay has a major impact on drug response measurements. However, the correlation between GSK and CCLE drug sensitivity data was still only fair (Spearman correlation < 0.51), implying that the drug sensitivity assay used is not the only source of inconsistency [5].
The primary goal of CCLE and CGP studies was to discover new associations between genomic and transcriptomic features of cancer cells and drug response that could advance our understanding of drug sensitivity and allow development of robust genomic predictors of response. Thus, we tested whether these associations were consistent across studies. We observed that, although we were able to validate the strongest associations identified in both studies, coupling consistent transcriptomic data with inconsistent drug sensitivity measurements resulted in inconsistent pharmacogenomic associations for most of the investigated drugs. Hence, any single study may not be able to identify robust predictors of drug response because a strong association observed in one study may not necessarily be evident in independent studies [5].
In vitro Cell-Based Drug Screening: What Can Affect Assay Consistency?
When considered independently, both the CCLE and CGP studies approached their experimental design, implementation, and data analysis with apparently adequate care and rigor; however, there were methodological differences between the two studies (Supplementary Information). Several factors could introduce variability in the results of in vitro cell-based assays. It is important to establish best practices on how the chemical compound collection is managed, maintained, and delivered to the assay plates for screening [18]. Critical factors include compound handling to ensure accuracy in the amount of the compound transferred to the master solution, maximal solubility in the solvent used (usually DMSO), optimal storage conditions to ensure compound integrity, and to minimize liquid loss through evaporation or leaching of the test compounds through the storage containers [19]. Because of such potential artifacts, the purity, integrity and concentration of the compound stock solutions should be verified prior to being used in the assay. Of the two studies, only CCLE reported pre-assay compound library validation. Furthermore, the type of the liquid transfer system can affect the accuracy of the amount of compound and cell culture stocks delivered to the recipient plates [20]. Tip-based serial dilution and dispensing could lead to larger IC50 values for some compounds, resulting in more than 100-fold underestimation of their potency compared with acoustic dispensing methods [21].
Minor variations in cell culture conditions including seeding density, plating efficiency, growth rate and cell cycle distribution may affect cell metabolism and drug responsiveness. The number of cells per well at the time of pharmacological testing also has a major impact on the dynamic range and sensitivity of the assay, and this variable depends both on the seeding density and on the grow rate of the cells. Several of these variables are affected by cell culture conditions including substrate, choice of cell culture medium (including relative concentrations of glucose, glutamine, and amino acids), and serum (final concentration, type, source, and batch effects). Other factors affecting the cell lines include their reference source, passage number, and the quality of the stock cultures used to seed the experiments. Even cell lines obtained from the same reference source may diverge enormously unless similar passages from the reference culture are used. Both the CCLE and CGP groups appropriately attempted to reduce the contribution of several of these variables, which would help control intra-study variability. However, these actions do not necessarily improve inter-study comparability.
Adaptive responses to the stress of drug treatment can occur within the transcriptome, proteome, metabolome, kinome, and methylome; some of these changes may be detected in hours or less of the initiation of exposure to a drug. Where rapid drug-induced changes in cells reflect common and/or evolutionarily conserved stress response pathways, key features that drive profile changes may appear qualitatively more similar than different across studies. Thus, it is likely that, provided the cell lines are of the same origin, the genomic data will appear broadly similar. If the same cell lines respond in the same way to the same drugs, selective features of the transcriptomes might be expected to exhibit at least qualitatively similar changes. These observations are broadly consistent with the findings of our comparative study [5].
The type of cytotoxicity assay used in assessing drug sensitivity could significantly impact the measured phenotypic response. Among the several assays compatible with HTS [22, 23], bioluminescent detection of adenosine triphosphate (ATP) is one of the most sensitive and reliable methods [24] and has been used extensively in HTS studies (Table 1). CCLE used an ATP detection cell proliferation assay, but CGP used two different cell viability assays, a fluorescent nucleic acids stain and a redox indicator dye to assess drug inhibition (for details see Supplementary Information). A recent study by Chan et al. directly compared cell viability assays based on quantifying total amount of nucleic acid using fluorescent DNA-binding dyes (similar to the SYTO 60 assay used in the CGP study) vs. ATP-dependent luminescence (CellTiter-Glo assay also used in the CCLE study) [25]. The study shows that the ATP-dependent luminescence assay is prone to underestimation of drug potency and efficacy, particularly for DNA synthesis-targeting agents [25]. The ATP-dependent luminescence and fluorescent DNA-binding assays are measuring different aspects of the drug response phenotype, and therefore it is not surprising that the assays show only moderate correlation in our comparative analysis [5]. Given the limitations of each assay, it has been suggested that multi-parameter testing, incorporating complementary cell-viability assays yields the most robust and informative phenotypic measures [23].
Table 1.
Comparison of available pharmacological assays for measuring cell viability.
| Cell viability assay |
Type of Assay | Pros | Cons | Suitable for HTS |
Used in* |
|---|---|---|---|---|---|
| Cell Titer Glo (Promega) |
Viability, Membrane integrity, ATP |
Non-toxic, does not require fixing or washing steps |
ATP could be reduced by metabolic interferences, leading to false positive results; underestimation of drug effect by DNA-targeting agents, where cells may be incapacitated but intact |
Yes | CCLE, GSK, FIMM, SU2C, NTP |
| Syto60 (Invitrogen) |
Proliferation; Fluorescent DNA stain |
Simple | requires fixation, washing; could stain DNA of intact but not-viable cells |
Yes | CGP |
| Bromodeoxyur idine-ELISA (CytoSelect, Cell BioLabs) |
Proliferation, DNA synthesis |
Only live and replicating cells can incorporate BrDU |
Assay is time consuming and cost prohibitive in high throughput format |
No | |
| DAPI | Proliferation, nucleic acids stain |
Simple | Could stain non viable cells if DNA is intact |
Yes | GSK |
| MTT, MTS, XTT |
Viability, Metabolic reduction of tetrazolium dye |
Simple colorimetric assay |
formazan product insoluble in aqueous media; secondary step required to solubilize it before optical detection |
No | NCI60 |
| SRB | Proliferation, anionic general biomass stain |
Simple Colorimetric assay; stable for extended periods; differentiates cell kill from growth inhibition |
requires fixation, washing steps | Yes | NCI60 |
| Resazurin (Cell Titer-Bue, Promega) |
Viability, cell permeable redox indicator |
Simple and inexpensive; more sensitive than tetrazolium assays |
Potentially cytotoxic; requires long incubation 1–4 h; could underestimate toxic effect |
Yes | |
| GF-AFC (Cell Titer-Fluor, Promega) | Viability; penetrates live cells where cytoplasmic cytopeptidase activity releaces AFC that fluoresces; Protease becomes inactive upon cell death |
Non-toxic, does not require fixing or washing steps; short incubation time < 1h |
potential underestimation of drug effect by DNA-targeting agents, where cells may be incapaciated but intact |
Yes | |
| Ruthenium Dye |
Proliferation; the fluoresecence of the dye is quenced by oxygen; proliferating cells reduce external oxygen and increase fluorescence |
Non-toxic, simple, rapid assay |
Narrow dynamic range; signal depends on metabolic activity |
Yes | |
| LDH (Cytotox-ONE, Promega) | Cytotoxity; released LDH is measured by enzymatically coupled reagent chemistry |
Reagents compatible with viable cells; quick incubation; cost effective |
Susceptible to background signal from serum sources of LDH in growth media or from compounds inhibiting LDH activity |
Yes |
CCLE: Cancer Cell Line Encyclopedia (Barretina et al, Nature 2012, 483:603-7); CGP: Cancer Genome Project (Garnett et al, Nature 2012, 483:570-5); FIMM: Institute for Molecular Medicine, Finland (Pemovskaet al, Cancer Discovery 2013, 3:1416-29); GSK: Glaxo Smith Kline Study (Greshock. et al, Cancer Res 2010, 70:3677-86); NCI60: National Cancer Institute (Shoemaker, Nature Rev Cancer 2006, 6:813-23); NTP: National Toxicology Program (Xia et al, Environ Health Perspect 2008, 116:284-91); SU2C: Lawrence Berkeley National Lab (Heiser et al, Genome Biol 2009, 10:R31)
Primary phenotypic responses measured in pharmacogenomic studies include dose response curves and two related measures of drug potency: the half maximal inhibitory concentration (IC50) and the area under the dose response curve (AUC). These measures can be estimated using many different laboratory assays and analytical methods – no standard “best practices” have been shown to be either highly reproducible or generally reflective of the response in humans [26]. The shape of the dose-response curve, and thus the estimates for both AUC and IC50, depends not only on the drug-cell line combination but also on the range of drug concentrations tested, which was not identical in the two studies. More broadly, the physiological definitions of AUC and IC50, originally intended to describe the mass-action law of a single-substrate competitive inhibition enzyme kinetics, [27] might not provide similarly meaningful measures of growth inhibition kinetics in whole cell systems. Cell population heterogeneity and cell-to-cell variability in drug response may affect the shape of the dose response curve, yielding shallow curves with reduced steepness or Hill slope [26]. An independent reanalysis of the two studies found that in the CCLE study a drug could be assessed both as effective or ineffective against a cell line depending on whether the assessment of drug sensitivity was based on an IC50 or AUC [6]. Thus, it may not be a simple task to obtain meaningful quantitative, and perhaps even qualitative, comparisons of responsiveness to many drugs across studies.
Standardization of In Vitro Cell-Based HTS
Reproducibility is a fundamental prerequisite for any scientific study, and this requirement becomes even more critical when experimental results impact medical decision-making. As large panels of genomically-annotated cancer cell lines (such as those in the CCLE and CGP studies) are being used to screen cancer drugs and guide treatment selection for patients whose cancers show similar genomic alterations, it is important to ensure the reproducibility of cell-line based drug sensitivity data from HTS assays. The unexpectedly low concordance in the results of the CCLE and CGP studies also illustrates a broader challenge. Scientifically appealing, novel high throughput analytical methods often become widely used before the factors that affect assay performance are well understood. Due to the substantial experimental costs, limited funding, and lower priority assigned to this type of technology assessment, parameters that influence the results may not be established and standardized until after widespread use of the method.
When DNA microarray analysis was first introduced in the 1990’s there was great excitement about the potential of genome-wide expression profiling. The initial euphoria soon evaporated as many studies failed to be replicated. In 1999, the Microarray Gene Expression Data Society (MGED) was formed to address the need for standardization in microarray experiments and the reporting of experimental data and metadata. The group produced the Minimal Information About a Microarray Experiment (MIAME) standard [28, 29] and, through a series of meetings and workshops, helped to catalyze development of experimental and data analysis standards. Data preprocessing tools such as dChip [30] and RMA [31] provided the means to compare large-scale assays, methods for removing batch effects [32, 33] helped to facilitate meta-analysis across experimental datasets, and approaches to creating robust biomarkers [34–37] have provided a framework to use expression data to make more reliable predictions of disease phenotypes.
It is clear that there is a need for a similar standardization of drug-response measurements in addition to the development of new, robust drug sensitivity assays that can both be replicated across studies and that are physiologically relevant for the treatment of human disease. Lack of standardization may limit the potential of high-throughput drug screening studies for discovering novel drug sensitivities. While gene expression and genome sequencing data may be reliable measures of the individual cell lines being screened, the lack of a robust phenotypic anchor for developing and validating predictive models can limit the use of data from HTS studies. A community-based effort, similarly to that undertaken by MGED, and an investment of resources to support the development of common standards to facilitate comparisons, would pay a huge dividend in establishing high-throughput screening as a valuable tool in drug discovery and could enable a host of robust and reproducible clinical and translational applications.
Community-wide Assessment for Standardization
Given the complexity of HTS assays, a community-wide consortium effort with multiple stakeholders from industry, academia, and government working together may be needed to reach consensus on best practices and strategies for objective quality assessment and inter-site reproducibility of drug sensitivity data. HTS assays are complex, multi-step processes with many options; however, the respective advantages, limitations, and inter-site reproducibility of the alternative protocols have not been adequately evaluated. Noise introduced at any step could seriously limit the robustness of an entire experiment. For the objective assessment of quality control of cell-based HTS assays, a community-wide consortium effort ("HTSQC") similar to the MAQC-I [37] could be invaluable.
We offer some initial ideas for further consideration. A common panel of cancer cell lines whose identity would be verified genomically, and a collection of standardized anticancer compounds could be used as common "reference materials" for assessing the performance of each participating HTS platform and testing laboratory, providing a common ground to support meaningful comparisons. The reference materials (cell lines and compounds) could be maintained under strict quality control and could be used with defined protocols at a centralized location before being distributed to testing laboratories. Specifying defined cultured media and conditions, number of passages, seeding densities, and duration of assays could help establish common assay protocols. Similarly, reagents required for a given HTS platform could be kept as consistent as possible among the testing laboratories.
The same HTS platform could be tested in at least three independent laboratories using the same reference materials and the same lots of reagents with multiple replicates in the same laboratory, allowing both inter- and intra-laboratory reproducibility, and cross-platform comparability to be assessed. All study protocols and experimental details would be recorded and submitted along with raw experimental data to designated data analysis teams for independent analysis, allowing variability among data analysis methods to be assessed.
Finally, members of the consortium should share and discuss the results transparently, and publish the results in a peer-reviewed journal as a consortium consensus. Raw data and experimental details should be deposited in a public repository such as PubChem, to allow the community to replicate, further evaluate, and improve HTS assays and data analysis approaches. Such a consortium effort would provide an estimate of the overall reproducibility of current HTS assays, develop best practices, and provide an upper limit of the robustness of measured drug response phenotypes. If executed properly, this project could provide critical information for industry, academia, and regulatory agencies to objectively assess the implications of the limits of consistency of HTS assay results in drug discovery and development.
Conclusion
While our evaluation suggests caution when considering the data generated from large-scale pharmacogenomic HTS studies, our results do not undermine the value of such studies. Indeed large-scale pharmacogenomic HTS studies hold great potential for generating robust genomic predictors of drug response. However this potential will not be fully realized until improvements are made in the design, application, and community-wide implementation of robust, standardized assays for measuring drug response phenotypes. Efforts should be made to promote transparency and availability of raw assay data for HTS studies. These data will be critical to identify the sources and magnitudes of experimental uncertainties, and for developing methods to reduce their impact. One proven path forward is the establishment of best practices and standardization of the critical steps in these assays through a collective effort. Such an approach could ensure that the data produced from large-scale efforts can be replicated over a broad range of conditions and compared successfully across multiple laboratories.
Supplementary Material
Definitions (Box).
Pharmacogenomics: the use of genetic information from populations to inform drug design and development or from individual patients to inform the clinical management of pharmacotherapy, including drug selection, dosing and analysis of drug toxicities. Here this term is used more broadly to include experimentation on model systems.
(Conventional) High-Throughput Screening: highly automated methodology that heavily relies on robotics, liquid handling devices and automated detection to quickly conduct millions of pharmacological tests, typically on specific protein targets
in vitro cell-based/pharmacogenomic HTS: extension of conventional HTS to include large panel of cancer cell lines to assess in vitro drug response.
IC50: half maximal inhibitory concentration as a measure of the effectiveness of a substance in inhibiting a specific biological or biochemical function. This quantitative measure is estimated from the drug dose-response curve.
AUC: Area under the drug dose-response curve.
DMSO: Dimethyl sulfoxide is an organosulfur compound with the formula (CH3)2SO. This colorless liquid is an important polar aprotic solvent that dissolves both polar and nonpolar compounds and is miscible in a wide range of organic solvents as well as water.
References
- 1.Bredel M, Jacoby E. Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat Rev Genet. 2004;5(4):262–275. doi: 10.1038/nrg1317. [DOI] [PubMed] [Google Scholar]
- 2.Drews J. Drug discovery: a historical perspective. Science. 2000;287(5460):1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 3.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haibe-Kains B, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang IS, et al. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac Symp Biocomput. 2014:63–74. [PMC free article] [PubMed] [Google Scholar]
- 7.America PRa.M.o. More Than 900 Medicines and Vaccines in Clinical Testing Offer New Hope in the Fight Against Cancer. Medicines in Development for Cancer. 2012 Available from: http://www.phrma.org/research/new-medicines. [Google Scholar]
- 8.Tan DS, et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer J. 2009;15(5):406–420. doi: 10.1097/PPO.0b013e3181bd0445. [DOI] [PubMed] [Google Scholar]
- 9.Yap TA, et al. Envisioning the future of early anticancer drug development. Nat Rev Cancer. 2010;10(7):514–523. doi: 10.1038/nrc2870. [DOI] [PubMed] [Google Scholar]
- 10.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Held MA, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer Discov. 2013;3(1):52–67. doi: 10.1158/2159-8290.CD-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macarron R, et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 14.Walters WP, Namchuk M. Designing screens: how to make your hits a hit. Nat Rev Drug Discov. 2003;2(4):259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 15.Melnick JS, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A. 2006;103(9):3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malo N, et al. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24(2):167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 17.Greshock J, et al. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70(9):3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 18.Matson SL, et al. Best practices in compound management for preserving compound integrity and accurately providing samples for assays. J Biomol Screen. 2009;14(5):476–484. doi: 10.1177/1087057109336593. [DOI] [PubMed] [Google Scholar]
- 19.McDonald GR, et al. Bioactive contaminants leach from disposable laboratory plasticware. Science. 2008;322(5903):917. doi: 10.1126/science.1162395. [DOI] [PubMed] [Google Scholar]
- 20.Marx V. Pouring over liquid handling. Nat Methods. 2014;11(1):33–38. doi: 10.1038/nmeth.2785. [DOI] [PubMed] [Google Scholar]
- 21.Ekins S, Olechno J, Williams AJ. Dispensing processes impact apparent biological activity as determined by computational and statistical analyses. PLoS One. 2013;8(5):e62325. doi: 10.1371/journal.pone.0062325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slater K. Cytotoxicity tests for high-throughput drug discovery. Curr Opin Biotechnol. 2001;12(1):70–74. doi: 10.1016/s0958-1669(00)00177-4. [DOI] [PubMed] [Google Scholar]
- 23.Niles AL, Moravec RA, Riss TL. In vitro viability and cytotoxicity testing and same-well multi-parametric combinations for high throughput screening. Curr Chem Genomics. 2009:333–341. doi: 10.2174/1875397300903010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crouch SP, et al. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods. 1993;160(1):81–88. doi: 10.1016/0022-1759(93)90011-u. [DOI] [PubMed] [Google Scholar]
- 25.Chan GK, et al. A simple high-content cell cycle assay reveals frequent discrepancies between cell number and ATP and MTS proliferation assays. PLoS One. 2013;8(5):e63583. doi: 10.1371/journal.pone.0063583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallahi-Sichani M, et al. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol. 2013;9(11):708–714. doi: 10.1038/nchembio.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T. Relationships between inhibition constants and fractional inhibition in enzyme-catalyzed reactions with different numbers of reactants, different reaction mechanisms, and different types and mechanisms of inhibition. Mol Pharmacol. 1974;10(2):235–247. [PubMed] [Google Scholar]
- 28.Ball CA, et al. Standards for microarray data. Science. 2002;298(5593):539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 29.Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8) doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolstad B, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 32.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 34.Prat A, et al. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135(1):301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haibe-Kains B, et al. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104(4):311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck AH, et al. Significance analysis of prognostic signatures. PLoS Comput Biol. 2013;9(1):e1002875. doi: 10.1371/journal.pcbi.1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium M, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.