Abstract
The interferon regulatory factor family member 8 (IRF8) regulates differentiation of lymphoid and myeloid lineage cells by promoting or suppressing lineage-specific genes. How IRF8 promotes hematopoietic progenitors to commit to one lineage while preventing the development of alternative lineages is not known. Here we report an IRF8-EGFP fusion protein reporter mouse that revealed previously unrecognized patterns of IRF8 expression. Differentiation of hematopoietic stem cells into oligopotent progenitors is associated with progressive increases in IRF8-EGFP expression. However, significant induction of IRF8-EGFP is found in granulocyte-myeloid progenitors (GMPs) and the common lymphoid progenitors (CLPs) but not the megakaryocytic-erythroid progenitors. Surprisingly, IRF8-EGFP identifies three subsets of the seemingly homogeneous GMPs with an intermediate level of expression of EGFP defining bipotent progenitors that differentiation into either EGFPhi monocytic progenitors or EGFPlo granulocytic progenitors. Also surprisingly, IRF8-EGFP revealed a highly heterogeneous pre-pro-B population with a fluorescence intensity ranging from background to 4 orders above background. Interestingly, IRF8-EGFP readily distinguishes true B cell-committed (EGFPint) from those that are non-committed. Moreover, dendritic cell progenitors expressed extremely high levels of IRF8-EGFP. Taken together, the IRF8-EGFP reporter revealed previously unrecognized subsets with distinct developmental potentials in phenotypically well-defined oligopotent progenitors, providing new insights into the dynamic heterogeneity of developing hematopoietic progenitors.
Introduction
Hematopoietic stem cells (HSCs) constantly differentiate into all blood cell lineages via distinct differentiation programs. Lineage specification and commitment are marked by timely activation of one set of transcription factors associated with downregulation of other set(s) of transcription factors important for alternate cell lineage potential. While early studies led to the proposal that the flow of intermediate cells within each lineage is fixed (1, 2), recent evidence suggests otherwise - that oligopotent progenitor differentiation is very “plastic”, especially when the host is stressed, by infection for example. This causes reprogramming of early lymphoid and myeloid progenitors leading to enhanced development of myeloid lineage cells but curbed production of lymphoid lineage cells(3-6).
The plasticity of hematopoietic differentiation has long been known and was recently confirmed at single cell level by Naik et al. using a novel “cellular barcoding” technique (7). The developmental heterogeneity of lineage progenitor cells has led not only to inconsistencies in identifying phenotypes of intermediate stage cells but also to difficulties in positioning the newly found precursors in the orderly progression of lineage differentiation pathways. For example, macrophages are thought to derive from myeloid progenitors whereas dendritic cells (DCs) are thought to develop from separate pathways originating from either CLPs or CMPs (1, 8-11). However, it was recently found that macrophage-DC progenitors (MDPs) with a phenotype of CD117+CX3CR1+ can give rise to both macrophages and DCs (12). These findings suggest that most, if not all, well-characterized progenitor populations are heterogeneous at the clonal level even though they appear to have a homogeneous phenotype by certain criteria.
Most of our current knowledge of how blood cells are made came from studies of transcription factors. One of a series of transcription factors modulating hematopoietic fate determination is IRF8, also known as ICSBP (interferon consensus sequence binding protein). IRF8 is expressed mostly in cells of the hematopoietic system. Microglial cells with a hematopoietic origin also express IRF8 (13, 14). Functional analyses revealed broad contributions of IRF8 to the regulation of myeloid and lymphoid lineage development. The levels of IRF8 transcripts are low in HSCs, but increased in yet poorly defined CLPs, MPs, and common DC progenitors (CDPs) (15, 16). IRF8 deficiency in mice causes disrupted development of monocytes and macrophages but increased differentiation of neutrophils (17). The numbers of several subtypes of DCs including plasmacytoid DCs (pDCs), CD8α+ DCs and CD103+ non-lymphoid tissue DCs are also greatly diminished in Irf8−/− mice (15, 18-23). In humans, a loss of function mutation of IRF8 also causes a monocytic and DC immunodeficiency (24).
While IRF8 expression is upregulated in both myeloid and lymphoid progenitors, as determined by conventional PCR methods on sorted bulk populations, little is known about how IRF8 participates in the distinct transcriptional programs that control lineage specification and commitment. Here, we created an IRF8-EGFP reporter mouse by a knockin of the EGFP sequence into the IRF8 stop codon that results in transcription and translation of an IRF8-EGFP fusion protein under the regulation of endogenous IRF8 regulatory elements. Our data revealed previously unappreciated expression patterns of IRF8 that help to explain the functions of IRF8 in distinct lineages of hematopoietic cells and to better understand the heterogeneity of early progenitors.
Materials and Methods
Mice
IRF8-EGFP fusion protein reporter mice were generated by Ozgene using a B6 germ line targeting strategy illustrated in Fig. 1. Mice were genotyped by PCR analysis of tail DNA using primers Wt IRF8 R (5'-CTGTCAGCTGACACAGAGTC-3'), IRF8 F (5'-TGTACCTCACACCAGAGACC-3') and IRF8 GFP R (5'-CGCTGAACTTGTGGCCGTTT-3'). C57BL/6J (B6) and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) congenic mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The use of mice in this study followed a protocol (LIG-16E) approved by NIAID Animal Care and Use Committee, a protocol (K052-LBG-07) approved by NIDDK Animal Care and Use Committee, and protocols approved by the WEHI animal and ethics committee.
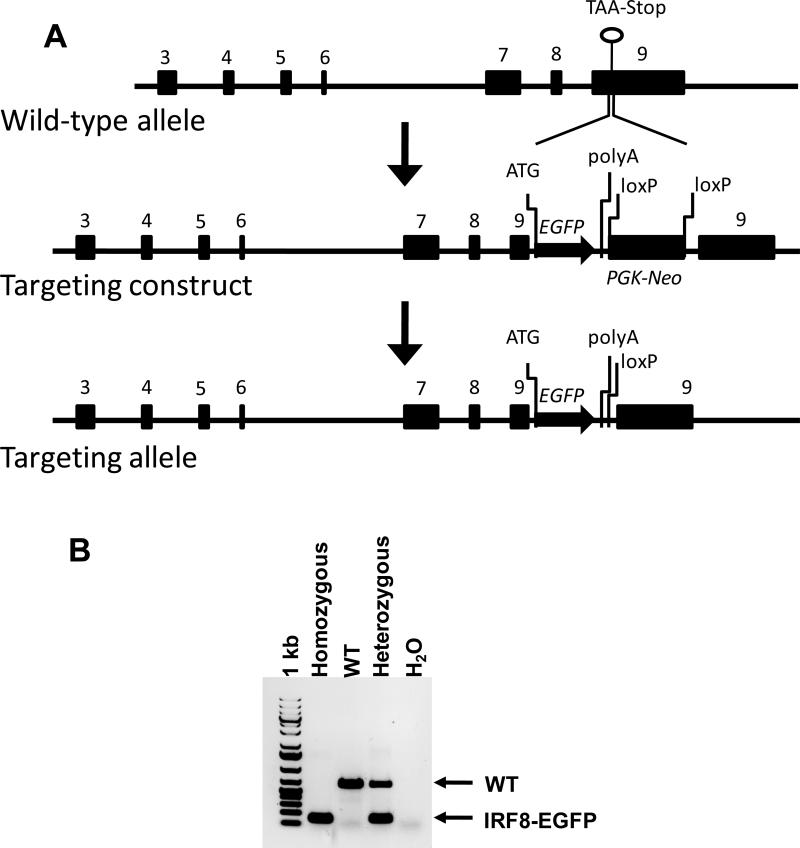
Fig. 1. Generation of the IRF8-EGFP reporter.
(A) A diagram of the targeting strategy used to replace the stop codon with an EGFP-PGK-Neo cassette. The PGK-Neo sequence was subsequently deleted by Cre-mediated excision. (B) The tail DNA of IRF8-EGFP mice was analyzed by PCR. The primers detected the knock-in fragment at 139 bp and the WT fragment at 642 bp.
Flow Cytometry
Cells were prepared and stained as previously reported (25). Monoclonal antibodies specific for cell surface markers are listed in Supplemental Table 1. Cells were analyzed using a LSR II analyzer (BD Biosciences) and FlowJo software. Dead cells were excluded by gating on cells negative for a viability dye (7AAD, propidium iodide, or fixable viability dye eFluor506). Doublets were excluded electronically by setting a SSC-A vs. FCS-W gate. For some experiments, cells were sorted by a FACSAria sorter (BD Biosciences). For intracellular staining, BM cells were first stained with antibodies for defining B cell subsets, followed by fixation and permeabilization with a Fix-and-Perm kit (Invitrogen). The cells were then stained with a polyclonal anti-IRF8 antibody (C-19x, Santa Cruz Biotechology) and a secondary Alexa Fluro546-labeled donkey anti-goat Ab (Invitrogen). The cells were analyzed by flow cytometry.
Cytomorphology
Cytocentrifuge preparations were stained with May-Grunwald-Giemsa stain before microscopic examination. Images were acquired using a Nikon Eclipse E600 microscope, 100x/1.3 NA oil objective with AxioCam Hrc and AxioVision 3.1 image acquisition software.
Clonogenic colony assays
Cultures were performed with sorted populations using semi-solid agar cultures in 1mL volumes of 0.3% agar in Iscove-modified Dulbecco medium containing 20% vol/vol newborn calf serum using cytokine stimuli as described (26), incubated for 7 days in a fully humidified atmosphere of 5% CO2 in air. Cultures were fixed, dried onto glass slides, and stained for acetylcholinesterase and then with luxol fast blue and hematoxylin and the number and type of colonies were determined by microscopic examination.
Immunization
Mice were immunized i.p. with 100 μg of NP-KLH (Biosearch) in alum. Splenocytes were analyzed 7 days later by flow cytometry.
Quantitative real time PCR (qPCR)
FACS-purified B cell subsets were extracted for RNA using a RNeasy Mini kit (Qiagen) including a DNA digestion step according to the manufacturer's instructions. Approximately 50 -100 ng of total RNA in 20 μl was reverse-transcribed with Superscript II reverse transcriptase (Invitrogen). One ng of cDNA was amplified in triplicate using an ABI Prism 7900HT SDS with the SYBR Green PCR Master Mix reagents (Applied Biosystems) and primers (Table 1). The housekeeping genes Gapdh and Hprt were amplified as internal controls. The relative RNA levels were calculated by 2−ΔΔCT algorithm (27).
Table 1.
Primer sequences used for qPCR.
| Pu.1 | CGGATGTGCTTCCCTTATCAAAC | 5′ |
| Pu.1 | TGACTTTCTTCACCTCGCCTGTC | 3′ |
| Ebfl | CCCCTCCAACTGCAGTAGCT | 5′ |
| Ebfl | GACCATGTTGGCTGGTGAGAA | 3′ |
| E2a | GCAACCTGAACCCCAAAGC | 5′ |
| E2a | ACCACGCCAGACACCTTCTC | 3′ |
| Pax5 | GCAGAGCGAGTCTGTGACAATG | 5′ |
| Pax5 | IGCIGIACIIIIGICCGAAIGAIC | 3′ |
| Cebpa | CCCCCAGTCAGACCAGAAAG | 5′ |
| Cebpa | CCCACAAAGCCCAGAAACCT | 3′ |
| Csflr | TTTTAAAAAACCCGTCCCAAACT | 5′ |
| Csflr | AGCCTTTGAGACTCTTGTCTTTTGA | 3′ |
| Rag2 | TCCTGCTTGTGGATGTGAAA | 5′ |
| Rag2 | GTGCCGAGTTTAATTCCTGG | 3′ |
| Statl | CTGAATATTTCCCTCCTGGG | 5′ |
| Statl | TCCCGTACAGATGTCCATGAT | 3′ |
| Csf2ra | CTTTCGTTGACGAAGCTCAG | 5′ |
| Csf2ra | GCTGGTTCAGGAGGATGATG | 3′ |
| Cebpb | GGCCCGGCTAGACAGTTAC | 5′ |
| Cebpb | GTTTCGGGACTTGATGCAAT | 3′ |
| Flt3 | AACIGGGCGICAICAIIIIC | 5′ |
| Flt3 | GTGAACAGAGAGGCCTGGAG | 3′ |
| Cx3crl | ATCCAGTTCAGGGAAGGAGG | 5′ |
| Cx3cr2 | AGACTGGGTGAGTGACTGGC | 3′ |
| Ifngrl | CAGCATACGACAGGGTTCAA | 5′ |
| Ifngrl | GATGCTGTCTGCGAAGGTC | 3′ |
| Ifngr2 | TGACGGCTCCCAAGTTAGAA | 5′ |
| Ifngr2 | CTGCTGCTCTGTGGGCTC | 3′ |
| Ifnarl | ACACTGCCCATTGACTCTCC | 5′ |
| Ifnarl | TTGGGTGCTACCCTCAGC | 3′ |
| Ifnar2 | CCACAAGACACAAGCTGAGG | 5′ |
| Ifnar2 | CAGAGGGGGATTCACGAGAC | 3′ |
| Stat2 | CAGGAACAGGCTGTCAAGGT | 5′ |
| Stat2 | CGCTTGGAGAATTGGAAGTT | 3′ |
| Irf9 | ACTCGGCCACCATAGATGAA | 5′ |
| Irf9 | TGAGCTAGAGGAGGGAGCTG | 3′ |
In vitro differentiation assay
Sort-purified B cells were cultured on an OP9 stromal cell layer (purchased from ATCC) with cytokines including IL-7 (10 ng/ml), Flt3L (200 ng/ml), SCF (50 ng/ml), GM-CSF (40 ng/ml), IL-4 (20 ng/ml), or IL-15 (10 ng/ml) for different periods of time. The cells were then analyzed by flow cytometry.
In vivo adoptive transfer assay
0.4-2×105 of sort-purified cells were injected i.v. into sublethally (500 rad) irradiated B6.CD45.1 mice. The mice were given antibiotics in their drinking water after irradiation. The splenocytes of recipients were stained and analyzed by flow cytometry 10 days later.
Statistical analysis
Student's t test was used to determine the statistical significance of the data. P<0.05 was considered to be statistically significant.
RESULTS
Generation of IRF8-EGFP fusion protein reporter mice
The stop codon at exon 9 of the mouse Irf8 locus was replaced with an EGFP sequence (Fig. 1A). The PGK-Neo selection cassette inserted into exon 9 next to the EGFP sequence was deleted from the mouse germline using Cre recombinase. The targeting strategy resulted in the expression of mRNA encoding an IRF8-EGFP fusion protein under the control of the endogenous Irf8 regulatory elements. A retroviral construct encoding essentially the same IRF8-EGFP fusion protein was previously used to study IRF8 function in infected cells or cell lines and was not found to cause unexpected effects due to the EGFP sequence (28). Genotyping of Irf8-EGFP knock-in mice identified heterozygous and homozygous mice (Fig. 1B). As expected, the IRF8-EGFP mice developed normally without differences from WT mice. Moreover, homozygous mice, which expressed twice as much IRF8-EGFP as heterozygous mice, were phenotypically indistinguishable from heterozygous and WT mice for the cellular distribution of lymphocytes and myeloid cells among different lymphoid organs (Table 2).
Table 2.
Cellular distribution of hematopoietic cells in wild-type and IRF8-EGFP mice
| BMa | Absolute cell numbers (x105) per femur (Mean ± SD) | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | CMPs | GMPs | Fr. A | Fr. B-C | Fr.D | Fr. E | Fr. F |
| WT | 0.15 ± 0.02 | 0.08 ± 0.02 | 0.24 ± 0.04 | 0.37 ± 0.22 | 3.08 ± 2.13 | 2.0 ± 1.12 | 2.53 ± 1.25 |
| IRF8- EGFPEgfp/+ | 0.18 ± 0.04 | 0.1 ± 0.02 | 0.29 ± 0.05 | 0.47 ± 0.33 | 2.41 ± 1.25 | 1.3 ± 0.56 | 2.73 ± 0.64 |
| IRF8- EGFPEgfp/Egfp | 0.15 ± 0.03 | 0.1 ± 0.03 | 0.22 ± 0.07 | 0.55 ± 0.13 | 3.15 ± 0.65 | 2.01 ± 0.48 | 3.66 ± 1.04 |
| Spleenb | Absolute cell numbers (x106) (Mean ± SD) | |||||
|---|---|---|---|---|---|---|
| Genotype | T cells | FO B cells | MZ B cells | DCs | N | M |
| WT | 18.3 ± 7.0 | 14.6 ± 4.0 | 1.7 ± 0.6 | 0.2 ± 0.1 | 0.7 ± 0.4 | 0.5 ± 0.3 |
| IRF8- EGFPEgfp/+ | 20.1 ± 7.8 | 12.5 ± 5.3 | 1.4 ± 0.7 | 0.2 ± 0.1 | 0.6 ± 0.3 | 0.4 ± 0.2 |
| IRF8- EGFPEgfp/Egfp | 23.6 ± 5.0 | 14.1 ± 2.0 | 1.6 ± 0.6 | 0.2 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.1 |
BM cells from indicated mice were stained and analyzed by flow cytometry. The gating schemes used for calculating each population were depicted in Fig. 2 and Fig. 4. Data represent 4 mice per group.
Splenocytes from indicated mice were stained and analyzed by flow cytometry. The cells were gated for T cells (CD4+), FO B cells (CD19+IgM+CD23+CD21lo/-), MZ B cells (CD19+IgM+CD23-CD21+), DCs (MHCII+CD11chi), neutrophils (N) (CD3-CD19-Gr- 1hiCD11b+) and monocytes (M) (CD3-CD19-Gr-1intCD11b+). Data represent 4 mice per group.
IRF8-EGFP expression distinguishes bipotent granulocyte-macrophage progenitors from more differentiated progeny
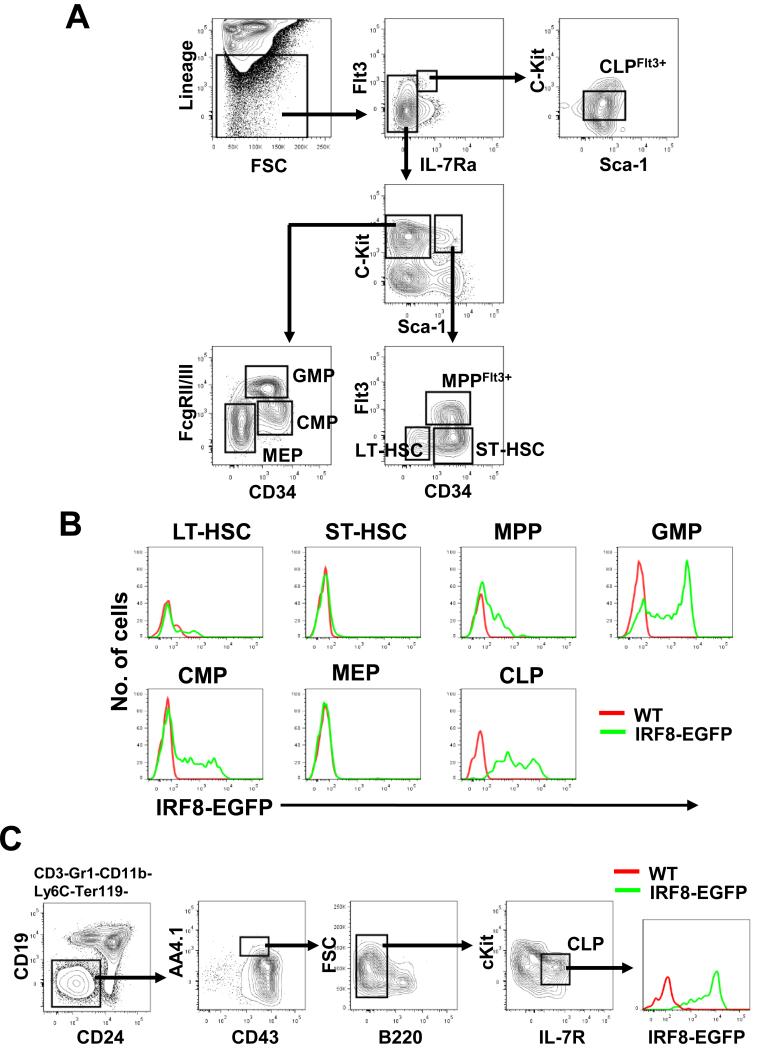
Previous analyses of Irf8 transcripts showed that expression of IRF8 increased after differentiation of HSCs into Lin−IL-7R−Sca-1−cKit+ myeloid progenitors and Lin−IL-7R+Sca-1+cKitlo CLPs (16). By using multicolor flow cytometry and a gating strategy of Pronk et al. (29), we detected IRF8-EGFP expression in early hematopoietic progenitors. As shown in Fig. 2 A and B, long-term (LT)- and short-term (ST)-HSCs expressed negligible levels of IRF8-EGFP (Fig. 2 B). Differentiation of HSCs into the MPP→CMP→GMP pathway coincided with sequential increases in IRF8-EGFP expression (Fig. 2 B). Nearly all Flt3+ CLPs were positive for IRF8-EGFP, whereas MEPs were negative (Fig. 2 B). Further characterization of CLPs by using the Hardy criteria (CD3−Gr1−CD11b−Ly6C−Ter119−CD19-CD24−AA4.1hiCD43+B220−cKitloIL7R+) (30) or the Weissman criteria (CD3−Gr1−CD11b-Ter119−CD19−CD27+Flt3+IL7R+B220−CD11c−) (31) revealed similar results (Fig. 2C and Supplemental Fig. 1A).
Fig. 2. IRF8-EGFP expression in BM hematopoietic progenitors.
(A) Gating schemes shown for the identification of LT-HSC, ST-HSC, MPP, GMP, CMP, MEP, and CLP. Lineage panel Abs included anti-CD3, B220, CD11b, Gr-1, and Ter119. Cells were gated on 7AAD− singlet cells. (B) Overlays are expression of IRF8-EGFP over WT controls. (C) IRF8-EGFP expression in CLPs defined by Rumfelt et al. (30). Data are representative of eight independent experiments.
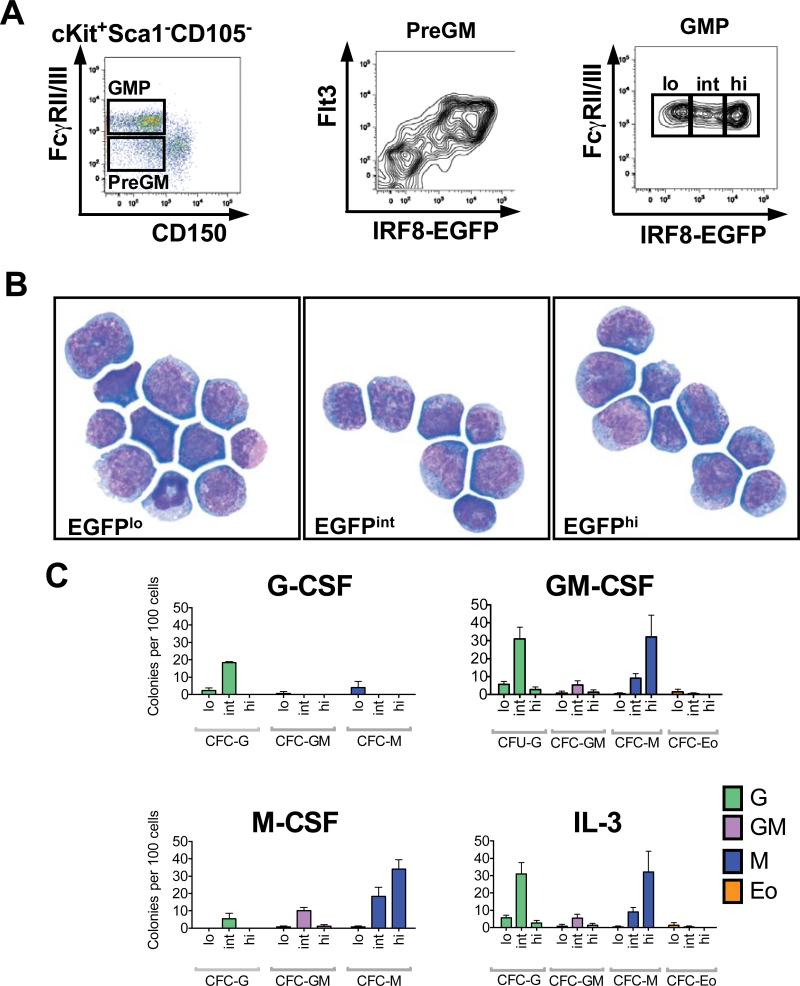
To study this further, we examined myeloid progenitor populations using an expanded range of markers to characterize IRF8-EGFP expression in the granulocyte-macrophage committed Lin−IL-7R−Sca-1−cKit+ CD150−FcγRII/III− pre-granulocyte-macrophage (PreGM) Flt3+ progenitors and multi-potential PreGM Flt3− progenitors (Fig. 3A, left panel), both of which lie within the CMP fraction that we had characterized previously (32, 33). Using this approach, we identified IRF8-EGFP expression in Flt3+ PreGMs, which are the first identifiable cells committed to forming GMPs (Fig. 3 A, middle panel), but not in the multi-potential Flt3− PreGMs that form GMPs as well as bipotential MEPs in vivo (32). These data suggest that IRF8 expression identifies the first point in the myeloid progenitor hierarchy associated with granulocyte-monocyte commitment from CMPs.
Fig. 3. IRF8-EGFP identifies specific myeloid PreGM and GMP populations.
(A) Gating schemes for identification of PreGM and GMP populations with IRF8-EGFP expression shown. (B) Cytomorphology of IRF8-EGFPlo/-, -EGFPint and -EGFPhi GMP populations sorted from Lin− BM cells of IRF8-EGFP mice. (C) Clonogenic colony forming assays of IRF8-EGFPlo/-, -EGFPint and -EGFPhi GMP populations cultured with 103 U/mL of G-CSF, GM-CSF, M-CSF, or IL-3. The numbers are colony forming units (CFU) of granulocyte (G), granulo-monocyte (GM), monocyte (M), and eosinophil (Eo) per 100 cells plated (means ± standard deviations of 3 biological replicates).
Given the range of IRF8-EGFP expression in GMPs (Fig. 2B), we then fractionated GMPs into 3 subpopulations based on IRF8-EGFPlo, IRF8-EGFPint and IRF8-EGFPhi expression (Fig. 3A, right panel) and examined their cytomorphology (Fig. 3B) and myeloid colony forming potential by semi-solid agar clonogenic colony assays using cytokine stimuli associated with specific cytokine receptor signaling, namely; granulocyte colony stimulating factor (G-CSF), macrophage colony stimulating factor (M-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF) and interleukin-3 (IL-3) (Fig. 3C).
Morphologically, all GMP populations were comprised primarily of promyelocytes, myelocytes and metamyelocytes but with IRF8-EGFPlo GMPs containing some more differentiated granulocyte ring forms and eosinophil progenitors as well (Fig. 3B). In semi-solid agar colony forming assays, the IRF8-EGFPint GMPs were capable of forming all myeloid colonies in vitro and on a per cell basis possessed the greatest granulocyte colony forming potential (CFC-G) in response to all cytokine stimuli (Fig. 3C). Interestingly, IRF8-EGFPhi GMPs were biased toward macrophage colony formation (CFC-M) after exposure to GM-CSF and M-CSF in particular while having virtually no response to G-CSF (Fig. 3C). In contrast, IRF8-EGFPlo GMPs did not possess significant colony forming potential to any of the cytokine stimuli when compared to the other GMP populations, consistent with this population being comprised primarily by more differentiated and no longer clonogenic granulocyte progenitors. Eosinophil colony forming potential (CFC-Eo) was minimal and basically seen only in cultures of IRF8-EGFPlo cells.
Taken together, these data demonstrate that GMPs are a heterogeneous collection of myeloid-committed progenitors, with intermediate IRF8 expression identifying GMPs that could contribute to all forms of myeloid colonies in vitro with the greatest potential to form CFC-G colonies. IRF8-EGFPhi cells have predominantly monocytic lineage potential whereas IRF8-EGFPlo GMPs appear to have lost most myeloid clonogenic potential.
IRF8-EGFP expression revealed heterogeneous pre-pro-B and homogeneous late B cell populations at the single cell level
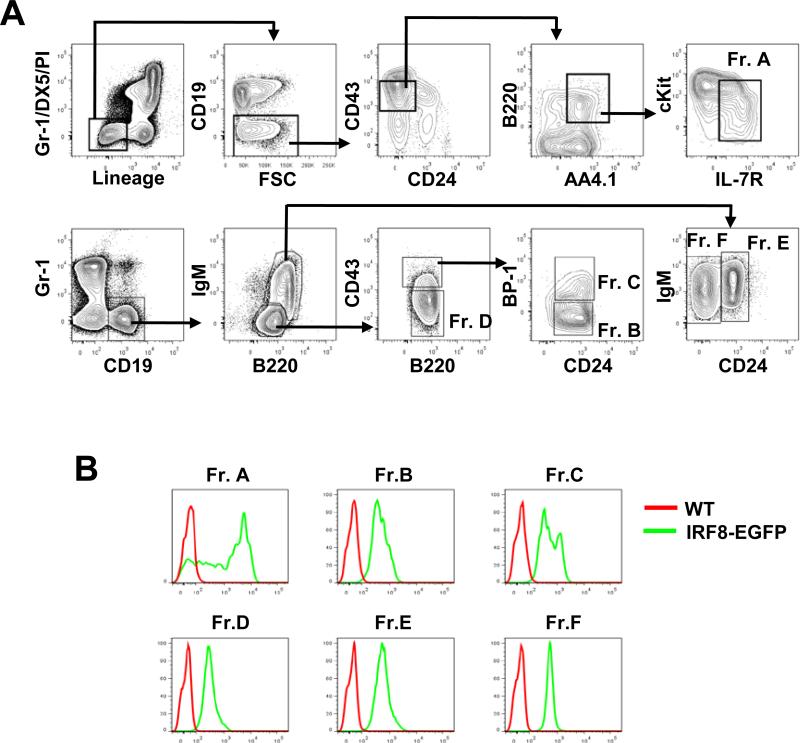
B cell lineage specification has been proposed to occur at the pre-pro-B cell stage (Hardy Fr. A), which is characterized by the generation of IgH D to JH gene rearrangements (34). The criteria used for phenotypic identification of pre-pro-B cells has been inconsistent in the literature, possibly due to the use of different markers and gating strategies. For example, the original definition of Fr. A was based on four markers: B220, CD43, CD24 and BP-1 (34). This was later expanded to more than 8 (B220, CD43, CD24, CD19, AA4.1, cKit, IL-7R, BP-1) as well as the lineage markers (30). Kincade's group defined pre-pro-B cells as DX5−Ly6C−IgM−B220+CD43+CD19−CD24− (35). More recently, Weissman's group redefined early stage B cells by using Ly6D (31). To accommodate these different nomenclatures for this B lineage subset, we combined markers used by the Hardy and Kincade groups to exclude granulocytes, monocytes, NK cells and T cells as well as erythroid and megakaryocytic lineage cells. Thus, the pre-pro-B cells were defined as CD19−CD43+CD24-B220+AA4.1+cKit+IL7R+DX5−CD3−Gr-1−CD11b−CD11c−Ly6C−Ter119− (Fig. 4A). As shown in Fig. 4B, the expression of IRF8-EGFP in Fr. A cells was strikingly heterogeneous with fluorescence intensity ranging from negative (background) to 104 above background. In contrast, cells in Fr. B through Fr. F expressed moderate and relatively homogeneous levels of IRF8-EGFP (Fig. 4B).
Fig. 4. IRF8-EGFP expression in BM B lineage cells.
(A) Gating schemes for identification of Hardy Frs. A, B, C, D, E and F, which represent pre-pro-B, early pro-B, late pro-B, pre-BII, immature and mature B cells, respectively. Lineage panel Abs contained anti-CD3, -CD11b, -CD11c, -Ly6C and -Ter119 Abs. (B) Overlays are expression of IRF8-EGFP in each subset over WT controls. Data are representative of nine independent experiments.
Using the Weissman nomenclature, which includes Ly6D to distinguish all-lymphoid progenitors (ALP), B-cell lineage progenitors (BLP) and pre-pro-B cells (31), we observed similar heterogeneity of IRF8-EGFP expression in Ly6D+ pre-pro-B cells (Supplemental Fig. 1A). Most recently, Medina et al. have shown that PDCA-1 could reduce pDC contamination from the pre-pro-B fraction (36). By including PDCA-1 as an additional marker, we evaluated the Hardy Fr. A for IRF8-EGFP expression. We found that PDCA-1 expression was heterogeneous in the Fr. A compartment. Excluding PDCA-1+ cells did not change the heterogeneous nature of IRF8-EGFP expression (Supplemental Fig. 1B). From this, we conclude that the pre-pro-B compartment is a mixture of cells with different levels of IRF8-EGFP expression.
Further analyses of peripheral B cells revealed consistently homogeneous and similar levels of IRF8-EGFP expression throughout the transitional to mature stages (Supplemental Fig. 2). In keeping with previous studies of IRF8 transcripts employing microarray and quantitative RT-PCR analyses (37), expression of IRF8-EGFP was significantly increased in germinal center B cells and decreased in plasma cells (Supplemental Fig. 2). In addition, neutrophils were negative for IRF8-EGFP whereas NK cells and macrophages expressed intermediate levels (Supplemental Fig. 2).
IRF8-EGFP expression distinguishes B cell-committed from non-committed pre-pro-B cells
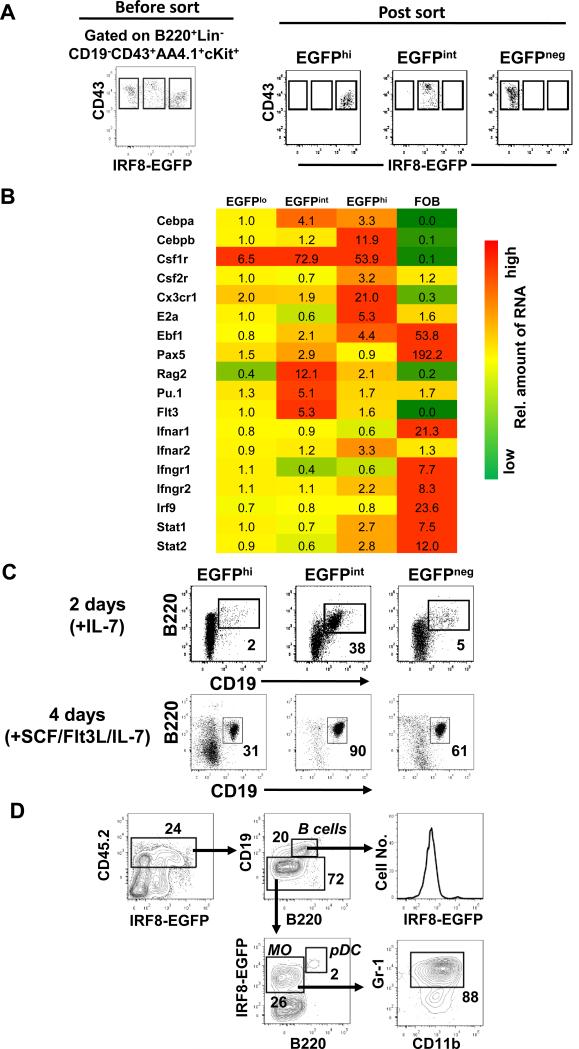
The finding that expression levels of IRF8-EGFP in the pre-pro-B cell compartment spanned 4 logs in fluorescence intensity prompted us to sort-purify EGFPneg, EGFPint and EGFPhi pre-pro-B cells and examine their B cell developmental potentials. First, we examined expression levels of a selected set of 18 genes that are considered to be “signature genes” representing lymphoid, myeloid and dendritic cell lineages as well as signaling activities of cytokine receptor pathways. As shown in Fig. 5A and B, EGFP neg, int and hi subsets were well distinguished by expression patterns of these genes. The lymphoid genes Pax5, Rag2, Pu.1 and Flt3 were enriched in the EGFPint subset, whereas the myeloid and dendritic progenitor representative genes Cebpb, Csf1r, Csf2r, and Cx3cr1 were over-expressed in the EGFPhi subset. Next, we evaluated the B cell developmental potential of these subsets in vitro under lymphoid permissive conditions. EGFPint pre-pro-B cells cultured in the presence of IL-7 gave rise to substantial numbers of CD19+ B cells within 48 hours, whereas EGFPneg and EGFPhi pre-pro-B cells did so at greatly reduced frequencies (Fig. 5C). After 4 days of culture, the generation of CD19+ B cells was significantly enriched in all subsets, but the frequency of B cells generated by EGFPint cells was still the highest (90%), followed by EGFPhi (60%) and EGFPneg (31%) (Fig. 5C). Finally, because the EGFPhi prepro-B cells expressed both lymphoid and myeloid progenitor genes (Fig. 5B), we tested the developmental potential of EGFPhi pre-pro-B cells by an adoptive transfer assay. Although EGFPhi pre-pro-B cells could yield a small number of B cells in vivo at 10 days following adoptive transfer, most of the progeny belonged to DC and/or monocyte lineages based on expression of markers including B220, CD19, IRF8-EGFP, Gr-1 and CD11b (Fig. 5D). Altogether, we conclude that B cell lineage specification and commitment is primarily associated with intermediate levels of IRF8-EGFP expression.
Fig. 5. The developmental potential of IRF8-EGFP+ pre-pro-B cells.
(A) Sort-purification of pre-pro-B cells with different levels of IRF8-EGFP. Lineage panel Abs included anti-CD3, -CD11b, -CD19, -Gr-1 and -Ter119. (B) Sort-purified cells were analyzed by real time qPCR for expression of indicated genes. Numbers are relative amounts of mRNA. Data is representative of two independent experiments with similar results. FOB, follicular B cells. (C) Sort-purified cells were cultured with IL-7 for 2 days (top panel) or with SCF, Flt3L and IL-7 for 4 days (bottom panel). The cells were then analyzed by flow cytometry. The numbers are percentages of cells falling in each gate. Data are representative of three independent experiments. (D) EGFPhi pre-pro-B cells sort-purified as in (A) were injected i.v. into sublethally irradiated CD45.1 congenic mice. The splenocytes of recipient mice were analyzed by flow cytometry 10 days later. Data are representative of 8 mice from three independent experiments. MO, monocyte-like cells. The numbers are percentages of cells falling in each gate.
DC progenitors express very high levels of IRF8-EGFP
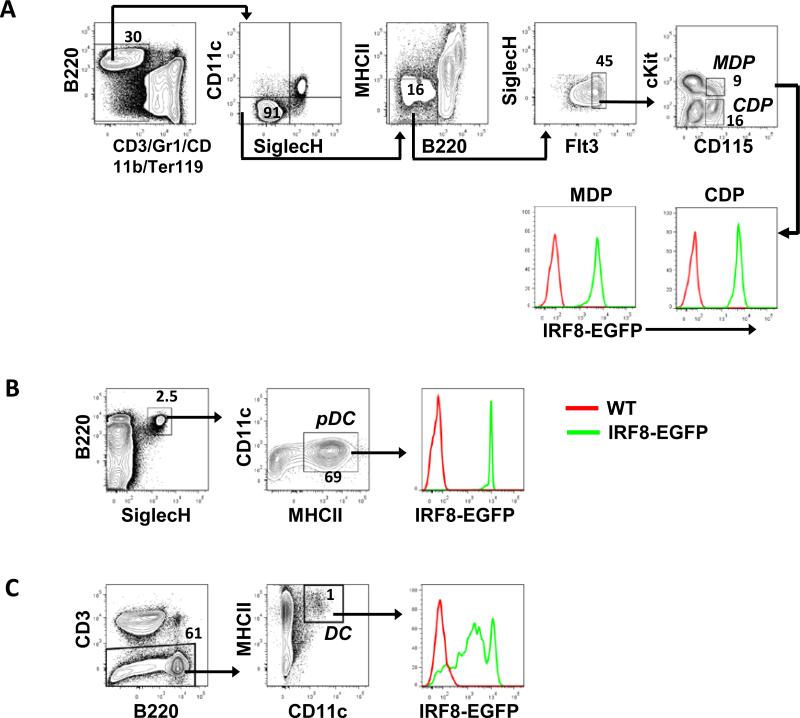
DCs are generated from lineage potential-restricted progenitors including MDPs and CDPs (38, 39). Studies of MDPs (Lin−CD11c−SiglecH−MHCII−Flt3+cKit+CD115+) and CDPs (Lin−CD11c−SiglecH−MHCII−Flt3+cKit−CD115+) revealed strikingly high and homogeneous expression of IRF8-EGFP (Fig. 6 A). BM pDCs (B220+SiglecH+CD11c+MHCII+) were also IRF8-EGFPhi (Fig. 6 B). By contrast, splenic DCs exhibited heterogeneous expression of IRF8-EGFP (Fig. 6 C). We conclude that DC lineage specification and commitment are associated with very high levels of IRF8-EGFP expression.
Fig. 6. IRF8-EGFP expression in DC progenitors and mature DCs.
(A and B) BM cells were stained and analyzed by flow cytometry. MDPs and CDPs (A) and pDCs (B) were identified as indicated. Overlays were IRF8-EGFP over WT controls. (C) Splenocytes were analyzed for IRF8-EGFP expression in DCs. Cells were gated on Viability dye eFluor506-negative singlets. The numbers are percentages of cells falling in each gate. Data are representative of 8 independent experiments.
T lineage cells express negligible levels of IRF8-EGFP
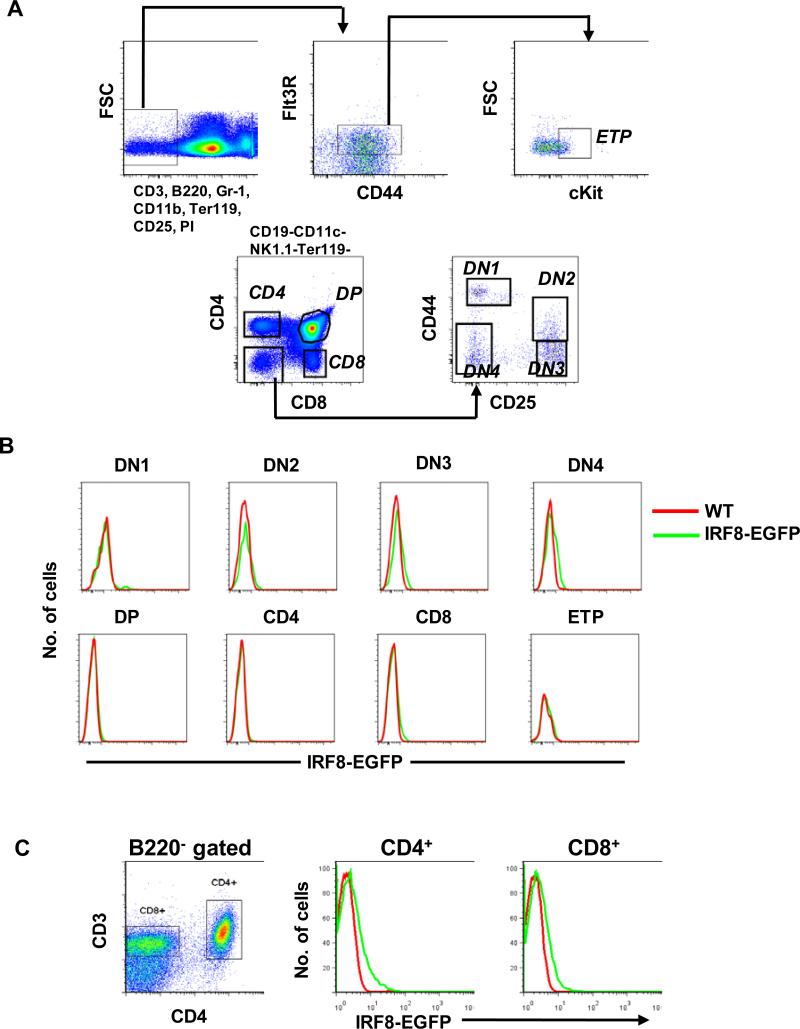
A previous report indicated that IRF8 is dispensable for development of T cells (17). To determine whether IRF8-EGFP was expressed by T cells, we examined thymic and splenic T lineage cells by flow cytometry. The overall levels of IRF8-EGFP expression in thymic T lineage cells including early T cell progenitors (ETPs) were negative to weak positive (Fig. 7 A and B). Most peripheral naïve T cells including CD4 and CD8 T cells were also negative for IRF8-EGFP expression (Fig. 7 C). The minimal levels of IRF8-EGFP expression in T lineage cells under steady state conditions argues that IRF8 is not required for thymic T cell development and homeostasis, consistent with the characteristics of IRF8 knockout mice (17).
Fig. 7. IRF8-EGFP expression in T cells.
(A) Gating schemes used to identify early T cell progenitors (ETP) and double-negative (DN) and double-positive (DP) populations in the thymus. (B) Overlays show expression of IRF8-EGFP over WT controls in thymocytes. (C) Splenocytes of WT and IRF8-EGFP mice were stained and analyzed by flow cytometry. All cells were gated on 7AAD− single cells. Data are representative of five independent experiments.
IRF8-EGFP expression is upregulated in activated B and T cells
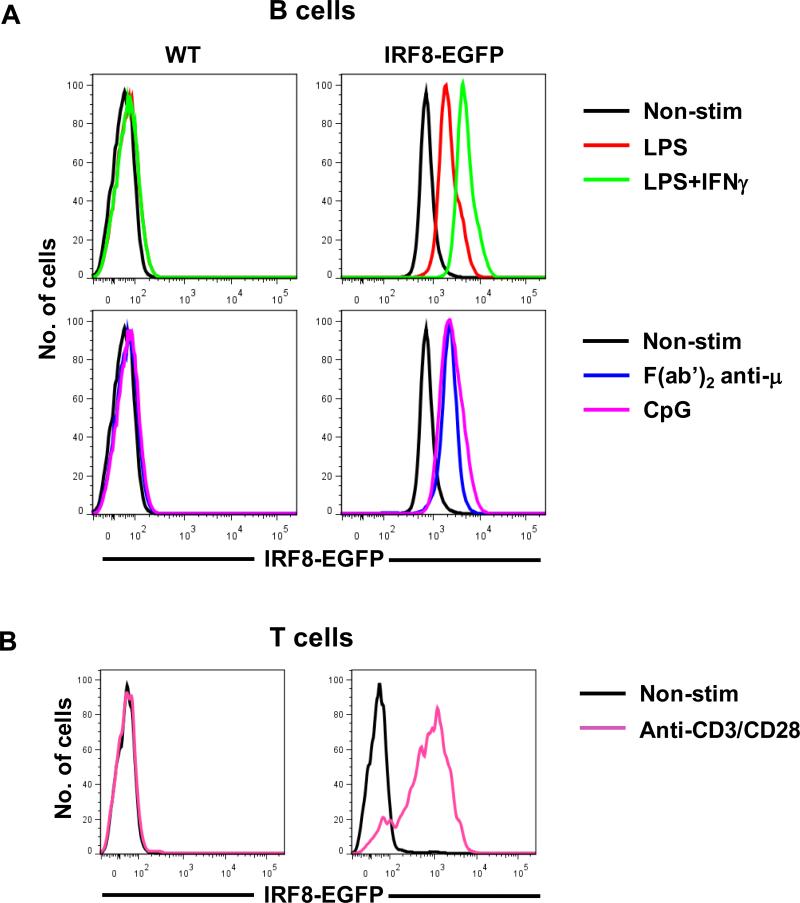
To determine if expression of IRF8-EGFP is altered in activated lymphocytes, we stimulated purified B and T cells with anti-B cell receptor (BCR), -T cell receptor (TCR), LPS, CpG and/or IFNγ and measured expression levels of IRF8-EGFP by flow cytometry. As shown in Fig. 8 A, expression of IRF8-EGFP was significantly upregulated in B cells stimulated with LPS, CpG, and anti-IgM antibodies. A synergistic effect on IRF8-EGFP expression was observed between LPS and IFNγ. Stimulation of T cells by coligation of the TCR and CD28 also dramatically enhanced expression of IRF8-EGFP (Fig. 8 B). We therefore conclude that expression of IRF8-EGFP, similar to IRF8 native proteins in stimulated B and T cells (40), is markedly upregulated in activated B and T cells.
Fig. 8.
Induction of IRF8-EGFP expression in B and T cells in vitro. (A) Splenic B cells from WT and IRF8-EGFP mice were purified and stimulated with LPS (20 μg/ml), IFNγ (10 ng/ml), anti-μ (10 μg/ml), or CpG (1 μg/ml) overnight. The cells were then analyzed by flow cytometry. (B) Splenocytes were cultured in the presence of anti-CD3 and anti-CD28 Dyna beads overnight. The cells were then stained with APC-labeled anti-CD4 Ab and analyzed by flow cytometry. Cells are gated on 7AAD−CD4+ cells. The data are representative of two experiments with similar results.
Discussion
Previous studies showing that IRF8-deficient mice exhibited broad defects in the development of a variety of cell types led to the concept that IRF8 might regulate gene programs facilitating cellular lineage differentiation. However, except for the evidence that IRF8 may extinguish gene programs for neutrophil fate and promote differentiation of the macrophage and the DC lineages (15, 41), little is known whether and how IRF8 could regulate myeloid vs. lymphoid lineage selection at different branch points. We now use an IRF8-EGFP reporter mouse to demonstrate that IRF8-EGFP exhibits different expression patterns in distinct progenitor cells of the myeloid and lymphoid lineages, correlating with previous studies showing that IRF8 is differentially expressed in distinct lineages of hematopoietic cells. Most importantly, IRF8-EGFP reporter mice revealed previously unappreciated heterogeneity in expression of IRF8 in some of the seemingly homogeneous populations of oligopotent progenitors including CMPs, GMPs and pre-pro-B cells.
The most prominent finding of this study is that the IRF8-EGFP reporter allele made it possible to accurately measure the levels of IRF8 protein expression, on a single cell level, in all hematopoietic cell types from the earliest adult progenitors to their terminally-differentiated progeny. The levels of IRF8-EGFP expression were initially low in HSCs but progressively increased along with HSC differentiation into two major branches of the hematopoietic tree, the myeloid and lymphoid lineages. The lack of IRF8-EGFP expression in the third branch, the megakaryocytic and erythroid lineages, is consistent with the idea that IRF8 has no role in promoting differentiation along these pathways. Surprisingly, expression of IRF8-EGFP was extremely heterogeneous in the well-defined and seemingly homogeneous GMPs with ~40% of these cells being negative for IRF8-EGFP expression. By using additional markers, we were able to define the IRF8-EGFP-positive and -negative subpopulation as being Flt3+ pre-GMs with restricted GMP potential and Flt3− pre-GMs with GMP, megakaryocytic and erythroid potential, respectively. We further identified three subsets of GMPs with varying levels of IRF8-EGFP expression and differentiation potential. Perhaps counter intuitively, the IRF8-EGFPint GMPs represented the true bipotent GMPs as they are readily primed for the granulocyte and macrophage lineages. In contrast, the IRF8-EGFPhigh GMPs significantly enriched for monocytic lineage forming progenitors, while the IRF8-EGFPneg GMPs were more mature granulocytic and eosinophilic progenitors that had lost significant CFU potential. While the driving force for IRF8 expression in GMP subsets is currently unknown, our finding that IRF8-EGFP subsets of GMPs responded differently to cytokine-driven differentiation in vitro (Fig. 3) supports the hypothesis that cytokines may play more selective (42) than instructive (43) roles in development of oligopotential progenitors.
Another striking finding of the IRF8-EGFP reporter is the heterogeneity of the prepro-B cell compartment. Since the first description of B220+CD19− pre-pro-B cells (Fr. A) by Hardy and colleagues (34), the B cell developmental potential of this population has been assessed by both in vitro and in vivo assays. The inconsistent observations in the literature have been largely attributed to contamination by other types of cells, such as DCs that express Ly6C (44) or PDCA-1 (36). In this study, we used stringent gating strategies revised by Hardy and colleagues (30) and identified three subsets with low, intermediate and high levels of IRF8-EGFP in this well-defined Fr. A compartment (Fig. 4). When tested under lymphoid permissible conditions, EGFPint Fr. A cells quickly differentiated into CD19+ B cells, suggesting that IRF8-EGFPint pre-pro-B cells are readily specified to the B cell lineage. While IRF8-EGFPneg Fr. A cells could be precursors of IRF8-EGFPint and -EGFPhi pre-pro-B cells, as suggested by studies of cells cultured with Flt3L, SCF, IL-7 and IL-15 (Supplemental Fig. 3A), most of IRF8-EGFPhi Fr. A cells appeared to be specified for the DC and/or monocyte lineages (Fig. 5 D). Because pDCs express B220 and very high levels (104 above background) of IRF8-EGFP (Fig. 6), it was possible that IRF8-EGFPhi pre-pro-B cells could be contaminated by a small number of pDCs that expressed low levels of Ly6C and were not excluded efficiently by gating. To exclude this possibility, we assessed PDCA-1 expression in Fr. A cells. PDCA-1+ pDCs have recently been shown to contaminate the analysis of pre-pro-B cells (36). Consistent with the results from using the Hardy and the Weissman gating strategies, gating on PDCA-1− Fr. A cells still yielded heterogeneous expression levels of IRF8-EGFP (Supplemental Fig. 1B). We believe that the existence of IRF8-EGFPhi subset in the Fr. A compartment could be a true nature of this population. A strong bias of IRF8-EGFPhi Fr. A cells to develop into DC-myeloid lineages as assessed by adoptive transfer assays (Fig. 5D) is consistent with the notion that some DCs bear rearranged Ig DHJH sequences (45, 46). This is consistent with the idea that DCs might be generated from different developmental pathways including at least the MDP-CDP pathway and the pre-pro-B pathway. Therefore, the IRF8-EGFP reporter mouse could be a valuable tool for investigating the nature of the pre-pro-B cell population.
Another intriguing finding of this study is that differentiation to the DC lineage, and pDC in particular, is associated with very high expression of IRF8. Both MDPs and CDPs are uniformly IRF8-EGFPhi. This suggests that commitment to the DC lineage is highly dependent on IRF8-regulated gene programs. Indeed, IRF8 deficient mice exhibit a complete blockade in the development of pDCs and CD8α+ cDCs (21, 22). While this study was in preparation for submission, Rosenbauer and colleagues reported a very similar finding in DC progenitors by using an IRF8-Venus reporter mouse (47).
As with any other EGFP reporter models, there is a concern of reporter efficiency during a dynamic differentiation process. On the one hand, a prolonged requirement for IRF8-EGFP to build up to a sufficient level for detection would under-report true IRF8 protein. On the other hand, a longer half-life of IRF8-EGFP would over-report true expression. There are several pieces of evidence that argue against a poor reporter efficiency of our model. First, the targeting strategy used to generate the IRF8-EGFP fusion protein reporter preserved all natural regulatory elements of the IRF8-EGFP locus (Fig. 1). Second, we previously showed that the Fr. A cells expressed at least 100 times more IRF8 transcripts than Fr. B cells measured by qPCR (16). Consistent with this finding, the mean fluorescence intensity of IRF8-EGFP of the bulk Fr. A population was 6 times higher than Fr. B cells (Supplemental Fig. 3B), a pattern well matched by messenger RNA expression. Analysis of IRF8 protein expression by intracellular staining with a polyclonal anti-IRF8 antibody in combination with cell surface markers revealed similar patterns of IRF8 protein expression (Supplemental Fig. 3C). It is worth noting that the IRF8-EGFP reporter is superior to all other available methods by revealing, at the single cell level, the heterogeneity of the Fr. A population. Third, similar to Fr. A, the GMP population is also heterogeneous for IRF8-EGFP expression (Fig. 2). If IRF8-EGFPlo GMPs were under-reported, prolonged culture would be expected to yield similar frequencies of progeny cells generated from IRF8-EGFPlo and IRF8-EGFPint GMPs. To the contrary, these GMP subsets exhibited distinct developmental potentials (Fig. 3). Fourth, IRF8 expression in rapidly cycling GCs is increased at both the RNA and protein levels (37, 48, 49). When GCs mature to PCs, IRF8 is rapidly downregulated (37, 50, 51). Consistent with this, the IRF8-EGFP reporter revealed the same patterns of expression in that GCs express higher levels and PCs express markedly lower levels of IRF8-EGFP than naïve B cells (Supplemental Fig. 2). Finally, the IRF8-EGFP reporter mouse has recently been demonstrated to faithfully reveal dynamic changes of IRF8 expression during progression of an experimental autoimmune encephalomyelitis disease (52) and in differentiating Langerhans cells (53). Altogether, the IRF8-EGFP reporter appeared to faithfully reflect true IRF8 expression in both physiological and pathological conditions.
In view of the data indicating that IRF8-EGFP could identify subsets of GMPs and pre-pro-B cells with distinct lineage potentials, we propose that there exists an IRF8 concentration-dependent mechanism for myeloid and lymphoid lineage specification and commitment. In this model, the development of macrophages and DCs requires very high levels of IRF8, whereas the generation of B lineage cells depends on intermediate levels. Minimal or no requirement of IRF8 is found for the development of megakaryocytic and erythroid cells as well as the T cell lineage in general. The phenotypes of IRF8-EGFP reporter mice described here and IRF8-deficient mice reported previously support this conclusion.
In addition to its function in regulating immune cell differentiation under the steady state conditions, IRF8 also plays important roles in the effector stages of innate and adaptive immune responses against a variety of microbial pathogens (54-59). This is at least partially achieved by IRF8-mediated transcriptional control of cytokine expression, including IL-12p40 and type I interferons (60). Because both B and T cells significantly upregulate IRF8-EGFP expression following in vitro stimulation with LPS, LPS plus IFNγ, anti-IgM (which stimulates B cells) or anti-CD3/CD28 (which stimulates T cells) (Fig. 8), the IRF8-EGFP reporter mice could be valuable for studying the functions of IRF8 in the settings of inflammatory responses such as the EAE (52). Furthermore, this IRF8-EGFP reporter should be useful for studying IRF8 functions in other cell types such as microglia and smooth muscle where IRF8 has been recently found to be present and function (14, 61-64).
Supplementary Material
Acknowledgments
We thank Mehrnoosh Abshari for assistance in cell sorting and Alfonso Macias in mouse colony maintenance.
Glossary
Abreviations used in this article: HSC, hematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythroid progenitor; MDP, macrophage-dendritic cell progenitor; CDP, common dendritic cell progenitor; DC, dendritic cell; CLP, common lymphoid progenitor; ETP, early T cell progenitor; BM, bone marrow.
Footnotes
Author Contributions: H.W. designed, directed and performed experiments, analyzed data and wrote the paper. M.Y., J.S., S. J., R.Y., S.M.A., A.P.N., D.M., and L.D. designed and performed experiments (A.P.N. wrote the paper); K.O., W.G.C., S.L.N. and H.C.M directed the research; H.C.M. steered the research, analyzed data and wrote the paper.
This work was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (NIAID) (H.W., J.S., H.C.M.), National Institute of Child Health and Human Development (NICHD) (K.O.), and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (M.Y., W.C.). A.P.N., L.D and D.M. were supported by a Program Grant (1016647) and Independent Research Institutes Infrastructure Support Scheme Grant (361646) from the Australian National Health and Medical Research Council, the Carden Fellowship (to D.M.) of the Cancer Council, Victoria; a Cure Cancer Australia/Leukaemia Foundation Australia Post Doctoral Fellowship and Lions Fellowship, Cancer Council of Victoria (to A.P.N.), and Victorian State Government Operational Infrastructure Support. S.L.N. was supported by an Australian Research Council Future Fellowship.
The authors have no conflicting financial interests.
References
- 1.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 2.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 3.King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nature reviews. Immunology. 2011;11:685–692. doi: 10.1038/nri3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, Moore J, Sieweke MH. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013 doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, Takatsu K, Kincade PW. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagaoka H, Gonzalez-Aseguinolaza G, Tsuji M, Nussenzweig MC. Immunization and infection change the number of recombination activating gene (RAG)-expressing B cells in the periphery by altering immature lymphocyte production. The Journal of experimental medicine. 2000;191:2113–2120. doi: 10.1084/jem.191.12.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naik SH, Perie L, Swart E, Gerlach C, van Rooij N, de Boer RJ, Schumacher TN. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 8.D'Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. The Journal of experimental medicine. 2003;198:293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manz MG, Traver D, Miyamoto T, Weissman IL, Akashi K. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 2001;97:3333–3341. doi: 10.1182/blood.v97.11.3333. [DOI] [PubMed] [Google Scholar]
- 10.Traver D, Akashi K, Manz M, Merad M, Miyamoto T, Engleman EG, Weissman IL. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, D'Amico A, Hochrein H, O'Keeffe M, Shortman K, Lucas K. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood. 2001;98:3376–3382. doi: 10.1182/blood.v98.12.3376. [DOI] [PubMed] [Google Scholar]
- 12.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi M, Wakayama K, Itoh A, Kawai K, Pleasure D, Ozato K, Itoh T. Interferon regulatory factor 8/interferon consensus sequence binding protein is a critical transcription factor for the physiological phenotype of microglia. Journal of neuroinflammation. 2012;9:227. doi: 10.1186/1742-2094-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda T, Tsuda M, Yoshinaga R, Tozaki-Saitoh H, Ozato K, Tamura T, Inoue K. IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype. Cell reports. 2012;1:334–340. doi: 10.1016/j.celrep.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker AM, Michael DG, Satpathy AT, Sciammas R, Singh H, Bhattacharya D. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood. 2012;119:2003–2012. doi: 10.1182/blood-2011-06-364976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC., 3rd IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC, 3rd, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 18.Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, Sher A. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 19.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, Nussenzweig M, Merad M. The origin and development of nonlymphoid tissue CD103+ DCs. The Journal of experimental medicine. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheller M, Foerster J, Heyworth CM, Waring JF, Lohler J, Gilmore GL, Shadduck RK, Dexter TM, Horak I. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood. 1999;94:3764–3771. [PubMed] [Google Scholar]
- 21.Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC, 3rd, Belardelli F, Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. The Journal of experimental medicine. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131–1135. doi: 10.4049/jimmunol.170.3.1131. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto M, Kato T, Hotta C, Nishiyama A, Kurotaki D, Yoshinari M, Takami M, Ichino M, Nakazawa M, Matsuyama T, Kamijo R, Kitagawa S, Ozato K, Tamura T. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PloS one. 2011;6:e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, Menon G, Trouillet C, McDonald D, Carey P, Ginhoux F, Alsina L, Zumwalt TJ, Kong XF, Kumararatne D, Butler K, Hubeau M, Feinberg J, Al-Muhsen S, Cant A, Abel L, Chaussabel D, Doffinger R, Talesnik E, Grumach A, Duarte A, Abarca K, Moraes-Vasconcelos D, Burk D, Berghuis A, Geissmann F, Collin M, Casanova JL, Gros P. IRF8 mutations and human dendritic-cell immunodeficiency. The New England journal of medicine. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Ye J, Arnold LW, McCray SK, Clarke SH. A VH12 transgenic mouse exhibits defects in pre-B cell development and is unable to make IgM+ B cells. J Immunol. 2001;167:1254–1262. doi: 10.4049/jimmunol.167.3.1254. [DOI] [PubMed] [Google Scholar]
- 26.Metcalf D, Di Rago L, Mifsud S. Synergistic and inhibitory interactions in the in vitro control of murine megakaryocyte colony formation. Stem Cells. 2002;20:552–560. doi: 10.1002/stem.200552. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, Ozato K. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14368–14373. doi: 10.1073/pnas.0504014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell stem cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. The Journal of experimental medicine. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes & development. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng AP, Kauppi M, Metcalf D, Di Rago L, Hyland CD, Alexander WS. Characterization of thrombopoietin (TPO)-responsive progenitor cells in adult mouse bone marrow with in vivo megakaryocyte and erythroid potential. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2364–2369. doi: 10.1073/pnas.1121385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. The Journal of experimental medicine. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tudor KS, Payne KJ, Yamashita Y, Kincade PW. Functional assessment of precursors from murine bone marrow suggests a sequence of early B lineage differentiation events. Immunity. 2000;12:335–345. doi: 10.1016/s1074-7613(00)80186-7. [DOI] [PubMed] [Google Scholar]
- 36.Medina KL, Tangen SN, Seaburg LM, Thapa P, Gwin KA, Shapiro VS. Separation of plasmacytoid dendritic cells from B-cell-biased lymphoid progenitor (BLP) and Pre-pro B cells using PDCA-1. PloS one. 2013;8:e78408. doi: 10.1371/journal.pone.0078408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, van der Zouwen B, Zhou JX, Lougaris V, Lipsky PE, Grammer AC, Morse HC., 3rd Regulation of the germinal center gene program by IFN regulatory factor 8 / IFN consensus sequence binding protein. The Journal of experimental medicine. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O'Keeffe M, Bahlo M, Papenfuss A, Kwak JY, Wu L, Shortman K. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nature immunology. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 39.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nature immunology. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 40.Nelson N, Kanno Y, Hong C, Contursi C, Fujita T, Fowlkes BJ, O'Connell E, Hu-Li J, Paul WE, Jankovic D, Sher AF, Coligan JE, Thornton A, Appella E, Yang Y, Ozato K. Expression of IFN regulatory factor family proteins in lymphocytes. Induction of Stat-1 and IFN consensus sequence binding protein expression by T cell activation. J Immunol. 1996;156:3711–3720. [PubMed] [Google Scholar]
- 41.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O'Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 42.Robb L. Cytokine receptors and hematopoietic differentiation. Oncogene. 2007;26:6715–6723. doi: 10.1038/sj.onc.1210756. [DOI] [PubMed] [Google Scholar]
- 43.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- 44.Miller JP, Izon D, DeMuth W, Gerstein R, Bhandoola A, Allman D. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. The Journal of experimental medicine. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, Buer J, Murphy KM, Reizis B, Jung S. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, Leder P, Sakaguchi N, Akashi K. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 47.Schonheit J, Kuhl C, Gebhardt ML, Klett FF, Riemke P, Scheller M, Huang G, Naumann R, Leutz A, Stocking C, Priller J, Andrade-Navarro MA, Rosenbauer F. PU.1 Level-Directed Chromatin Structure Remodeling at the Irf8 Gene Drives Dendritic Cell Commitment. Cell reports. 2013 doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Cattoretti G, Shaknovich R, Smith PM, Jack HM, Murty VV, Alobeid B. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 49.Shin DM, Lee CH, Morse HC., 3rd IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PloS one. 2011;6:e27384. doi: 10.1371/journal.pone.0027384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez A, Pittaluga S, Rudelius M, Davies-Hill T, Sebasigari D, Fountaine TJ, Hewitt S, Jaffe ES, Raffeld M. Expression of the interferon regulatory factor 8/ICSBP-1 in human reactive lymphoid tissues and B-cell lymphomas: a novel germinal center marker. Am J Surg Pathol. 2008;32:1190–1200. doi: 10.1097/PAS.0b013e318166f46a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, Morse HC., 3rd IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188–3194. doi: 10.4049/jimmunol.0803693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida Y, Yoshimi R, Yoshii H, Kim D, Dey A, Xiong H, Munasinghe J, Yazawa I, O'Donovan MJ, Maximova OA, Sharma S, Zhu J, Wang H, Morse HC, 3rd, Ozato K. The Transcription Factor IRF8 Activates Integrin-Mediated TGF-beta Signaling and Promotes Neuroinflammation. Immunity. 2014;40:187–198. doi: 10.1016/j.immuni.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chopin M, Seillet C, Chevrier S, Wu L, Wang H, Morse HC, 3rd, Belz GT, Nutt SL. Langerhans cells are generated by two distinct PU.1-dependent transcriptional networks. The Journal of experimental medicine. 2013;210:2967–2980. doi: 10.1084/jem.20130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alter-Koltunoff M, Goren S, Nousbeck J, Feng CG, Sher A, Ozato K, Azriel A, Levi BZ. Innate immunity to intraphagosomal pathogens is mediated by interferon regulatory factor 8 (IRF-8) that stimulates the expression of macrophage- specific Nramp1 through antagonizing repression by c-Myc. The Journal of biological chemistry. 2008;283:2724–2733. doi: 10.1074/jbc.M707704200. [DOI] [PubMed] [Google Scholar]
- 55.Fehr T, Schoedon G, Odermatt B, Holtschke T, Schneemann M, Bachmann MF, Mak TW, Horak I, Zinkernagel RM. Crucial role of interferon consensus sequence binding protein, but neither of interferon regulatory factor 1 nor of nitric oxide synthesis for protection against murine listeriosis. The Journal of experimental medicine. 1997;185:921–931. doi: 10.1084/jem.185.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hein J, Kempf VA, Diebold J, Bucheler N, Preger S, Horak I, Sing A, Kramer U, Autenrieth IB. Interferon consensus sequence binding protein confers resistance against Yersinia enterocolitica. Infection and immunity. 2000;68:1408–1417. doi: 10.1128/iai.68.3.1408-1417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ko J, Gendron-Fitzpatrick A, Splitter GA. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J Immunol. 2002;168:2433–2440. doi: 10.4049/jimmunol.168.5.2433. [DOI] [PubMed] [Google Scholar]
- 58.Scharton-Kersten T, Contursi C, Masumi A, Sher A, Ozato K. Interferon consensus sequence binding protein-deficient mice display impaired resistance to intracellular infection due to a primary defect in interleukin 12 p40 induction. The Journal of experimental medicine. 1997;186:1523–1534. doi: 10.1084/jem.186.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turcotte K, Gauthier S, Malo D, Tam M, Stevenson MM, Gros P. Icsbp1/IRF-8 is required for innate and adaptive immune responses against intracellular pathogens. J Immunol. 2007;179:2467–2476. doi: 10.4049/jimmunol.179.4.2467. [DOI] [PubMed] [Google Scholar]
- 60.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 61.Horiuchi M, Wakayama K, Itoh A, Kawai K, Pleasure D, Ozato K, Itoh T. Interferon regulatory factor 8/interferon consensus sequence binding protein is a critical transcription factor for the physiological phenotype of microglia. Journal of neuroinflammation. 2012;9:227. doi: 10.1186/1742-2094-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, Muller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, Rosenbauer F, Prinz M. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nature neuroscience. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 63.Minten C, Terry R, Deffrasnes C, King NJ, Campbell IL. IFN regulatory factor 8 is a key constitutive determinant of the morphological and molecular properties of microglia in the CNS. PloS one. 2012;7:e49851. doi: 10.1371/journal.pone.0049851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang SM, Gao L, Zhang XF, Zhang R, Zhu LH, Wang PX, Tian S, Yang D, Chen K, Huang L, Zhang XD, Li H. Interferon regulatory factor 8 modulates phenotypic switching of smooth muscle cells by regulating the activity of myocardin. Molecular and cellular biology. 2014;34:400–414. doi: 10.1128/MCB.01070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.