Abstract
Background
Over the past several decades, advances in lung cancer research and practice have led to refinements of histological diagnosis of lung cancer. The differential use and subsequent alterations of non-specific morphology codes, however, may have caused artifactual fluctuations in the incidence rates for histologic subtypes, thus biasing temporal trends.
Methods
We developed a multiple imputation (MI) method to correct lung cancer incidence for non-specific histology using data from the Surveillance, Epidemiology, and End Results (SEER) Program during 1975–2010.
Results
For adenocarcinoma in men and squamous in both genders, the change to a increasing trend around 2005, after more than ten years of decreasing incidence, is apparently an artifact of the changes in histopathology practice and coding system. After imputation, the rates remained decreasing for adenocarcinoma and squamous in men, and became constant for squamous in women.
Conclusions
As molecular features of distinct histologies are increasingly identified by new technologies, accurate histological distinctions are becoming increasingly relevant to more effective 'targeted' therapies, and therefore, are important to track in patients. However, without incorporating the coding changes, the incidence trends estimated for histologic subtypes could be misleading.
Impact
The MI approach provides a valuable tool for bridging the different histology definitions, thus permitting meaningful inferences about the long-term trends of lung cancer by histological subtype.
Keywords: SEER, ICD-O-3, Non-specific morphology codes, Missing data, Ridge-penalized logistic regression
INTRODUCTION
Lung cancer is the leading cause of cancer death in women and men in the United States. On average, only approximately 15% of newly diagnosed cases survive for five years or longer (1). Histologically, lung cancers are classified as small-cell and non-small-cell (NSC) carcinoma (2). The latter is usually further divided into squamous-cell carcinoma, adenocarcinoma, and large-cell carcinoma. Within the NSC category, etiologic and morphologic differences by histology have been recognized, but in the past, treatment and prognosis were considered relatively homogeneous for different histologies of the same stage. Emerging data now increasingly identifies subsets of adenocarcinoma (3) and squamous histologies (4) with specific genetic alterations. For example, the epidermal growth factor receptor (EGFR) protein overexpression and activating EGFR mutations, associated with responsiveness to EGFR therapies (tyrosine kinase inhibitors) (5, 6), are almost exclusively found in adenocarcinoma histology. Similarly, echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) rearrangements are also more common in adenocarcinoma and these mutations indicate responsiveness to another therapeutic agent, crizotinib (7). As we move into the future, clinical strategy for tumor management will be determined by molecular studies of the tumors and their underlying mutations (8). Inherited variation in lung cancer that has been identified may eventually have therapeutic implications in terms of efficacy and side-effects. The recent results from the National Lung Screening Trial further suggested that histology might be attributable to the differential computed tomography (CT) screening efficiency (9). As the broader implications of histologic classification are becoming increasingly relevant to screening, treatment, prognosis, and etiology, so will the examination of temporal trends separately for each subtype.
Cancer registry data collected by the National Cancer Institute (NCI)’s Surveillance Epidemiology and End Results (SEER) Program has been a primary source of data for providing national trends of lung cancer incidence and mortality (10). SEER registries have been coding cancer histology according to the International Classification of Diseases for Oncology (ICD-O). In the 1990s, pathologists tended not to report NSC carcinomas with specificity since their treatments and prognoses were considered similar, thus an increasing number of cases are coded with 8010 (carcinoma, NOS) since 1980. In recognition of this trend, 8046 (NSC carcinoma), was added into ICD-O-3 in 2001 to group cases that could not be classified beyond the exclusion of small-cell. Collectively, the percentage of cases coded with 8010 or 8046 increased dramatically, from 5% in 1982 to over 22% in 2005 (11). Some of these cases could have been derived from one of the specific histologic subtypes, which would have subsequently reduced their incidence rates. However, this increasing use of non-specific codes did not continue. In light of the advances in cancer research and therapy, increasingly NSC cases have been diagnosed with more histologic specificity (12) over the last few years, which may have driven up the rates for squamous or adenocarcinoma. Such differential use of nonspecific morphology codes could bias the estimated temporal trends of histologic subtypes and complicate interpretations. Appropriate statistical adjustments are necessary to improve the quality of inferences using the authoritative cancer registry data, which otherwise has been compromised by the unavoidable limitations imposed by the imperfect earlier classification system.
Multiple imputation (MI) has been shown to be a useful approach for handling measurement or coding changes for settings both in the presence (13–16) and absence (17) of calibration data (observations that are measured in all measurement scales or coding systems). When a calibration data (usually on a random subsample) is available, one can generate plausible values in all measurement scales from an imputation model and analyze the imputed data using the preferred scale. For the issue associated with the change in the use of nonspecific morphology codes, i.e. 8010 and 8046, two types of calibration data could be useful for correcting coding inconsistency. The first type comprises cancer cases that are originally assigned to a nonspecific code, but are updated with a specific code through reexamination. This type of data provides information about the association between nonspecific and specific histologies that one can use to recover the missing histology for all cases with a nonspecific code. Because nonspecific codes no longer exist in the imputed data, the trend analysis of incidence by histology is valid (provided that the imputation model is correct). The second type consists of cancer cases coded in multiple classification systems. Using this data as a bridge, one can convert data from one system to another. Although non-specific codes still exist, temporal comparisons of imputed histology in any classification system is valid because coding consistency is maintained. However, neither type of calibration data could be easily obtained due to practical reasons, such as budget constraints and the lack of diagnostic data sources. Thus, this problem becomes a missing data issue where the specific histologies for cases with a nonspecific morphology code are missing and an assumption about the association between the missing specific histology and observed data (18–20) is required. We make a reasonable assumption that for cancer cases with similar tumor, treatment, survival, patients’ demographic characteristics, the distribution of nonspecific and specific histology is similar. Based on this assumption, we developed a MI approach using the sequential regression imputation method (SRMI) (21) to redistribute cases without specific histology to one of specific subtypes, thus correcting the biased estimates of incidence rates.
Materials and Methods
Data Sources
We selected 522,416 malignant lung cancer cases diagnosed from 1975 to 2010 from the SEER 9 registries database (including Atlanta, Connecticut, Detroit, Hawaii, Iowa, New Mexico, San Francisco-Oakland, Seattle-Puget Sound, and Utah). We created six histologic categories according to the most recent NCI’s SEER Cancer Statistics Review (1) and they are small-cell carcinoma (8041–8045), squamous and transitional-cell carcinoma (8051–8052, 8070–8084, 8120–8131), adenocarcinoma (8050, 8140–8149, 8160–8162, 8190–8221, 8250–8263, 8270–8280, 8290–8337, 8350–8390, 9400–8560, 8570–8576, 8940–8941), large-cell carcinoma (8011–8015), other NSC carcinoma (8020–8022, 8030–8040, 8090–8110, 8150–8156, 8170–8175, 8180, 8230–8231, 8240–8249, 8340–8347, 8561–8562, 8580–8671), and other specified and unspecified types (8680–8713, 8800–8912, 8990–8991, 9040–9044, 9120–9136, 9150–9252, 9370–9373, 9540–9582, 8720–8790, 8930–8936, 8950–8983, 9000–9030, 9060–9110, 9260–9365, 9380–9539, 8000–8005). We singled out 8010 and 8046 from these categories, for which we performed statistical adjustments. We excluded cases with other specified and unspecified types because their incidence is not likely to be affected by the recent change in coding system. We also excluded cases which were not histologically confirmed or with unknown histologic confirmation status, because their diagnoses tended to be inaccurate and lacked specificity. The final sample size for this analysis is 470,326.
Data Analysis
We treated the cases with 8010 or 8046 as missing data that we dealt with by MI (22). This MI approach took each case with missing histology and imputed it with a specific histologic subtype. Cases coded with 8010 were imputed with one of the five carcinoma subtypes, i.e. small-cell, squamous, adenocarcinoma, large-cell, and other NSC. For cases coded with 8046, the imputation was limited to one of the NSC subtypes, i.e. excluding small-cell. This process was repeated independently 10 times to create 10 completed datasets to account for imputation uncertainty. Age-adjusted incidence rates (using the 2000 U.S standard population in 19 age groups) were estimated from each completed data set in the same way as using the original data set, thus producing 10 sets of estimates. We then combined these estimates to produce MI estimates. For a single incidence rate, the MI point estimate was the average of 10 imputed data estimates. The associated standard error was calculated by combining the average of the squared standard errors of the 10 estimates and the variance of the 10 rate estimates (22). Joinpoint linear regression models (23) were used to fit connected linear trends on a log scale with up to four joinpoints using the Joinpoint regression program version 3.5.0 developed by the NCI. Annual percentage change (APC) with a corresponding 95% confidence interval (CI) was calculated to describe each joined trend.
Imputation Method
The non-specific histological diagnoses are highly likely to have nonrandom characteristics. For example, patients may not merit further histological diagnostic procedures because they have diseases too advanced to permit curative surgery (i.e. Stage 3B or greater) or because their medical status preclude surgery or other modalities with curative intent. When surgery is not a clinical option, obtaining adequate tissue to establish a histologic subtype may be impossible and, in this circumstance, clinicians may elect to forgo further histologic classification. Therefore, we considered using the information that is predictive of histology and the missingness of specific histology to recover the incomplete specific histology. We assumed the missingness is random conditional on this information, and this assumption has been shown to be reasonable in most practical situations (24, 25).
Specifically, we selected the covariates to be included in the imputation model following the principle of reducing missing data bias in a statistical analysis (26). Socio-demographic covariates include age, gender, race, Hispanic origin, nativity, and marital status. Covariates describing tumor characteristics and treatment include tumor size (27), grade, stage, survival time, and receipt of cancer-directed surgery. Certain therapies have shown to be more responsive in some histologic subtypes, thus making them important predictors. However, such information can only be made available for patients 65 years and older through the linked SEER-Medicare database (28) for 1991 and later. Considering the lack of analytics tools to handle the dynamics of the availability and access to particular regimen over time and patients’ age, we did not include more detailed treatment variables in the model. We also did not include lymph node involvement in the final model because it is highly collinear with stage. We included a nominal variable of nine SEER registries to reflect the variability among registries in the use of nonspecific morphology codes. Cancer diagnosis year was entered into the models as a nominal variable (instead of a continuous variable) to relax the temporal assumption about the intervariable relationships. Smoking and socioeconomic deprivation are also strongly predictive of histology (29), but they are not routinely collected in SEER. To substitute, we used county-level smoking prevalence estimates obtained from the Model-based Small Area Estimates Projects of NCI (http://sae.cancer.gov/) (30), and poverty prevalence estimates from the 2000 U.S. Census Bureau (31).
Because missing histology cannot be imputed for cases that are associated with missing covariates using simple regression-based imputation approaches, we developed an algorithm using SRMI technique to deal with multivariate missing data with arbitrary missing patterns. Specifically, SRMI fits a conditional model for each variable at a time on the remaining variables sequentially for multiple rounds to achieve convergence. The form of conditional model depends on the type of variable imputed. Our algorithm offers two new capacities beyond what is available in existing SRMI-based imputation packages, such as IVEware (http://www.isr.umich.edu/src/smp/ive/) and MICE (http://cran.r-project.org/web/packages/mice/index.html). First, for imputing binary data (categorical variables with more than two levels can be expressed as a series of nested dummy variables), we used ridge-penalized logistic regressions (32, 33) to improve imputation precision in the presence of binary outcome with skewed distribution and highly correlated covariates (34). The standard approach for imputing missing binary data is usually based on a logistic regression model (21, 35). However, the adequacy of logistic models could highly depend upon the extent to which the binary outcome is balanced and there is an absence of collinearity. In the presence of either condition or both at the same time, logistic regression coefficients may still be unbiased, but the precision could be very low, which could lead to poorly imputed data. The proposed approach improves the imputation by estimating a penalized log likelihood to obtain coefficients estimates with minimum prediction errors. Optimizing the penalty parameters is critical and usually requires intensive cross-validation studies (36). We follow the simplified approach proposed by Yu (34) and obtain the optimized parameters directly from the data by estimating the unrestricted log likelihood. The remaining steps are similar to those when standard logistic models are used (21). Second, we added a module to impute discrete right-censored survival data. For the data we chose for this study, over 25% of survival time was censored because the patient was still alive at the end of study or died from other causes. Because both survival and censoring are highly correlated with histology as well as other covariates such as age, stage, tumor size, and grade, it is problematic to use relatively simple approaches, such as the indicator method where censoring is taken care of by including a censoring indicator (37, 38). The proposed method applies the MI principle to impute the censored time with a plausible future survival time. Specifically, to generate the imputed values, we first aggregate continuous survival time (in month) into several meaningful categories and sort them in an increasing order of survival. We then define an imputing risk set for each censored case as the cases with observed survivals no shorter than the censoring time. Using data from this imputing risk set, we finally estimate the predictive conditional distributions of survival categories, from which we randomly draw a value to be the imputed survival. Note that the possible value of an imputed survival is always equal to or longer than the censoring time category. This is reasonable because a censored case could only die at a later time in its own survival category or be still alive and die at a future category, but not die at a past category. This imputation process starts with censored cases in the first survival category and cycles through all categories to complete one imputed survival data. Because the survival is now a discrete variable, we estimate its predictive conditional distribution using nested ridge-penalized logistic models similar to what we have outlined for categorical data. Furthermore, to deal with the inconsistency in stage definitions over time, we conducted the imputation separately for 1975–1982, 1983–1987, and 1988–2010, so that staging is comparable within each period.
Simulation Study
To explore information recovery from the MI in estimating the distribution of histology, we generated a simulated dataset from the analysis data with only complete observations included (n=10,659). We considered a situation similar to the main analysis where histology is missing at random and the probability of the induced missingness is determined by a logistic regression model with the coefficients estimated using the analysis data. The rate of induced missing data was 8.4% (the observed missing rate was 10.0% for the portion of data with all covariates observed). Twenty imputed datasets were generated using the proposed approach and the standard logistic regression method respectively.
The ridge-penalized logistic regression model outperformed the standard logistic regression model in recovering the missing information based on the Akaike information criterion (AIC) (ridge-penalized method: AIC=31,008 and standard method: AIC=31,065). The imputed distributions of histology obtained using the proposed method were similar to the complete data distribution (with absolute difference less than 2% in estimating the percentage of cases in each histology and gender group). We also calculated the overlap probability (39) to evaluate how much the associated 95% CI estimated from the imputed and complete data overlap. Suppose (Limp, Uimp) and (Lcom, Ucom) are the 95% confidence intervals for estimating p, the percentage of adenocarcinoma among men, using the imputed and complete data respectively. The probability overlap in the CIs for p is , where fimp and fimp are the distributions of p computed under the imputed and complete data respectively. Note that fimp could take a different form of distribution depending on the type of statistics for which one wish to obtain estimates, but fimp is always t-distributed according to Rubin’s rules (22). I takes value 0.95 if two CIs overlap perfectly and 0 if they do not overlap at all. A large value in I suggests that the imputed data highly maintains the analytical properties of the complete data. This measure provides more information than a simple comparison of two point estimates by also considering the standard errors. Estimates with large standard errors might still have a high confidence interval overlap even if their point estimates differ considerably from each other because the CI will increase with the standard error of the estimate. In this simulation study, most overlap probabilities (for estimating the distributions of cases by histology and gender) were over 0.8, which suggested a very strong agreement, with a few exceptions in which the probabilities were around 0.75, which still suggested a strong agreement. These evaluation results provided strong evidences for model adequacy in the proposed method.
RESULTS
Table 1 shows the distribution of histologic categories by histology confirmation status. Ninety percent of cases are histologically confirmed. Among the cases that are not- confirmed and the cases for which the confirmation status is unknown, 8010 accounts for about 50% of the total, whereas 8046 only accounts for less than 2%. Possible explanation for the differential use of 8010 and 8046 could be that the latter is mainly used when histological diagnosis, although not quite specific, exists, and the former is also used when the diagnosis is not available.
Table 1.
The numbers and percentages of lung cancer cases by histologic type and histological confirmation status, SEER 9*, 1975–2010.
| Overall | Histological Confirmation Status (Column %) |
|||
|---|---|---|---|---|
| (n=522,416 (100.0%)) |
Confirmed (n=470,326 (90.0%)) |
Not confirmed (n=38,657 (7.4%)) |
Unknown (n=13,433 (2.6%)) |
|
| Small-cell Carcinoma | 14.4 | 15.7 | 1.7 | 3.4 |
| Non-small Cell (NSC) Carcinoma | 68.5 | 75.2 | 7.0 | 8.9 |
| Squamous | 22.6 | 24.8 | 1.9 | 2.3 |
| Adenocarcinoma | 32.1 | 35.3 | 3.1 | 4.0 |
| Large-cell | 5.6 | 6.2 | 0.3 | 0.6 |
| Other specified NSC | 3.1 | 3.4 | 0.2 | 0.3 |
| 8046 (NSC carcinoma) | 5.1 | 5.5 | 1.5 | 1.7 |
| 8010 (carcinoma, NOS) | 12.5 | 7.6 | 61.5 | 43.0 |
| Other specified and unspecified types | 4.6 | 1.4 | 29.8 | 44.7 |
Note:
The SEER 9 registries include Atlanta, Connecticut, Detroit, Hawaii, Iowa, New Mexico, San Francisco-Oakland, Seattle-Puget Sound, and Utah.)
NSC=Non-small Cell; NOS=Not otherwise specified.
Table 2 shows the distributions of lung cancer cases by histology and selected covariates. All covariates are closely associated with histology. Men and older patients were more likely to be diagnosed with squamous type. Squamous and adenocarcinoma tumors tended to be more well-differentiated than large cell and other specific NSC tumors. Squamous and large cell tumors tended to be larger at diagnosis. Small-cell tumors were likely detected at a later stage (61.6%) as compared to other types. In contrast, tumors of squamous and adenocarcinoma types tended to be detected at early stage. There are also a few notable differences in the use of nonspecific codes across registries. For example, a lower use of 8046 (15.2% in 8046 compared to the overall percentage of 20.8%) is observed in Detroit, and a higher use of both 8010 (16.9% compared to the overall percentage of 15.0%) and 8046 (19.9%) is observed in Seattle. The use of nonspecific code is also slightly higher for cases not reported by a hospital (2.8% in 8010 and 2.9% in 8046 compared to the overall percentage of 1.8%). These variables are also predictive to the use of nonspecific morphology codes. As we expected, tumors without specific histological diagnosis tended to be less well-differentiated, diagnosed at a late stage, had shorter survivals, and were less likely to to be candidates for surgery.
Table 2.
Distribution of histologically confirmed lung cancer cases by histology and selected covariates, SEER 9*, 1975–2010
| Overall | Small- cell |
Squamous | Adeno- carcinoma |
Large- cell |
Other specified NSC |
8010 (carcinoma, NOS) |
8046 (NSC carcinoma) |
||
|---|---|---|---|---|---|---|---|---|---|
| Overall | 463,609 (100.0%) |
73,994 (100.0%) |
116,775 (100.0%) |
166,006 (100.0%) |
29,123 (100.0%) |
15,914 (100.0%) |
35,954 (100.0%) |
25,843 (100.0%) |
|
| Age | <50 Yrs | 6.5 | 5.4 | 4.1 | 7.8 | 8.5 | 14.6 | 6.3 | 5.7 |
| 50–<60 Yrs | 17.9 | 19.5 | 15.2 | 19.1 | 20.6 | 19.7 | 16.4 | 16.6 | |
| 60–<70 Yrs | 32.4 | 35.3 | 33.5 | 31.5 | 33.0 | 30.5 | 30.4 | 26.8 | |
| 70–<80 Yrs | 31.2 | 30.3 | 34.9 | 29.6 | 28.4 | 25.9 | 32.3 | 32.7 | |
| >=80 Yrs | 12.0 | 9.4 | 12.3 | 11.9 | 9.5 | 9.4 | 14.6 | 18.2 | |
| Sex | Male | 59.8 | 56.1 | 71.4 | 53.4 | 63.0 | 53.2 | 62.1 | 55.8 |
| Race | White | 84.0 | 88.2 | 83.3 | 83.1 | 84.6 | 86.1 | 79.5 | 83.0 |
| Black | 10.4 | 7.8 | 12.1 | 9.9 | 11.2 | 9.5 | 12.6 | 11.2 | |
| Other | 5.5 | 4.0 | 4.5 | 6.9 | 4.1 | 4.1 | 7.7 | 5.8 | |
| Missing | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.2 | 0.1 | |
| Ethnicity | Non-Hispanic | 2.7 | 2.4 | 2.4 | 3.0 | 2.6 | 3.3 | 2.6 | 3.8 |
| Marital Status | Single | 9.0 | 8.2 | 8.8 | 9.0 | 8.2 | 10.7 | 3.4 | 3.7 |
| Married | 58.2 | 57.2 | 59.0 | 59.0 | 60.4 | 59.2 | 9.2 | 12.2 | |
| Sep/Div/Wid | 29.7 | 31.6 | 29.1 | 29.0 | 28.4 | 27.1 | 56.1 | 51.1 | |
| Missing | 3.1 | 3.0 | 3.1 | 3.1 | 3.0 | 3.0 | 31.3 | 33.0 | |
| Nativity | Native-born | 81.0 | 86.1 | 83.1 | 77.9 | 84.9 | 72.8 | 83.7 | 74.2 |
| Foreign-born | 8.2 | 7.0 | 7.9 | 9.0 | 8.1 | 7.2 | 9.0 | 8.0 | |
| Missing | 10.8 | 7.0 | 9.1 | 13.1 | 7.0 | 20.1 | 7.3 | 17.8 | |
| Data Source | Non-hospital | 1.8 | 1.6 | 1.5 | 1.8 | 1.1 | 1.9 | 2.8 | 2.9 |
| Grade | Grade 1 | 4.0 | 0.1 | 4.1 | 7.8 | 0.2 | 4.9 | 0.2 | 0.2 |
| Grade 2 | 13.1 | 0.7 | 24.8 | 18.2 | 0.5 | 2.6 | 0.6 | 1.8 | |
| Grade 3 | 27.4 | 5.9 | 36.0 | 30.6 | 21.8 | 8.0 | 39.5 | 30.5 | |
| Grade 4 | 27.4 | 44.7 | 2.2 | 1.8 | 48.3 | 41.9 | 2.8 | 2.4 | |
| Missing | 42.3 | 48.7 | 32.9 | 41.7 | 29.3 | 42.7 | 57.0 | 65.0 | |
| Tumor Size | <2cm | 8.3 | 4.6 | 5.9 | 12.2 | 5.3 | 17.1 | 4.2 | 8.1 |
| 2-<3cm | 10.9 | 6.3 | 9.1 | 15.0 | 8.8 | 12.4 | 7.4 | 11.6 | |
| 3-<4cm | 10.2 | 6.4 | 10.2 | 12.3 | 9.6 | 8.4 | 8.4 | 11.9 | |
| 4-<5cm | 8.1 | 5.7 | 9.1 | 8.4 | 8.3 | 6.0 | 7.1 | 10.4 | |
| >=5cm | 19.6 | 17.9 | 24.4 | 15.7 | 23.0 | 14.7 | 18.8 | 28.2 | |
| Missing | 43.0 | 59.0 | 41.3 | 36.4 | 45.0 | 41.4 | 54.1 | 29.8 | |
| Stage | Localized | 19.7 | 7.9 | 24.7 | 23.8 | 16.5 | 33.0 | 11.4 | 11.9 |
| Regional | 27.8 | 24.7 | 36.2 | 25.6 | 30.0 | 21.7 | 21.4 | 23.1 | |
| Distant | 46.5 | 61.6 | 31.7 | 46.1 | 47.1 | 40.3 | 56.1 | 62.0 | |
| Missing | 6.0 | 5.9 | 7.4 | 4.4 | 6.4 | 5.0 | 11.1 | 3.1 | |
| Surgery | Performed | 27.4 | 5.9 | 31.9 | 38.6 | 25.6 | 43.6 | 11.7 | 10.3 |
| Not perf’d | 69.1 | 89.2 | 64.3 | 58.8 | 69.0 | 52.5 | 83.4 | 89.4 | |
| Missing | 3.5 | 4.9 | 3.9 | 2.6 | 5.4 | 3.8 | 4.9 | 0.3 | |
| Survival | <1 Yr | 43.8 | 53.7 | 39.9 | 38.7 | 52.1 | 37.0 | 54.6 | 45.5 |
| 1-<2 Yrs | 11.2 | 15.5 | 11.4 | 9.9 | 10.7 | 7.3 | 10.1 | 10.3 | |
| 2-<3 Yrs | 3.7 | 3.1 | 4.1 | 4.0 | 3.4 | 2.3 | 3.1 | 3.2 | |
| >=3 Yrs | 16.8 | 6.7 | 18.1 | 22.1 | 14.4 | 30.8 | 9.0 | 9.7 | |
| Censored | 24.7 | 21.0 | 26.5 | 25.4 | 19.4 | 22.7 | 23.2 | 31.3 | |
| SEER 9 Registry | SMS | 15.3 | 13.2 | 13.5 | 16.6 | 17.9 | 15.0 | 16.0 | 17.6 |
| Connecticut | 16.2 | 15.9 | 15.4 | 17.3 | 15.4 | 15.7 | 16.9 | 14.5 | |
| Detroit | 20.8 | 21.3 | 23.0 | 19.8 | 20.9 | 21.2 | 20.8 | 15.2 | |
| Hawaii | 4.0 | 3.4 | 3.6 | 4.6 | 2.5 | 3.6 | 4.4 | 4.0 | |
| Iowa | 13.6 | 15.5 | 15.6 | 12.7 | 10.5 | 13.6 | 11.7 | 11.6 | |
| New Mexico | 4.5 | 4.9 | 4.5 | 4.2 | 4.2 | 3.9 | 4.3 | 5.6 | |
| Seattle | 15.0 | 15.3 | 13.8 | 15.1 | 11.5 | 15.1 | 16.9 | 19.9 | |
| Utah | 2.8 | 2.8 | 2.9 | 2.7 | 2.8 | 4.6 | 2.4 | 2.6 | |
| Atlanta | 7.9 | 7.7 | 7.8 | 6.9 | 14.4 | 7.3 | 6.7 | 9.1 | |
| % Below poverty | 0-<5 | 1.7 | 1.8 | 1.7 | 1.9 | 1.2 | 1.6 | 1.8 | 1.6 |
| 5-<10 | 54.9 | 55.6 | 52.7 | 56.3 | 52.2 | 56.2 | 53.8 | 57.5 | |
| 10-<20 | 41.7 | 40.7 | 43.9 | 40.4 | 44.7 | 40.8 | 42.7 | 38.6 | |
| >=20 | 1.7 | 1.9 | 1.7 | 1.4 | 1.9 | 1.4 | 1.7 | 2.2 | |
| % Current smoker (mean) | 21.4 | 21.7 | 21.9 | 21.1 | 20.9 | 21.5 | 21.4 | 21.4 | |
Notes: All two-way associations are significant at .001 level.
The SEER 9 registries include Atlanta, Connecticut, Detroit, Hawaii, Iowa, New Mexico, San Francisco-Oakland, Seattle-Puget Sound, and Utah.)
NSC=Non-small Cell; NOS=Not otherwise specified; SMS=San Francisco-Oakland; Seattle=Seattle- Puget Sound; Atlanta=Atlanta Metropolitan.
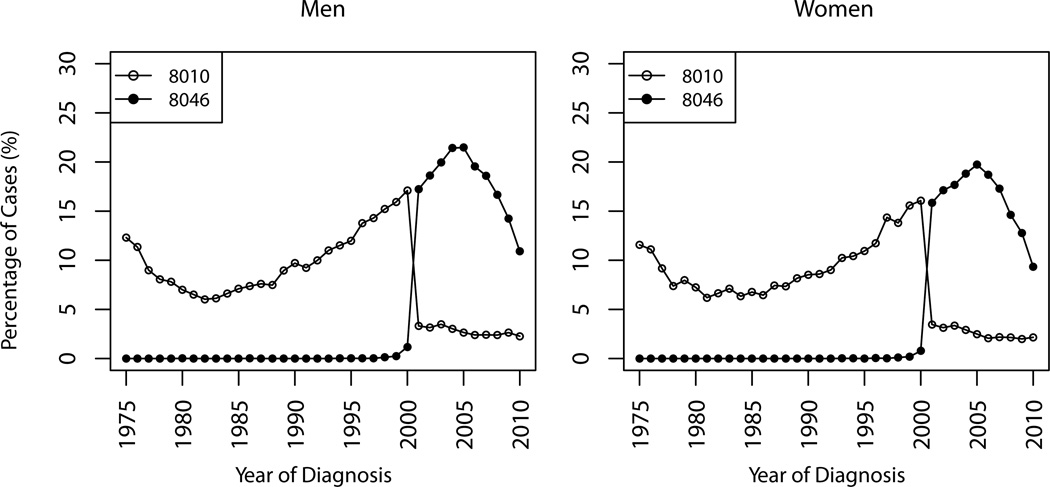
Figure 1 shows the percentages of cases coded with 8046 and 8010 by year of diagnosis for men and women separately. The temporal distributions are similar for both genders. The percentage of cases coded with 8010 had increased from 1982 until the introduction of 8046 into ICD-O-3 in 2001, when it dropped to around 3%. There seems to be a smooth compensation between 8010 and 8046 in 2001, which suggests that 8010 and 8046 are probably used interexchangebly in practice.
Figure 1.
Percentages of histologically confirmed lung cancer cases coded as 8010 and 8046, SEER 9, 1975–2010.
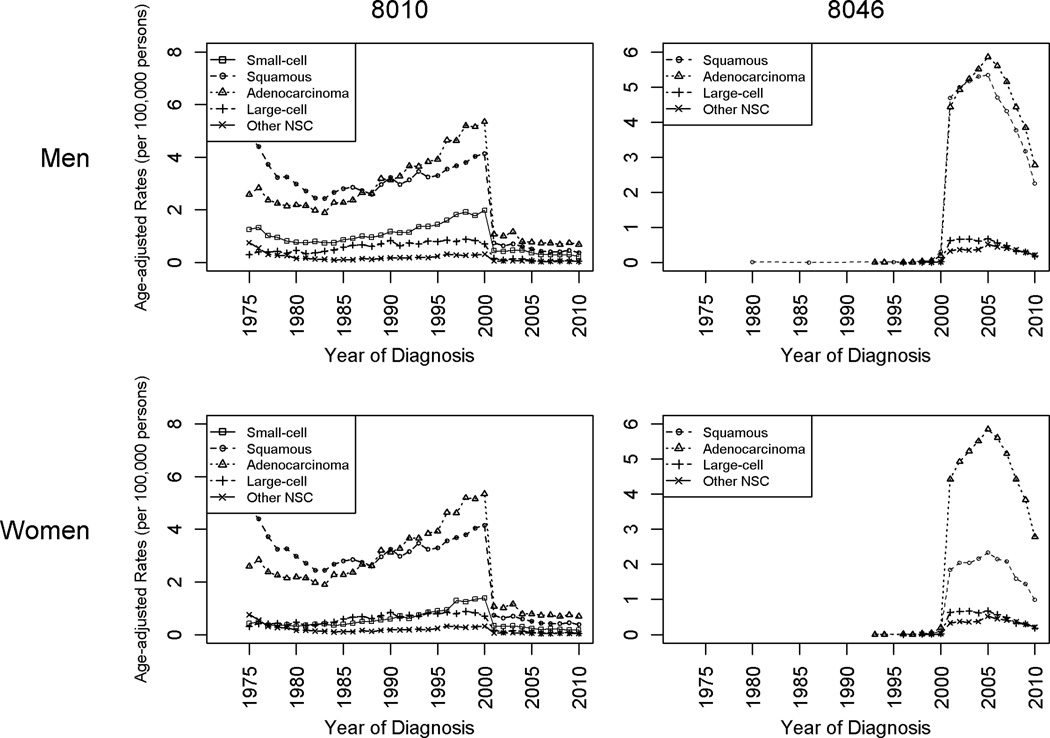
Figure 2 shows the rates of incidence by imputed histology among cases coded with 8010 or 8046. Overall, the amount of imputed histology differs by histologic subtype and year. For both 8010 and 8046, the rates of incidence raised by imputation were greatest for adenocarcinoma and squamous. For both histologic subtypes, the rates followed an n-shaped pattern over the most recent 15 years. Small-cell was the third-most raised category, although only contributed from imputing 8010 cases, and the amount of increases was relatively stable over time.
Figure 2.
Imputed incidence rates by histologic subtype and gender, histologically confirmed cases that were originally coded as 8010 or 8046, SEER 9, 1975–2010.
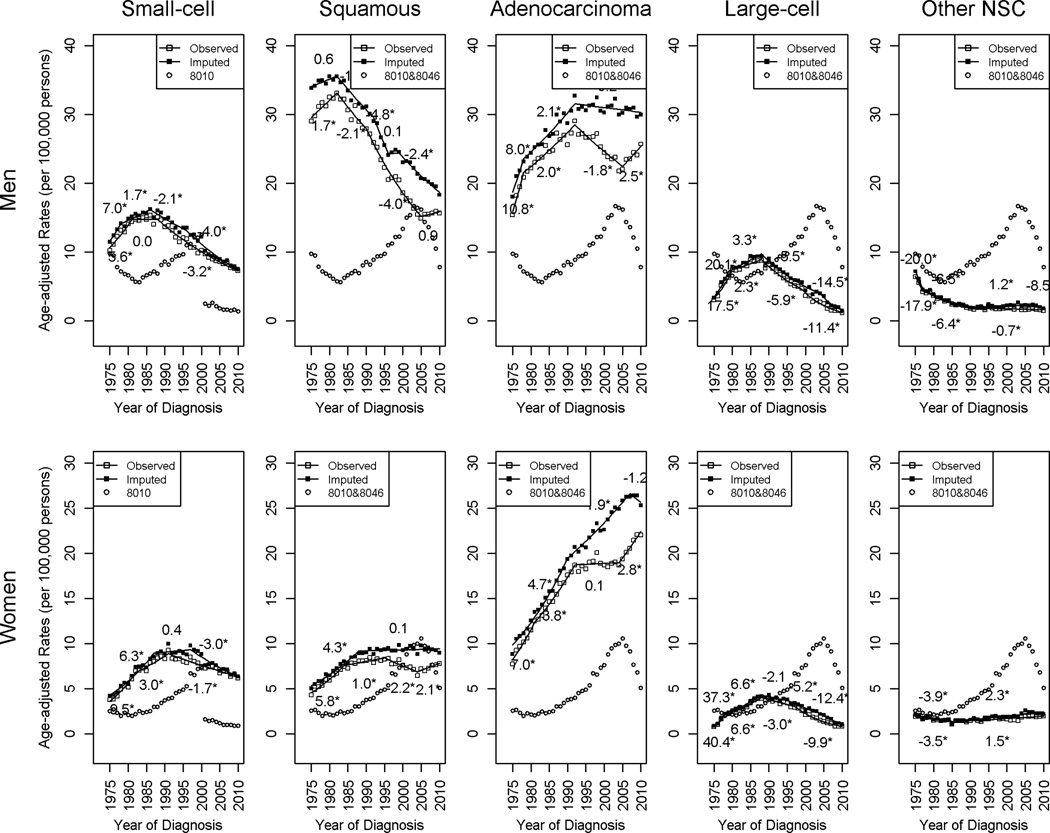
Figure 3 compares the before and after imputation temporal trends in age-adjusted incidence rate of lung cancer by histology for men and women separately (see Table 3 for detailed results of the joinpoint trends analysis.) The numbers listed over (imputed) or under (original) each segment represents the APC for that portion of the trend and an asterisk indicates a statistically significant trend at 0.05 level. The rates for 8010 (small-cell type) and 8010 and 8046 combined (NSC subtypes) are also included in these plots to help examine how cases are distributed by the imputation procedure.
Figure 3.
Observed and imputed incidence rates by histologic subtype and gender, histologically confirmed malignant cancer cases, SEER 9, 1975–2010.
Table 3.
Joinpoint analysis for histologically confirmed malignant lung cancers by imputation status, gender, and histology, SEER 9*, 1975–2010
| Trend 1 | Trend 2 | Trend 3 | Trend 4 | Trend 5 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Years | APC (95% C.I.) |
Years | APC (95% C.I.) |
Years | APC (95% C.I.) |
Years | APC (95% C.I.) |
Years | APC (95% C.I.) |
||
| Men | |||||||||||
| Small-cell | Original | 1975–1981 | 5.6 (3.8,7.4) | 1981–1988 | 0.0 (−1.4,1.5) | 1988–2010 | −3.2 (−3.4,−3.0) | ||||
| Imputed | 1975–1978 | 7.0 (0.3,14.2) | 1978–1986 | 1.7 (0.2,3.2) | 1986–1996 | −2.1 (−3.0,−1.1) | 1996–2010 | −4.0 (−4.5,−3.5) | |||
| Squamous | Original | 1975–1982 | 1.7 (0.6,2.7) | 1982–1990 | −2.1 (−3.1,−1.2) | 1990–2005 | −4.0 (−4.3,−3.6) | 2005–2010 | 0.9 (−1.0,2.8) | ||
| Imputed | 1975–1982 | 0.6 (−0.1,1.3) | 1982–1992 | −1.8 (−2.3,−1.4) | 1992–1996 | −4.8 (−7.2,−2.2) | 1996–1999 | 0.1 (−5.2,5.6) | 1999–2010 | −2.4 (−2.8,−2.0) | |
| Adenocarcinoma | Original | 1975–1978 | 10.8 (4.3,17.6) | 1978–1992 | 2.0 (1.5,2.5) | 1992–2005 | −1.8 (−2.3,−1.3) | 2005–2010 | 2.5 (0.7,4.3) | ||
| Imputed | 1975–1978 | 8.0 (2.6,13.7) | 1978–1992 | 2.1 (1.7,2.6) | 1992–2010 | −0.2 (−0.4,0.0) | |||||
| Large-cell | Original | 1975–1980 | 17.5 (12.1,23.2) | 1980–1988 | 2.3 (0.3,4.4) | 1988–1999 | −5.9 (−7.0,−4.8) | 1999–2010 | −11.4 (−12.9,−10.0) | ||
| Imputed | 1975–1979 | 20.1 (12.4,28.3) | 1979–1988 | 3.3 (1.7,4.9) | 1987–2004 | −5.5 (−6.1,−4.9) | 2004–2010 | −14.5 (−18.0,−10.9) | |||
| Other specific NSC | Original | 1975–1977 | −17.9 (−28.7,−5.3) | 1977–1990 | −6.4 (−7.5,−5.3) | 1990–2010 | −0.7 (−1.3,−0.1) | ||||
| Imputed | 1975–1977 | −20.0 (−31.1,−7.2) | 1977–1990 | −6.5 (−7.6,−5.5) | 1990–2007 | 1.2 (0.4,2) | 2007–2010 | −8.5 (−17.7,1.6) | |||
| Women | |||||||||||
| Small-cell | Original | 1975–1982 | 9.5 (7.4,11.6) | 1982–1991 | 3.0 (1.9,4.2) | 1991–2010 | −1.7 (−2.0,−1.5) | ||||
| Imputed | 1–75–1987 | 6.3 (5.3,7.4) | 1987–1997 | 0.4 (−0.8,1.6_ | 1997–2010 | −3.0 (−3.6,−2.3) | |||||
| Squamous | Original | 1975–1984 | 5.8 (4.8,6.8) | 1984–1995 | 1.0 (0.3,1.6) | 1995–2004 | −2.2 (−3.1,−1.4) | 2004–2010 | 2.1 (0.8,3.5) | ||
| Imputed | 1975–1988 | 4.3 (3.7,4.9) | 1988–2010 | 0.1 (−0.1,0.3) | |||||||
| Adenocarcinoma | Original | 1975–1981 | 7.0 (5.1,9.0) | 1981–1992 | 3.8 (3.2,4.5) | 1992–2004 | 0.1 (−0.3,0.5) | 2004–2010 | 2.8 (1.8,3.8) | ||
| Imputed | 1975–1990 | 4.7 (4.3,5.0) | 1990–2007 | 1.9 (1.6,2.1) | 2007–2010 | −1.2 (−3.6,1.2) | |||||
| Large-Cell | Original | 1975–1978 | 40.4 (24.1,59.0) | 1978–1988 | 6.6 (5.2,7.9) | 1988–1997 | −3.0 (−4.3,−1.7) | 1997–2010 | −9.9 (−10.7,−9.1) | ||
| Imputed | 1975–1978 | 37.3 (20.8,56.2) | 1978–1988 | 6.6 (5.2,8.0) | 1988–1995 | −2.1 (−4.1,0.1) | 1997–2004 | −5.2 (−6.6,−3.7) | 2004–2010 | −12.4 (−15.3,−9.4) | |
| Other specific NSC | Original | 1975–1985 | −3.5 (−5.3,−1.6) | 1985–2010 | 1.5 (1.1,1.9) | ||||||
| Imputed | 1975–1985 | −3.9 (−5.7,−2.1) | 1985–2010 | 2.3 (1.8,2.7) | |||||||
Notes:
The SEER 9 registries include Atlanta, Connecticut, Detroit, Hawaii, Iowa, New Mexico, San Francisco-Oakland, Seattle-Puget Sound, and Utah.) APC=Annual Percent Change
The imputation adjustment affected the incidence trends differently for each histologic subtype. For small-cell in both genders, the original and imputed trends are similar. For squamous cell cancer in both genders and adenocarcinoma in men, the trends showed a similar pattern overall from 1970 to early 1990’s before and after imputation. From early 1990’s to 2005, the decreasing trends also remained unchanged after imputation, but the pace of decline slowed. After 2005, the increasing trends based on the original data had been replaced by the steady continuations of earlier decreasing trends for squamous and adenocarcinoma in men, a constant trend for squamous in women, after imputation. For adenocarcinoma in women, the trends, before and after imputation, exhibited similar patterns overall before early 1990’s. From 1992 to 2007, the plateau followed by an increasing trend started in 2004 changed to a continuously increasing trend after imputation. It is also worth noting that the imputed rates showed a nonsignificant decreasing tendency during the most recent 3 years starting in 2007. For large cell cancer and cancer in other specified NSC type, the imputed rates were similar to the original rates and the imputation did not change the overall trends.
To rule out the possibility that changes in trends may be due to the absence of cases that are not histologically confirmed or have missing confirmation status, we conducted a sensitivity analysis on all cases. The imputation affected the trends similarly (see Supplementary Figure 1 and Supplementary Table 1 for detailed results on the rates and jointpoint analysis), which suggests that excluding these cases does not affect the overall findings and conclusions.
DISCUSSION
In cancer surveillance data collections, it is common for the morphological classification systems to change to reflect the contemporary pathology practice. Hence, the data often comprise cancer cases coded one way at one time and others a different way at another time. When classification systems differ in coding histology, temporal inferences by histologic subtype can be misleading and difficult to interpret. Without access to a calibration data to inform the underlying distribution of histology among cases coded without specificity or the association in histology between editions of classification systems, we carefully developed a MI approach to correct for biases in statistical inferences about temporal trends of lung cancer incidence based on the MAR assumption.
Although this assumption is not empirically testable, we argue that MAR is reasonable in our setting because we have identified and included into the imputation models an extensive set of auxiliary variables that can explain the missingness of specific histology, e.g. receipt of cancer-directed surgery, and that are correlates of histology, e.g. the stage, grade, and size of a tumor, as well as patient survival. Other important variables that could enhance the MAR assumption plausibility are patients’ smoking status and socioeconomic status (40, 41), for which we substituted county-level estimates at 2000 (pooled estimates from 2000 to 2003 for smoking) from the decennial census because they are not routinely collected in SEER. Although such estimates are not available for every diagnosis year, we believe the ranking of a county in smoking prevalence or poverty level relative to the rest of the country remains relatively unchanged over time. The potential confounding between smoking status and poverty (40) is not likely a cause for concern in our analysis because both are aggregate measures and neither is a strong predictor to histology after conditional on other patient-level information.
Ensuring the plausibility of MAR assumption imposed two modeling challenges of handling a large number of variables with missing data and a general missing data pattern, which often cannot be adequately addressed by simple imputation methods (21). The proposed MI approach based on SRMI is particularly suitable to this complex situation because of its flexibility in specifying and fitting conditional distributions. The search for refined ridge-penalized logistic regression imputation models is necessary because the standard SRMI approach (based on logistic regressions) might be inadequate in handling a categorical outcome with a skewed distribution (e.g. certain histology categories only contain 3% to 6% of cases) and correlated covariates (e.g. stage and survival). The simulation study demonstrated the adequacy and prediction benefits of the proposed semi-parametric models.
The amount of lung cancer cases lacking specific histologic subtypes was predominantly associated with the year of diagnosis, which reflected the evolution of SEER coding algorithms and recent changes in diagnostic practice. The imputation raised the incidence rates across the entire study period for both genders and histology subgroups. However, the magnitudes of the elevations varied. Of the various histologic subtypes, the most impacted were squamous and adenocarcinoma, on which the most pronounced impacts occurred during the last decade. This result further supports our hypothesis that 8010 and 8046 are mainly used to group cases which could have been coded as either adenocarcinoma or squamous type if more coding information were extracted and available to support detailed histological coding. For both subtypes, the decreasing trends from early or mid 1990’s to 2005, had persisted, although at a slower pace. The increasing trends after 2005 are apparently an artifact of this coding change and imprecision in histopathologic classification, which, after imputation, became a continuation of earlier decreasing trends. The sensitivity analyses including cases that are not histologically confirmed or have missing histological confirmation information showed similar results.
We classified lung cancers according to a schema developed based on Travis et al. (42) and earlier versions of ICD-Os. WHO recently published a revised version of the histological grouping for lung cancers (43). Different histological classification systems have been used in practice, for example, the recently published classification schema by the International Agency for Research on Cancer of the WHO (43) in 2007. The differences between this new classification and the one used in this research are summarized in Supplementary Table 2. Because the groupings of the most frequently used morphologic codes are consistent between the two schemas, we suspect that the effect of using this alternative schema on the inferences of incidence trends is noticeable for the histologic subtypes that we investigated in this research.
In summary, molecular, genetic, and etiologic features are increasingly associated with histology distinctions (3, 4, 44). Progress in linking molecular features to morphology will facilitate mechanistic understanding and further characterization of the molecular and genetic features specific to histologic subtypes in lung cancer. These considerations, along with the emergence of targeted therapies within specific histologic subtypes especially adenocarcinoma, clearly indicates that accurate population tracking of trends by lung cancer histology will be increasingly important in the future, and that the MI technique applied in this study can help refine these trends. Planned data collections for bridge data in the future will further enhance the quality of data augmented by MI.
Supplementary Material
Footnotes
The authors do not have any conflicts of interest.
Contributor Information
Mandi Yu, Division of Cancer Control and Population Sciences, National Cancer Institute.
Eric J. Feuer, Division of Cancer Control and Population Sciences, National Cancer Institute
Kathleen A. Cronin, Division of Cancer Control and Population Sciences, National Cancer Institute
Neil E. Caporaso, Division of Cancer Epidemiology and Genetics, National Cancer Institute
REFERENCES
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, J Ruhl, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review. Bethesda, MD: National Cancer Institute; 1975–2010. http://seer.cancer.gov/csr/1975_2010/, based on November 2012 SEER data submission, posted to the SEER web site, April 2013. [Google Scholar]
- 2.Lamb D. Histological classification of lung cancer. Thorax. 1984;39:161–165. doi: 10.1136/thx.39.3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J, Chatterjee N, Rotunno M, Wang Y, Pesatori AC, Consonni D, et al. Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2:131–139. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Husain H, Rudin CM. ALK-targeted therapy for lung cancer: ready for prime time. Oncology. 2011;25:597–560. [PubMed] [Google Scholar]
- 8.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Tsao A, et al. The BATTLE rrial:personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinsky P. Board of Scientific Advisor & National Cancer Advisory Board. Bethesda, Maryland: National Cancer Institute; 2013; National Lung Screening Trial (NLST) subset analysis. [Google Scholar]
- 10.Jemal A, Simard E, Dorell C, Noone A, Markowitz L, Kohler B, et al. Annual report to the nation on the status of cancer, 1975–2009, reaturing the burden and trends in HPV-associated cancers and HPV vaccination coverage levels. J Natl Cancer Inst. 2013;105:175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surveillance, Epidemiology, and End Results (SEER) Program ( www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 9 Regs Research Data, Nov 2011 Sub (1975–2010) <Katrina/Rita Population Adjustment> - Linked To County Attributes - Total U.S., 1969–2010 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, released April 2013, based on the November 2012 submission. [Internet]
- 12.Travis W, Brambilla E, Noguchi M, Nicholson A, Geisinger K, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8:381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 13.Cole SR, Chu H, Greenland S. Multiple-imputation for measurement-error correction. Int J Epidemiol. 2006;35:1074–1081. doi: 10.1093/ije/dyl097. [DOI] [PubMed] [Google Scholar]
- 14.Durrant GB, Skinner C. Using missing data methods to correct for measurement error in a distribution function. Surv Methodol. 2006;32:25–36. [Google Scholar]
- 15.Schenker N, Parker JD. From single-race reporting to multiple-race reporting: Using imputation methods to bridge the transition. Stat Med. 2003;22:1571–1587. doi: 10.1002/sim.1512. [DOI] [PubMed] [Google Scholar]
- 16.Thomas N, Raghunathan TE, Schenker N, Katzo MJ, Johnson CL. An evaluation of matrix sampling methods using data from the National Health and Nutrition Examination Survey. Surv Methodol. 2006;32:217–232. [Google Scholar]
- 17.Burgette LF, Reiter JP. Nonparametric Bayesian multiple imputation for missing data due to mid-study switching of measurement methods. J Am Stat Assoc. 2012;107:439–449. [Google Scholar]
- 18.Anderson WF, Katki HA, Rosenberg PS. Incidence of breast cancer in the United States: current and future trends. J Natl Cancer Inst. 2011;103:1397–1402. doi: 10.1093/jnci/djr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howlader N, Noone A, Yu M, Cronin K. Use of imputed population-based cancer registry data as a method of accounting for missing information: application to estrogen receptor status for breast cancer. Am J Epidemiol. 2012;176:347–356. doi: 10.1093/aje/kwr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.sLittle RJA, Rubin DB. Statistical analysis with missing data. Hoboken, New Jersey: John Wiley & Sons, Inc; 2002. [Google Scholar]
- 21.Raghunathan TE, Lepkowski JM, van Hoewyk J, Solenberger P. A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv Methodol. 2001;27:85–95. [Google Scholar]
- 22.Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York: Wiley & Sons; 1987. [Google Scholar]
- 23.Kim H-J, Fay MP, Feuer EJ, Midthune DN. Permutation tests for joinpoint regression with applications to cancer rates. Stat Med. 2000;19:335–351. doi: 10.1002/(sici)1097-0258(20000215)19:3<335::aid-sim336>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.David M, Little RJA, Samuhel ME, Triest RK. Alternative methods for CPS income imputation. J Am Stat Assoc. 1986;81:29–41. [Google Scholar]
- 25.Rubin DB, Stern HS, Vehovar V. Handling "don't know" survey responses: the case of the slovenian plebiscite. J Am Stat Assoc. 1995;90:822–828. [Google Scholar]
- 26.Little RJA. Missing-data adjustments in large surveys. J Bus Econom Statist. 1988;6:287–296. [Google Scholar]
- 27.Lin P-Y, Chang Y-C, Chen H-Y, Chen C-H, Tsui H-C, Yang P-C. Tumor size matters differently in pulmonary adenocarcinoma and squamous cell carcinoma. Lung Cancer. 2010;67:296–300. doi: 10.1016/j.lungcan.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Warren JLKC, Schrag D, Bach PB, Riley GF. Overview of the SEER-Medicare Data: Content, Research Applications, and Generalizability to the United States Elderly Population. Med Care. 2002;40:IV-3-18. doi: 10.1097/01.MLR.0000020942.47004.03. [DOI] [PubMed] [Google Scholar]
- 29.Thun MJ, Lally CA, Calle EE, Heath CW, Flannery JT, Flanders WD. Cigarette smoking and changes in the histopathology of lung cancer. J Natl Cancer Inst. 1997;89:1580–1586. doi: 10.1093/jnci/89.21.1580. [DOI] [PubMed] [Google Scholar]
- 30.Small Area Estimates for Cancer Risk Factors & Screening Behaviors. National Cancer Institute, DCCPS, Statistical Methodology & Applications Branch, released May 2010 ( sae.cancer.gov). Underlying data provided by Behavioral Risk Factor Surveillance System ( http://www.cdc.gov/brfss/) and National Health Interview Survey ( http://www.cdc.gov/nchs/nhis.htm). [Internet]
- 31.U.S. Census Bureau. Census 2000, Summary File 3, Table QT-P35; using American FactFinder. http://factfinder2.census.gov [Internet]
- 32.Le Cessie S, van Houwelingen JC. Ridge Estimators in Logistic Regression. Appl Statist. 1992;41:191–201. [Google Scholar]
- 33.Schaefer R, Roi L, Wolfe R. A ridge logistic estimator. Commun Stat-Theor M. 1984;13:99–113. [Google Scholar]
- 34.Yu M. Disclosure risk assessments and control. Ann Arbor: University of Michigan; 2008. [Google Scholar]
- 35.SAS Institute Inc. SAS/STAT 9.2 User's Guide. Cary, NC: 2008. [Google Scholar]
- 36.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data mining, Inference, and Prediction. second ed. New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 37.Greenland S, Finkle W. A critical look at methods for handling missing covariates in epidemiologic regression analyses. Am J Epidemiol. 1995;142:1255–1264. doi: 10.1093/oxfordjournals.aje.a117592. [DOI] [PubMed] [Google Scholar]
- 38.van der Heijden G, Donders A, Stijnen T, Moons K. Imputation of missing values is superior to complete case analysis and the missing-indicator method in multivariable diagnostic research: A clinical example. J Clin Epidemiol. 2006;59:1102–1109. doi: 10.1016/j.jclinepi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Karr AF, Kohnen CN, Oganian A, Reiter JP, Sanil AP. A framework for evaluating the utility of data altered to protect confidentiality. Amer Statist. 2006;3:224–232. [Google Scholar]
- 40.Menvielle G, Boshuizen H, Kunst A, Dalton S, Vineis P, Bergmann M, et al. The role of smoking and diet in explaining educational inequalities in lung cancer incidence. J Natl Cancer Inst. 2009;101:321–330. doi: 10.1093/jnci/djn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett VA, Davies EA, Jack RH, Mak V, Møller H. Histological subtype of lung cancer in relation to socioeconomic deprivation in South East England. BMC Cancer. 2008;8:139. doi: 10.1186/1471-2407-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer. 1995;75:191–202. doi: 10.1002/1097-0142(19950101)75:1+<191::aid-cncr2820751307>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Curado MPEB, Shin HR, Storm H, Ferlay J, Heanue M, Boyle P, editors. Cancer Incidence in Five Continents. IX. Lyon, France: IARC; 2007. [Google Scholar]
- 44.Rotunno M, Yu K, Lubin JH, Consonni D, Pesatori AC, Goldstein AM, et al. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 2009;4:e5652. doi: 10.1371/journal.pone.0005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.