Abstract
The 24-h rhythmic production of melatonin by the pineal gland is essential for coordinating circadian physiology. Melatonin production increases at night in response to the release of norepinephrine from sympathetic nerve processes which innervate the pineal gland. This signal is transduced through G-protein-coupled adrenergic receptors. Here, we found that the abundance of regulator of G-protein signaling 2 (RGS2) increases at night, that expression is increased by norepinephrine and that this protein has a negative feedback effect on melatonin production. These data are consistent with the conclusion that RGS2 functions on a daily basis to negatively modulate melatonin production.
Keywords: Melatonin, Pineal gland, Regulator of G-protein signaling, RGS2, Norepinephrine
1. Introduction
Daily rhythms characterize essentially all physiological functions in animals [1]. Disruption of normal 24-h rhythms in man can cause sleep disorders and a wide range of metabolic and psychiatric diseases [2,3]. An essential element in the circadian system is melatonin, which is produced by the pineal gland and acts to optimize circadian biology. The 24-h pattern in melatonin production and release is controlled by the suprachiasmatic nucleus (SCN), the central circadian clock in mammals [4]. Time-of-day information from SCN is transmitted to the pineal gland by a neural circuit which includes central and peripheral neural structures, terminating in sympathetic fibers.
At night, neural signals from the SCN elicit norepinephrine release from these fibers into the pineal perivascular space; norepinephrine acts through G-protein-coupled adrenergic receptors to increase cAMP. This increases melatonin production by increasing the activity of the penultimate enzyme in melatonin synthesis, arylalkylamine N-acetyltransferase (Aanat), which acetylates serotonin to produce N-acetylserotonin, the melatonin precursor. In rodents, the increase in Aanat activity reflects in part cAMP-dependent induction of Aanat transcription, in addition to phosphorylation [4].
Here we have focused on a critical element of the regulation of melatonin production, signaling through G-proteins. G-Protein signaling is activated by binding of GTP; hydrolysis of bound GTP by intrinsic GTPase activity of the Gα subunit inactivates G-protein signaling. This hydrolysis is accelerated by GTPase activating proteins (GAPs), and through this mechanism, GAPs accelerate G-protein deactivation, thereby downregulates the signaling. The regulator of G-protein signaling (RGS) family of proteins act as GAPs and accelerate the otherwise slow intrinsic GTPase activity. Transcription of some RGS family members is dynamically regulated [5], and RGS2 proteins provide feedback regulation of G-protein signaling [6,7]. Here we describe the expression of RGS family members in the rat pineal gland, and report that one negatively regulates melatonin production.
2. Materials and methods
2.1. Animals and tissue collection
Pineal glands were prepared from Sprague Dawley rats (female, 180–250 g; Taconic Farms Inc., Germantown, NY), that had been entrained to a 14:10 light:dark (L:D) cycle for at least one week. Six pineal glands were pooled and used for RGS family member expression screening; the day pool was obtained at Zeitgeber time (ZT) 7 and the night pool at ZT19. Euthanasia at ZT19 was done under a dim red light. Animal use and care protocols were approved by local ethical review and were in accordance with National Institutes of Health guidelines.
2.2. Western blotting
Protein was extracted from pineal glands for quantification of daily or induced changes in RGS2 levels; and, from pineal cells to quantify levels of virus-directed induction of RGS2. Protein extraction was done in radio immunoprecipitation assay buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). The lysate was centrifuged (15,000×g, 30 min, 4 °C); protein concentration of the supernatant was determined by the bicinchoninic acid assay method (Pierce, Rockford, IL, USA). Proteins (10 µg/lane) were resolved using 12% bis-tris SDS–polyacrylamide gel electrophoresis in 3-(N-morpholino) propanesulfonic acid (MOPS) buffer, and electroblotted onto Immobilon-P (Millipore, Billerica, MA, USA) membrane in NuPAGE transfer buffer. The membranes were incubated for 1 h in Odyssey Blocking buffer (Li-Cor, Lincoln, NE, USA) before overnight incubation with a polyclonal chicken anti-full-length RGS2 (GW22245F, Sigma Aldrich, St. Louis, MO, USA; 1:800 dilution) and monoclonal mouse anti-β-actin (A5441, Sigma Aldrich; 1:5000 dilution) in Tris-buffered saline with 0.1% Tween20. After washing (Tris-buffered saline with 0.1% Tween20, 5 min, 3 times), membranes were incubated (Tris-buffered saline, 5% non-fat milk, 0.1% Tween20 and 0.02% SDS, 1 h, room temperature) with two secondary antisera: anti-mouse IgG (IRDye 800CW, #827-08364, Li-Cor) and anti-chicken IgG (IRDye 680LT, #926-68028, Li-Cor). Blots were visualized and band density of immunopositive bands was determined with an Odyssey Infrared Imaging System (Li-Cor); RGS2 and β-actin immunopositive bands were of the predicted size. The RGS2 signal was normalized to β-actin. The normalized RGS2 values from three biological samples were used to determine mean intensity.
2.3. Pineal cell culture
Pineal cells were prepared as described previously [8]; animals were euthanized between ZT4 and ZT7. Approximately 100,000 cells were added to each well of a poly-d-lysine coated 96-well plate (Corning, Tewksbury, MA, USA). Pineal cells were cultured for 48 h, and then incubated in serum-free medium for 24 h prior to and during drug treatments.
2.4. Virus infection
A replication-defective adenovirus encoding RGS2 was provided by Peter Chidiac (University of Western Ontario, Canada) [9]; β-galactosidase-encoding adenovirus (Ad-CMV-beta-gal, Vector BioLabs, Philadelphia, PA, USA) was used as a control. After 24 h of incubation in complete media, pineal cells were infected by addition of adenovirus (24 h). The virus was used at a multiplicity of infection (MOI) of 40 virus particles per cell, unless otherwise stated. Subsequently, cells were maintained in serum-free medium for 24 h prior to drug treatment.
2.5. mRNA quantification by qPCR
RNA from the pineal gland or pineal cells was extracted and treated with DNase using RNeasy Micro Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. RNA quantity and quality was checked by UV absorbance using a NanoDrop 1000 (Thermo Scientific, Wilmington DE, USA). cDNA was synthesized from 1 µg total RNA by SuperScript III reverse transcriptase (Life Technologies, Gland island, NY, USA) using random hexamers at 50 °C for 50 min. qRT-PCR was done using SYBR Green qPCR Mastermix (Qiagen) in a LightCycler 480 (Roche, Indianapolis, IN, USA) as described elsewhere [10]. Molecule specific primer sets that span at least one intron were designed by Primer-Blast software, as listed in Supplementary Table 1. The PCR cycling was: 95 °C for 10 min and then 45 cycles for 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. Product specificity was initially confirmed by agarose gel electrophoresis and sequencing, and melting curve analysis during every qRT-PCR thereafter. For each molecule, 10-fold serial dilutions of each internal standard (10 fM to 1 nM) were used to generate a standard curve. mRNA values were normalized to the abundance of Gapdh, and results are shown based on three independent experiments.
2.6. Melatonin and N-acetylserotonin determination
A published LC/MS/MS method was used [10].
2.7. cAMP assay
cAMP in culture media was detected by the cAMP-Glo Assay (Promega, Madison, WI, USA) following manufacturer’s instructions.
2.8. Data presentation and statistical analysis
All data are presented as the mean of three independent experimental results ± standard error of the mean. Statistical analyses were performed with Prism 5 for Windows (GraphPad software, La Jolla, CA, USA). Statistical tests used are given in the text or in the figure legends.
3. Results
3.1. Transcript levels of RGS family members in the pineal gland
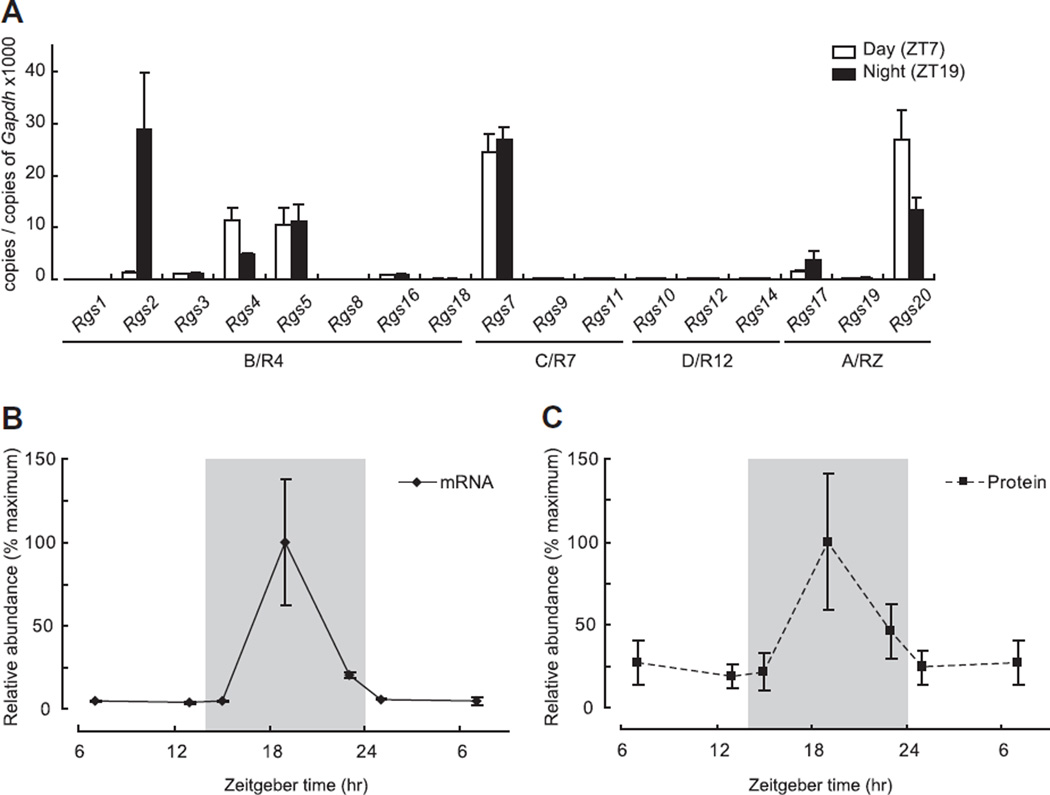
Expression levels of 17 RGS family members were examined by qRT-PCR of RNA extracted from pineal glands obtained during the day and night. Data are grouped according to subfamily membership [11] (Fig. 1A). Except for the D/R12 family, expression of at least one member of all subfamilies was detected. In the C/R7 subfamily, Rgs7 was the only highly expressed member; expression did not exhibit a day/night difference. In the A/RZ subfamily, Rgs20 was highly expressed, with highest levels occurring at ZT7. Three members of the B/R4 subfamily, Rgs2, Rgs4 and Rgs5 were highly expressed, and expression of the first two exhibited a night/day difference. The expression pattern of Rgs2 was notable because it exhibited a ~20-fold increase at night (ZT7 vs ZT19; P < 0.05, two-tailed t-test, n = 3 each), consistent with the day/night difference reported based on microarray analysis [12].
Fig. 1.
Transcript levels of RGS family members in pineal glands obtained during the day or night. (A) Transcript levels of 17 RGS members in pineal gland either at day (Zeitgeber time; ZT7, white box) or night (ZT19, black box) are shown. Rgs2, Rgs4 and Rgs20 exhibited significantly different expression levels between ZT7 and ZT19 (ZT7 vs ZT19 of Rgs2: 1.3 ± 0.12 vs 28.9 ± 6.28; Rgs4: 11.4 ± 1.32 vs 4.7 ± 0.23; Rgs20: 26.9 ± 3.27 vs 13.4 ± 1.28, P < 0.05, two-tailed t-test, n = 3 each). Note Rgs2 was the only molecule that had higher expression at ZT19. (B and C) Levels of Rgs2 mRNA or RGS2 protein expression are shown relative to their peak at ZT19. Gray shading indicates time when lights were off (night time), and the expression value at ZT7 is plotted twice for presentation. For further details see Section 2.
3.2. The Rgs2 transcript and RGS2 protein exhibit similar 24-h profiles
The abundance of the Rgs2 mRNA was determined at 6 time points throughout a 24-h period (Fig. 1B). The peak in Rgs2 expression occurred at ZT19. Expression was no more than 20% of the peak value at other times. The Rgs2 expression pattern generally resembles that of Aanat and of a group of transcripts known to be controlled by the SCN and to peak at night (Dio2, Fosl2, Pde4b, Pde10a, Crem, Dusp1 [12]). Western blot analysis indicated that RGS2 protein exhibited similar dynamics (Fig. 1C, Supplementary Fig. 1).
3.3. Norepinephrine regulates Rgs2 transcript and protein abundance
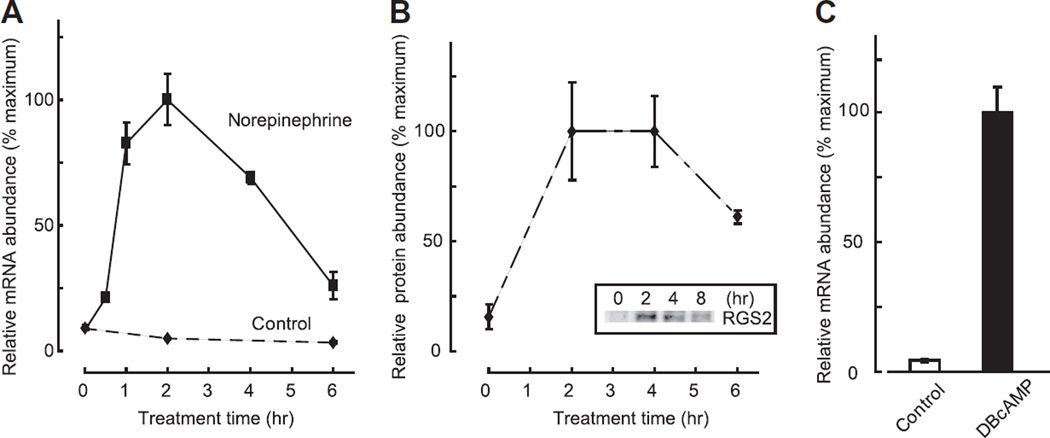
To determine if the night time increase in Rgs2 expression could be reproduced in vitro by norepinephrine treatment, isolated pineal cells were treated with 1 µM norepinephrine. Rgs2 mRNA increased immediately following norepinephrine application; the response peaked within 2 h and decreased gradually thereafter to ~30% of peak value by 6 h (Fig. 2A). Also, RGS protein was found to increase within 2 h (Fig. 2B). However, the subsequent decrease in RGS2 protein was delayed about 2 h relative to that of Rgs2 mRNA.
Fig. 2.
Increase of Rgs2 transcripts and RGS2 protein following treatment with norepinephrine or a cAMP analog. (A) Rgs2 transcription was induced by 1 µM norepinephrine application, and reached its peak 2 h after stimulation. Significant induction of Rgs2 was found 1 to 4 h after stimulation (P < 0.05, one-way ANOVA vs expression level at 0 h, n = 3 each). (B) RGS2 protein increased within 2 h by treatment with 1 µM norepinephrine (P < 0.05, one-way ANOVA vs expression level at 0 h, n = 3 each). Inset shows a representative Western blot result of RGS2 protein expression following norepinephrine stimulation. (C) Application of the cAMP analog, dibutyryl–cAMP, induced transcription of Rgs2 (Control vs DBcAMP, 4.3 ± 0.55% vs 100.0 ± 9.41%; P < 0.05, two-tailed t-test, n = 3 each). For further details see Section 2.
3.4. cAMP controls Rgs2 transcript abundance
This rapid increase of Rgs2 mRNA is consistent with reports of a similar response after carbacohol or geldanamycin treatment in cultured cell lines, or after seizure in brain tissues [7,13,14]. It has also been shown that an upstream cAMP-response element controls Rgs2 expression regulation [15]. In the pineal gland, norepinephrine acts through cAMP to increase expression of many genes, which in some cases appears to involve a cAMP-responsive element (Aanat, Dio2, Fosl2, Crem, Dusp1). To examine if cAMP might regulate Rgs2 expression in the pineal gland, we tested the effect of 1 mM dibutyryl-cAMP (DB-cAMP), a soluble cAMP analog and found that a 6-h treatment increased Rgs2 transcript abundance by >20-fold (Fig. 2C). This supports the conclusion that norepinephrine acts through cAMP to control the abundance of the Rgs2 transcript in pineal cells.
3.5. RGS2 inhibits adrenergic stimulation of N-acetylserotonin and melatonin production
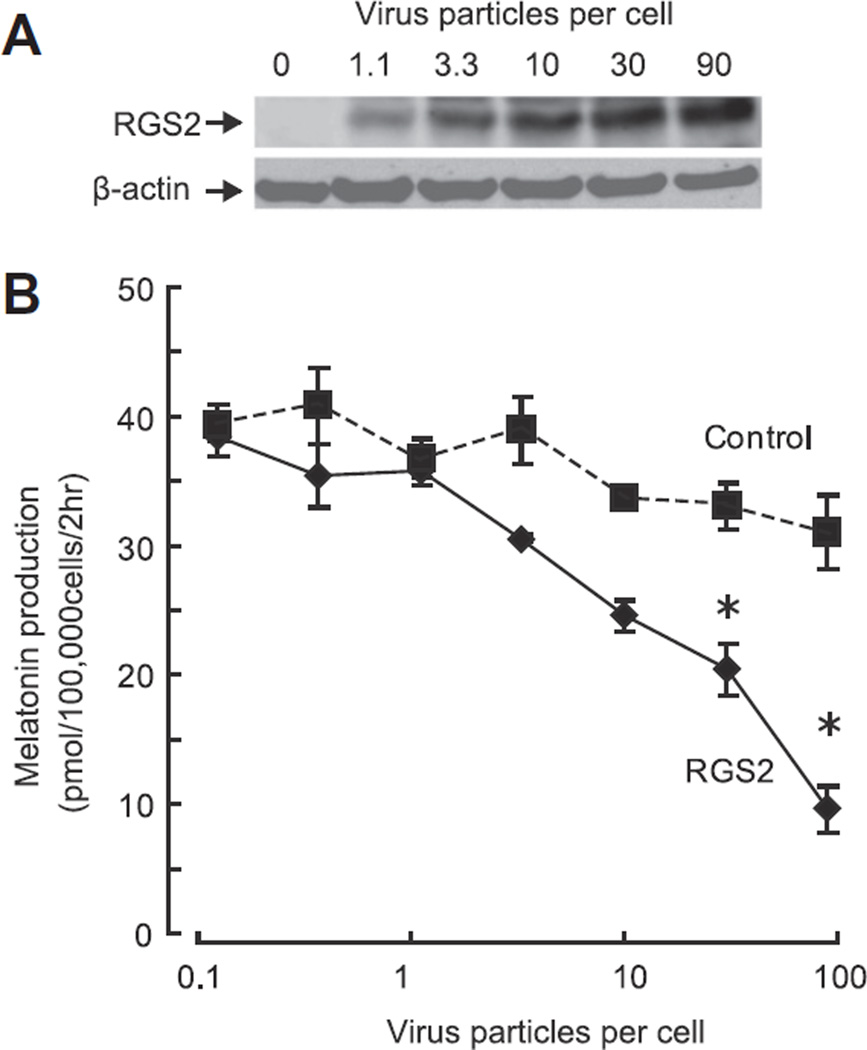
Based on the findings that norepinephrine elevates RGS2 protein in the pineal gland and that RGS2 protein is known to inhibit G-protein signaling, we hypothesized that overexpression of RGS2 would inhibit G-protein signaling in this tissue. To examine this, RGS2 protein was overexpressed by use of an adenovirus vector (Fig. 3A).
Fig. 3.
RGS2 inhibits melatonin production in a dose-dependent manner. (A) Western blot of cell lysates infected with serial titers of RGS2-coding adenovirus; 1.1 to 90 virus particles per cell (MOI). The increase in pineal RGS2 protein was titer-dependent. (B) Virus titer-dependent decrease of melatonin production in pineal cells. Although no significant decrease of melatonin was found up to 10 virus particles per cell, a significant and titer-dependent decrease by RGS2 was found at 30 and 90 virus particles per cell (P < 0.05, one-way ANOVA, Control vs RGS2 infected cells at same virus concentration, n = 3 each). For further details see Section 2.
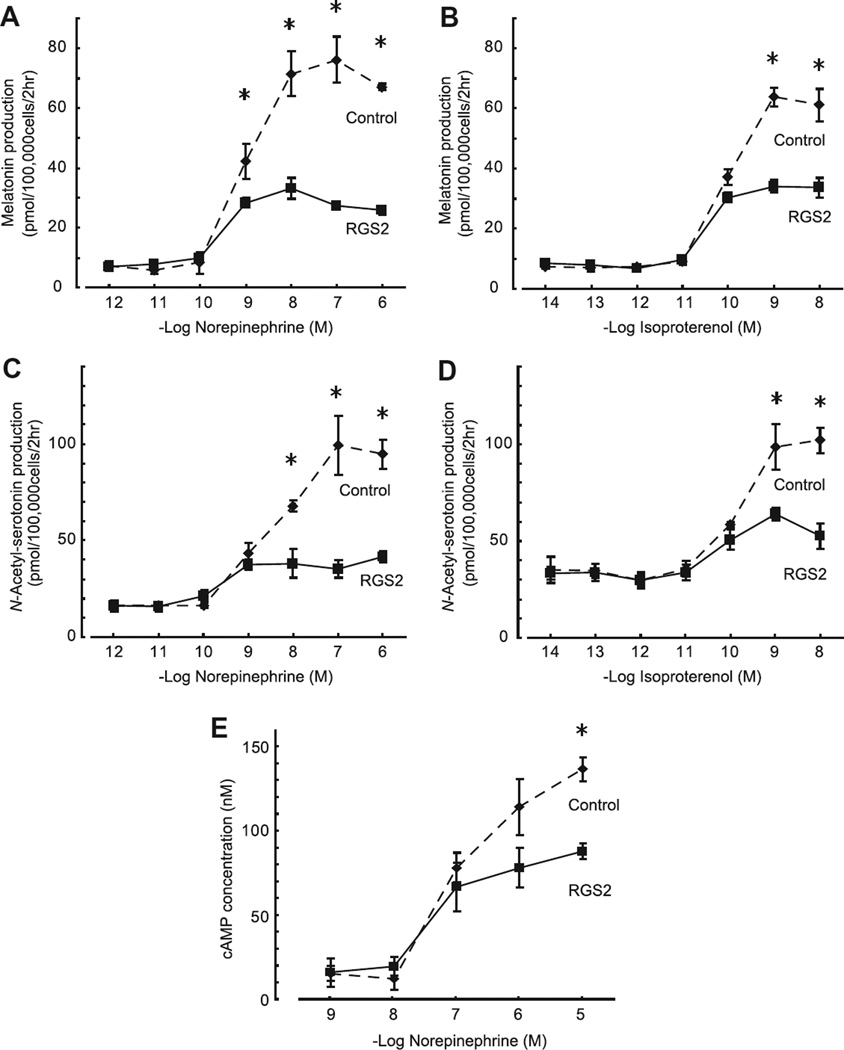
RGS2 overexpression suppressed the norepinephrine-dependent increase of melatonin production, as reflected in an increase in medium melatonin (Fig. 3B). This finding was extended by using a range of concentrations of norepinephrine, which revealed that RGS2 reduced by 64% the maximal level of medium melatonin in the presence of norepinephrine (Fig. 4A, Control vs RGS2; 76.1 ± 4.47 vs 27.3 ± 0.17 pmol, P < 0.05, one-way ANOVA at 100 nM norepinephrine, n = 3 each). In addition, it was found that RGS2 overexpression decreased levels of the melatonin precursor, N-acetylserotonin by approximately 50% (Fig. 4C). This suggested to us that RGS2 acts by decreasing the acetylation of serotonin by AANAT.
Fig. 4.
RGS2 inhibits norepinephrine- and isoproterenol-dependent elevation of N-acetylserotonin, melatonin and cAMP in pinealocyte culture media. Pineal cells were stimulated by a range of concentrations of norepinephrine (A and C) or isoproterenol (B and D). Significant inhibition of melatonin and N-acetylserotonin production by RGS2 occurred at concentrations of 10−9–10−6 and 10−8–10−6 M norepinephrine, respectively (A and C, P < 0.05, one-way ANOVA, Control vs RGS2-infected cells at the same concentration of norepinephrine, n = 3 each). Significant inhibition of both melatonin and N-acetylserotonin production by RGS2 was found at the concentrations of 10−9–10−8 M isoproterenol (B and D, P < 0.05, one-way ANOVA, Control vs RGS2-infected cells at the same concentration of isoproterenol, n = 3 each). (E) Norepinephrine stimulation of cAMP content of media was decreased in cultures of cells in which RGS2 was overexpressed (P < 0.05, one-way AVNOVA, Control vs RGS2-infected cells at 10−5 µM norepinephrine, n = 3 each). For further details see Section 2.
Norepinephrine is a mixed α- and β-adrenergic agonist and elicits maximal stimulation of the pineal gland in a dual receptor mechanism in which β-adrenergic stimulation of adenylyl cyclase is essential; this is enhanced by α-adrenergic elevation of intracellular Ca2+ and translocation of protein kinase C [16]. In studies in which RGS2 was overexpressed, we found that this suppressed melatonin production in cells treated with isoproterenol, a selective β-adrenergic agonist by 47% (Fig. 4B, Control vs RGS2; 63.8 ± 1.77 vs 34.0 ± 1.25 pmol, P < 0.05 one-way ANOVA at 1 nM isoproterenol, n = 3 each). Similar effects were seen with medium N-acetylserotonin (Fig. 4D). These findings indicate that RGS2 inhibits melatonin production by inhibiting β-adrenergic signaling.
3.6. RGS2 suppresses cAMP production
To determine if cAMP production was reduced by RGS2, we measured medium cAMP concentrations after 15 min of norepinephrine stimulation of pineal cells in which GRS was overexpressed (Fig. 4E). This revealed that RGS2 decreased the cAMP amount in media when cells were treated with 10 µM norepinephrine (Fig. 4E, Control vs RGS2; 136.5 ± 7.04 vs 87.7 ± 4.75 nM, P < 0.05 one-way ANOVA at 10 µM norepinephrine, n = 3 each). This provides evidence that cAMP production by pineal cells is reduced by RGS2.
3.7. RGS2 suppresses the adrenergic induction of Aanat and other adrenergic cAMP-inducible genes
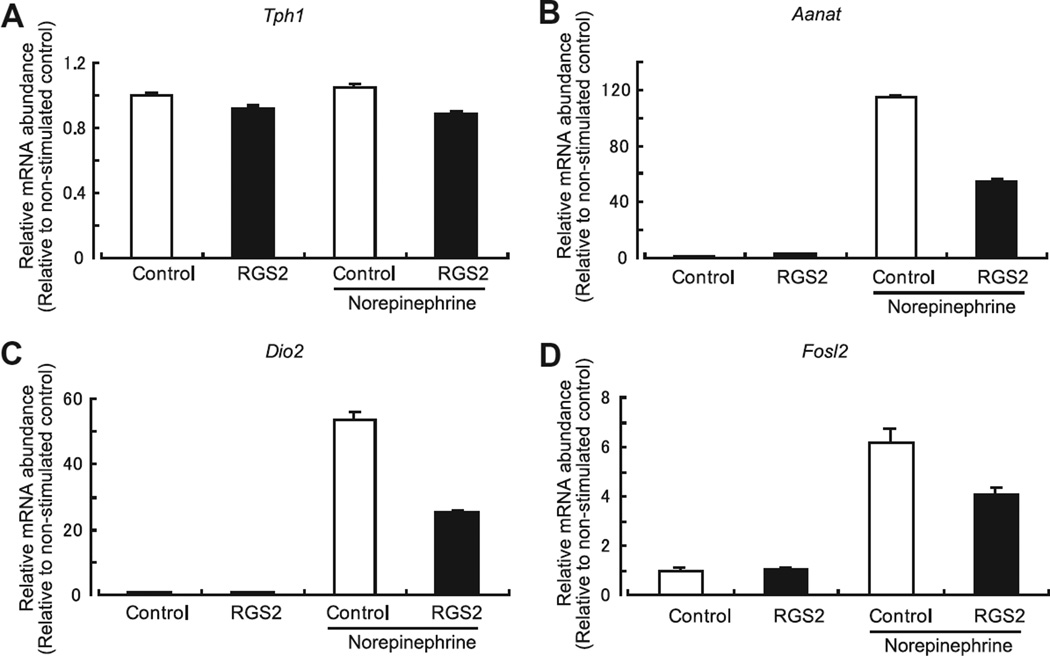
Based on the findings that RGS2 inhibited N-acetylserotonin and cAMP production, it is likely that inhibition of RGS2 reflects in part a decrease in AANAT activity. This might occur as a result of a decrease in Aanat transcription which in turn would result in a decrease in protein and enzyme activity. Analysis of Aanat mRNA indicated that RGS2 suppressed by approximately 50% the norepinephrine stimulation of Aanat mRNA (Fig. 5B, Control vs RGS2; P < 0.05, one-way ANOVA, n = 3 each). In the same experiment, we found that the expression levels of Tph1, the first enzyme in the tryptophan to melatonin pathway did not change as a function of RGS2 infection in control or norepinephrine-treated cells, providing indication that the effect of viral infection was selective.
Fig. 5.
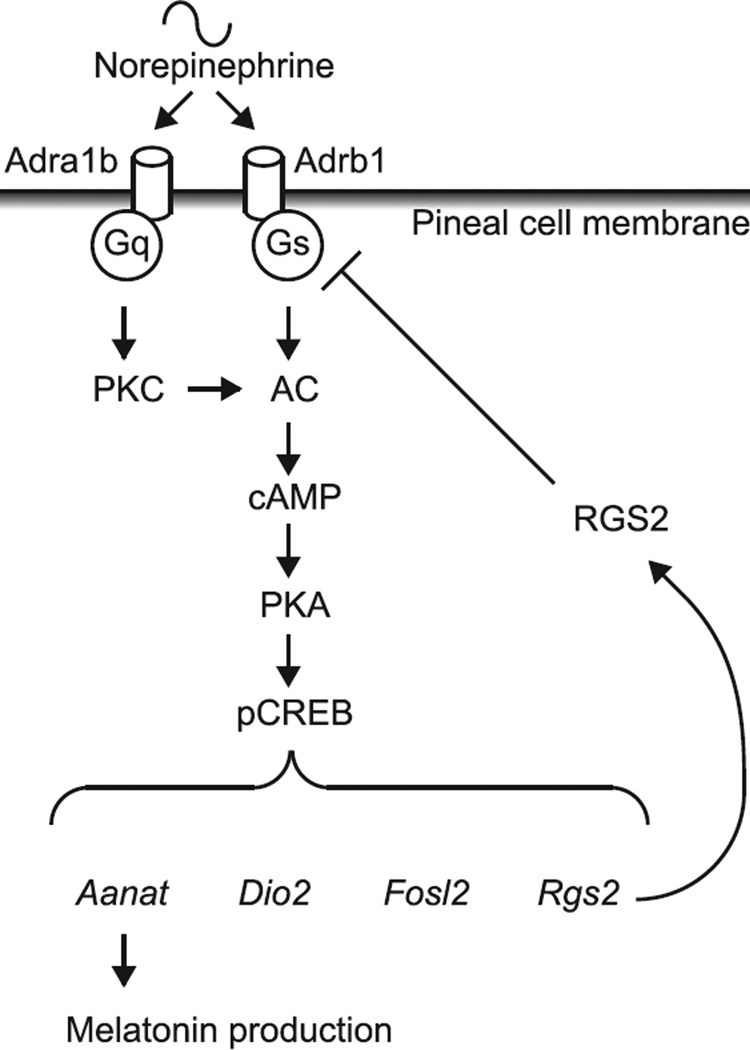
RGS2 inhibits the norepinephrine-dependent increase of the abundance of Aanat, Dio2 and Fosl2 transcripts. The effect of RGS2 overexpression on the abundance of several transcripts was determined following by a 2-h application of 1 µM norepinephrine. Norepinephrine did not increase the abundance of Tph1 mRNA (A). However, a greater than 5-fold increase was observed for Aanat (B), Dio2 (C) and Fosl2 (D). Increased transcript levels in RGS2-infected cells was decreased to 47.7% for Aanat, to 47.1% for Dio2, and to 66.0% for Fosl2 (P < 0.05, one-way ANOVA, n = 3 each) compared to norepinephrine-stimulated control cells. For further details see Section 2. Fig. 6. Schematic presentation of negative regulation of melatonin synthesis by RGS2. RGS2 is induced by the daily release of norepinephrine which binds to α1band β1-adrenergic receptors and activates adenylyl cyclase. The resulting increase in cAMP leads to the transcription of several genes including Rgs2. Translated RGS2 protein inhibits G-protein signaling thereby attenuating cAMP-mediated transcription.
It was also found that RGS2 regulates other transcripts regulated by norepinephrine–cAMP signaling [12], including Dio2 and Fosl2 (Fig. 5C and D). Expression of both was inhibited by RGS2 overexpression, thereby providing evidence of broad effects of RGS2 on adrenergic stimulation of gene expression in the pineal gland.
4. Discussion
The results presented in this report provide the first evidence that pineal signal transduction is regulated at the G-protein level through a member of the RGS family proteins. The finding that norepinephrine controls the abundance of RGS2 supports the conclusion that a negative feedback loop exists in which stimulation of pineal cells by norepinephrine elevates RGS2 which in turn attenuates adrenergic stimulation, as outlined in Fig. 6. Analyses of melatonin and N-acetylserotonin indicate that RGS2 impacts melatonin production at the first step in the serotonin → N-acetylserotonin → melatonin pathway, serotonin acetylation by AANAT.
Fig. 6.
Schematic presentation of negative regulation of melatonin synthesis by RGS2. RGS2 is induced by the daily release of norepinephrine which binds to α1b- and β1-adrenergic receptors and activates adenylyl cyclase. The resulting increase in cAMP leads to the transcription of several genes including Rgs2. Translated RGS2 protein inhibits G-protein signaling thereby attenuating cAMP-mediated transcription.
The mechanism through which RGS2 inhibits serotonin acetylation appears to involve inhibition of Aanat transcription, based on our results. Moreover, in view of the evidence that cAMP mediates norepinephrine regulation of RGS2 and that norepinephrine elevates pineal cAMP, we believe that RGS2 inhibits Aanat induction through a suppressive effect on cAMP production, as in other tissues [17]. This conclusion is supported by the finding that RGS2 also inhibits induction of other cAMP-regulated genes in the pineal gland, including Dio2 and Fosl2. Accordingly, it is not only reasonable to suspect that RGS2 is acting on melatonin synthesis by suppressing cAMP-dependent induction of Aanat, but that suppression of cAMP might also result in a decrease in cAMP-dependent phosphorylation of AANAT, which would in turn decrease enzyme activity because posttranslational phosphorylation promotes AANAT stability and activity [4].
The evidence that RGS2 suppresses induction of two other adrenergic–cAMP regulated genes in the pineal gland points to the likelihood that RGS2 broadly impacts pineal biology by inhibiting the adrenergic–cAMP regulation of hundreds of genes in this tissue [12], thereby playing a broad governing role.
As such, RGS2 joins a growing group of proteins that contribute to negative feedback of adrenergic stimulation of pineal function. This group includes PDE4B, which like RGS2 is (1) induced by norepinephrine acting through a cAMP mechanism and (2) suppresses norepinephrine–cAMP signaling [18]. In the case of PDE4B and perhaps PDE10A, the mechanism involves increased cAMP degradation. In addition, another mechanism that represses pineal cAMP-directed transcription involves Snf1-lk/SIK induction [19].
Early work on signal transduction in the pineal gland clearly established that mechanisms exist to control daily change of pineal sensitivity to adrenergic signals [20]. It would appear that the phenomenon of subsensitivity can now be explained on a molecular level in part by the daily induction of Rgs2, Pde4b, Pde10a, and Snf1-lk/SIK, acting through several negative feedback mechanisms focusing on cAMP signaling to govern the magnitude of the response of the pinealocyte to norepinephrine.
We found that two other highly expressed RGS family members, Rgs4 and Rgs20, exhibited ~2-fold higher expression in the day time. The antiphase relationship to the dynamics of Rgs2 raises the possibility that these molecules might also modulate G-protein signaling, and modify those of RGS2. Although Rgs2 and Rgs4 belong to the same B/R4 subfamily, opposite transcriptional regulation has been reported [14]. Rgs20 is a member of A/RZ subfamily and mainly targets Gαz [21]; therefore, interference in the action of Rgs2 is unlikely because it primarily targets Gαs and Gαq [11].
Besides the regulation of G-proteins, RGS2 acts through other mechanisms to regulate signaling pathways. Direct interactions with adenylyl cyclase III and V have been reported [22,23]; however, neither is highly expressed in the pineal gland (unpublished data). Also, emerging data has described RGS2 regulation of transient receptor potential (TRP) ion channels, specifically TRPV6 [24]; although it is expressed at low levels in the pineal gland (unpublished data), other members of this family which are highly expressed might be RGS2 targets.
In conclusion, the results of our studies indicate that RGS2 is expressed on a 24-h schedule in the pineal gland by a norepinephrine– cAMP mechanism; and, that this constitutes a negative feedback mechanism that suppresses effects of norepinephrine, including melatonin production and induction of several genes. This suggests that RGS2 acts broadly to govern G-protein signaling in the pineal gland, and therefore RGS2 might have a suppressive role in the daily pattern of melatonin production. Moreover, a similar 24-h cAMP-RGS2 feedback mechanism may function in other tissues to influence daily changes in cAMP-driven gene expression, and malfunction of this mechanism can lead to disease.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NICHD. We express appreciation to Peter Chidiac for providing the RGS2 adenovirus and valuable scientific input.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.febslet.2013.03.
References
- 1.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin DB. Influence of sleep-wake and circadian rhythm disturbances in psychiatric disorders. J. Psychiatry Neurosci. 2000;25:446–458. [PMC free article] [PubMed] [Google Scholar]
- 3.Morris CJ, Yang JN, Scheer FA. The impact of the circadian timing system on cardiovascular and metabolic function. Prog. Brain Res. 2012;199:337–358. doi: 10.1016/B978-0-444-59427-3.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein DC. Arylalkylamine N-acetyltransferase: “the Timezyme”. J. Biol. Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- 5.Kach J, Sethakorn N, Dulin NO. A finer tuning of G-protein signaling through regulated control of RGS proteins. Am. J. Physiol. Heart Circ. Physiol. 2012;303:H19–H35. doi: 10.1152/ajpheart.00764.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 7.Ingi T, et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J. Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho AK, Ogiwara T, Chik CL. Thapsigargin modulates agoniststimulated cyclic AMP responses through cytosolic calcium-dependent and - independent mechanisms in rat pinealocytes. Mol. Pharmacol. 1996;49:1104–1112. [PubMed] [Google Scholar]
- 9.Zou MX, Roy AA, Zhao Q, Kirshenbaum LA, Karmazyn M, Chidiac P. RGS2 is upregulated by and attenuates the hypertrophic effect of alpha1-adrenergic activation in cultured ventricular myocytes. Cell. Signal. 2006;18:1655–1663. doi: 10.1016/j.cellsig.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Clokie SJ, Lau P, Kim HH, Coon SL, Klein DC. MicroRNAs in the pineal gland: miR-483 regulates melatonin synthesis by targeting arylalkylamine N-acetyltransferase. J. Biol. Chem. 2012;287:25312–25324. doi: 10.1074/jbc.M112.356733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 12.Bailey MJ, et al. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J. Biol. Chem. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS. Second messengers regulate RGS2 expression which is targeted to the nucleus. Biochim. Biophys. Acta. 2001;1541:201–211. doi: 10.1016/s0167-4889(01)00144-6. [DOI] [PubMed] [Google Scholar]
- 14.Song L, Jope RS. Cellular stress increases RGS2 mRNA and decreases RGS4 mRNA levels in SH-SY5Y cells. Neurosci. Lett. 2006;402:205–209. doi: 10.1016/j.neulet.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z, Liu D, Liu S, Calderon L, Zhao G, Turk J, Guo Z. Identification of a cAMP-response element in the regulator of G-protein signaling-2 (RGS2) promoter as a key cis-regulatory element for RGS2 transcriptional regulation by angiotensin II in cultured vascular smooth muscles. J. Biol. Chem. 2011;286:44646–44658. doi: 10.1074/jbc.M111.265462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanecek J, Sugden D, Weller J, Klein DC. Atypical synergistic alpha 1- and beta-adrenergic regulation of adenosine 3′,5′-monophosphate and guanosine 30,50-monophosphate in rat pinealocytes. Endocrinology. 1985;116:2167–2173. doi: 10.1210/endo-116-6-2167. [DOI] [PubMed] [Google Scholar]
- 17.Nunn C, Zou MX, Sobiesiak AJ, Roy AA, Kirshenbaum LA, Chidiac P. RGS2 inhibits beta-adrenergic receptor-induced cardiomyocyte hypertrophy. Cell. Signal. 2010;22:1231–1239. doi: 10.1016/j.cellsig.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Bailey MJ, Ho AK, Moller M, Gaildrat P, Klein DC. Daily rhythm in pineal phosphodiesterase (PDE) activity reflects adrenergic/3′,5′-cyclic adenosine 5′-monophosphate induction of the PDE4B2 variant. Endocrinology. 2007;148:1475–1485. doi: 10.1210/en.2006-1420. [DOI] [PubMed] [Google Scholar]
- 19.Kanyo R, Price DM, Chik CL, Ho AK. Salt-inducible kinase 1 in the rat pinealocyte: adrenergic regulation and role in arylalkylamine N-acetyltransferase gene transcription. Endocrinology. 2009;150:4221–4230. doi: 10.1210/en.2009-0275. [DOI] [PubMed] [Google Scholar]
- 20.Romero JA, Axelrod J. Pineal beta-adrenergic receptor: diurnal variation in sensitivity. Science. 1974;184:1091–1092. doi: 10.1126/science.184.4141.1091. [DOI] [PubMed] [Google Scholar]
- 21.Barker SA, Wang J, Sierra DA, Ross EM. RGSZ1 and Ret RGS: two of several splice variants from the gene RGS20. Genomics. 2001;78:223–229. doi: 10.1006/geno.2001.6659. [DOI] [PubMed] [Google Scholar]
- 22.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. Identification of RGS2 and type V adenylyl cyclase interaction sites. J. Biol. Chem. 2003;278:15842–15849. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 23.Sinnarajah S, et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 24.Schoeber JP, Topala CN, Wang X, Diepens RJ, Lambers TT, Hoenderop JG, Bindels RJ. RGS2 inhibits the epithelial Ca2+ channel TRPV6. J. Biol. Chem. 2006;281:29669–29674. doi: 10.1074/jbc.M606233200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.