Abstract
The transcription factor HIF1α is implicated in the development of clear cell renal cell carcinoma (ccRCC). Although HIF1α was initially believed to be essential for ccRCC development, recent studies hypothesize an oncogenic role for HIF2α in ccRCC, but a tumor suppressor role for HIF1α [1], leading to uncertainty as to the precise roles of the different HIF transcription factors in this disease.
Using evidence available from studies with human ccRCC cell lines, mouse xenografts, murine models of ccRCC, and human ccRCC specimens, we evaluate the roles of HIF1α and HIF2α in the pathogenesis of ccRCC. We present a convergence of clinical and mechanistic data supporting an important role for HIF1α in promoting tumorigenesis in a clinically important and large subset of ccRCC. This indicates that current understanding of the exact roles of HIF1α and HIF2α is incomplete and that further research is required to determine the diverse roles of HIF1α and HIF2α in ccRCC.
Keywords: Clear cell renal cell carcinoma, Cancer stem cells, Hypoxia inducible factor 1α, Kidney cancer, Carcinogenesis, Review
REVIEW
Clear Cell Renal Cell Carcinoma and Von Hippel-Lindau Disease
Approximately 75–85% of all kidney cancers are clear cell renal cell carcinomas (ccRCC). Known for many years to be hypervascular tumors, it was not until the discovery and characterization of the Von Hippel-Lindau (VHL) gene that researchers gained a better understanding of the pathogenesis of ccRCC. Families with VHL disease have a high incidence of ccRCC that occurs as a consequence of VHL inactivation. However, most patients with sporadic ccRCC acquire a single deleted or inactivated VHL allele in normal kidney proximal tubule cells and over time, loss of heterozygosity occurs and both VHL alleles become inactivated. This is believed to lead to the development of sporadic renal ccRCCs [2].
The VHL protein is a member of the E3 ubiquitin ligase complex that reduces the levels of the transcription factors HIF1α and HIF2α during normoxia [3–5]. Both HIF1α and HIF2α normally function in part to transcriptionally activate target genes in response to hypoxic conditions. In the normal rat kidney, hypoxia results in high levels of HIF1α protein in proximal and distal tubules and in collecting ducts, while high HIF2α protein levels are found in peritubular endothelial cells and in renal fibroblasts [5]. However, in the absence of active VHL protein, as is the case in the majority of ccRCCs, the VHL-associated proteolysis of HIF1α and HIF2α that occurs in the absence of hypoxia is lost, leading to constitutive activity of HIF1α and HIF2α independent of the oxygen level ([6, 7]; for review [8] ). Whereas the VHL protein normally functions as an E3 ubiquitin ligase that targets HIF1α, studies show that VHL may have other functions, such as in metabolism and inflammation, as judged by various studies in model organisms in addition to mice [9].
The loss of VHL tumor suppressor function and the resulting loss of regulated HIF degradation in ccRCC cells results in the increased expression of several proteins transcriptionally activated by HIFα that are involved in angiogenesis, such as vascular endothelial growth factor (VEGF) and platelet-derived growth factor B chain (PDGF-B). The increased expression of VEGF in ccRCCs explains the vascularity of these tumors, and directly led to the development of a variety of therapies that specifically target the VEGF pathway. Currently, sunitinib, pazopanib, sorafenib, and axitinib, all small molecule inhibitors of receptor tyrosine kinases, including the VEGF receptor, are in use for the treatment of advanced ccRCC [10]. The humanized monoclonal antibody (bevacizumab) that recognizes and inactivates VEGF, a HIF target gene, is also widely used to treat advanced ccRCC [11, 12].
Two other small molecular weight drugs approved to treat ccRCC, temsirolimus and everolimus, act by inhibiting the mammalian target of rapamycin (mTOR) [13]. mTOR consists of two enzymatically active complexes, mTOR complex 1 (mTORC1) and mTORC2 [14]. Activation of mTOR complexes leads to the stimulation of ribosomal translation of various mRNAs, including the translation of HIF1α message, whereas inhibition of mTOR results in decreased HIF1α translation [15]. Thus, the successful treatment of ccRCC today involves direct and indirect targeting of the HIF pathway, though it is becoming clear that significant intratumoral heterogeneity exists within primary and metastatic ccRCCs in the same patient, and this heterogeneity makes successful eradication of ccRCC more difficult [16].
The Roles of HIF1α and HIF2α in Human Clear Cell Renal Cell Carcinoma
Over the past 10 years, numerous researchers have studied the roles of the VHL target genes HIF1α and HIF2α in renal carcinogenesis (for review [17]). Many of these studies directly implicate the overexpression of HIF1α as a critical factor in ccRCC tumorigenesis. In contrast, others have reported that HIF2α is more tumorigenic than HIF1α in ccRCC [1, 18], as well as implicating HIF1α as a tumor suppressor in ccRCC [1]. We recently developed transgenic mouse models that specifically express either a mutated, constitutively active HIF1α or HIF2α in mouse proximal tubule cells, the normal progenitor cells of ccRCC (see below). In these models, we observed the development of ccRCC in mice expressing constitutively active HIF1α but not in mice expressing constitutively active HIF2α [19, 20]. These results have led us to critically re-examine the evidence for the specific roles of HIF1α and HIF2α in human renal clear cell carcinogenesis.
Cell and Animal Model Data
There are numerous, somewhat contradictory reports concerning the results of HIF1α and HIF2α overexpression and/or shRNA knockdown in tumor cell lines and xenograft models of human tumor cell proliferation. Xu et al [21] demonstrated that the silencing of HIF1α in the human RCC lines Caki-1 and OS-RC-2 inhibited growth in cell culture and inhibited tumorigenicity in tumor xenograft experiments in athymic mice. In another xenograft model, the apoptosis repressor with a caspase recruitment domain ARC gene was shown to be activated directly by HIF1α at the transcriptional level in human renal cell carcinoma cell lines. Loss of expression of ARC led to a great reduction in RCC proliferation in SCID mice in vivo [22], indicating that this HIF1α target gene regulates the growth of human RCC cells. The data from these two publications implicate HIF1α in driving RCC cell proliferation. In contrast, the knockdown of HIF2α prevented the growth of renal tumors in numerous xenograft models, whereas HIF1α knockdown did not prevent the growth of tumors in xenografts [23–28]. Conversely, overexpression of HIF2α also caused increased growth rates of tumors in xenografts, but not in cell culture experiments [29].
It is important to note, however, that in all of these experiments only the proliferation of kidney tumor cells was examined, and not the actual process of tumor development. The striking differences observed in these cell culture and xenograft assays with respect to HIF1α and HIF2α actions on cancer cell proliferation, i.e. the contradictory results obtained by various laboratories, are not easily explained and may be related to the particular human tumor cell lines used in the assays [18, 21–28].
Murine Models of Kidney Cancer with Constitutive HIF1α Expression
Currently, few mouse models of RCC possess the major features of human RCC (for review [30–32]). Attempts to recapitulate human kidney carcinogenesis by inactivation of the human tumor suppressor gene VHL have not been successful [33–38]. Clear cells, renal cysts, and tumors characteristic of human RCC are not generally seen in these models, but why these VHL animal models lack the ability to mimic the human disease is not understood. Other RCC models have long latency times [39, 40]. Models of adenomatous polyposis coli (APC) deficiency in mice lead to RCC, but this occurs over an extended period and is associated with activation of the β-catenin pathway, an event that is not common in human RCC [41]. Currently, murine xenograft models using human RCC-derived cell lines or human ccRCC explants are used to test new drugs for efficacy (e.g. [42–46]).
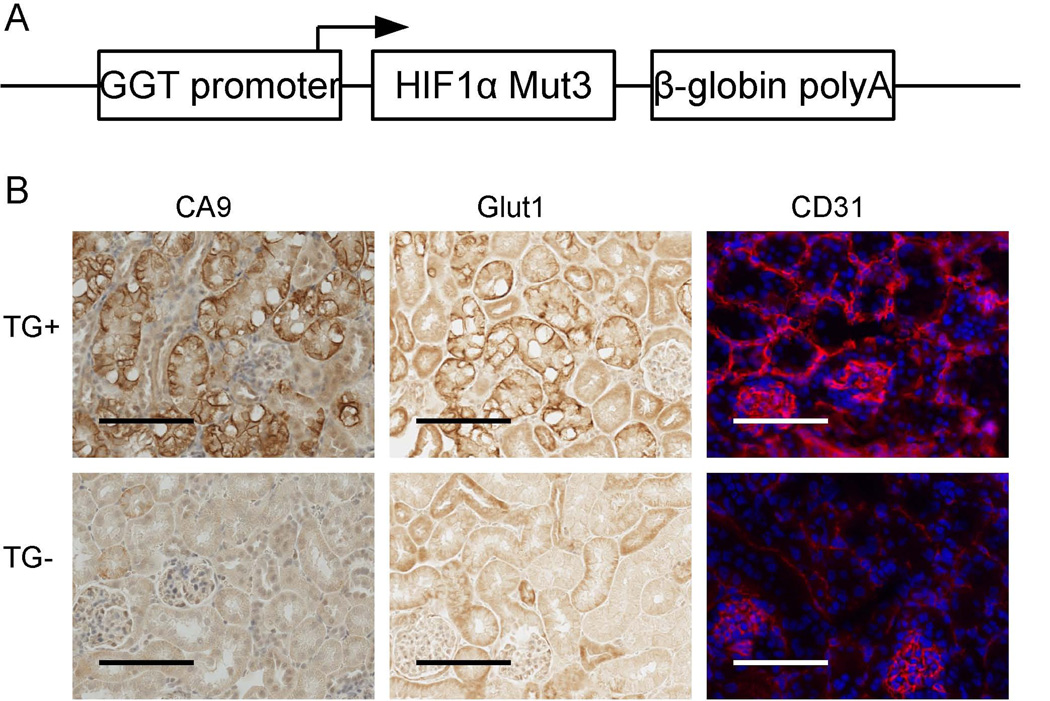
We recently developed the TRACK (transgenic cancer of the kidney) murine model of kidney cancer in which constitutively active HIF1α is expressed specifically in renal proximal tubules, and this expression drives tumorigenesis that recapitulates many pathological and molecular features of early human ccRCC [20] (Fig. 1). For instance, expression of CA9, Glut1, and CD31 (as a marker of angiogenesis) is highly elevated in TRACK kidneys, and these proteins are similarly highly expressed in human ccRCC [47, 48]. In contrast, we and another research group have shown that constitutive HIF2α expression specifically in the proximal tubules does not lead to neoplastic transformation in ccRCC [19, 49]. Furthermore, we showed by genome-wide RNA-seq that the transcripts expressed in the mice that constitutively express HIF2α in their kidneys do not resemble the transcripts highly expressed in human ccRCC samples assessed in Oncomine, a cancer array database [19, 50]. Crossing the TRACK mice with the mice that express constitutively active HIF2α in their kidneys did not make the tumorigenic phenotype more severe; the double transgenic TRACK-HIF2α-mut3 mice resembled mice expressing only the TRACK-HIF1α-mut3 gene [19]. Therefore, taken together, the transgenic animal models of kidney cancer are most consistent with HIF1α having a major role in driving ccRCC tumorigenesis.
Figure 1. (A). Simplified construction map of the linearized γ-HIF1α-M3 plasmid used to create the TRACK mouse.
The expression of the triple mutant (P402A, P564A, N803A) HIF1α ORF is driven by the proximal tubule specific type 1 gamma-glutamyl transpeptidase (GGT) promoter. (B). Immunostaining of CA-9, Glut-1 and CD31 in TRACK kidney proximal tubule cells. Increased expression levels of CA-9, Glut-1 and CD31 proteins are shown as stronger immunohistochemistry (CA-9, Glut-1) or immunofluorescence (CD31, red) signals in the TRACK TG+ mouse compared to the TG- mouse. Scale bars represent 100µm.
Protein Expression Data in Human RCCs
When tumor specimens are examined for HIF1α and HIF2α protein expression, generally nuclear expression is expected since both HIF1α and HIF2α are transcription factors that should act in the nucleus. Numerous studies have examined the expression of HIF1α and HIF2 α by immunostaining in ccRCCs (Table 1) [28, 51–62]. Although the definition of high HIF1α expression varied among studies, on average HIF1α protein was detected in 70% (range 17% to 97%) of ccRCC specimens. In a number of studies, HIF1α expression was associated with an inferior survival [55, 56, 61, 63]. Schultz et al. [61] showed that HIF1α levels were significantly higher in primary and metastatic ccRCCs compared with benign tissues (P<0.0001), and that tumor size and HIF1α expression were independent predictors of both reduced disease free survival and tumor progression in primary ccRCC. In another recent report, the highest level of HIF1α expression in ccRCCs was associated with the worst prognoses [60]. Studies have also examined both HIF1α and HIF2α in the same tissue samples. Gordan et al. [28] reported HIF immunostaining patterns that separated ccRCCs into three distinct classes: no HIF protein detected (“VHL WT,” 12%), both HIF1α and HIF2α detected (61%), and HIF2α only detected (27%). Their data suggested that high HIF expression was inversely correlated with VHL expression. In another recent study, HIF1αHIGH/HIF2αLOW renal tumors had a worse overall survival compared with HIF1αLOW/HIF2αLOW tumors [51]. Thus, numerous analyses of human ccRCC specimens show that HIF1α is highly expressed in the majority of primary renal tumors, either alone or with HIF2α. Sato et al [54] recently reported expression of HIF1α protein by immunostaining in 84% of 106 primary surgical ccRCC specimens.
Table 1. Evaluation of HIF1α and HIF2α protein expression in renal cell carcinoma samples.
% positive indicates the percentage of tumor samples in which HIF1α or HIF2α protein expression was detected. % negative indicates the percentage of tumor samples in which both HIF1α and HIF2α protein were not detected. All studies used immunohistochemistry to evaluate protein expression unless otherwise specified in the remarks. For studies in which the % positive was not specified, details regarding their findings are noted in the remarks. Information regarding how the different studies determined positivity and negativity is also included in the remarks. The table is divided into three sections: the top section cites publications in which both HIF1α and HIF2α were assayed; the middle section includes publications in which only HIF1α was assessed; the bottom section includes one publication in which only HIF2α was assayed.
| Reference | Number of patients |
% HIF1α positive |
% HIF2α positive |
% HIF negative |
Remarks |
|---|---|---|---|---|---|
| Biswas, et al, [51] | 106 | 97 | 99 | Positivity was determined from intensity of staining.and percentage of cells stained positive (histoscore), For HIF1α. the histoscore cutoff >2 was used, and for HIF2α. the histoscore cutoff >0 was used. |
|
| Chintala, et al. [52] | 88 | 41 | 79 | 13 | HIF1α positive samples have staining in more than 66% of nuclei. HIF2α positive samples have an average of 15% of nuclei stained positive. |
| Gordan, et al. [28] | 57 | 51 | 88 | 12 | Positive samples have more than 40% of nuclei stained positive. |
| Nyhan, et at. [53] | 17 | 88 | 100 | 0 | Positive samples have expression of HIF1/2α as determined by Western blot analysis. |
| Sato, et at. [54] | 106 | 84 | 73 | 7 | Positive samples have low moderate or high expression 0f HIF1/2α. |
| Reference |
Number of patients |
% HIF1α positive | Remarks | ||
| Di Cristofano, et al. [55] | 136 | 95 | Positive samples have cytoplasmic staininq in more than 50% of cells. |
||
| Dorević, et al. [56] | 94 | Not specified | Immunoreactivity was evaluated as a percentage of nuclear or cytoplasmic positivity. The median value for positive nuclei is 47.1 and for positive cytoplasm is 12.9. |
||
| Klatte, et al. [57] | 308 | 17 | Positive samples have specific nuclear staining in more than 35% of cells. |
||
| Lidgren, et al. 2005[58] | 66 | 67 | Positive samples have high HIF1α expression as determined by Western blot analysis. |
||
| Lidgren, et al. 2006 [59] | 176 | 58 | Positive samples have medium or strong HIF1α staining. | ||
| Medina Villaamil, et al. [60] | 80 | 73 | HIF1α positive samples have strong staining (in an average of 80% of cells) or moderate positivity. | ||
| Schultz, et al. [61] | 176 | Not specified | Mean H-score (0–300) was calculated by taking into account the intensity of staining and the percentage of positive cells. For primary tumors, H-score mean is 76.2 and for metastatic ccRCC. H-score mean is 99.E. Benign tissue has an H-score mean of 0.6. |
||
| Reference |
Number of patients |
% HIF2α positive | Remarks | ||
| Kroeger, et al. [62] | 300 | 97 | Positive samples have > 32% cells with HIF2α staining in the nuclei. |
||
In a TMA (tissue microarray) study of 308 ccRCC patient specimens, high HIF2α nuclear (N) (cutoff >32%) expression correlated with smaller tumor sizes (p=0.002) and lower Fuhrman grades (p=0.044), whereas tumors with high cytoplasmic (C) HIF2α staining had a higher frequency of positive lymph nodes (p=0.004), distant metastases (p=0.021), and higher Fuhrman grades (p<0.0001). The localization of HIF2α in the cytoplasm rather than the nucleus associated with increased ccRCC severity, indicating a complex and poorly understood role for HIF2α in the cytoplasm in ccRCC [62].
Of note, HIF1α expression correlates significantly with the “clear” histological subtype of renal cell carcinoma (p<0.01) [60], possibly because HIF1α (and not HIF2α) increases the expression of lipin1, a phosphatidate phosphatase that catalyzes the last step in triglyceride biosynthesis [64]. Triglycerides form lipid droplets, the major neutral lipid stores in cells, and these lipid droplets cause the “clear” phenotype in human ccRCC. HIF1α activation also increases glutamine-dependent lipid synthesis in tumors by reducing the activity of the enzyme α-ketoglutarate dehydrogenase via SIAH-targeted ubiquitination; no effects of HIF2α on this enzyme were presented in this study [65].
Gene Expression and HIF Mutation/Deletion in Human RCCs
Genome wide chromatin immunoprecipitation, coupled with next generation sequencing (ChIPseq), has been used to identify HIF1α and HIF2α regulated gene networks [66]. Of the high stringency genomic regions identified for HIF1α (359 sites) and HIF2α (301 sites) in MCF-7 cells, only 157 are common to both HIF1α and HIF2α [66].
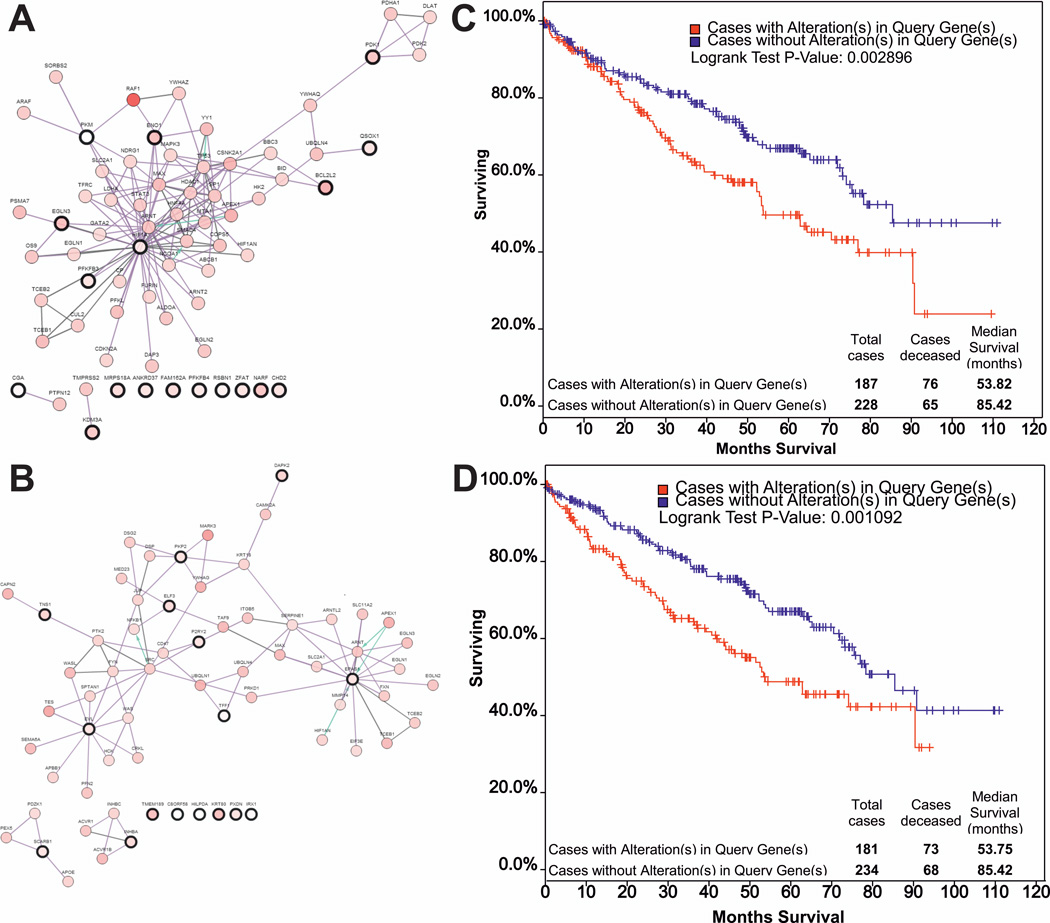
Analysis of the published renal cancer data from The Cancer Genome Atlas (TCGA) research network [67] shows that HIF1α and HIF2α mRNA expression is altered in 3% and 5% of patients, respectively, and that these genes are mutated in <1% of patients. However, an analysis of the expression in the same renal cancer cohort of putative HIF1α and HIF2α regulated gene targets [66] demonstrates that HIF1α (Figure 2A) and HIF2α (Figure 2B) regulate distinct networks which are both associated with poorer overall survival (Figure 2C, D). It is important to note that in the TCGA patients [67] only mRNA levels and no protein expression data are available. Furthermore, in these TCGA data the expression of HIF1α and HIF2α transcripts is compared to the average gene expression in kidney tumor tissue rather than to the levels of HIF1α and HIF2α in the normal kidney; this suggests that HIF1α and HIF2α levels are even higher in tumor tissue relative to the levels in normal kidney tissue. Of importance, mRNA expression data do not reflect the levels of HIF protein, which is regulated primarily postranscriptionally by protein degradation.
Figure 2. The cBio database was used to compare expression and clinical correlation in renal cancer of putative HIF1α and HIF-2α/EPAS1 target genes [66].
HIF1α (A) (RSBN1,CHD2,PFKFB3,QSCN6,PKM2,CGA,BCL2L2,EGLN3,MRPS18A,PFKFB4,PDK1,ANKRD 37,C3orf28, ZFAT1,ENO1,NARF,JMJD1A/KAT3A), and HIF2α (B) (TFF1, HIG2,TNS1, P2RY2,KRT80, EVL,C8orf58, PXDN,INHBA,IRX1,Kua,ELF3,SCARB1,PKP2,DAPK2) target genes form distinct networks. Kaplan-Meier survival estimates for HIF1α (C) and HIF-2α/EPAS1 (D) target gene expression are associated with poorer overall survival.
The identities of protein partners shown to interact with HIF1α and HIF2α also differ, supporting distinct roles for the HIF1α and HIF2α protein complexes in normal physiology and in carcinogenesis. For example, the protein interactions summarized in NCBI (http://www.ncbi.nlm.nih.gov/gene/) (Figure 3) indicate that HIF1α can interact with multiple, distinct epigenetic regulatory proteins, including the coactivators KAT2B/pCAF, NCoA1/SRC-1, and NCoA2/SRC-2/TIF2. In addition, multiple histone deacetylases (HDACs), which are typically associated with transcriptional repression, interact with HIF1α. This suggests that HIF1α can have both positive and repressive effects on transcriptional regulation. Interestingly, while there is evidence that HIF2α can interact with the mediator transcriptional complex and the CBP histone acetyltransferase [68], there is little overlap in known protein interactions of HIF1α and HIF2α in the diverse cell lines studied (Figure 3).
Figure 3. Comparison of HIF1α and HIF2α/(EPAS1) protein interactions.
HIF1α and HIF2α protein interactions reported in the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene/) were compared. HIF1α and HIF2α have 90 and 74 unique interaction partners respectively, with only 31 partners common to both. This suggests that the composition and function of HIF1α and HIF2α complexes may differ.
Is the HIF1α Gene Deleted in a High Proportion of Human ccRCCs?
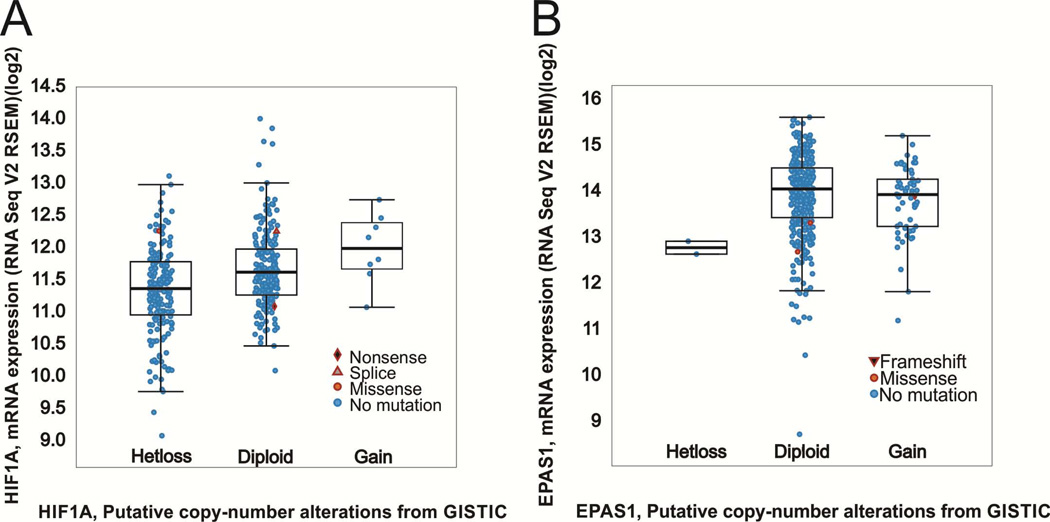
The report by Shen et al [1] implicating “HIF1α as a 14q kidney cancer suppressor gene” has been widely quoted in the literature, but these and related data [69] have not been carefully analyzed and compared to other published integrated molecular analyses of ccRCC. The deletions reported in Shen et al. [1] and Beroukhim et al. [69] in ccRCC specimens are at 14q31.1. It was also recently reported that “loss of chromosome 14q, associated with loss of HIF1α, occurs frequently in ccRCC,” with SNP (single nucleotide polymorphism) arrays indicating loss of 14q24.3 (encompassing NRXN3 and ∼462 other genes) in 45% of patients [67]. However, HIF1α maps at 14q23.2 (chr14:62162117-62214976) whereas NRXN3 is located at chr14:79745682-80334633 (HG19/GRCh37), some 17 million nucleotides distant. Moreover, RNAseq analysis of ccRCC specimens [67] identified a transcriptional hub related to HIF1α/ARNT that is believed to contribute to the metabolic changes in ccRCC. Furthermore, RNAseq data suggest that HIF1α expression persists at level within the range of diploid HIF1α tumors in most ccRCC specimens affected by HIF1α loss of heterozygosity (Fig. 4) [67]. Consistent with these data [67] showing HIF1α mRNA expression, Sato et al. [54] detected HIF1α protein in 84% of primary ccRCC specimens. Sato et al. [54] do not report any major deletions at chromosome 14q in their patient specimens.
Figure 4. HIF1α and HIF2α mRNA levels.
(A) Expression analysis of ccRCC specimens from [62] indicates that HIF1 α expression generally persists in patients harboring HIF1 α loss of heterozygosity within the range of diploid HIF1 α expression. (B) Expression of HIF2α/EPAS1 is provided for comparison.
A genetic polymorphism in the human HIF1α gene, C1772T, is associated with overexpression of HIF1α and a poorer outcome in a subgroup of breast cancer patients [70]. The same SNP, C1772T, was found at a higher frequency in ccRCC patients than in healthy subjects [71].
Collectively, the data summarized here indicate both expression of HIF1α protein in the majority of ccRCC specimens and a pro-oncogenic role for HIF1α in ccRCC. Therefore, the putative tumor suppressor function for HIF1α in ccRCC [1, 67] should be reconsidered.
HIF1α and HIF2α Have Specific Functions in Normal Stem Cell Physiology
Although the HIFs are both induced under hypoxic stress conditions, the genes that HIF1α and HIF2α regulate can overlap or be entirely unique. Many HIF target genes are involved in mediating the change from oxidative to aerobic glycolytic metabolism [72]. HIFs not only have a major role in cancer cells by driving this Warburg effect, but indeed, HIF1α and HIF2α display specific effects on the functions of various types of normal stem cells. For instance, the level of HIF1α was shown to be critical for hematopoietic stem cell (HSC) maintenance in the bone marrow, in part because HIF1α regulates anaerobic glycolysis in these cells [73, 74]. HIF1α maintains anaerobic glycolysis in normal HSCs by activation of the transcription of pyruvate dehydrogenase kinase (PDK) isoforms 2 and 4, which prevent pyruvate from entering the TCA cycle [74]. In contrast, HIF2α knockdown prevents the repopulating ability of human CD34+ umbilical cord blood cells [75].
HIF1α−/− mouse embryos die by E11 and display major defects in cardiovascular development, neural tube malformations, and massive cell death within mesenchymal cell populations [76]. HIF2α−/− mouse embryos die by E16.5 from reduced levels of noradrenaline and a slow heart rate (bradycardia) [77]. In contrast to the HIF1α−/− embryos, HIF2α−/− embryos have no morphological defects and display normal vascular development, which indicates that HIF2α does not play an essential role in vasculogenesis or angiogenesis during development [77]. ARNT−/− mice die in utero between E9.5 and E10.5, and display neural tube closure defects, forebrain hypoplasia, delayed rotation of the embryo, placental hemorrhaging, and visceral arch abnormalities [78]. Indeed, these knockout mice display the most extensive defects because without ARNT, both HIF1α and HIF2α are inactive [78].
Specific Functions of HIF1α in Carcinogenesis, Tumor Progression, and the Generation of Cancer Stem Cells
In addition to the specific effects of HIF1α and HIF2α in various types of normal stem cells, HIF1α can mediate changes associated with processes that occur both during carcinogenesis and in cancer progression. For instance, HIF1α mediates the activation of histone deacetylase 3 (HDAC3) in both epithelial and mesenchymal cells, ultimately resulting in epithelial-mesenchymal transition associated with both repression of the gene E-cadherin and loss of polarity of epithelial cells [79]. Another example is the regulation of HIF1α transcription by Sirt6; Sirt6, a member of the sirtuin family of proteins, can act as a tumor suppressor via its role in the transcriptional repression of the HIF1α gene. In the absence of Sirt6, specifically HIF1α is transcriptionally activated to a high level and this directly leads to neoplastic transformation without any additional, secondary mutations in the cells [80, 81]. The transcription factor YY1 is involved in stabilizing HIF1α under hypoxic conditions in tumors, and inhibition of YY1 suppressed the proliferation of metastatic cancer cells, potentially via a reduction in HIF1α levels [82]. While in many types of cells HIF1α protein associates with polymerized microtubules to traffic to the nucleus, the connection between microtubules and HIF1α is lost in RCC [83]. As a result, microtubule-targeting drugs do not impair HIF1α nuclear activity and transcription in RCC, which can potentially explain the lack of clinical activity of microtubule inhibitors in RCC patients.
Another example of the specific association of HIF1α and oncogenesis is from a recent study by Xiang et al [84], which showed that the drug ganetespib, an inhibitor of heat shock protein 90, lowered the level of HIF1α, but not HIF2α protein, and also inhibited the subsequent expression of the HIF1α target genes involved in tumor progression in a mouse model of triple-negative breast cancer. A HIF1α high expression signature is found in human triple-negative breast cancer [85] and in human head and neck squamous cell carcinomas [86]. In another intriguing study, Conley et al [87] demonstrated that anti-angiogenic drugs such as sunitinib and bevacizumab can increase the numbers of cancer stem cells in a human breast cancer xenograft model by generating hypoxic conditions. These hypoxic conditions lead to a massive increase in expression of HIF1α in the ALDEFLUOR™+ cancer stem cell population, whereas HIF2α is not detectable in this population. Furthermore, a major pathway of breast cancer stem cell self-renewal, the Wnt/β-catenin signaling pathway, was reported to be activated by HIF1α in embryonic stem cells cultured under hypoxic conditions [88]. In glioma cells, knockdown of HIF1α impairs the ability of these glioma cells to form tumor spheres [89]. In glioma and glioblastoma the expression of both HIF1α and HIF2α stimulates cancer stem cell growth ([90, 91]; for review [92]). Thus, large numbers of studies have specifically implicated HIF1α in carcinogenesis, tumor cell proliferation, cancer progression, and in the generation of larger cancer stem cell populations.
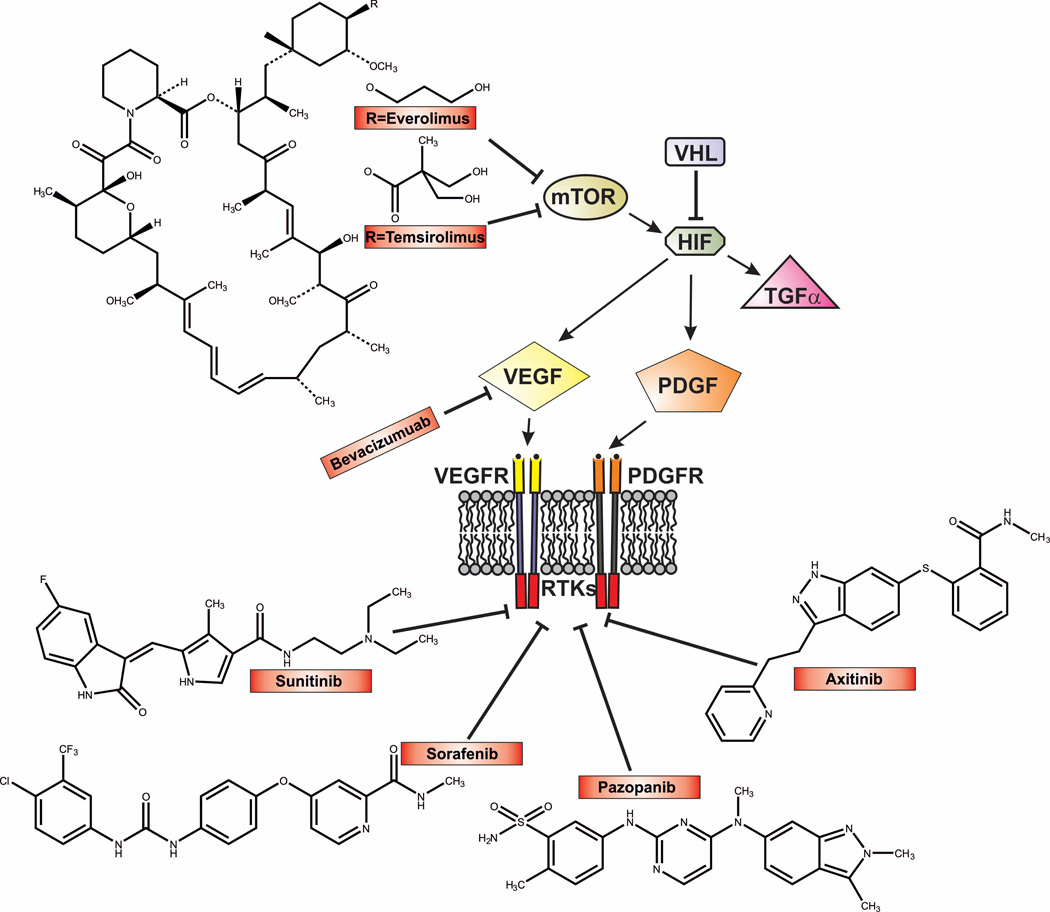
Clinical Data and New Treatment Strategies for ccRCC
As discussed earlier, the major drugs currently used to treat late stage ccRCC are temsirolimus, everolimus, bevacizumab, and the tyrosine kinase inhibitors that act of VGFR (Fig. 5). mTOR consists of two enzymatically active complexes, mTOR complex 1 (mTORC1) and mTORC2 [14]. Activation of mTOR complexes leads to the stimulation of ribosomal translation of various mRNAs, including the translation of HIF1α message, which is governed by mTORC1, and the translation of HIF2α mRNA, which is regulated by mTORC2 [93]. Temsirolimus competitively inhibits mTORC1 kinase, indicating that HIF1α translation should be reduced to a greater extent than HIF2α translation by this drug (Fig. 5).
Figure 5. Current FDA approved drugs used for the treatment of ccRCC and their mechanisms of action.
Researchers are investigating strategies to target other HIF1α target genes for treatment of HIF overexpressing tumors, such as ccRCC, rather than blocking tumor angiogenesis, which is and has been the focus for drug development [10]. For example, IL-11, a member of the IL-6 family of cytokines, is a HIF1α target that, when silenced, significantly abrogates the ability of hypoxia to increase anchorage-independent growth and results in reduced tumor growth in xenograft models [94]. Thus, targeting inhibition of IL-11 with small molecules could be a useful approach. In work on diabetic nephropathy, the drug fasudil, a rho-kinase inhibitor, promoted HIF1α degradation [95, 96]. By inhibiting expression of HIF1α target genes in the kidney via reduction in HIF1α expression, fasudil may also inhibit ccRCC. In a breast cancer model, the oncogene HER2/neu required HIF1α for anoikis resistance, anchorage independence, and 3D culture growth. Researchers also demonstrated that in this model ErbB2 overexpression in cells stabilized HIF1α under normoxic conditions and that HIF1α is a major downstream effector of ErbB2 actions [97]. Therefore, inhibitors of ErbB2 could potentially be tested in ccRCC because such inhibitors could reduce HIF1α actions downstream of ErbB2. ErbB2 is expressed in human ccRCC [98].
The protein XBP1 drives tumorigenicity in triple negative breast cancer by forming a transcription complex with HIF1α, and this transcription signature is associated with a poor prognosis. Thus, inhibition of XBP1 activity could form a new treatment strategy for ccRCC, as XBP1 is moderately overexpressed in ccRCC in the Oncomine database [99].
Mitochondrial autophagy is induced by hypoxia in normal mouse embryo fibroblasts in a HIF1α dependent process that requires the expression of BNiP3, beclin1, and ATG5; mitochondrial autophagy is necessary to prevent increased reactive oxygen species and cell death during long-term hypoxia [100]. In ccRCC high expression of HIF1α in the presence of either hypoxia or normoxia (in the absence of VHL) could also lead to mitochondrial autophagy as a strategy for cell survival. Blocking such autophagy could be a useful strategy to promote ccRCC death by generating increased reactive oxygen species. However, STF-62247 is a drug that stimulates autophagy and inhibits RCC growth in xenograft models [101]. Autophagy is a complex process and the VHL protein enhances the expression of LC3C, a HIF-regulated LC3B paralog, that suppresses ccRCC growth [102]. This suppression of ccRCC growth by LC3C occurs by protection of RCC cells from the loss of LC3B-mediated autophagy [102]. Thus, targeting mitochondrial or cell autophagy programs could be a viable strategy for ccRCC treatment in the future.
Conclusions
Clear cell renal cell carcinoma remains a major health concern despite advances over the past 10 years in understanding the genetic pathways leading to overexpression of the VEGF axis and the development of a variety of therapies that target these pathways. Our understanding of the precise roles of the HIF transcription factors, HIF1α and HIF2α, in the development and progression of ccRCC continues to evolve. Unrepressed expression of HIF1α through VHL loss was initially proposed to be an initiating oncogenic event leading to ccRCC development. This concept was subsequently challenged by the idea that genetic loss of HIF1α was a key event in ccRCC evolution, leaving HIF2α as a major driver of renal cell neoplastic transformation. However, as reviewed here, a large number of studies, including transgenic mouse models, expression analyses of human tumors, and refined analyses of large data sets profiling ccRCCs recently made available call into question the dismissal by some in the field of HIF1α as a critical protein necessary and possibly sufficient to initiate development of ccRCC. These and other studies also indicate that ccRCC is a highly heterogeneous disease, with both intra- and inter-tumor heterogeneity. Collectively, the data reviewed here indicate that HIF1α plays a key role in promoting the tumor phenotype in a major subset of ccRCCs and that therapeutic strategies aimed at targeting HIF1α and the genes it affects should continue to be explored.
KEY MESSAGES.
The TRACK mouse ccRCC model with constitutively active HIF1α but not HIF2α expressed in proximal tubules develops RCC.
HIF1α protein is expressed in the majority of human ccRCC specimens.
Elevated HIF1α in ccRCC correlates with a worse prognosis.
Many publications do not support a tumor suppressor role for HIF1α in ccRCC.
HIF1α, but not HIF2α, is expressed in some types of cancer stem cells.
ACKNOWLEDGMENTS
This was supported by the University of Nottingham (NPM) and by Weill Cornell funds. DM and LF are supported by NCI T32-CA062948.
We thank Dr. Paraskevi Giannakakou for critically reading this manuscript.
Footnotes
DISCLOSURE STATEMENT
The authors report nothing to disclose.
REFERENCES
- 1.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, Kaelin WG. Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1:222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 4.Miller F, Kentsis A, Osman R, Pan ZQ. Inactivation of VHL by tumorigenic mutations that disrupt dynamic coupling of the pVHL.hypoxia-inducible transcription factor-1alpha complex. J Biol Chem. 2005;280:7985–7996. doi: 10.1074/jbc.M413160200. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberger C, Mandriota S, Jürgensen JS, Wiesener MS, Hörstrup JH, Frei U, Ratcliffe PJ, Maxwell PH, Bachmann S, Eckardt KU. Expression of hypoxia-inducible factor-1alpha and −2alpha in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13:1721–1732. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 6.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 7.Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- 8.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu T. Complex cellular functions of the von Hippel-Lindau tumor suppressor gene: insights from model organisms. Oncogene. 2012;31:2247–2257. doi: 10.1038/onc.2011.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 11.Oladipupo S, Hu S, Kovalski J, Yao J, Santeford A, Sohn RE, Shohet R, Maslov K, Wang LV, Arbeit JM. VEGF is essential for hypoxia-inducible factor-mediated neovascularization but dispensable for endothelial sprouting. Proc Natl Acad Sci U S A. 2011;108:13264–13269. doi: 10.1073/pnas.1101321108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes DF. Bevacizumab treatment for solid tumors: boon or bust? JAMA. 2011;305:506–508. doi: 10.1001/jama.2011.57. [DOI] [PubMed] [Google Scholar]
- 13.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Hollaender N, Urbanowitz G, Berg WJ, Kay A, Lebwohl D, Ravaud A, Group R-S. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 14.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 15.Verheul HM, Salumbides B, Van Erp K, Hammers H, Qian DZ, Sanni T, Atadja P, Pili R. Combination strategy targeting the hypoxia inducible factor-1 alpha with mammalian target of rapamycin and histone deacetylase inhibitors. Clin Cancer Res. 2008;14:3589–3597. doi: 10.1158/1078-0432.CCR-07-4306. [DOI] [PubMed] [Google Scholar]
- 16.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keith B, Johnson RS, Simon MC. HIF1αlpha and HIF2αlpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. Activation of HIF2α in kidney proximal tubule cells causes abnormal glycogen deposition but not tumorigenesis. Cancer Res. 2013;73:2916–2925. doi: 10.1158/0008-5472.CAN-12-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. Generation of a Mouse Model of Von Hippel-Lindau Kidney Disease Leading to Renal Cancers by Expression of a Constitutively Active Mutant of HIF1αlpha. Cancer Res. 2011;71:6848–6856. doi: 10.1158/0008-5472.CAN-11-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu K, Ding Q, Fang Z, Zheng J, Gao P, Lu Y, Zhang Y. Silencing of HIF-1alpha suppresses tumorigenicity of renal cell carcinoma through induction of apoptosis. Cancer Gene Ther. 2010;17:212–222. doi: 10.1038/cgt.2009.66. [DOI] [PubMed] [Google Scholar]
- 22.Razorenova OV, Castellini L, Colavitti R, Edgington LE, Nicolau M, Huang X, Bedogni B, Mills EM, Bogyo M, Giaccia AJ. The Apoptosis Repressor with a CARD Domain (ARC) is a Direct HIF1 Target Gene and Promotes Survival and Proliferation of VHL Deficient Renal Cancer Cells. Mol Cell Biol. 2013 doi: 10.1128/MCB.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 24.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF2αlpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 1: E83. 2003 doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol Cancer Res. 2004;2:89–95. [PubMed] [Google Scholar]
- 26.Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 27.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK, Keith B, Simon MC, Nathanson KL. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas S, Troy H, Leek R, Chung YL, Li JL, Raval RR, Turley H, Gatter K, Pezzella F, Griffiths JR, Stubbs M, Harris AL. Effects of HIF-1alpha and HIF2αlpha on Growth and Metabolism of Clear-Cell Renal Cell Carcinoma 786-0 Xenografts. J Oncol 2010: 757908. 2010 doi: 10.1155/2010/757908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook JD, Walker CL. The Eker rat: establishing a genetic paradigm linking renal cell carcinoma and uterine leiomyoma. Curr Mol Med. 2004;4:813–824. doi: 10.2174/1566524043359656. [DOI] [PubMed] [Google Scholar]
- 31.Nanus DM, Walker CL. In: Experimental Models of Renal Cancer. Raghavan D, Scher HI, Leibel SA, Lange P, editors. Philadelphia: Principles and Practice of Genitourinary Oncology Lippincott-Raven; 1997. pp. 779–785. [Google Scholar]
- 32.Kaelin WG., Jr. Treatment of kidney cancer: insights provided by the VHL tumor-suppressor protein. Cancer. 2009;115:2262–2272. doi: 10.1002/cncr.24232. [DOI] [PubMed] [Google Scholar]
- 33.Frew IJ, Thoma CR, Georgiev S, Minola A, Hitz M, Montani M, Moch H, Krek W. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J. 2008;27:1747–1757. doi: 10.1038/emboj.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleymenova E, Everitt JI, Pluta L, Portis M, Gnarra JR, Walker CL. Susceptibility to vascular neoplasms but no increased susceptibility to renal carcinogenesis in Vhl knockout mice. Carcinogenesis. 2004;25:309–315. doi: 10.1093/carcin/bgh017. [DOI] [PubMed] [Google Scholar]
- 36.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol. 2009;11:994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 38.Mathia S, Paliege A, Koesters R, Peters H, Neumayer HH, Bachmann S, Rosenberger C. Action of hypoxia-inducible factor in liver and kidney from mice with Pax8-rtTA-based deletion of von Hippel-Lindau protein. Acta Physiol (Oxf) 2013;207:565–576. doi: 10.1111/apha.12058. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59:1206–1211. [PubMed] [Google Scholar]
- 40.Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/-) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole AM, Ridgway RA, Derkits SE, Parry L, Barker N, Clevers H, Clarke AR, Sansom OJ. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO Mol Med. 2010;2:472–486. doi: 10.1002/emmm.201000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammers HJ, Verheul HM, Salumbides B, Sharma R, Rudek M, Jaspers J, Shah P, Ellis L, Shen L, Paesante S, Dykema K, Furge K, Teh BT, Netto G, Pili R. Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: evidence from a xenograft study. Mol Cancer Ther. 2010;9:1525–1535. doi: 10.1158/1535-7163.MCT-09-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kedar D, Baker CH, Killion JJ, Dinney CP, Fidler IJ. Blockade of the epidermal growth factor receptor signaling inhibits angiogenesis leading to regression of human renal cell carcinoma growing orthotopically in nude mice. Clin Cancer Res. 2002;8:3592–3600. [PubMed] [Google Scholar]
- 44.Morais C, Healy H, Johnson DW, Gobe G. Inhibition of nuclear factor kappa B attenuates tumour progression in an animal model of renal cell carcinoma. Nephrol Dial Transplant. 2010;25:1462–1474. doi: 10.1093/ndt/gfp673. [DOI] [PubMed] [Google Scholar]
- 45.Touma SE, Goldberg JS, Moench P, Guo X, Tickoo SK, Gudas LJ, Nanus DM. Retinoic acid and the histone deacetylase inhibitor trichostatin a inhibit the proliferation of human renal cell carcinoma in a xenograft tumor model. Clin Cancer Res. 2005;11:3558–3566. doi: 10.1158/1078-0432.CCR-04-1155. [DOI] [PubMed] [Google Scholar]
- 46.Yi Y, Mikhaylova O, Mamedova A, Bastola P, Biesiada J, Alshaikh E, Levin L, Sheridan RM, Meller J, Czyzyk-Krzeska MF. von Hippel-Lindau-dependent patterns of RNA polymerase II hydroxylation in human renal clear cell carcinomas. Clin Cancer Res. 2010;16:5142–5152. doi: 10.1158/1078-0432.CCR-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semenza GL. Cancer-stromal cell interactions mediated by hypoxia-inducible factors promote angiogenesis, lymphangiogenesis, and metastasis. Oncogene. 2013;32:4057–4063. doi: 10.1038/onc.2012.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schietke RE, Hackenbeck T, Tran M, Günther R, Klanke B, Warnecke CL, Knaup KX, Shukla D, Rosenberger C, Koesters R, Bachmann S, Betz P, Schley G, Schödel J, Willam C, Winkler T, Amann K, Eckardt KU, Maxwell P, Wiesener MS. Renal tubular HIF-2α expression requires VHL inactivation and causes fibrosis and cysts. PLoS One. 2012;7:e31034. doi: 10.1371/journal.pone.0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biswas S, Charlesworth PJ, Turner GD, Leek R, Thamboo PT, Campo L, Turley H, Dildey P, Protheroe A, Cranston D, Gatter KC, Pezzella F, Harris AL. CD31 angiogenesis and combined expression of HIF-1α and HIF-2α are prognostic in primary clear-cell renal cell carcinoma (CC-RCC), but HIFα transcriptional products are not: implications for antiangiogenic trials and HIFα biomarker studies in primary CC-RCC. Carcinogenesis. 2012;33:1717–1725. doi: 10.1093/carcin/bgs222. [DOI] [PubMed] [Google Scholar]
- 52.Chintala S, Najrana T, Toth K, Cao S, Durrani FA, Pili R, Rustum YM. Prolyl hydroxylase 2 dependent and Von-Hippel-Lindau independent degradation of Hypoxia-inducible factor 1 and 2 alpha by selenium in clear cell renal cell carcinoma leads to tumor growth inhibition. BMC Cancer. 2012;12:293. doi: 10.1186/1471-2407-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyhan MJ, El Mashad SM, O'Donovan TR, Ahmad S, Collins C, Sweeney P, Rogers E, O'Sullivan GC, McKenna SL. VHL genetic alteration in CCRCC does not determine deregulation of HIF, CAIX, hnRNP A2/B1 and osteopontin. Cell Oncol (Dordr) 2011;34:225–234. doi: 10.1007/s13402-011-0029-5. [DOI] [PubMed] [Google Scholar]
- 54.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 55.Di Cristofano C, Minervini A, Menicagli M, Salinitri G, Bertacca G, Pefanis G, Masieri L, Lessi F, Collecchi P, Minervini R, Carini M, Bevilacqua G, Cavazzana A. Nuclear expression of hypoxia-inducible factor-1alpha in clear cell renal cell carcinoma is involved in tumor progression. Am J Surg Pathol. 2007;31:1875–1881. doi: 10.1097/PAS.0b013e318094fed8. [DOI] [PubMed] [Google Scholar]
- 56.Dorević G, Matusan-Ilijas K, Babarović E, Hadzisejdić I, Grahovac M, Grahovac B, Jonjić N. Hypoxia inducible factor-1alpha correlates with vascular endothelial growth factor A and C indicating worse prognosis in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2009;28:40. doi: 10.1186/1756-9966-28-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klatte T, Seligson DB, Riggs SB, Leppert JT, Berkman MK, Kleid MD, Yu H, Kabbinavar FF, Pantuck AJ, Belldegrun AS. Hypoxia-inducible factor 1 alpha in clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:7388–7393. doi: 10.1158/1078-0432.CCR-07-0411. [DOI] [PubMed] [Google Scholar]
- 58.Lidgren A, Hedberg Y, Grankvist K, Rasmuson T, Vasko J, Ljungberg B. The expression of hypoxia-inducible factor 1alpha is a favorable independent prognostic factor in renal cell carcinoma. Clin Cancer Res. 2005;11:1129–1135. [PubMed] [Google Scholar]
- 59.Lidgren A, Hedberg Y, Grankvist K, Rasmuson T, Bergh A, Ljungberg B. Hypoxia-inducible factor 1alpha expression in renal cell carcinoma analyzed by tissue microarray. Eur Urol. 2006;50:1272–1277. doi: 10.1016/j.eururo.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 60.Medina Villaamil V, Aparicio Gallego G, Santamarina Caínzos I, Valladares-Ayerbes M, Antón Aparicio LM. Searching for Hif1-α interacting proteins in renal cell carcinoma. Clin Transl Oncol. 2012;14:698–708. doi: 10.1007/s12094-012-0857-4. [DOI] [PubMed] [Google Scholar]
- 61.Schultz L, Chaux A, Albadine R, Hicks J, Kim JJ, De Marzo AM, Allaf ME, Carducci MA, Rodriguez R, Hammers HJ, Argani P, Reuter VE, Netto GJ. Immunoexpression status and prognostic value of mTOR and hypoxia-induced pathway members in primary and metastatic clear cell renal cell carcinomas. Am J Surg Pathol. 2011;35:1549–1556. doi: 10.1097/PAS.0b013e31822895e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kroeger N, Seligson DB, Signoretti S, Yu H, Magyar CE, Huang J, Belldegrun AS, Pantuck AJ. Poor prognosis and advanced clinicopathological features of clear cell renal cell carcinoma (ccRCC) are associated with cytoplasmic subcellular localisation of Hypoxia inducible factor-2α. Eur J Cancer. 2014 doi: 10.1016/j.ejca.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 63.Minardi D, Lucarini G, Filosa A, Milanese G, Zizzi A, Di Primio R, Montironi R, Muzzonigro G. Prognostic role of tumor necrosis, microvessel density, vascular endothelial growth factor and hypoxia inducible factor-1alpha in patients with clear cell renal carcinoma after radical nephrectomy in a long term follow-up. Int J Immunopathol Pharmacol. 2008;21:447–455. doi: 10.1177/039463200802100225. [DOI] [PubMed] [Google Scholar]
- 64.Mylonis I, Sembongi H, Befani C, Liakos P, Siniossoglou S, Simos G. Hypoxia causes triglyceride accumulation by HIF-1-mediated stimulation of lipin 1 expression. J Cell Sci. 2012;125:3485–3493. doi: 10.1242/jcs.106682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014;19:285–292. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Network CGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen R, Xu M, Hogg RT, Li J, Little B, Gerard RD, Garcia JA. The acetylase/deacetylase couple CREB-binding protein/Sirtuin 1 controls hypoxia-inducible factor 2 signaling. J Biol Chem. 2012;287:30800–30811. doi: 10.1074/jbc.M111.244780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG, Jr., Signoretti S. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim HO, Jo YH, Lee J, Lee SS, Yoon KS. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep. 2008;20:1181–1187. [PubMed] [Google Scholar]
- 71.Lessi F, Mazzanti CM, Tomei S, Di Cristofano C, Minervini A, Menicagli M, Apollo A, Masieri L, Collecchi P, Minervini R, Carini M, Bevilacqua G. VHL and HIF-1α: gene variations and prognosis in early-stage clear cell renal cell carcinoma. Med Oncol. 2014;31:840. doi: 10.1007/s12032-014-0840-8. [DOI] [PubMed] [Google Scholar]
- 72.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 73.Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 75.Rouault-Pierre K, Lopez-Onieva L, Foster K, Anjos-Afonso F, Lamrissi-Garcia I, Serrano-Sanchez M, Mitter R, Ivanovic Z, de Verneuil H, Gribben J, Taussig D, Rezvani HR, Mazurier F, Bonnet D. HIF-2α protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell. 2013;13:549–563. doi: 10.1016/j.stem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 76.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 79.Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC, Teng SC, Wu KJ. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43:811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 80.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via HIF1αlpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu S, Kasim V, Kano MR, Tanaka S, Ohba S, Miura Y, Miyata K, Liu X, Matsuhashi A, Chung UI, Yang L, Kataoka K, Nishiyama N, Miyagishi M. Transcription factor YY1 contributes to tumor growth by stabilizing hypoxia factor HIF-1α in a p53-independent manner. Cancer Res. 2013;73:1787–1799. doi: 10.1158/0008-5472.CAN-12-0366. [DOI] [PubMed] [Google Scholar]
- 83.Carbonaro M, Escuin D, O'Brate A, Thadani-Mulero M, Giannakakou P. Microtubules regulate hypoxia-inducible factor-1α protein trafficking and activity: implications for taxane therapy. J Biol Chem. 2012;287:11859–11869. doi: 10.1074/jbc.M112.345587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang L, Gilkes DM, Chaturvedi P, Luo W, Hu H, Takano N, Liang H, Semenza GL. Ganetespib blocks HIF-1 activity and inhibits tumor growth, vascularization, stem cell maintenance, invasion, and metastasis in orthotopic mouse models of triple-negative breast cancer. J Mol Med (Berl) 2013 doi: 10.1007/s00109-013-1102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunkel J, Vaittinen S, Grénman R, Kinnunen I, Irjala H. Prognostic markers in stage I oral cavity squamous cell carcinoma. Laryngoscope. 2013;123:2435–2441. doi: 10.1002/lary.23888. [DOI] [PubMed] [Google Scholar]
- 87.Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, Clouthier SG, Wicha MS. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazumdar J, O'Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Méndez O, Zavadil J, Esencay M, Lukyanov Y, Santovasi D, Wang SC, Newcomb EW, Zagzag D. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol Cancer. 2010;9:133. doi: 10.1186/1476-4598-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, Pollack IF, Park DM. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28:3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 91.Seidel S, Garvalov BK, Wirta V, von Stechow L, Schänzer A, Meletis K, Wolter M, Sommerlad D, Henze AT, Nistér M, Reifenberger G, Lundeberg J, Frisén J, Acker T. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 92.Philip B, Ito K, Moreno-Sánchez R, Ralph SJ. HIF expression and the role of hypoxic microenvironments within primary tumours as protective sites driving cancer stem cell renewal and metastatic progression. Carcinogenesis. 2013;34:1699–1707. doi: 10.1093/carcin/bgt209. [DOI] [PubMed] [Google Scholar]
- 93.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Onnis B, Fer N, Rapisarda A, Perez VS, Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J Clin Invest. 2013;123:1615–1629. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matoba K, Kawanami D, Okada R, Tsukamoto M, Kinoshita J, Ito T, Ishizawa S, Kanazawa Y, Yokota T, Murai N, Matsufuji S, Takahashi-Fujigasaki J, Utsunomiya K. Rho-kinase inhibition prevents the progression of diabetic nephropathy by downregulating hypoxia-inducible factor 1α. Kidney Int. 2013;84:545–554. doi: 10.1038/ki.2013.130. [DOI] [PubMed] [Google Scholar]
- 96.Turcotte S, Desrosiers RR, Béliveau R. HIF-1alpha mRNA and protein upregulation involves Rho GTPase expression during hypoxia in renal cell carcinoma. J Cell Sci. 2003;116:2247–2260. doi: 10.1242/jcs.00427. [DOI] [PubMed] [Google Scholar]
- 97.Whelan KA, Schwab LP, Karakashev SV, Franchetti L, Johannes GJ, Seagroves TN, Reginato MJ. The oncogene HER2/neu (ERBB2) requires the hypoxia-inducible factor HIF-1 for mammary tumor growth and anoikis resistance. J Biol Chem. 2013;288:15865–15877. doi: 10.1074/jbc.M112.426999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomasson M, Hedman H, Ljungberg B, Henriksson R. Gene expression pattern of the epidermal growth factor receptor family and LRIG1 in renal cell carcinoma. BMC Res Notes. 2012;5:216. doi: 10.1186/1756-0500-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, Mai J, Shen H, Hu DZ, Adoro S, Hu B, Song M, Tan C, Landis MD, Ferrari M, Shin SJ, Brown M, Chang JC, Liu XS, Glimcher LH. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. The Journal of biological chemistry. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, Czyzyk-Krzeska MF. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]