Abstract
Just when you thought that you had heard it all about autophagy - the conserved cellular process that mediates turnover of cellular constituents in the lysosomes - studies keep coming out highlighting new types of autophagy, new functions for autophagy or even new autophagy-independent roles for the proteins associated with this process. The field of immunology has been riding the autophagic wave since the beginning of its revival; first due to its role in the host defense against pathogens, and more recently through the better understanding of the unique characteristics and functions of different autophagic pathways in immune cells. Here, we describe some of these new functions that are tightening the connection between autophagy and acquired or innate immunity and their malfunctioning with age.
Introduction
The identification of the molecular players that participate in autophagy, the process that mediates the degradation cellular components in lysosomes, and the numerous associations established between autophagy malfunctioning and human disease – including those of immune origin– were a major push for the revival of autophagy in the late 90’s [1]. However, autophagy is far from fading in the scientific stardom, and it keeps instead emerging as hot topic because of its multifunctional nature and reciprocal functional interactions with important cellular processes [2]. Immunology has not been an exception to this extended autophagy honeymoon. Immunologist became initially interested in the role of autophagy in the cellular defense against pathogens [3] (xenophagy, see Box 1). Soon it became evident that the interplay between autophagy and innate immunity was more complex, which made it necessary to analyze specialized functions of autophagy in different cell populations of the immune system [4]. Current research in innate and adaptive immunity has also embraced some of the newly elucidated roles for autophagy, such as its contribution to the cellular energetic balance, remodeling of the proteome or unconventional secretion [5-7]. In some instances, research in Immunology has been the driving force in new discoveries in autophagy. For example, Crohn’s disease was one of the first human diseases linked to mutations in an autophagy gene [8], and studies on phagocytosis discovered new autophagy-independent functions of autophagy related proteins (Atg) [9]. Here, we review recent findings that have kept autophagy in the headline of Immunology news and comment on the implications of the newly identified roles of autophagy in the regulation of the adaptive immune response and its decline with age.
Box 1. Autophagic pathways: the basics.
Three basic autophagic pathways co-exist in mammals:
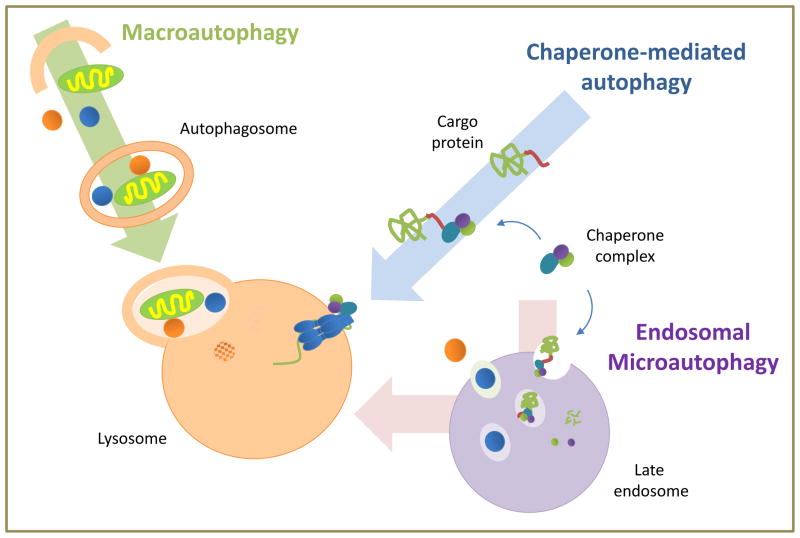
Macroautophagy: the gene products (Atg proteins) that participate in macroautophagy act sequentially in the assembly of lipid and proteins from different sources [10] to form a double membrane around cargo identified by receptor proteins that sequesters it from the cytosol. These double membrane vesicles (autophagosomes) use the microtubule tracks to encounter and fuse with lysosomes, where luminal hydrolases degrade the cargo [1].
Macroautophagy can occur “in bulk” – when cargo is sequestered in a random manner – or selectively – when the cargo is identified through interactions between cargo-receptors and structural components of the autophagosome. Examples of selective macroautophagy include mitophagy (mitochondria), lipophagy (lipid droplets), ribophagy (ribosomes), aggregophagy (aggregosomes) or xenophagy (extracellular pathogens). In the case of xenophagy [3], the endocytic or phagocytic vesicles used for pathogen internalization rather than fusing with lysosomes are identified in the cytosol by the autophagy machinery that delivers them to lysosomes. Other pathogens find their way out of the internalizing phagocytic vesicles into the cytosol where they become autophagy substrates.
Microautophagy: Cargo sequestration in vesicles is also the first step in microautophagy, but in this case, vesicles form from the invagination of the limiting membrane of lysosomes (in yeast microautophagy [12]) or late endosomes (in endosomal microautophagy [15]). These cargo-containing vesicles then pinch off into the lumen for degradation (Fig. 1).
Chaperone-mediated autophagy (CMA): In CMA, client proteins are selectively identified by a cytosolic chaperone that delivers them to the lysosomal membrane [11]. Once bound to the integral lysosomal membrane protein (LAMP-2A), the substrate protein unfolds and reaches the lumen through a LAMP-2A-enriched translocation complex.
Autophagy: what is new?
Analysis of the way in which autophagic cargo is delivered to the lysosomes has revealed the co-existence in most mammalian cells of three different autophagic pathways (Fig. 1): macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). The steps, effectors and regulators of these pathways have been extensively reviewed elsewhere and are also summarized in Box 1 [1,2,10-12].
Figure 1. Schematic of the main autophagic pathways mammalian cells.
Macroautophagy and chaperone-mediated autophagy deliver cytosolic cargo directly to lysosomes for degradation. In the case of endosomal microautophagy, cytosolic proteins are internalized in single membrane vesicles into late endosomes and undergo degradation in this compartment or in lysosomes upon endosome/lysosome fusion.
New themes on autophagy mechanisms
Several recent findings in the autophagy field have gotten considerable attention. One of them is the existence of shared components among autophagic pathways which could underlie the basis of the compensatory activation of one autophagic pathway when another fails [13]. For example, hsc70 targets cytosolic proteins to CMA, endosomal microautophagy or macroautophagy depending on their folding state [14,15]. A second interesting finding is the fact that macroautophagy can proceed without some of the Atg proteins previously considered essential (i.e. Beclin, Atg5 or Atg7) [16,17]. The challenge now is to determine whether these unconventional variants are alternatives to canonical autophagy or if they coexist. Also worth mentioning is the now well-accepted idea that regulation of autophagy is not linear but that instead inputs from multiple pathways converge and often co-modulate autophagy [18]. Lastly, the recognition that autophagy-related proteins have functions independent of the autophagic process is also gaining momentum [7].
Autophagy functions
The roles of autophagy in cellular quality control and cellular defense have been actively researched but some of the “old” functions have also been revisited. For example, although the early connections between autophagy and the cell’s energetics were focused on the contribution of autophagy to the replenishment of the intracellular pool of free amino acids through protein breakdown, recent studies support that macroautophagy also contributes to sustain a positive energetic cellular balance through mobilization of intracellular lipid [6] and glycogen stores [19]. CMA also contributes to the regulation of cellular energetics by selectively degrading key enzymes involved in essential metabolic pathways [20]. The interplay between autophagy and cell death has also been revised in recent years, resulting in a stronger support for autophagy as a pro-survival mechanism, except for very specific scenarios mostly in invertebrate models. The early labeling of autophagy as a programmed type of cell death originated from the observation of autophagosomes in dying cells. However, later studies demonstrated that higher autophagosome content was often not a signature of increased autophagy but rather reflected the cell’s inability to degrade them, and therefore an expression of incomplete autophagy. These studies of the new and revisited functions of autophagy have also made an impact on our current understanding of the role of autophagy in the immune response.
Autophagy and the adaptive immune response
Studies in recent years have firmly established macroautophagy as a crucial component of the innate immune response [4]. Macroautophagy aids in the elimination of pathogens, but its role extends beyond that to include delivery of endogenous or exogenous molecules to intracellular compartments, modulation of the activity of the inflammasome, control of cytokine secretion and regulation of phagocytosis [21-25]. Though most of these functions are a consequence of the activation of conventional autophagy, non-autophagy functions of Atg proteins, such LC3-associated-phagocytosis, also participate in the orchestration of the innate immune response [9,22]. The complex circuitries that regulate autophagy in macrophages and other phagocytic cells and the full spectrum of the functions regulated by autophagy in those cells have been the object of comprehensive recent reviews [4]. Here, we focus in the less well-studied functions that different forms of autophagy exert to modulate the adaptive immune response.
Autophagy and antigen presentation
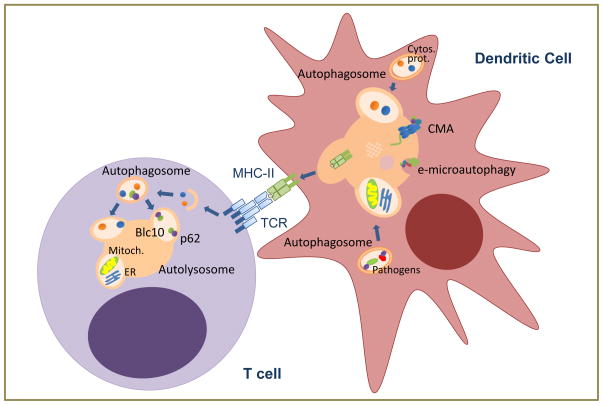
Autophagy has emerged as an important mechanism of cross-delivery of intracellular pathogen’s antigens to MHC-class II loading compartments (Fig. 2) [26-28]. Mice with macroautophagy-deficient macrophages become more susceptible to Mycobacteria infection and display increased bacterial burden, which has been attributed to defective pathogen killing and unregulated inflammatory response [29,30]. However, it is also likely that inefficient antigen presentation prevents adequate T cell activation and contributes to the deficient global response to Mycobacteria. Several recent findings underscore the contribution of macroautophagy to the development of an efficient adaptive immune response in vivo. For example, the protective efficiency of yellow fever vaccination depends on the ability of the virus to induce autophagy in dendritic cells [31], and likewise, chemical activation of autophagy potentiates immunization with BCG [32]. Furthermore, although the association of mutations in Atg16L or in NOD2 with inflammatory bowel disease has been explained on the basis of defective function of Panneth and goblet cells in the intestinal mucosa [8,33], it has also been reported that NOD2 activation regulates macroautophagy-mediated presentation of bacterial antigens and that, interestingly, dendritic cells from Crohn’s disease patients with NOD2 mutations fail to upregulate MHC-II presentation in response to NOD2 ligands [34].
Figure 2. Autophagic functions in adaptive immunity.
In dendritic cells, cytosolic proteins or those derived from intracellular pathogens can be delivered to be loaded into MHC-II through macroautophagy, CMA or endosomal microautophagy (e-microautophagy) to activate T cells. Whereas basal macroautophagy in T cells regulates organelle homeostasis (including mitochondria and endoplasmic reticulum), activation-induced macroautophagy in response to TCR engagement degrades cytosolic components to respond to increased metabolic demand and to modulate specific signaling pathways.
Beside pathogens, self-proteins can also be delivered through macroautophagy for MHC-II loading, implicating this pathway in the regulation of tolerance. In this respect, preferential involvement of autophagy in the presentation of citrullinated peptides, frequently associated with autoimmunity, has been proposed, as the presentation of this peptides was inhibited by silencing Atg5 in B lymphoma cells [35]. The involvement of macroautophagy in the regulation of tolerance is not restricted to peripheral presentation of self-peptides but it extends to regulate thymocyte selection. Thymic epithelial cells use macroautophagy to present self-peptides and shape the TCR repertoire. Interestingly, nude mice receiving an Atg5-deficient thymic graft develop autoimmunity, supporting that macroautophagy participates also in negative selection [36]. Recent analyses suggest that autophagy becomes increasingly important to mediate thymocyte deletion when concentrations of antigen are low, which is likely the case for many tissue-restricted genes expressed in thymic epithelial cells [37].
It is not only macroautophagy that regulates antigen presentation (Fig. 2). Silencing of LAMP-2A, the CMA receptor at the lysosomal membrane, decreases loading of a constitutively expressed cytosolic protein in B lymphoblastoid cells [38]. Moreover, delivery of cytosolic proteins to late endosomal compartments in a manner that resembles microautophagy occurs in dendritic cells [15]. Cytosolic proteins are recognized by hsc70 that then binds to specific lipids in the endosomal membrane. Proteins eventually access the endosome lumen in small vesicles that form by invagination of the endosome membrane in a process mediated by SCRT complex proteins [15].
Autophagy and lymphocyte function
T cells display basal autophagy that controls organelle homeostasis [39,40]. Consequently, T cells deficient in essential Atg proteins accumulate defective mitochondria and present higher levels of ROS and increased cell death [40]. Defective mitochondrial homeostasis likely underlies the reduced number of peripheral T cells observed in mice bearing Atg-deficient T cells or those with a T cell-specific deletion of Vps34, a PI3 kinase class III required for autophagy initiation [41-43]. Interestingly, old mice with Vps34-deficient T cells develop intestinal inflammation that correlates with decreased suppressive function of regulatory T cells [41]. How the absence of Vps34 affects regulatory T cell development or function and whether macroautophagy plays a role in these processes remains unknown.
Macroautophagy is also activated in response to TCR engagement (Fig. 2), and cells with defective macroautophagy respond poorly to activation and show reduced levels of proliferation and cytokine production [42,44]. The specific roles of activation-induced macroautophagy in T cells and the signaling pathways that regulate it are not yet fully characterized, but it is likely that basal and activation-induced autophagy might serve different purposes. For example, autophagosome cargo shifts from mainly organelles to mostly soluble cytosolic components as cells transition from a resting to an activated state [44]. Furthermore, two specific roles for macroautophagy in activated T cells have been described: providing substrates to adapt to an increased metabolic demand; and modulating NFκB activation through the p62-mediated selective degradation of Bcl10 (Fig. 2) [44,45].
Less is known about the role of autophagy in B cells. Studies using bone marrow chimeras showed that Atg5-deficiency led to reduced numbers of peripheral B cells, with specific defects in B1 populations and partial blockage of pro-B to pre-B cell differentiation [46]. Autophagy has also been shown to be necessary to fulfill the metabolic demands of activated plasma cells and ensure survival of short- and long-lived plasma cells to maintain an efficient antibody response in vivo [47].
Autophagy and aging
Malfunctioning of macroautophagy and CMA occurs in many organs and tissues with age [48,49]. Activation of macroautophagy has proven necessary for the extension of life-span observed in worms and flies after specific mutations or caloric restriction [50,51]. Macroautophagy is also necessary to sustain normal life-span, as mice with compromised autophagy in individual systems all show shorter life-span. More excitingly mice overexpressing one of the essential Atg proteins display a noticeable increase in lifespan [52]; and genetic correction of the age-dependent decline on CMA in liver has proven sufficient to prevent the functional decline of this organ in old rodents [53]. Interestingly, most interventions that improve life- and health-span, such as caloric restriction, or treatment with rapamycin, resveratrol, spermidine or metformin, also activate autophagy [54].
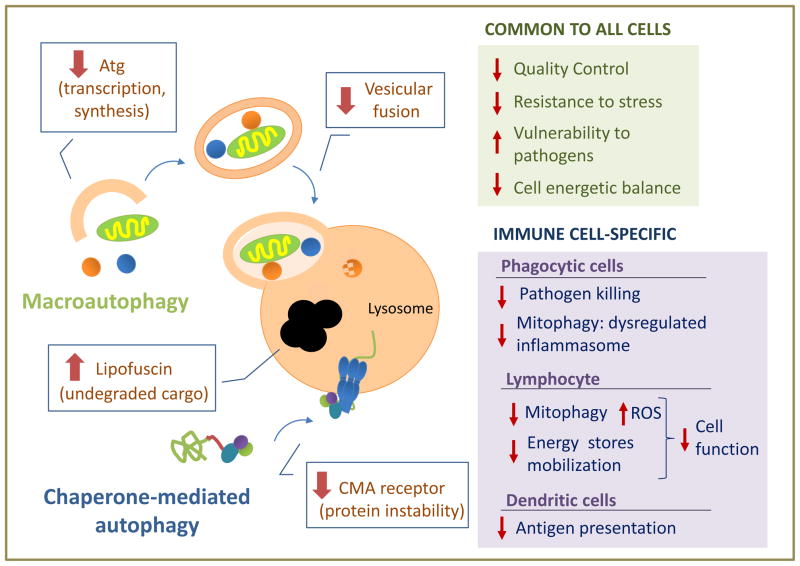
The mechanisms behind the autophagic malfunctioning in aging are still poorly understood (Fig. 3). For CMA, reduced stability of the lysosomal receptor for this pathway is responsible for the CMA decline in many organs [48]. In the case of macroautophagy, changes in Atg proteins at the transcriptional or protein level have been reported, but whether they are the primary defect behind the age-dependent failure requires additional clarification [54]. Besides primary defects, changes in autophagy with age can also be reactive to disease conditions, where autophagy may be activated to assure cell survival. However, this initial compensatory activation may be followed by failure of this pathway as the disease condition becomes chronic.
Figure 3. Age-related alteration in autophagy and its cellular consequences.
The scheme depicted the most common mechanisms that contributed to malfunctioning of macroautophagy and chaperone-mediated autophagy with age. Boxes on the right summarized the main consequences that could follow autophagic malfunctioning with age separating those common to all cells (top) from those specific for immune cells (bottom).
Malfunctioning of autophagy in old organism could contribute to loss of cellular function and often to cell death by limiting the cell’s ability to sustain a healthy proteome and organelles, making them more vulnerable to cellular stressors and pathogens, and rendering them incapable to adapt to energetically demanding conditions (Fig. 3) [54].
Dysfunction of the autophagic systems may also be behind some cell- and tissue-specific functions that fail with age. Decreased autophagy in phagocytic cells may not only impair their ability to kill pathogens but also cause dysregulated activation of the inflammasome due to deficient mitophagy, with the subsequent increase in the production of inflammatory cytokines, likely contributing to the age-associate inflammaging phenotype. Indeed, activation of the NLPR3 inflammasome responds to mitochondrial ROS production and the release of oxidized mitochondrial DNA [55,56], and macroautophagy has been shown to play an essential role in regulating this process [57,58].
It was recently reported that CD8+ T cells isolated from old healthy humans presented significantly lower levels of basal macroautophagy [59]. How this dysfunction affects T cell responses and the mechanisms that may account for the reduced autophagy in T cells with age are still unknown. However, one possible mechanism might involve deficient mitophagy by analogy to reports of reduced mitophagy with age in other cell types [60]. It would be then expected that decreased mitophagy would result in ROS accumulation, leading to altered T cell responses and increased susceptibility to stress. In addition, the T cell energetic balance may also be compromised as a result of the autophagic failure with age. In fact, a recent study found that T cells isolated from patients with rheumatoid arthritis, which usually present characteristics of premature aging, failed to properly activate autophagy and were unable to utilize internal cell energy sources [61].
Although little is still known on how age affects autophagy in dendritic cells, a recent study reported that oxidized proteins accumulate in the endosomal compartment of dendritic cells from old mice, which correlates with decreased macroautophagy in those cells (Fig. 3). As a consequence, aged dendritic cells cannot efficiently present antigen and activate T cells [62]. Interestingly, in vivo administration of an antioxidant was able to restore the ability of aged mice to present antigen and elicit T cell responses, suggesting that therapeutic interventions aimed at restoring autophagy or palliating the effects of decreased autophagy with age might be a valid approach to improve immune function in the elderly [62].
Concluding remarks and a promising future
The challenge in the study of autophagy in immune cells originates in part by the fact that conventional regulatory mechanisms do not always apply, and that the autophagic process often fulfills very specific needs different from other organs and systems. The recent emphasis in “exceptions” in the autophagic process, the identification of novel autophagic functions and the better characterization of different autophagic pathways has provided a fertile ground for the future expansion of research on autophagy in immunity. Future efforts should be directed at elucidating how each autophagic pathway regulates specific processes, such as antigen presentation, cell activation or memory generation.
Autophagy is being flagged as an anti-aging mechanism and the growing number of successful attempts to improve health-span through modulation of autophagy has provided momentum to the search for more effective and specific chemical modulators of autophagy. A better characterization of the changes with age in autophagic pathways in immune cells, their time- course and whether the mechanisms behind this failure are universal or cell-type specific are needed before novel autophagic modulators can be considered for use in the prevention of immunosenescence or the restoration of the immune function in aging.
Highlights.
Different forms of autophagy play key roles regulating innate and adaptive immunity
New functions of autophagy are reshaping the understanding of how autophagy modulates immunity
An age-associated dysregulation of autophagy in immune cells may underlie immunosenescence
Acknowledgments
Research in our groups is supported by grants from the National Institutes of Health, the Glenn Foundation and a generous gift from Robert and Renee Belfer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5:427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 6••.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. First evidence of the contribution of macroautophagy to lipid metabolism by direct mobilization and breakdown of lipid droplets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. Demonstrated that the risk allele of ATG16L1 identified in Crohn’s patients leads to altered autophagy in the intestinal epithelium and disrupted function of Panneth cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. (and 22) First characterization of non-autophagy related functions of Atg proteins in the regulation of innate immunity. [DOI] [PubMed] [Google Scholar]
- 10.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. Journal of Cell Biology. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. Demonstrates cross-talk between macroautophagy and chaperone-mediated autophagy. Cells respond to blockage of CMA by upregulating macroautophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 15••.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. Identification of endosomal microautophagy as a new mechanism of delivery of intracellular proteins to the endosomal compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu CT, Zhu J, Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3:663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 18.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonilla DL, Bhattacharya A, Sha Y, Xu Y, Xiang Q, Kan A, Jagannath C, Komatsu M, Eissa NT. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity. 2013;39:537–547. doi: 10.1016/j.immuni.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. (See 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWt, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. (and 27-28) Initial identification of macroautophagy as a mechanism of delivery of intracellular proteins for presentaion on MHC-II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. (See 26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. (See 26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A. 2012;109:E3168–3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, Park Y, Jones DP, Chappert P, Davoust J, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–317. doi: 10.1126/science.1246829. (and 32) Describe how modulation of macroautophagy can induce increase antigen presentaion and potentiate vaccine efficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. (See 31) [DOI] [PubMed] [Google Scholar]
- 33.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 Inflammasome Orchestrates the Colonic Host-Microbial Interface by Regulating Goblet Cell Mucus Secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. Defines that the induction of macroautopahgy enhances presentation of pathogen antigens in dendiritc cells. [DOI] [PubMed] [Google Scholar]
- 35.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208:2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. (and 37) First evidence that macroautophagy in thymic epithelial cells regulates antigen presentation and hence thymocyte positive and negative selection. [DOI] [PubMed] [Google Scholar]
- 37••.Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med. 2013;210:287–300. doi: 10.1084/jem.20122149. (See 36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. Defines a essential role of macroautophagy in mitochondrial turnover in T cells. [DOI] [PubMed] [Google Scholar]
- 41.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagomez D, Cover TL, Zong WX, Zhang J, Van Kaer L. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013;190:5086–5101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. Initial characterization of the involvement of macroautiphagy in the regulation of T cell activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. First evidence of the contribution of macroautophagy to the T cell energetic balance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. Characterization of a novel role of selective macroautophagy in the regulation of NFκB activation in T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 47•.Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. Initial characterization of the role of macroautophagy in the regulation of plasma cell responses in vivo. [DOI] [PubMed] [Google Scholar]
- 48•.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. Identification of the molecular defect that leads to reduced CMA in aging. [DOI] [PubMed] [Google Scholar]
- 49.Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci. 2001;56:B288–293. doi: 10.1093/gerona/56.7.b288. [DOI] [PubMed] [Google Scholar]
- 50.Hansen M, Chandra A, Mitic L, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. First genetic evidence linking macroautophagy and longevity. [DOI] [PubMed] [Google Scholar]
- 52.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. First example of the beneficial effect of genetic restoration of autophagy in mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 55••.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. First report that clearly shows that the NLR3 inflammasome is activated by mitochnodrial ROS. [DOI] [PubMed] [Google Scholar]
- 56.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. (and 58) Initial evience that associates defective mitophagy with increased inflammasome activation. [DOI] [PubMed] [Google Scholar]
- 58••.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. (See 57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, Wade-Martins R, McMichael A, Klenerman P, Simon AK. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–689. doi: 10.4161/auto.18935. Presents evidence of reduced macroautophagy with age in human T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira, Gutkind S, Daniels MP, Komatsu M, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210:2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Cannizzo ES, Clement CC, Morozova K, Valdor R, Kaushik S, Almeida LN, Follo C, Sahu R, Cuervo AM, Macian F, et al. Age-related oxidative stress compromises endosomal proteostasis. Cell Rep. 2012;2:136–149. doi: 10.1016/j.celrep.2012.06.005. First evidence of the association of increased oxidative stress and reduced macroautophagy activity in dendritic cells with age and the implications for antigen presentation. [DOI] [PMC free article] [PubMed] [Google Scholar]