Abstract
The third variable region (V3) of HIV-1 gp120 plays a key role in viral entry into host cells; thus, it is a potential target for vaccine design. Human monoclonal antibody (mAb) 447–52D is one of the most broadly and potently neutralizing anti-V3 mAbs. We further characterized the 447–52D epitope by determining a high-resolution crystal structure of the Fab fragment in complex with a cyclic V3 and interrogated the antigen–antibody interaction by a combination of site-specific mutagenesis, isothermal titration calorimetry (ITC) and neutralization assays. We found that 447–52D’s neutralization capability is correlated with its binding affinity and at 25 °C the Gibbs free binding energy is composed of a large enthalpic component and a small favorable entropic component. The large enthalpic contribution is due to (i) an extensive hydrogen bond network, (ii) a π–cation sandwiching the V3 crown apex residue Arg315, and (iii) a salt bridge between the 447–52D heavy chain residue AspH95 and Arg315. Arg315 is often harbored by clade B viruses; thus, our data explained why 447–52D preferentially neutralizes clade B viruses. Interrogation of the thermodynamic signatures of residues at the antigen binding interface gives key insights into their contributions in the antigen–antibody interaction.
The third variable loop (V3) of gp120 is a promising AIDS vaccine target because it mediates HIV-1’s contact with coreceptors CCR4 or CXCR5 (1, 2). It is very immunogenic and readily accessible to antibodies (3). Passive immunization with anti-V3 antibodies also shows protection in nonhuman primate studies (4, 5). V3 is generally 35 amino acids in length, beginning with disulfide bond between Cys at position 296 (Cys296) and Cys331 (HXB2 numbering (6)). It can be divided into three structural regions: the disulfide linkage at the base in the gp120 core, the distal crown region of about 13 amino acids which projects ~20 Å from the core, and the flexible stem region between the base and the crown (7, 8). The epitopes of most known human anti-V3 mAbs have been mapped to the crown region of V3. Many of these mAbs have been carefully characterized by functional and structural methods. Recent structural studies have demonstrated that although the crown often forms a β-hairpin structure, it can be further divided into three distinct subregions: the band, the circlet, and the arch (8). The arch at the center of the V3 sequence contains the highly conserved 4-residue motif of gp120 residues 312–315, often composed of the sequence GPGR for clade B or GPGQ for nonclade B viruses. The circlet at the middle of the crown has a hydrophobic face and a hydrophilic face; the hydrophobic face contains two highly conserved isoleucine residues (Ile307 and Ile309). The band consists of the often positively charged residues 304 and 305 at the N-terminus and the highly conserved Tyr318 at the C-terminus. Among these subregions of V3 are four conserved structural elements: the arch, the band, the hydrophobic face of the circlet, and the peptide backbone. Anti-V3 antibodies that target these conserved structural elements are broadly reactive (8). Structural studies have also revealed that there are two general modes of antigen recognition for the human V3 mAbs: the ladle mode and the cradle mode. The ladle mode, typified by mAb 447–52D, is one where the antigen binding site is shaped like a soup ladle (9). The bowl of the ladle binds the arch of the V3 crown while the handle, formed by a long CDRH3 in the case of 447–52D, interacts with the N-terminal half of the V3 crown by main-chain interactions. Conversely, the cradle-binding mode, typified by mAbs 2219, is one where the antigen binding site is shaped like a cradle and the V3 crown sinks into the cradle, often burying the hydrophobic face of the circlet (8).
Although mAb 447–52D (IgG3, λ) has somatic mutation frequencies of only 5.4% and 2.5% for VH and VL, respectively (10), it harbors a sophisticated antigen binding site. 447–52D has a long CDRH3 of 20 amino acids (Kabat definition) that forms a β-hairpin structure standing tall at one side of the antigen binding site (the handle of the ladle). The antigen binding side β strand of the CDRH3, decorated with 5 consecutive tyrosines (residues 100G–100K), makes main chain interactions with the N-terminal strand of the V3 crown. There is a shallow pocket at the base of CDRH3 that can be considered as the bowl of the ladle. On one side of the bowl, two tryptophans, residue 91 of the light chain (TrpL91) and TrpL96, form a wall that packs against the GPG turn of the V3 arch. On the other side of the bowl, TyrH100J and TrpH33 of the heavy chain form perfect π–cation stacking geometry that sandwiches the guanidinium group of Arg315 of V3. In addition, the side chain of Arg315 of V3 forms a salt bridge with the side chain of AspH95. This highly specific binding of the Arg315 side chain with a π–cation stacking and a salt bridge has led to speculation that Arg315 is the signature motif of the epitope of 447–52D (11). This might be the cause of 447–52D’s preference for neutralizing clade B viruses over nonclade B viruses (12).
Although investigated by us and other groups, analysis of the structure-function relationship of the antigen binding site of 447–52D still presents unanswered questions. For example, although the crystal structures of the antigen binding fragments (Fabs) of 447–52D in complex with V3 peptides of clade B and nonclade B sequences have been obtained (9, 10), the role of the C-terminal region of V3 crown hairpin in the antigen antibody interaction was not clear because the residues after 316 had poor electron densities. The C-terminus was later extended to residue 319 in another crystal structure (13), but the diffraction data set was epitaxially twinned and the structure was not sufficiently refined. There are also some inaccuracies of the placement of key residue Arg315 of the V3 loop and water molecules within the antigen binding site (9, 13). Most importantly, the contribution to antigen binding of the individual residues cannot be determined from the structure alone because residues at the antigen–antibody interface do not contribute equally to the affinity and neutralization function of the antibody. Instead, studies have indicated that the residue’s contribution to the antibody function is not uniformly distributed along the binding interface; rather, the majority of the contribution is often localized to a subset of residues (14–16).
To study the contribution of individual residues, a structure-guided computational approach has been used (17). However, it cannot accurately predict the affinity (kD) and Gibbs free binding energy (ΔG) from atomic structures. Purely functional studies are also limited in that they lack the precision at the single residue level (12, 18). Therefore, the best approach is to interrogate the contribution of individual residues at the molecular level by sensitive binding assays guided by computational and structural studies. SPR (19), ELISA (20), and FRET(21) assays have also been used to measure affinity with high sensitivity, but these techniques cannot determine a complete thermodynamic signature. Isothermal titration calorimetry (ITC) is a unique technique that can not only provide the accurate measurement of the binding affinities but also sensitively quantitate the thermodynamic signatures of individual residues at a specific temperature (22, 23). Here we present a detailed interrogation of the antigen binding site of 447–52D using a combination of mutagenesis, neutralization assays using pseudoviruses, and determination of affinity by ITC.
MATERIALS AND METHODS
Fab Production
Human mAb 447–52D was produced as previously described (3, 24). Briefly, peripheral blood mononuclear cells derived from HIV-1-infected individuals were transformed with Epstein–Barr virus, and the cells reactive with a 23-mer V3MN peptide were fused with heteromyeloma cells SHM-D33 (25). The resulting hybridomas were cloned by limiting dilution to monoclonality. mAb 447–52D was purified from culture supernatants on a Protein G affinity column (GE Healthcare).
Fab 447–52D was produced by papain digestion. Papain (Worthington) in 100 mM sodium acetate pH 5.5 was activated with 10 mM cysteine and incubated with mAb at a 1:20 molar ratio for 4 h at 37 °C. Papain was then deactivated using iodoacetamide, and the Fab was then isolated by removing the Fc fragment and undigested IgG with a Protein G column. The Fab was further purified by size exclusion chromatography after dialysis overnight against 50 mM sodium acetate pH 5.5, 100 mM NaCl.
Fv Production
To produce the variable region (Fv) molecules we first created single chain Fv (scFv) constructs. Genes encoding variable fragments of the heavy and light chains of mAb 447–52D were synthesized in a single chain connected by a flexible linker, LVPRGSGGGGLVPRGS (26), with 2 thrombin cleavage sites and a C-terminal poly-His tag (Supporting Information Figure S1). The scFv were cloned into the pET30a kanamycin resistant vector. Select mutations were made by PCR-driven overlap extension primers. Resulting plasmids were then transformed into competent E. coli BL-21 cells (Rosetta-DE3) and protein overexpression was induced with IPTG in an LB medium with added antibiotics. scFv molecules were found in the insoluble fraction of the cell lysate. Inclusion bodies were isolated by centrifugation (15 000 rpm, 30 min, 4 °C) and dissolved in a denaturing buffer (6 M guanidine HCl, 50 mM Tris pH 7.5, 20 mM EDTA, 20 mM DTT) at 4 °C overnight. Aggregates were isolated by centrifugation (18 000 rpm, 4 °C, 30 min), and denatured proteins were filtered through a 0.45 μm filter and stored at −80°C.
To refold the scFv and generate functional Fv molecules, scFv protein was denatured into 600 mL to a final concentration of 0.08 mg/mL in 2.4 M guanidine HCl, 50 mM Tris pH 7.5 and dialyzed into 50 mM Tris pH 7.5, 150 mM NaCl, and 10% glycerol for 24 h at 4 °C. The protein was then dialyzed into 50 mM Tris pH 7.5, 150 mM NaCl, and 5% glycerol for 48 h. Refolded scFv protein was captured by Ni-NTA sepharose beads (Qiagen). The immobilized protein was washed (20 mM imidazole pH 8.0, 50 mM Tris pH 7.5, 150 mM NaCl) and eluted (500 mM imidazole pH 8.0, 50 mM Tris pH 7.5, 150 mM NaCl). Soluble refolded scFv protein was digested with thrombin for 1 h at 37 °C to create soluble Fv molecules. Fv was separated from thrombin by incubation with heparin sulfate beads (4 °C, 30 min) and further purified by size exclusion chromatography on a Superdex 75 column in 25 mM Tris pH 7.5 and 100 mM NaCl (Supporting Information Figure S2).
Crystallization, Data Collection, Structure Determination, and Refinement
Commercially synthesized V3MNCYCLIC 14mer peptide (Biomatik, >99% purity) with amino acid sequence CRIHIGPGRAFYTC was dissolved in water to 10 mg/mL and added at a 5:1 molar ratio to purified 447–52D Fab. The mixture was concentrated to ~20 mg/mL for crystallization. Initial crystals were obtained by hanging drop vapor diffusion over reservoirs containing 1.6 M ammonium sulfate and 0.1 M Tris pH 7.5 as previously described (9, 10, 13). Crystals were optimized further by streak seeding. Selected crystals were briefly soaked in their respective mother liquor with additional 30% glycerol (v/v) prior to being flash frozen in liquid nitrogen. X-ray diffraction data were collected at beamline X6A, National Synchrotron Light Source, Brookhaven National Laboratory. All data sets were processed using the HKL2000 Package, (27) and structures were solved by molecular replacement using MolRep (28) with the 447–52D Fab/V3(W2RW020) structure (Protein Data Bank entry 3GHB) as the search model. Alternating cycles of real space fitting and restrained refinement for each model were carried out in Coot (29) and Refmac5 (30). Structural figures were generated using PyMOL (31).
Isothermal Titration Calorimetry (ITC)
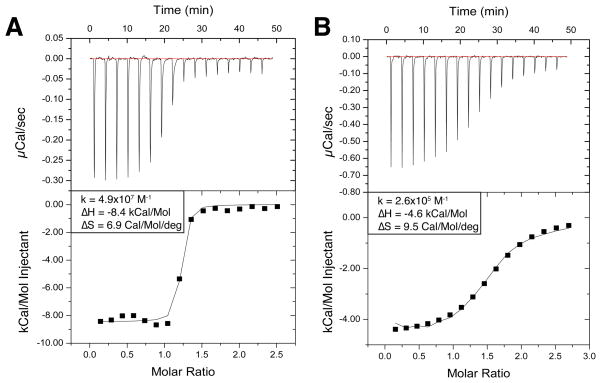
ITC experiments were performed using an ITC200 system (Micro-Cal) (32) as previously described by Bradshaw et al. (33), at 25 °C in a buffer composed of 25 mM Tris pH 7.5 and 100 mM NaCl. Fvs were dialyzed against this buffer overnight before the experiment. The peptides were dissolved in the final dialysis buffer. In a typical binding experiment to characterize the Fv-V3 interactions, 300 mM V3 peptide was injected in 16 aliquots of 2.43 μL into a 280 μL sample cell containing 30 μM Fv. In all cases, data were corrected for the heat of dilution and fitted using a nonlinear least-squares routine using a single-site binding model with Origin for ITC version 7.0383 (MicroCal). The reported uncertainty in all thermodynamic parameters is the standard deviation of at least three experiments. Raw data for representative experiments are included in Figure 2 and Supporting Information Figures S5–S11.
Figure 2.
Representative ITC raw data (top) and integrated isotherms (bottom) of (A) Fv 447–52D wild type and (B) Fv 447–52D TrpH33Ala mutant titrated with V3MN peptide. Panel A shows raw data (μCal/sec) and integrated isotherm (kcal/M V3) for wild type 447–52D Fv binding to a V3MN peptide. From the isotherm we are able to directly observe thermodynamic properties (ΔH, kD) and determine other thermodynamic parameters (ΔS, ΔG) based on the basic equations of thermodynamics (ΔG = ΔH − TΔS and ΔG = −RT ln kD) (49). The steep slope of the Fv WT and V3MN isotherm coincides with a nanomolar binding affinity. Contrastingly, the slope of the isotherm for the 447–52D Fv TrpH33Ala mutant and V3MN (B) is much shallower, indicating a micromolar binding affinity. The data represented in panels A and B (as well as all others discussed here) are representative of at least 3 replicates.
Neutralization Assays with TZM-bl Target Cells
The wild type Fv construct, its five mutants (TrpL91Ala, TrpL96Ala, TrpH33Ala, TyrH100JThr, AspH95Asn), IgG 447–52D (positive control), and parvovirus B19-specific mAb 1418 (negative control) were tested for neutralizing activity against SF162 based pseudovirus (psV) using the TZM-bl cell line as target cells. The psV carried Env from SF162 and backbone vector from strain NL43 lacking the Env region. Fvs were tested with seven 2-fold serial dilutions starting at concentrations of 2–7 mg/ml. Each Fv dilution was preincubated with the psV; subsequently, each Fv/psV mixture was incubated for 1 h with TZM-bl cells expressing CD4, CXCR4, and CCR5 at 37 °C. Pseudovirus infectivity was determined by measuring the luciferase activity in the cell lysates. The reduction of infectivity was expressed as percent neutralization by comparing the enzyme activity, as relative light units (RLU), in the presence of antibody versus the absence of antibody (34, 35). The IC50 values were determined by the standard method of least-squares(36). The IC50 values were analyzed by one-way ANOVA analysis using GraphPad Prism (version 5.04 for Windows, GraphPad Software, La Jolla, CA, www.graphpad.com) (Supporting Information Figure S4).
RESULTS AND DISCUSSION
High-Resolution Structure of Fab 447–52D in Complex with a Cyclic V3 Peptide
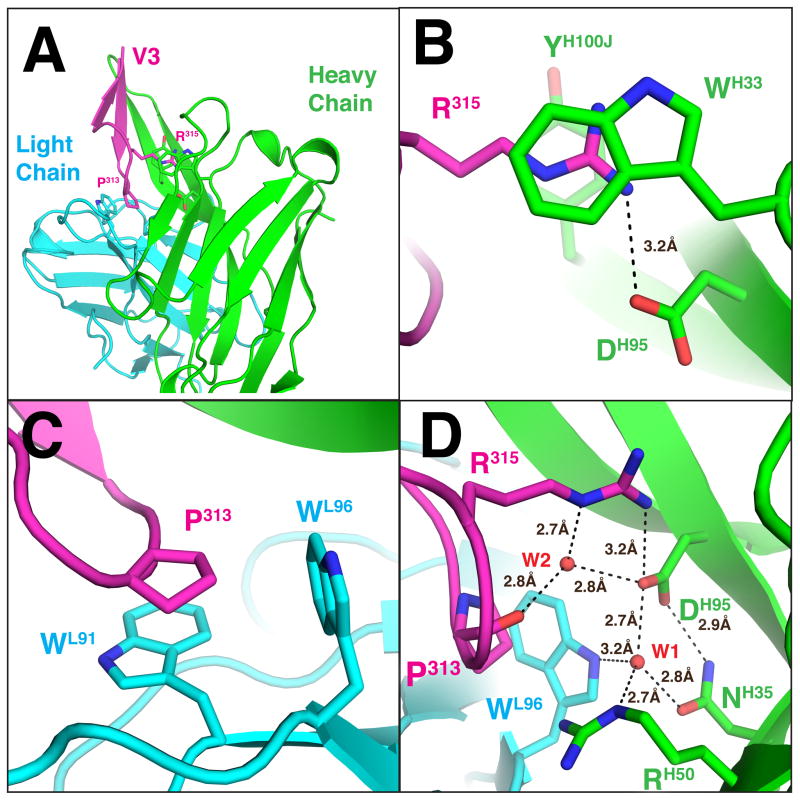
First, we sought to obtain a high-quality structure of the 447–52D–epitope complex that would result in an accurate visualization of its antigen–antibody interaction. We determined a crystal structure of Fab 447–52D in complex with a cyclic V3 peptide with the amino acid sequence of CRIHIGPGRAFYTC (derived from HIV-1 strain MN) and refined it to 1.8 Å resolution with an Rwork/Rfree of 0.187/0.219 (Figure 1, Supporting Information Table S1). All 14 amino acids in the cyclic peptide were visualized with good electron densities. The C-terminal region of the V3 crown forms a β strand (Figure 1A), antiparallel with the N-terminal strand of the V3 crown. Together with the two β strands of CDRH3 of 447–52D, they formed a continuous four-strand β sheet (Figure 1A). Our structure also permitted the accurate placement of water molecules within the antigen binding site: the epsilon nitrogen of residue Arg315 from the epitope was at a good hydrogen bonding distance to one of the water molecules (W2; Figure 1B,D). This placement of the side chain of Arg315 allowed it to have a better stacking with TyrH100J and TrpH33 of the heavy chain of 447–52D.
Figure 1.
(A) Structure of Fab 447–52D (only the Fv portion is shown) where the light and heavy chains are colored cyan and green, respectively; the V3 peptide is colored magenta. Several residues that are important for antibody–antigen interaction, including Pro313 and Arg315 of V3, TrpL91 and TrpL96 of the light chain, and TrpH33, AspH95, and TyrH100J of the heavy chain, are illustrated in stick representation. (B) Side chain of Arg315 of V3 interacts with TyrH100J (background) and TrpH33 (foreground) through the π–cation stacking and forms a salt bridge with AspH95. (C) Details of the interaction between Pro313 of the V3 epitope and TrpL91 and TrpL96 of 447–52D light chain. (D) Water mediated hydrogen bond network at the antigen binding site. Note that Arg315 and AspH95 of the π–cation stacking/salt-bridge are further coordinated by the water molecules.
Consistent with the previous data, we observed four key antigen–antibody interactions of 447–52D: (i) a π–cation stacking between the side chains of the aromatic residues TyrH100J and TrpH33 and Arg315 of the V3 loop (Figure 1B), (ii) a salt bridge between the cationic Arg315 of the V3 loop and the anionic AspH95 of 447–52D (Figure 1B), (iii) van der Waals interactions of Pro313 of the V3 loop with TrpL91 in a parallel orientation and with TrpL96 in an orthogonal orientation (Figure 1C), and finally (iv) main chain interactions of the N-terminal portion of the V3 crown with the CDRH3 of 447–52D (Figure 1A). There are six main chain mediated hydrogen bonds between the N-terminal β strand of the V3 crown and the C-terminal β strand of CDRH3 of 447–52D (Supporting Information Table S2). These four key interactions account for the contacts between 447–52D and V3.
The accurate placement of the C-terminal strand of the V3 crown allowed us to visualize its hydrophobic core in the context of 447–52D binding. However, the hydrophobic core in this complex structure is on the opposite side of the hairpin in comparison with the V3 crowns in complex with a family of mAbs (mAbs 1006–15D, 2219, 2557, etc.) (8, 37, 38) encoded by the VH5–51 germline genes, which is preferentially used by anti-V3 crown human antibodies and accounts for 35% of isolated anti-V3 human mAbs (37). In detail, when visualized from the perspective of the C-terminal strand, the hydrophobic core of this structure is located to the left of the V3 crown in the 447–52D structure. However, in the case of structures of anti-V3 antibodies encoded by VH5–51 genes, the hydrophobic core is located to the right of the crown. This observation suggests the hydrophobic core can flip-flop between the two sides of the V3 crown. The capacity for accommodating two such drastically different conformations might play a role in the entropic masking of the V3 crown, hiding this functionally important motif from human immune responses (39).
Generation of Fv Fragment of mAb 447–52D and Mutations of Its Antigen Binding Site
To further understand the antigen–antibody interaction of 447–52D and interrogate the thermodynamic contributions of individual residues, we studied the Fv fragment of 447–52D which solely consists of the terminal Ig domain that is responsible for antigen binding; in the case of 447–52D, these are residues 1–113 of heavy chain and residues 1–109 of the light chain (Supporting Information Figures S1 & S2). The Fv fragment was used because we had previously noticed that the Fab fragment of a mAb can adopt two very different elbow angles in a crystal (40). The potential flexibility of the elbow of a Fab fragment would increase the noise of a thermodynamic measurement that could obscure the sensitivity of measurements. Therefore, the Fv was the preferred molecule for our thermodynamic studies.
We then created a panel of Fv 447–52D and V3 mutants that would allow us to interrogate the contribution of individual residues to the antigen–antibody interaction. We divided the mutations of Fv 447–52D into 3 groups. The first, TyrH100J to Thr or TrpH33 to Ala, would destroy the π–cation sandwich interaction with Arg315 of the V3 epitope. We mutated TyrH100J to Thr to preserve the polarity of this position, because it is likely the mutated residue would still be exposed to the solvent upon V3 binding. The second, AspH95 to Asn, would remove this side chain’s charge and prevent the formation of the salt bridge with the side chain of Arg315. The third, TrpL91 to Ala or TrpL96 to Ala, would alter the contact with Pro313 of the GPG turn of the V3 arch. These mutant Fv molecules were all generated with the same procedures as the wild type Fv 447–52D and they all eluted from size exclusion chromatography similarly to the wild type (WT) Fv. We also created mutations in the V3 loop: Arg315 to Gln, which interrupts the specific π–cation and the salt bridge, and His308 to Arg, which is often associated with nonclade B viruses.
Functional Analysis of Fv 447–52D and Its Mutants by Neutralization
We tested the neutralization of the wild type Fv 447–52D and the panel of its mutants by a TZM-bl assay against a SF162 pseudovirus (36, 41). We first quantitated the neutralization capability of Fv 447–52D in comparison with that of the native 447–52D IgG molecule (Supporting Information Figure S3). Fv 447–52D is able to neutralize SF162 pseudoviruses but with a 230-fold molar increase in IC50 in comparison with that of IgG 447–52D. The weakening of neutralization capability of the Fv molecule may be partly explained by the difference in stoichiometry between IgG (two binding sites) and Fv (one binding site). Additionally, Fv molecules lack the constant domains of the IgG that may play an additional role in augmenting neutralization function. We then tested all the Fv mutants in the TZM-bl assay against the SF162 pseudovirus. The results indicated that: (i) Mutation of each of the aromatic residues that sandwich Arg315, TrpH33Ala, and TyrH100JThr, resulted in a 3.4- and 1.2-fold decrease in neutralization capability, respectively. (ii) Mutation AspH95Asn, which prevented the formation of the salt bridge with Arg315, resulted in a 2.2-fold decrease in neutralization capability. Finally, (iii) mutation of each of the aromatic residues that interact with Pro313, TrpL91Ala, and TrpL96Ala resulted in a 4.0- and 2.6-fold decrease in neutralization capability, respectively. All Fv mutants’ IC50 values are higher than that of the wild type Fv (Table 1, Supporting Information Figure S4).
Table 1.
Neutralization and ITC Dataa
| Mutation | IC50 | kD |
|---|---|---|
| WT | 19 μM (± 0.6) | 0.023 μM (± 0.001) |
| 447–52D YH100JT | 23 μM (± 6) | 1.3 μM (± 0.5) |
| 447–52D WH33A | 68 μM (± 5) | 3.9 μM (± 0.7) |
| 447- 52D DH95N | 41 μM (± 4) | 3.9 μM (± 0.1) |
| 447–52D WL91A | 76 μM (± 5) | 5.4 μM (± 0.3) |
| 447–52D WL96A | 50 μM (± 4) | 1.8 μM (± 0.5) |
| V3 R315Q | ND | 5.1 μM (± 0.7) |
| V3 H208R | ND | 0.057 μM (± 0.003) |
| V3 Cyclic | ND | 0.016 μM (± 0.002) |
IC50 values and error were obtained from two or three replicate experiments of Fv 447–52D WT or mutants against SF162 pseudovirus in a TZM-bl cell assay and determined by standard methods (36, 41). kD values and error were obtained from three or four replicated experiments of Fv and V3 molecules via Isothermal Titration Calorimetry.
Binding Affinity Measured by ITC
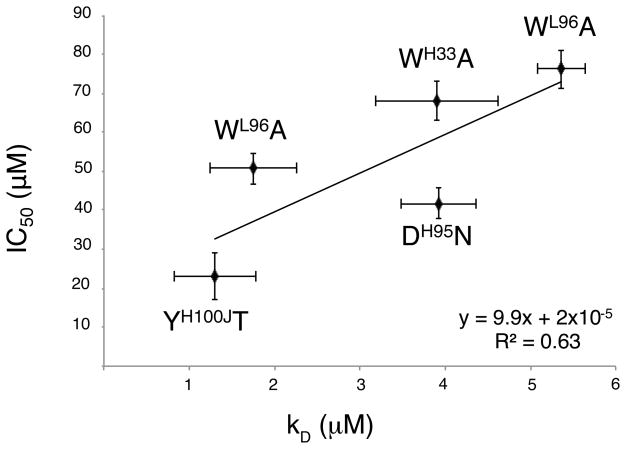
To determine if changes in neutralization capability of Fv 447–52D correlate with changes in antigen binding affinity, we measured the binding affinities of the wild type Fv 447–52D and its mutants with a 14-mer V3 peptide (KRIHIGPGRAFYTT) by ITC (Figure 2). We found that (i) mutations TrpH33Ala and TyrH100JThr, which destroy the sandwiching of Arg315, resulted in a 56- and a 170-fold decrease in affinity, respectively (Figure 2B and Supporting Information Figure S5B); (ii) mutation AspH95Asn which removed the salt bridge with Arg315 resulted in a sizable 170-fold decrease in affinity (Supporting Information Figure S6B); and finally, (iii) mutations TrpL91Ala and TrpL96Ala, which altered the interaction with Pro313 resulted in a 230- and a 76-fold decrease in affinity, respectively (Figures S7B and S8B). We found that there was a slight correlation between neutralization function and binding affinity, i.e., mutations of residues that resulted in large changes of neutralization capability also resulted in large changes of binding affinity and vice versa (Figure 3, Table 1). This is consistent with a previous study of IgG 447–52D using SPR and ELISA (19). Such correlation has also been noted by a recent study on an MPER (gp41) directed mAb 2F5 (42). However, studies on mAb b12 have shown that there was not a clear correlation between binding and neutralization (43).
Figure 3.
The neutralization activity (IC50) of each mutant Fv of 447–52D is plotted against its affinity (kD) (data from Table 1). Note that tighter binding (lower kD) results in greater neutralization activity (lower IC50) with a correlation R2 = 0.63.
We also measured the affinity of the wild type Fv 447–52D binding to V3 peptides mutated at two positions: (i) V3 with Arg315Gln mutation resulted in a GPGQ motif (KRIHIGPGQAFYTT) (Supporting Information Figure S9B) or (ii) V3 with His308Arg mutation (KRIRIGPGRAFYTT) (Supporting Information Figure S10B). Residues His308 and Arg315 of the V3 crown are often associated with clade B viruses. When compared to the wild type V3 with MN sequence (KRIHIGPGRAFYTT), we observed that (i) a 230-fold decrease in binding to an Arg315Gln peptide and (ii) a 2.5-fold decrease in binding to a His308Arg peptide. Thus 447–52D antigen binding affinity is much more contingent on Arg315, explaining the limited neutralization ability of mAb 447–52D for nonclade B viruses which often contain Gln315.
Finally, we also measured the binding affinity of the wild type Fv 447–52D to a cyclic V3 peptide (CRIRIGPGRAFYTC) used in our crystallographic study (Supporting Information Figure S11B). We observed a very modest 1.5-fold increase in binding affinity of the cyclic peptide. The very similar affinities of the linear and the cyclic V3 with Fv 447–52D indicates that the cyclization of the V3 crown did not affect much of its interaction with mAb 447–52D (44).
Thermodynamic Signature of the Antigen Binding Site of 447–52D
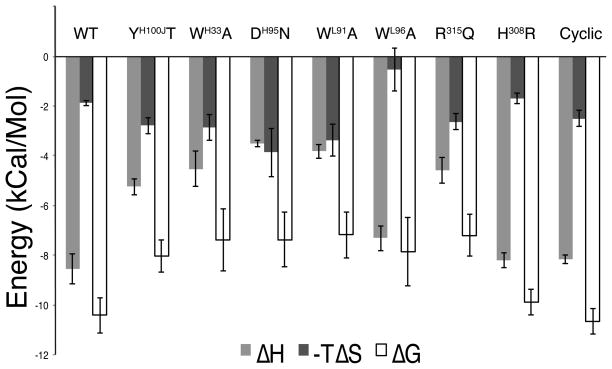
Our ITC measurements have also provided the thermodynamic signature of each antigen antibody interaction: the enthalpic (ΔH) and entropic (ΔS) contributions to the Gibbs free binding energy (ΔG = ΔH − TΔS) at a specific temperature (25 °C). The wild type Fv 447–52D binding to V3 peptide KRIHIGPGRAFYTT gave a ΔG value of −10.42 kcal/mol with an enthalpic contribution of −8.54 kcal/mol and an entropic contribution of −1.88 kcal/mol (Figures 2A and 4, Supporting Information Table S3). Thus the majority of the binding energy is derived from the enthalpy (ΔH) of the interaction. This sizable enthalpic contribution may indicate a large number of specific contacts formed when the antibody is complexed with the V3 epitope. These could include the nine hydrogen bonds between the V3 epitope and the heavy chain atoms (Supporting Information Table S2) and the hydrogen bond network mediated by the two solvent molecules at the base of the binding pocket (the bowl of the ladle) (Figure 1D). The salt bridge between AspH95 and Arg315 and the latter’s π–cation stacking with TyrH100J and TrpH33 likely also contribute significantly to this enthalpy (see below). However, there is also an unfavorable enthalpic contribution, probably due to the peptide-displacing solvent molecules that make contact (hydrogen bonds) with the unbound paratope. When the peptide binds, these contacts are lost, which is an unfavorable contribution to the Gibbs free binding energy. The peptide is able to make new contacts with the paratope, which is a favorable contribution to the Gibbs free binding energy. The net favorable enthalpy observed for the Fv–peptide interaction indicates that the favorable contacts made upon peptide binding outweigh unfavorable loss of contacts between the paratope and solvent.
Figure 4.
Enthalpic (ΔH) and entropic (ΔS) contributions of individual amino acid side chains to the Gibbs free binding energy (ΔG) are shown for the wild type (WT) interaction and each of the mutated Fv and V3 molecules. All the enthalpic contributions to binding of mutated molecules are less than the wild type. Most entropic contributions to binding of mutated molecules are more than the wild type, with the exceptions of Fv with the mutation TrpH96Ala and V3 with the mutation of His308Arg. Each of these parameters is derived from raw thermodynamic binding data (Supporting Information Table S3).
Interestingly, the V3–Fv 447–52D interaction also had a favorable entropic contribution (−TΔS) of −1.88 kcal/mol. The ordering of the flexible V3 crown peptide upon binding likely contributes unfavorably to the Gibbs free binding energy. However, this unfavorable contribution is perhaps overcome by a larger favorable contribution derived from expelling solvent molecules present at the surface of the unbound antigen binding site. Assuming each hydrogen bond donor/acceptor would originally bind one water molecule, there would be at least nine water molecules expelled from the binding site upon antigen binding. In addition, there is a pocket of 460 Å3 at the bottom of the antigen binding site (the bowl of the ladle). It is possible that water molecules self-assembled in this largely hydrophilic pocket would be expelled upon V3 binding, creating an additional favorable entropic contribution to the binding energy ΔG.
We determined the thermodynamic signatures of binding events for mutants of Fv 447–52D and V3 in comparison with the binding of the wild type Fv and V3 at 25 °C (Figure 4, Supporting Information Table S3). All five mutants of Fv 447–52D had reduced enthalpic contributions to their Gibbs free energy, which could be interpreted as reductions of side chain specific interactions made by the mutants. Mutation TyrH100JThr resulted in a substantial enthalpic reduction of 3.3 kcal/mol, indicating the important role TyrH100J in the π–cation interaction. TrpH33Ala resulted in an even larger enthalpic reduction of 4.02 kcal/mol. Thus, the indole group of TrpH33 contributed more to the π–cation interaction than the tyrosyl group of TyrH100J. Mutation Arg315Gln in the V3 loop resulted in a large enthalpic reduction of 3.96 kcal/mol. AspH95Asn also resulted in a large enthalpic reduction of 5.04 kcal/mol. These two large enthalpic reductions hint at the importance of the AspH95–Arg315 salt bridge to antigen binding. Mutation His308Arg resulted in a small enthalpic reduction of 0.35 kcal/mol, suggesting that if specific contacts are made at this position, the side chain of either a His or an Arg at position 308 can bridge them similarly. Mutation TrpL91Ala resulted in an enthalpic reduction of 4.72 kcal/mol, which is comparable to that of the mutation TrpH33Ala. TrpL96Ala resulted in only a small enthalpic reduction of 1.22 kcal/mol; this is understandable because it has the smallest contact area with the epitope among all the residues we mutated (Supporting Information Table S4).
All the mutants had more favorable entropic contributions than the wild type with the exceptions of TrpL96Ala and His308Arg. Mutations Tyr100JThr and TrpH33Ala resulted in comparable increases in favorable entropy of 0.91 kcal/mol and 0.98 kcal/mol, respectively. The entropic increases of these mutations were likely derived from the increased flexibility of the Arg315 side chain of V3 when the π–cation sandwiching is no longer complete. Mutation Arg315Gln resulted in a 0.75 kcal/mol increase in favorable entropy, which could reflect the imperfect fit of the shorter Gln within the 447–52D binding site. Similarly, AspH95Asn and TrpL91Ala had comparable increases in favorable entropy of 1.99 kcal/mol and 1.49 kcal/mol, respectively. These increases in entropy possibly resulted from increased flexibility of the complexed V3 arch in comparison with that of the wild type. Mutation His308Arg resulted in a very small decrease in favorable entropy of 0.19 kcal/mol, likely because the side chain of Arg308 has more entropy than that of His308. Interestingly, mutation TrpL96Ala resulted in a large reduction in favorable entropy of 1.34 kcal/mol. This mutation created a 137.9 Å3 cavity lining by hydrophobic residues LeuL34, PheL98 and MetH100L in the antigen binding site of 447–52D (Supporting Information Figure S12). This cavity could potentially trap solvent molecules that will not be liberated into the free solvent upon V3 binding, resulting in a less favorable entropic contribution. The important role of water bridging wild type interactions absent in a site-specific mutant was also observed in analysis of antihen egg white lysozyme (HEL) antibody D1.3 in complex with HEL(45).
By the investigation of the thermodynamics signatures of residues at the antigen binding surface of 447–52D we observed that its epitope binding energy is dominated by a large enthalpic contribution. This enthalpic contribution is derived from hydrogen bonds, a π–cation interaction, and most importantly the salt bridge between prototypic clade B residue Arg315 and AspH95 of 447–52D. The π–cation sandwich surrounding Arg315, together with the salt bridge, created an almost perfect binding environment for the long, positively charged arginine side chain. Mutation of any of these three residues resulted in a substantial reduction in the enthalpic contribution to binding and affinity. These data explain why 447–52D strongly prefers to neutralize viruses with the GPGR motif at the V3 crown. Interestingly, several charged residues line the surface of the binding site and there are two stably bound solvent molecules. These water molecules are likely a part of the antigen binding site because they are also present in the structure of 447–52D in complex with GPGQ V3 (9). Using water molecules to bridge interactions is a common mechanism of increasing affinity of antibodies during affinity maturation (46). The two highly conserved residues of the hydrophobic core, Ile307 and Ile309, are largely exposed when the epitope is bound. This is a very different binding topology from VH5–51 anti-V3 crown mAbs, which bind the V3 crown using the cradle mode and Ile307 and Ile309 are buried at the center of the cradle. This hydrophobic-binding-dominated antigen binding can tolerate variation on the antigen side so long as the mutations are also hydrophobic. On the other hand, hydrophilic interactions, like the hydrogen bonds and salt bridge discussed in this manuscript, are more specific such that they require particular residue functional groups to have a specific geometry and distance. This type of antibody likely requires a long time affinity maturation; thus, it is difficult to be elicited by vaccination.
The favorable entropic contribution to the Gibbs free binding energy of 447–52D differs from some other anti-gp120 antibodies which have a net unfavorable entropic contribution, such as VRC01 and PG9 (47, 48). This unfavorable entropy is derived from the limitation of gp120 conformation flexibility due to binding by antibodies such as VRC01 that will anchor gp120 molecules at a particular conformation, incurring an entropic penalty. Quaternary epitope neutralizing antibodies, such as PG9, may also stabilize the conformation of gp120 upon binding. Vaccination to induce these antibodies would have to overcome this very unfavorable entropic contribution with larger, favorable enthalpic contributions composed of many specific molecular interactions. To be able to make these large numbers of interactions, these antibodies have special characteristics including long CDRH3 and large numbers of somatic mutations. These types of antibodies will be challenging to elicit by immunization. Therefore, our AIDS vaccine development efforts should focus on generating immune responses similar to antibodies whose binding does not incur a large entropic penalty.
Supplementary Material
Acknowledgments
Special thanks to personnel at the beamline X6A of NSLS at Brookhaven National Laboratories for X-ray data collection, Dr. Steve Hubbard for ITC, Dr. Luzia Mayr and Dr. Sandra Cohen for neutralization assays, and Dr. Arthur Nadas for useful discussions.
Funding
This work was supported by a Graduate Research Fellowship to A.K. from the National Science and Engineering Research Council of Canada, and NIH grants AI100151 and HL059725.
ABBREVIATIONS
- HIV
human immunodeficiency virus
- mAb
monoclonal antibody
- gp120
glycoprotein 120
- V3
third variable region
- ITC
isothermal titration calorimetry
- CCR4
CC chemokine receptor 4
- CXCR5
C-X-C chemokine receptor type 5
- CDRH3
complementarity determining region 3 of the heavy chain
- SPR
surface plasmon resonance
- ELISA
enzyme-linked immunosorbent assay
- FRET
fluorescence resonance energy transfer
- Fab
antigen binding fragment
- Fv
variable domain
- scFv
single chain variable domain
Footnotes
Accession Codes
Atomic coordinates of Fab 447–52D in complex with V3MNCYCLIC have been deposited to RCSB Protein Data Band (PDB ID: 4M1D).
Author Contributions
The manuscript was written with contributions from all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Additional figures and tables supplied as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 2.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 3.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. Journal of immunology (Baltimore, Md: 1950) 1993;150:635–643. [PubMed] [Google Scholar]
- 4.Prince AM, Reesink H, Pascual D, Horowitz B, Hewlett I, Murthy KK, Cobb KE, Eichberg JW. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res Hum Retroviruses. 1991;7:971–973. doi: 10.1089/aid.1991.7.971. [DOI] [PubMed] [Google Scholar]
- 5.Watkins JD, Siddappa NB, Lakhashe SK, Humbert M, Sholukh A, Hemashettar G, Wong YL, Yoon JK, Wang W, Novembre FJ, Villinger F, Ibegbu C, Patel K, Corti D, Agatic G, Vanzetta F, Bianchi S, Heeney JL, Sallusto F, Lanzavecchia A, Ruprecht RM. An Anti-HIV-1 V3 Loop Antibody Fully Protects Cross-Clade and Elicits T-Cell Immunity in Macaques Mucosally Challenged with an R5 Clade C SHIV. PLoS ONE. 2011;6:e18207. doi: 10.1371/journal.pone.0018207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratner L, Fisher A, Jagodzinski LL, Mitsuya H, Liou RS, Gallo RC, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Research and Human Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 7.Huang Cc. Structure of a V3-Containing HIV-1 gp120 Core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong X-P. Conserved structural elements in the V3 crown of HIV-1 gp120. Nature Structural & Molecular Biology. 2010;17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 9.Burke V, Williams C, Sukumaran M, Kim S-S, Li H, Wang X-H, Gorny MK, Zolla-Pazner S, Kong X-P. Structural Basis of the Cross-Reactivity of Genetically Related Human Anti-HIV-1 mAbs: Implications for Design of V3-Based Immunogens. Structure/Folding and Design. 2009;17:1538–1546. doi: 10.1016/j.str.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural Rationale for the Broad Neutralization of HIV-1 by Human Monoclonal Antibody 447–52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Cardozo T, Swetnam J, Pinter A, Krachmarov C, Nadas A, Almond D, Zolla-Pazner S. Worldwide distribution of HIV type 1 epitopes recognized by human anti-V3 monoclonal antibodies. AIDS Res Hum Retroviruses. 2009;25:441–450. doi: 10.1089/aid.2008.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Research and Human Retroviruses. 2004;20:1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon AK, Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structure determination of an anti-HIV-1 Fab 447–52D–peptide complex from an epitaxially twinned data set. Acta Crystallographica Section D Biological Crystallography. 2008;64:792–802. doi: 10.1107/S0907444908013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lise S, Buchan D, Pontil M, Jones DT. Predictions of Hot Spot Residues at Protein-Protein Interfaces Using Support Vector Machines. PLoS ONE. 2011;6:e16774. doi: 10.1371/journal.pone.0016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosdidier S, Fernandez-Recio J. Identification of hot-spot residues in protein-protein interactions by computational docking. BMC Bioinformatics. 2008;9:447. doi: 10.1186/1471-2105-9-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Zhao B, Cui Z, Gan J, Sakharkar MK, Kangueane P. Identification of hot spot residues at protein-protein interface. Bioinformation. 2006;1:121–126. doi: 10.6026/97320630001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Pan D, Shen Y, Jin N, Liu H, Yao X. Understanding the molecular mechanism of the broad and potent neutralization of HIV-1 by antibody VRC01 from the perspective of molecular dynamics simulation and binding free energy calculations. Journal of molecular modeling. 2012:1–11. doi: 10.1007/s00894-012-1450-z. [DOI] [PubMed] [Google Scholar]
- 18.Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors Determining the Breadth and Potency of Neutralization by V3-Specific Human Monoclonal Antibodies Derived from Subjects Infected with Clade A or Clade B Strains of Human Immunodeficiency Virus Type 1. Journal of Virology. 2006;80:7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanCott TC, Bethke FR, Polonis VR, Gorny MK, Zolla-Pazner S, Redfield RR, Birx DL. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. Journal of immunology (Baltimore, Md: 1950) 1994;153:449–459. [PubMed] [Google Scholar]
- 20.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal Structures of Human Immunodeficiency Virus Type 1 (HIV-1) Neutralizing Antibody 2219 in Complex with Three Different V3 Peptides Reveal a New Binding Mode for HIV-1 Cross-Reactivity. Journal of Virology. 2006;80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennison SM, Stewart SM, Stempel KC, Liao H-X, Haynes BF, Alam SM. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. Journal of virology. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 23.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Current opinion in structural biology. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 24.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. Journal of Virology. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng NN, Lam KS, Riera FC, Kaplan HS. Construction and testing of mouse--human heteromyelomas for human monoclonal antibody production. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:7308–7312. doi: 10.1073/pnas.80.23.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Song L, Kim M, Holmes MA, Kraft Z, Sellhorn G, Reinherz EL, Stamatatos L, Strong RK. Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity. Journal of Virology. 2010;84:1076–1088. doi: 10.1128/JVI.02113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallographica Section D: Biological Crystallography. 2009;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D: Biological Crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL molecular graphics system 2002 [Google Scholar]
- 32.Wiseman TT, Williston SS, Brandts JFJ, Lin LNL. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 33.Bradshaw JM, Waksman G. Calorimetric Investigation of Proton Linkage by Monitoring both the Enthalpy and Association Constant of Binding: Application to the Interaction of the Src SH2 Domain with a High-Affinity Tyrosyl Phosphopeptide †. Biochemistry. 1998;37:15400–15407. doi: 10.1021/bi9814991. [DOI] [PubMed] [Google Scholar]
- 34.Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. Anti-V3 Monoclonal Antibodies Display Broad Neutralizing Activities against Multiple HIV-1 Subtypes. PLoS ONE. 2010;5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of Virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorny MK, Wang X-H, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. Preferential use of the VH5–51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Molecular Immunology. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang X-H, Williams C, O'Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong X-P. Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5–51/VL Lambda Genes Define a Conserved Antigenic Structure. PLoS ONE. 2011;6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PWIH, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 40.Gorny MK, Pan R, Williams C, Wang XH, Volsky B, O’Neal T, Spurrier B, Sampson JM, Li L, Seaman MS, Kong XP, Zolla-Pazner S. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals. Virology. 2012;427:198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 42.Guenaga J, Wyatt RT. Structure-guided alterations of the gp41-directed HIV-1 broadly neutralizing antibody 2F5 reveal new properties regarding its neutralizing function. PLoS Pathog. 2012;8:e1002806. doi: 10.1371/journal.ppat.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13950–13955. doi: 10.1073/pnas.0606312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dall’Acqua W, Goldman ER, Lin W, Teng C, Tsuchiya D, Li H, Ysern X, Braden BC, Li Y, Smith-Gill SJ, Mariuzza RA. A mutational analysis of binding interactions in an antigen–antibody protein-protein complex. Biochemistry. 1998;37:7981–7991. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- 46.Cauerhff A, Goldbaum FA, Braden BC. Structural mechanism for affinity maturation of an anti-lysozyme antibody. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3539–3544. doi: 10.1073/pnas.0400060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freire E, Murphy KP, Sanchez-Ruiz JM, Galisteo ML, Privalov PL. The molecular basis of cooperativity in protein folding. Thermodynamic dissection of interdomain interactions in phosphoglycerate kinase. Biochemistry. 1992;31:250–256. doi: 10.1021/bi00116a034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.