Abstract
Purpose
Endocrine therapies include aromatase inhibitors and the selective oestrogen receptor (ER) down-regulator fulvestrant. This study aimed to determine if the reported efficacy of fulvestrant over anastrozole, and high- over low-dose fulvestrant, reflect distinct transcriptional responses.
Experimental design
Global gene expression profiles from ERα-positive breast carcinomas before and during pre-surgical treatment with fulvestrant (n=22) or anastrozole (n=81), and corresponding in vitro models, were compared. Transcripts responding differently to fulvestrant and oestrogen (E) deprivation were identified and integrated using gene ontology (GO), pathway and network analyses to evaluate their potential significance.
Results
The overall transcriptional response to fulvestrant and E-deprivation was correlated (r=0.61 in pre-surgical studies, r=0.87 in vitro), involving down-regulation of E-regulated and proliferation-associated genes. The transcriptional response to fulvestrant was of greater magnitude than E-deprivation (slope=0.62 in pre-surgical studies, slope=0.63 in vitro). Comparative analyses identified 28 genes and 40 GO categories affected specifically by fulvestrant. Seventeen fulvestrant-specific genes, including CAV1/2, SNAI2 and NRP1, associated with ERα, androgen receptor (AR) and TP53, in a network regulating cell cycle, death, survival, and tumour morphology. Eighteen genes responding differently to fulvestrant specifically predicted anti-proliferative response to fulvestrant, but not anastrozole. Transcriptional effects of low-dose fulvestrant correlated with high-dose treatment, but were of lower magnitude (ratio=0.29).
Conclusions
The transcriptional response to fulvestrant has much in common with E-deprivation, but is stronger with distinctions potentially attributable to arrest of E-independent ERα activity and involvement of AR signalling. Genes responding differently to fulvestrant may have predictive utility. These data are consistent with the clinical efficacy of fulvestrant versus anastrozole and higher dosing regimens.
Keywords: Breast cancer, oestrogen, estrogen, oestrogen receptor, estrogen receptor, oestrogen deprivation, aromatase inhibitor, fulvestrant
INTRODUCTION
Endocrine therapies abrogate oestrogenic signalling through distinct mechanisms, impeding oestrogen (E) synthesis or transcriptional activity of oestrogen receptor alpha (ERα). Aromatase inhibitors (AIs), e.g. anastrozole, cause profound post-menopausal E-suppression and are used as first-line neo-adjuvant, adjuvant and metastatic therapies. Selective oestrogen receptor modulators (SERMs), e.g. tamoxifen, exert partial agonist activity. In contrast, the selective ER down-regulator (SERD) fulvestrant (Faslodex®, AstraZeneca) is a pure anti-oestrogen, inhibiting receptor dimerisation, nuclear uptake, E-response element binding and accelerating ERα degradation (1-5).
Fulvestrant is licensed for post-menopausal progression/relapse on first-line endocrine therapy (6-8). Low-dose fulvestrant (250mg/28 days) provides comparable disease outcome to anastrozole following first-line tamoxifen in metastatic disease (6-8), with utility after progression on AIs (9-11). As first-line therapy in metastatic or locally advanced disease, low-dose fulvestrant provides comparable disease outcome to tamoxifen (12). High-dose treatment (500mg on day 0, 14, 28, monthly thereafter) further improved progression-free (13) and overall survival (14) in the COmparisoN of Faslodex In Recurrent or Metastatic breast cancer (CONFIRM) trial. The Fulvestrant fIRst-line Study comparing endocrine Treatments (FIRST), found an improved time to progression with the high-dose compared to anastrozole (1mg/day) in advanced disease (15, 16). In the Neo-adjuvant Endocrine therapy for Women with Estrogen-Sensitive Tumours (NEWEST) trial of locally advanced disease, high-dose fulvestrant showed greater suppression of ERα, progesterone receptor (PgR), the proliferation marker Ki67 and radiological response than low-dose (17). These data show significant differences between fulvestrant dosing schedules and a mechanism of action which is different to, and may circumvent complete cross-resistance with, SERMs and AIs. The molecular mechanisms which underpin these clinically important differences are incompletely understood.
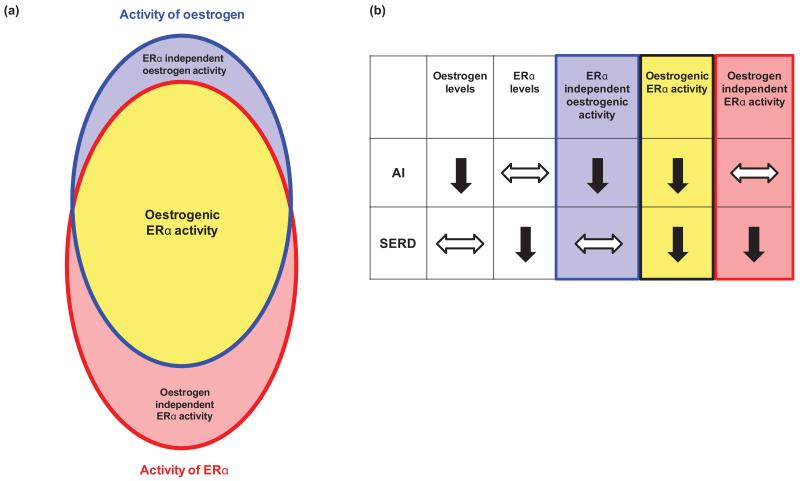
The transcriptional response to fulvestrant differs from SERMs, with the latter up-regulating particular E-regulated genes (ERGs) (18). In addition to more complete ERG antagonism, fulvestrant exclusively down-regulates numerous cell cycle, proliferation, and DNA synthesis genes in vitro (19), and some E-suppressed genes are up-regulated by fulvestrant and not tamoxifen (20). The transcriptional response to AIs and fulvestrant has not previously been compared and may be pertinent to the clinical utility of fulvestrant as an alternative, sequential or combination therapy. The potential for difference is supported by their contrasting effects on E and ERα. The interaction between E and ERα underpins classical oestrogenic signalling which is susceptible to both AIs and fulvestrant. Contemporary models also include activities which do not require interaction and may involve either E or ERα independently (21-25) (Figure 1a). Such non-classical activities might be affected selectively by AIs and fulvestrant respectively (Figure 1b). In vitro, the greater anti-proliferative effect of fulvestrant (26), has been attributed to continued ERα activity following E-withdrawal, with hypersensitivity to residual E and/or E-independent interactions between ERα and growth factor pathways. ERα has recently been shown to retain genomic binding activity following E-withdrawal and drive a CDK4/E2F-dependent transcriptional program. Such ligand-independent ERα activity has particular relevance to de novo and acquired AI resistance, where ERα is frequently expressed and fulvestrant may remain effective (9, 10, 27).
Figure 1.
(a) Summary of E-dependent ERα signalling (yellow) and the potential independent activities of E (blue) and ERα (red). (b) Comparison of the impact of AIs and SERDs on classical and non-classical oestrogenic signalling, illustrating activities targeted by both agents (yellow) and those affected specifically by AIs (blue) or fulvestrant (red), reduced activity is indicated by downward arrows (solid black) and no change is indicated by horizontal arrows (black outline).
In this study, global gene expression profiles from pre-surgical studies of fulvestrant or anastrozole, and corresponding in vitro models, were assessed. The primary objective was to compare and contrast transcriptional responses. Secondary objectives included evaluating the biological response to low- and high-dose fulvestrant and the extent to which transcriptional consequences were attributable to ERα depletion.
MATERIALS & METHODS
Pre-surgical study of fulvestrant
Pre- and on-treatment (four-week) core biopsies stored at −20°C in RNA-later (Qiagen, Sussex, UK) were available from NEWEST (clinicaltrials.gov/show/NCT00093002) (17), Supplementary Figure S1. This phase-II study recruited post-menopausal women with untreated, potentially operable, locally advanced, ERα-positive, primary invasive cancer ≥2 cm. No data were available for HER2 status. Randomisation was to low- (250mg/28 days) or high-dose (500mg on day 0, 14, 28, monthly thereafter) fulvestrant. The on-treatment biopsy was taken prior to the day 28 dose of fulvestrant in both arms of the study. RNA was extracted with RNeasy, assessed using an Agilent Bioanalyser (Santa Clara, CA, USA) and rejected if RNA integrity number <5. Following exclusions, 22 high-dose and 16 low-dose pre-/on-treatment pairs were available, Supplementary Figure S1.
Pre-surgical study of anastrozole
Pre- and on-treatment (two- and sixteen-week) core biopsies were available from post-menopausal women receiving anastrozole monotherapy (1mg/day) within a randomised phase-II neo-adjuvant trial of anastrozole alone or with gefitinib in early disease (clinicaltrials.gov/show/NCT00255463) (28). This subgroup constitutes the Functional Aromatase Inhibitor Molecular Study (FAIMoS) (29), Supplementary Figure S1b/c. Following exclusions, 81 two-week and 18 sixteen-week pairs were available, Supplementary Figure S1. Written informed consent was obtained from each subject and investigations performed after approval by a local institutional review board.
In vitro modelling of fulvestrant or E-deprivation
MCF7 cells (ATCC, Manassas, VA, USA), were cultured in phenol red-free RPMI-1640, 10% fetal bovine serum (FBS) (Gibco® Life Technologies), and 1nM 17β-oestradiol (E2). Cells were stripped of steroids for 48 hours in phenol red-free RPMI with 10% dextran-coated charcoal-stripped FBS (DCC). Cells were seeded into six well plates at a density of 3×105 cells/well for 24 hours. Monolayers were: (i) harvested at this stage, i.e. following 72 hours of E-deprivation (modelling AI), (ii) treated for 48 hours with 0.1nM E2 in DCC (modelling baseline), or (iii) treated for 48 hours with 10nM fulvestrant and 0.1nM E2 (modelling fulvestrant). Experiments were conducted in triplicate and RNA extracted using RNeasy (Qiagen, Sussex, UK).
Microarray-based global gene expression profiling
RNA was quantified, amplified, labelled and hybridized onto Expression BeadChips (Illumina, San Diego, CA, USA). Samples were processed with the following BeadChips: (i) FAIMoS - HumanWG-6v2, (ii) NEWEST - HumanWG-6v3, (iii) bridging study of ten high-dose pairs from NEWEST and ten two-week pairs from FAIMoS - HumanHT-12v4, and (iv) in vitro samples - HumanHT-12v4.
Global gene expression analysis
Raw expression data were extracted with BeadStudio, transformed by variance-stabilising transformation (VST) and normalised using Robust Spline Normalisation (RSN) in the Lumi package in Bioconductor. Probes were excluded if they were not present in any samples (detection p-value>1%). Microarray data are publicly available (29-32). Expression data and annotation files, HumanHT-12_V4_0_R2_15002873_B, HumanWG-6_V3_0_R3_11282955_A, HumanWG-6_V2_0_R4_11223189_A), were imported into Partek® Genomics Suite™ (PGS, 6.6_6.12.0531, Partek Incorporated, MO, USA).
Class comparison of pre- and on-treatment clinical samples employed two-way analysis of variance (ANOVA). Treatment status (i.e. pre- or on-treatment) was considered a categorical variable with fixed effect (as assignment represents all conditions of interest). Pre- and on-treatment samples were paired according to their patient identifier, which was considered a random effect variable which encompassed inter-patient variability (given that patients represent a random sample of all possible patients). Transcripts differentially expressed between fulvestrant-treated, E-deprived and control conditions in vitro were identified by ANOVA. A false discovery rate (FDR) of 5% was used to correct for multiple testing.
Analytical strategy
Matched Illumina probe identifiers were used to enable valid comparisons between BeadChips. All detected probes from HumanWG-6v3 (n=22550) were used to evaluate fulvestrant dosing regimens. Comparisons between NEWEST and FAIMoS used detected probes common to HumanWG-6v3 and HumanWG-6v2 (n=15051), followed by an unpaired t-test of treatment-induced alterations. Detected probes common to HumanHT-12v4, HumanWG-6v3, and HumanWG-6v2 (n=11122) were assessed in vitro. Biological interpretation involved: (i) identification of functional groupings from the Gene Ontology (GO) database, by GO ANOVA in PGS, and (ii) network analyses with Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, CA, USA).
Technical validation of microarray findings for selected genes
Expression of two selected genes, CAV1 and SNAI2, was assessed by quantitative real-time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) of the same RNA preparations used for expression profiling of high-dose fulvestrant (n=20) and two-week anastrozole (n=31) treated patients. TaqMan® assays (Applied Biosystems) were used to quantify CAV1 (Hs0971716_m1) and SNAI2 (Hs0950344_m1), which were normalised to FKBP15 (Hs0391480_m1) and PUM1 (Hs0982775_m1).
ESR1 knockdown in MCF7 cells
MCF7 cells were seeded into DCC at a density of 7×104 cells/well in 12 well plates. After 24 hours monolayers were transfected with 50nM of siRNA targeting ESR1 (ON-TARGETplus 003401, Dharmacon, ThermoFisher, UK), or non-targeting siRNA using DharmaFECT 3 reagent. Media (0.1nM E2 in DCC) was replenished the following day and cells were cultured for 24 hours. TaqMan assays were used to quantify ESR1 (Hs01046818_m1), and selected genes: BRI3 (Hs0854645_g1), CAV1, CAV2 (Hs0184597_m1), CCDC34 (Hs0293234_m1), CYP26B1 (Hs01011223_m1), C9orf140 (Hs0746788_s1), FNTA (Hs0357739_m1), GTSE1 (Hs0212681_m1), HMGN4 (Hs01549435_m1), LRP8 (Hs0182998_m1), NRP1 (Hs0826128_m1), NSUN2 (Hs0214829_m1), RBMS1 (Hs0377856_m1), RECQL4 (Hs01548660_g1), SEPP1 (Hs01032845_m1), SLC7A5 (Hs0185826_m1), SNAI2 and STRA13 (Hs0414534_m1), which were normalised to FKBP15.
RESULTS
Transcriptional response to high- and low-dose fulvestrant
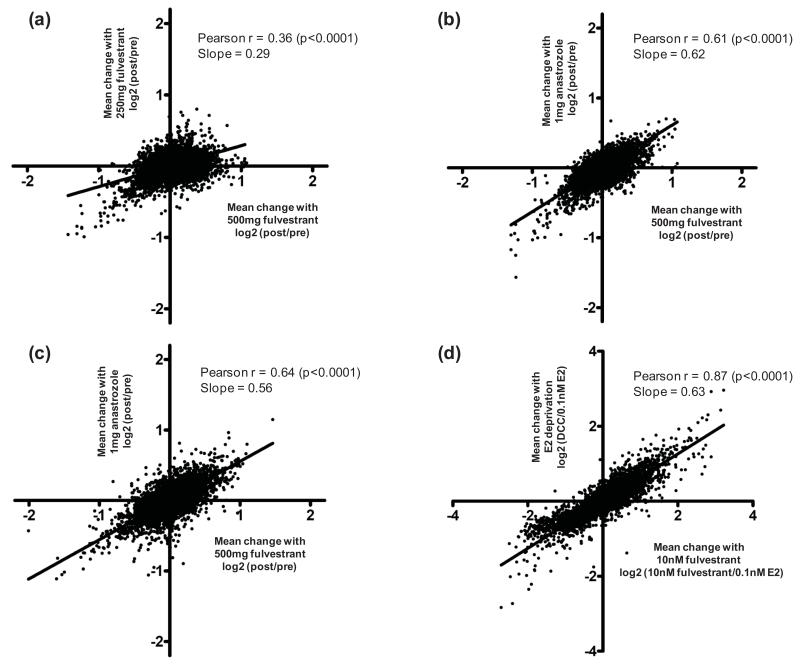
The overall transcriptional response to low-dose fulvestrant was significantly correlated with that to high-dose (Pearson r=0.36, p<0.0001), albeit of lesser magnitude (slope=0.29, Figure 2a, Deming linear regression), supporting a quantitative difference between the dosing schedules. None of the alterations in gene expression induced by low-dose treatment were statistically significant after multiple testing correction (FDR<0.05). However, down-regulation of individual proliferation-associated (e.g. AURKA) or E-regulated (e.g. PGR, PDZK1 and GREB1) genes reached significance (uncorrected p<0.05), Supplementary Table S1. In contrast, 2210 transcripts were significantly affected (977 up-regulated and 1233 down-regulated, FDR<0.05) in the high-dose cohort. Further comparative analyses were undertaken with only high-dose treated patients to avoid the potential for false-negativity by inclusion of those receiving low-dose fulvestrant.
Figure 2.
Scattergrams illustrating the overall transcriptional response to fulvestrant or E-deprivation: (a) Low-dose compared to high-dose fulvestrant, (b) High-dose fulvestrant compared to two weeks of anastrozole, (c) Bridging study of high-dose fulvestrant compared to two-weeks of anastrozole, and (d) MCF7 cells modelling fulvestrant treatment and E-deprivation.
Similarities in the transcriptional response to fulvestrant and E-deprivation
The overall transcriptional response to anastrozole and high-dose fulvestrant in pre-surgical studies was significantly correlated (Pearson r=0.61, p<0.0001, Figure 2b/c), as were those of E-deprivation and fulvestrant in vitro (Pearson r=0.87, p<0.0001, Figure 2d). In both settings, E-regulated genes (e.g. PDZK1, PGR, GREB1 and TFF1) were significantly down-regulated by E-deprivation and fulvestrant, full listings are provided in Supplementary Tables S2-4.
Proliferation-associated genes were also down-regulated by E-deprivation (e.g. TOP2A, CDCA5, CDC20, CCNB2, AURKA, and E2F2) and fulvestrant (e.g. TOP2A, CCNA2, CCNB2, CCND1, CDCA5, CDCA7, CDCA8, CDC2, CDC20, CDC25C, AURKA and POLE), detailed in Supplementary Tables S2-4. Proliferation-associated GO sets, pathways and networks were prominent in transcripts affected by both treatments (Supplementary Tables S5-8). Ki67 staining was comparably reduced in samples from patients receiving anastrozole (geometric mean of post-/pre-treatment expression=24.8% after two weeks, n=69) and fulvestrant (19.6%, following high-dose treatment, n=22).
Differences in the transcriptional response to fulvestrant and E-deprivation
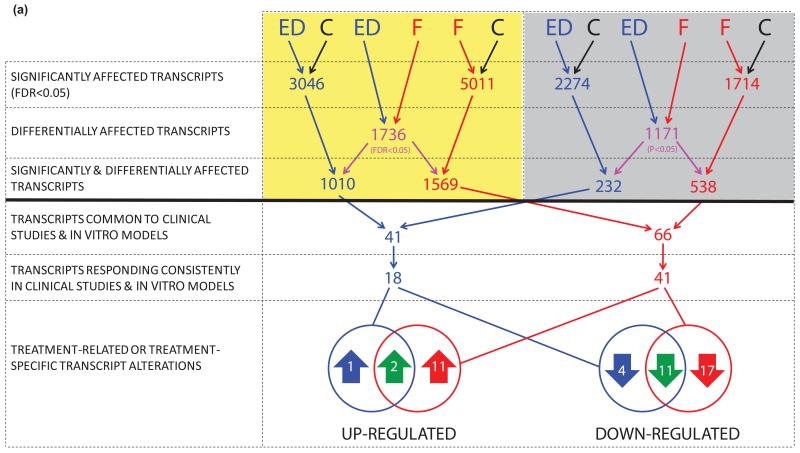
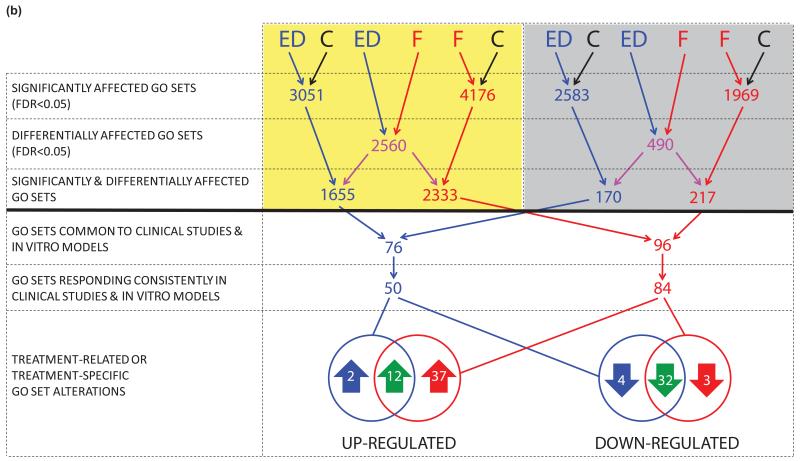
The overall transcriptional response to high-dose fulvestrant was of greater magnitude than anastrozole in pre-surgical studies (slope=0.62, Figure 2b). This difference was supported by the bridging study (slope=0.56, Figure 2c), excluding batch separation as a potential explanation, and consistent with the greater impact of fulvestrant compared to E-deprivation in vitro (slope=0.63, Figure 2d). To determine if any transcripts were affected differently by the agents, rigorous comparisons were undertaken following the scheme illustrated in Figure 3a. Treatment-related transcripts were identified that met the following criteria: (i) expression changed significantly after treatment in clinical samples and corresponding in vitro models (FDR<0.05), (ii) expression was affected differently by the two agents in vitro (discovery set, FDR<0.05) and in pre-surgical studies (validation set, uncorrected p<0.05), and (iii) changes in expression were consistent in clinical samples and corresponding in vitro models (direction of change and relative treatment effect). Transcript sets from Figure 3a are detailed in Supplementary Table S2.
Figure 3.
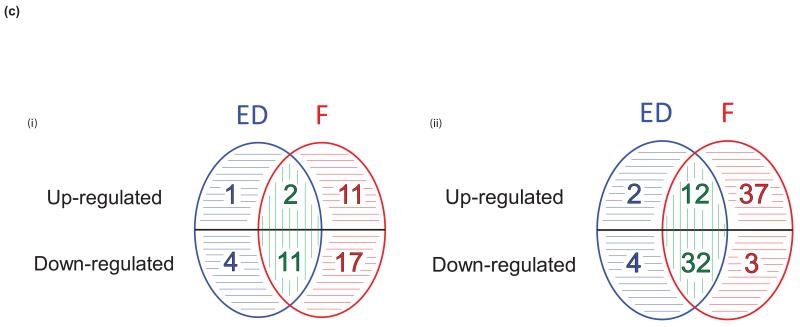
Summary of analytical strategy with (a) resultant transcripts and (b) Gene Ontology (GO) functional gene sets. Key: In vitro models (yellow panel), pre-surgical studies (grey panel), control conditions in vitro / pre-treatment clinical samples (C, black), E-deprivation (ED, blue), Fulvestrant (F, red), ED and F (green). Transcripts and GO sets resulting from comparative analyses are detailed in Web Appendix 1. (c) Summary of (i) transcripts and (ii) GO functional gene sets affected by E-deprivation (ED, blue), fulvestrant (F, red), or both (green), with treatment-specific and quantitative differences indicated by horizontal and vertical stripes respectively.
Fulvestrant-related transcripts (n=41, 13 up-regulated and 28 down-regulated) and E-deprivation-related transcripts (n=18, 3 up-regulated and 15 down-regulated) were then assessed for whether differences were treatment-specific or quantitative (Figure 3a). Of the 15 genes down-regulated by E-deprivation, four (KCNK6, KCNK15, RASGRP1 and TUBA3E) were not similarly affected by fulvestrant, while for the 28 down-regulated by fulvestrant, 17 were not similarly affected by E-deprivation (e.g. GTSE1, LRP8, NSUN2 and STRA13, p=0.005 for the comparison of those not similarly down-regulated, i.e. 4 vs. 17, McNemar test); 11 genes were down-regulated significantly more by fulvestrant than E-deprivation (e.g. CCDC34, RECQL4, SLC7A5 and SAPCD2). Of the three genes significantly up-regulated by E-deprivation, only one (DCXR) was not similarly affected by fulvestrant, while for the 13 up-regulated by fulvestrant, 11 were not similarly affected by E-deprivation (e.g. CAV1, CAV2, SNAI2 and NRP1, p=0.004 for the comparison of those not similarly up-regulated); 2 transcripts (e.g. ZMAT3) were up-regulated significantly more by fulvestrant than E-deprivation. Treatment-related and -specific alterations are summarised in Figure 3c (i) and detailed in Supplementary Table S9.
To assess whether transcript differences might be sufficient to influence distinct biological processes, differentially affected GO sets were identified in the same manner (Figure 3b, detailed in Supplementary Table S2). Fulvestrant down-regulated 32 GO sets significantly more than E-deprivation. Common down-regulated sets were predominantly proliferation-related (e.g. DNA helicase activity, microtubule motor activity and spindle assembly, Supplementary Table S10). Of the 14 GO sets up-regulated by E-deprivation, two were not similarly affected by fulvestrant, while for the 49 up-regulated by fulvestrant, 37 were not similarly affected by E-deprivation (e.g. apoptotic signalling pathway, and negative regulation of: ERK1/2 cascades, TGFβ receptor signalling, canonical Wnt receptor signalling and epithelial cell proliferation, p<0.001 for the comparison of those not similarly up-regulated). Some GO sets specifically up-regulated by fulvestrant pertained to fulvestrant-specific up-regulated genes (e.g. caveolae assembly and CAV1/CAV2). Fulvestrant up-regulated 12 GO sets significantly more than E-deprivation (e.g. negative regulation of MAPK cascade). Treatment-related and -specific alterations in GO sets are summarised in Figure 3c (ii) and detailed in Supplementary Table S10.
Pathway and network analysis of fulvestrant-specific genes
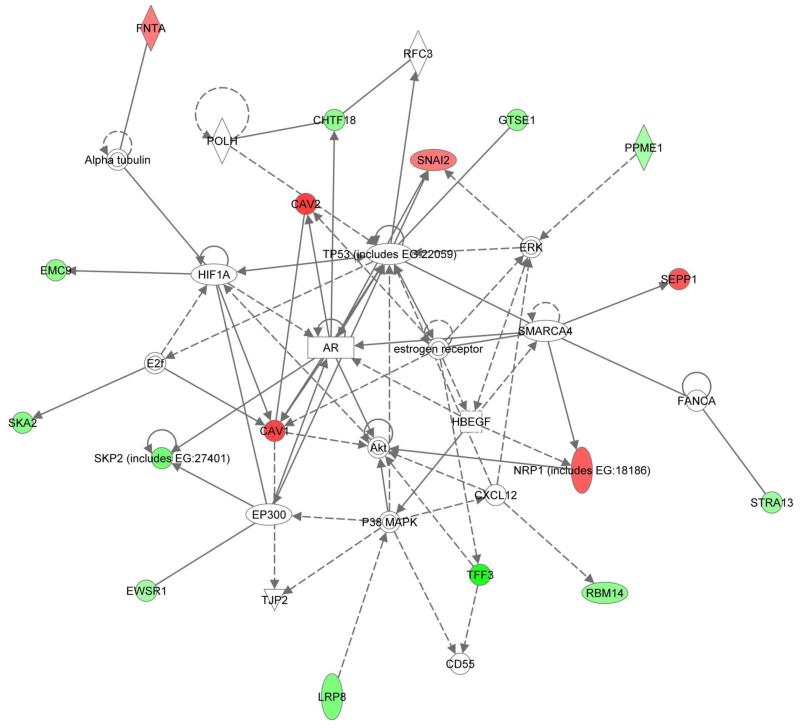
Canonical pathways associated with fulvestrant-specific genes (n=28, 11 up-regulated and 17 down-regulated) included signalling through heterotrimeric G-proteins (β-/γ-subunits) and E-mediated S-phase entry (Table 1). One principal network (IPA score=40) included 17/28 fulvestrant-specific genes with functions related to cell cycle, death, survival, and tumour morphology (Figure 4). Six fulvestrant-specific up-regulated genes were present, with CAV1/2, SNAI2 and NRP1 forming a core group associated directly and/or indirectly with the focal points of ERα, androgen receptor (AR) and TP53. Members of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) family and the serine/threonine protein kinase (AKT) were prominent. Eleven fulvestrant-specific down-regulated genes occupied the network periphery.
Table 1.
Ingenuity based biological interpretation of genes specifically affected by fulvestrant in pre-surgical studies and in vitro models: (a) Network functions and (b) Canonical pathways.
| (a) | ||
|---|---|---|
| Canonical Pathways | p-value | Genes |
| G Beta Gamma Signaling | 9.120E-03 | CAV1,CAV2 |
| Estrogen-mediated S-phase Entry | 3.890E-02 | SKP2 |
| Antiproliferative Role of TOB in T Cell Signaling | 4.169E-02 | SKP2 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 6.761E-02 | SKP2 |
| Semaphorin Signaling in Neurons | 8.128E-02 | NRP1 |
| (b) | |||
|---|---|---|---|
| Molecules in Network | Score | Focus Molecules | Top Functions |
| Akt, Alphatubulin, AR, CAV1, CAV2, CD55, CHTF18, CXCL12, E2f, EMC9, EP300, ERK, estrogen receptor, EWSR1, FANCA, FNTA, GTSE1, HBEGF, HIF1A, LRP8, NRP1 (includes EG:18186), P38 MAPK, POLH, PPME1, RBM14, RFC3, SEPP1, SKA2, SKP2 (includes EG:27401), SMARCA4, SNAI2, STRA13, TFF3, TJP2, TP53 (includes | 40 | 17 | Cell Cycle, Cell Death and Survival, Tumor Morphology |
| RBMS1, SUZ12 | 2 | 1 | Embryonic Development, Cancer, Skeletal and Muscular Disorders |
| CD63, SYTL4 | 2 | 1 | Cell Morphology, Endocrine System Development and Function, Nervous System Development and Function |
| FBL, NOP56 | 2 | 1 | RNA Post-Transcriptional Modification, Hereditary Disorder, Neurological Disease |
| AGTR1, NSUN2, PMS2 | 2 | 1 | Cancer, Gastrointestinal Disease, Hereditary Disorder |
Figure 4.
Network analysis of genes specifically affected by fulvestrant. Up- and down-regulated transcripts are indicated in red and green respectively. Direct and indirect relationships are indicated by solid and interrupted lines respectively.
To determine if this functional alliance of fulvestrant-specific genes was specifically induced by, and attributable to, congruent response to fulvestrant, their pre-treatment expression was assessed. Fulvestrant-related genes correlated significantly with one another at baseline; up-regulated genes were co-expressed, as were down-regulated genes, and these two groups were already inversely correlated, implicating a pre-existing regulatory system. Fulvestrant down-regulated genes correlated directly, whereas those up-regulated correlated inversely, with baseline AURKA and ESR1 (summarised in Supplementary Figure S2 and detailed in Supplementary Table S11).
Treatment-related gene alterations and anti-proliferative response
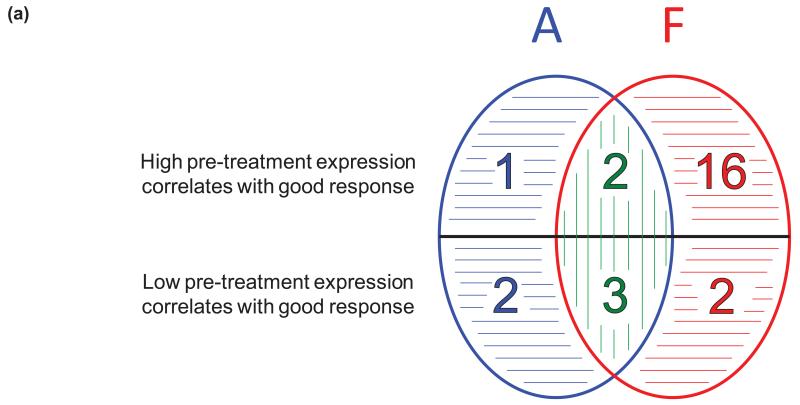
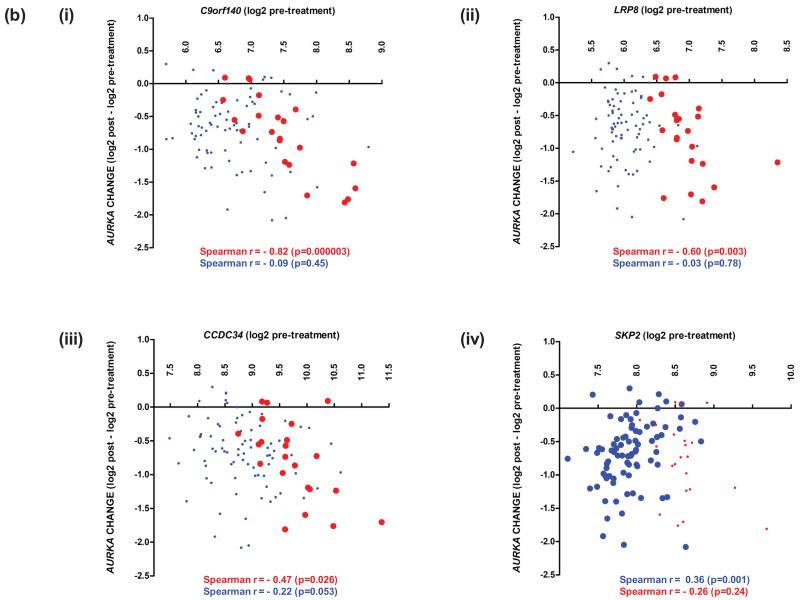
To determine if baseline expression of treatment-related genes (n=46) was associated with anti-proliferative response, their pre-treatment levels were correlated with change in AURKA expression. Twenty-six genes correlated with response to anastrozole or high-dose fulvestrant (uncorrected p<0.05), summarised in Figure 5a and detailed in Supplementary Table S12. Twenty-three fulvestrant-related genes correlated with response to high-dose fulvestrant, with 5/23 (C9orf140, POLD2, SAC3D1, ZMYND19 and STRA13) also doing so in the low-dose treated cohort (Supplementary Table S12c). Notably, 18/23 were not associated with response to anastrozole. Fulvestrant response was specifically associated with low pre-treatment expression of two genes (SNAI2 and NRP1) found to be up-regulated by fulvestrant, and high expression of 16 genes found to be down-regulated by fulvestrant; of these C9orf140, LRP8 and CCDC34 had predictive significance independent of pre-treatment AURKA expression (p<0.05), Figure 5b (i)-(iii).
Figure 5.
(a) Summary of treatment-related genes for which pre-treatment expression correlates significantly with treatment-induced changes in expression of the proliferation-associated gene AURKA. Anastrozole (A, blue), fulvestrant (F, red), or both (green), with agent-specific and quantitative differences indicated by horizontal and vertical stripes respectively. (b) Scattergrams illustrating the correlation between change in AURKA expression and three fulvestrant-specific genes (i)-(iii), and one E-deprivation-specific gene (iv), with predictive significance (p<0.05) independent of pre-treatment AURKA expression in NEWEST (red) and FAIMoS (blue).
Pre-treatment expression of five genes had predictive significance for both agents and their correlation was invariably stronger with response to fulvestrant. Anastrozole response was specifically correlated with three genes, of which KCNK15 was E-deprivation-related and SKP2 had predictive significance independent of pre-treatment AURKA expression, Figure 5b (iv). Further evaluation of the independence of the predictive performance of agent-specific genes was not conducted because of limited numbers.
Validation of differentially affected transcripts
qRT-PCR of clinical samples confirmed significant up-regulation of CAV1 (1.87 fold-increase, p=0.0095) and SNAI2 (1.88 fold-increase, p=0.0005) by fulvestrant, whereas changes induced by anastrozole were not significant. The differential impact of the two agents on treatment-related transcripts (n=46) was consistent between the parent and bridging studies (Pearson r=0.93, p<0.0001, slope=0.97). Fulvestrant-related transcripts (n=41) were concordantly affected, albeit to a lesser extent, following low-dose treatment (Pearson r=0.77, p<0.0001, slope=0.23). Genes which were found to respond differently to four weeks of fulvestrant and two weeks of anastrozole, were evaluated in a sub-group of patients receiving 16 weeks of anastrozole therapy. There was no significant change in any of the 11 transcripts specifically up-regulated by fulvestrant, or 11 of the 17 transcripts specifically down-regulated (Supplementary Table S13). This argues against the possibility that duration of treatment may have biased the identification of alterations in gene expression in favour of fulvestrant.
ESR1 knockdown in MCF7 cells
Knockdown of ESR1 invariably down-regulated the expression of genes which were found to be differentially down-regulated by fulvestrant (e.g. LRP8 and GTSE1) and up-regulated the expression of some genes which were differentially up-regulated by fulvestrant (e.g. SNAI2 and SEPP1), but not others (e.g. CAV1 and RBMS1). The majority of genes responded concordantly to fulvestrant and ESR1 knockdown (Supplementary Figure S3).
DISCUSSION
This study compared, for the first time, transcriptional profiles from breast cancer in situ following fulvestrant or anastrozole and corresponding in vitro models. The robust integrative strategy avoids the identification of spurious genes in clinical samples which may reflect increasing proportions of stroma with treatment response (33). The approach taken may discard alterations inadequately modelled in vitro, including those dependent upon three-dimensional structure, stromal interactions and hypoxia, but focuses on those genes which may be subjected to functional interrogation in model systems. The hallmark molecular responses to interrupted oestrogenic signalling, including suppression of ERGs and proliferative markers, followed both fulvestrant and E-deprivation. However, distinguishing features were apparent. Firstly, the overall transcriptional response to fulvestrant was of greater magnitude. Secondly, differences were not distributed uniformly across the transcriptome, but were most marked in a relatively limited cohort of genes.
A small number of differentially affected genes were specific to E-deprivation, potentially attributable to ERα-independent E-activity (34). Most were fulvestrant-specific and remained unaffected by extended anastrozole treatment, raising the possibility of regulation by unliganded ERα. Both fulvestrant and E-deprivation abrogate E-dependent ERα activity, but only fulvestrant, by virtue of ERα depletion, antagonises E-independent ERα activity, including cross-talk with growth factor pathways (26). The greater and differential transcriptional response to fulvestrant may be attributable to the arrest of E-independent ERα activity that is unaffected by E-deprivation (27). This greater anti-oestrogenic effect is consistent with the greater efficacy of fulvestrant in studies comparing the agents as first-line therapy in advanced disease (16) and in its sequential utility after AI relapse (9, 10). Incompletely overlapping transcriptional responses are also consistent with the reported efficacy of combination therapy with fulvestrant and anastrozole (35).
Fulvestrant-specific genes associated in networks with ERα, AR and TP53. ERα activity can be influenced by cross-talk with AR signalling (36), which may exert anti-oestrogenic/anti-proliferative effects in ERα-positive breast cancer, while having contrasting roles in ERα-negative tumours (37). The discovery of a subset of DNA binding elements and pioneer factors common to both receptors (38) raises the possibility that activity of AR-dependent networks may be influenced by fulvestrant-induced loss of the ERα transcriptional program. Transcripts specifically up-regulated by ERα-depletion, including CAV1/2 and SNAI2, are associated with AR signalling and the biology of ERα-negative and basal-like breast cancer (39-42). CAV1 encodes caveolin-1, the principal constituent of specialised membrane invaginations called caveolae. Caveolin-1/-2 are widely expressed and may co-localise. Caveolae have diverse functions, including: vesicular trafficking, lipid homeostasis, sub-cellular partitioning and integrating the activity of signalling molecules. Caveolin-1 may facilitate non-genomic/extra-nuclear and ligand-independent ERα activity (42, 43). SNAI2 encodes SLUG, a transcription factor implicated in breast cancer progression, nodal involvement and metastasis (44, 45). Expression is associated with epithelial-mesenchymal-transition, E-cadherin down-regulation and stem cell-associated gene expression (46, 47). NRP1, encoding neuropilin-1, a co-receptor for semaphorins and vascular endothelial growth factor (VEGF), also has links with stem cell phenotype (48) and poor prognosis (49).
Pre-treatment expression of fulvestrant-related genes may be influenced by, and reflect, inherent tumour proliferation and/or oestrogenicity. The possible agent-specific predictive utility of the differentially affected genes identified warrants further study. This may indicate whether such genes contribute mechanistically to the action of the two agents, are inconsequentially associated with treatment, and/or have external validity as predictive biomarkers. The impact of ESR1 knockdown on the expression of genes differentially affected by fulvestrant supports ERα destabilisation as the mechanism underpinning particular fulvestrant-induced alterations. The identification of such genes may facilitate the comparative pharmacology of SERDs in development. This study also provides the first comparison of transcriptional responses to fulvestrant dosing regimens, and supports the efficacy of low-dose treatment (8-10). The greater transcriptional impact of high-dose treatment is consistent with pharmacokinetic models predicting five-fold greater plasma concentrations on day 28 (50), and the increased efficacy observed in the NEWEST (17) and CONFIRM trials (13, 14).
Limitations of this study include in vitro modelling using a single cell line, which is also PIK3CA mutated, and expression profiling across different BeadChip versions which reduced the number of comparable probes. Potential confounding factors also include better patient compliance with treatment regimens in favour of fulvestrant given its mode of administration (although good E-suppression was found in the AI treated patients – data not shown) and duration of treatment; extended anastrozole treatment may have induced further changes in gene expression. Non-randomised comparisons may also be influenced by selection-bias, with differences in baseline patient and tumour characteristics between pre-surgical studies with incompletely overlapping entry criteria, and differences such as details in sample taking, storage and ethnicity of the populations. Sample sizes may have also restricted the statistical power to identify treatment-induced changes.
In conclusion, the molecular response to fulvestrant has much in common with E-deprivation, but is stronger with distinctions potentially attributable to arrest of E-independent ERα activity and involvement of AR signalling. Genes responding differently to fulvestrant may have specific predictive utility. These data are consistent with the efficacy of first-line fulvestrant versus anastrozole in advanced disease, combination therapy in the metastatic setting, sequential utility after AI relapse, and higher dosing regimens.
Supplementary Material
Statement of translational relevance.
Aromatase inhibitors (AIs) are first-line post-menopausal agents for oestrogen receptor alpha (ERα)-positive breast cancer. However, there is considerable response heterogeneity and women frequently relapse. Oestrogen (E)-deprivation does not completely arrest ERα-activity and transactivation of the unliganded receptor may continue through cross-talk with growth factor pathways. In contrast to AIs, the selective ER down-regulator fulvestrant also abrogates ligand-independent ERα-activity. The benefit of fulvestrant as an alternative, combination or sequential therapy to AIs has been reported, but molecular mechanisms underpinning its relative efficacy remain unclear and biomarkers for patient selection are lacking. This study demonstrates, for the first time, that the overall transcriptional response to fulvestrant is of greater magnitude than E-deprivation, consistent with its clinical efficacy and more complete blockade of oestrogenic signalling. Using a robust integrative approach, we identify a subset of genes differentially affected by fulvestrant which comprise distinct biological networks, correlate with anti-proliferative response and have potential utility as predictive biomarkers for fulvestrant.
ACKNOWLEDGEMENTS
Dr Scott Brouilette and Dr Jörg Mages provided technical support and assistance with Partek® Genomics Suite™.
Financial support: This work was funded by the Mary-Jean Mitchell Green Foundation and AstraZeneca. Prof. Mitch Dowsett and Dr Lesley-Ann Martin are supported by Breakthrough Breast Cancer and NHS funding to the Royal Marsden NIHR Biomedical Research Centre. Mr Neill Patani is funded by a Medical Research Council Clinical Research Training Fellowship.
Footnotes
Conflict of interest: The authors declare no competing interests that could be perceived as prejudicing the impartiality of this study.
REFERENCES
- 1.Parker MG. Action of “pure” antiestrogens in inhibiting estrogen receptor action. Breast Cancer Res Treat. 1993;26:131–7. doi: 10.1007/BF00689686. [DOI] [PubMed] [Google Scholar]
- 2.Dauvois S, White R, Parker MG. The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J Cell Sci. 1993;106(Pt 4):1377–88. doi: 10.1242/jcs.106.4.1377. [DOI] [PubMed] [Google Scholar]
- 3.Fawell SE, White R, Hoare S, Sydenham M, Page M, Parker MG. Inhibition of estrogen receptor-DNA binding by the “pure” antiestrogen ICI 164,384 appears to be mediated by impaired receptor dimerization. Proc Natl Acad Sci U S A. 1990;87:6883–7. doi: 10.1073/pnas.87.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson JFRGE, Cheung KL, Pinder S, Ellis IO, Wakeling A. Clinical efficacy of fulvestrant and effects on estrogen receptor levels during first-line endocrine treatment of patients with advanced breast cancer. Breast Cancer Res Treat. 2004;88:S236–S7. [Google Scholar]
- 5.Wakeling AE, Dukes M, Bowler J. A potent specific pure antiestrogen with clinical potential. Cancer Res. 1991;51:3867–73. [PubMed] [Google Scholar]
- 6.Robertson JF, Osborne CK, Howell A, Jones SE, Mauriac L, Ellis M, et al. Fulvestrant versus anastrozole for the treatment of advanced breast carcinoma in postmenopausal women: a prospective combined analysis of two multicenter trials. Cancer. 2003;98:229–38. doi: 10.1002/cncr.11468. [DOI] [PubMed] [Google Scholar]
- 7.Osborne CK, Pippen J, Jones SE, Parker LM, Ellis M, Come S, et al. Double-blind, randomized trial comparing the efficacy and tolerability of fulvestrant versus anastrozole in postmenopausal women with advanced breast cancer progressing on prior endocrine therapy: results of a North American trial. J Clin Oncol. 2002;20:3386–95. doi: 10.1200/JCO.2002.10.058. [DOI] [PubMed] [Google Scholar]
- 8.Howell A, Robertson JF, Quaresma Albano J, Aschermannova A, Mauriac L, Kleeberg UR, et al. Fulvestrant, formerly ICI 182,780, is as effective as anastrozole in postmenopausal women with advanced breast cancer progressing after prior endocrine treatment. J Clin Oncol. 2002;20:3396–403. doi: 10.1200/JCO.2002.10.057. [DOI] [PubMed] [Google Scholar]
- 9.Ingle JN, Suman VJ, Rowland KM, Mirchandani D, Bernath AM, Camoriano JK, et al. Fulvestrant in women with advanced breast cancer after progression on prior aromatase inhibitor therapy: North Central Cancer Treatment Group Trial N0032. J Clin Oncol. 2006;24:1052–6. doi: 10.1200/JCO.2005.04.1053. [DOI] [PubMed] [Google Scholar]
- 10.Perey L, Paridaens R, Hawle H, Zaman K, Nole F, Wildiers H, et al. Clinical benefit of fulvestrant in postmenopausal women with advanced breast cancer and primary or acquired resistance to aromatase inhibitors: final results of phase II Swiss Group for Clinical Cancer Research Trial (SAKK 21/00) Ann Oncol. 2007;18:64–9. doi: 10.1093/annonc/mdl341. [DOI] [PubMed] [Google Scholar]
- 11.Chia S, Gradishar W, Mauriac L, Bines J, Amant F, Federico M, et al. Double-blind, randomized placebo controlled trial of fulvestrant compared with exemestane after prior nonsteroidal aromatase inhibitor therapy in postmenopausal women with hormone receptor-positive, advanced breast cancer: results from EFECT. J Clin Oncol. 2008;26:1664–70. doi: 10.1200/JCO.2007.13.5822. [DOI] [PubMed] [Google Scholar]
- 12.Howell A, Robertson JF, Abram P, Lichinitser MR, Elledge R, Bajetta E, et al. Comparison of fulvestrant versus tamoxifen for the treatment of advanced breast cancer in postmenopausal women previously untreated with endocrine therapy: a multinational, double-blind, randomized trial. J Clin Oncol. 2004;22:1605–13. doi: 10.1200/JCO.2004.02.112. [DOI] [PubMed] [Google Scholar]
- 13.Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R, et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594–600. doi: 10.1200/JCO.2010.28.8415. [DOI] [PubMed] [Google Scholar]
- 14.Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R, et al. Final analysis of overall survival for the Phase II CONFIRM trial: fulvestrant 500 mg versus 250 mg. Cancer Res. 2012;72:90s. [Google Scholar]
- 15.Robertson JF, Llombart-Cussac A, Rolski J, Feltl D, Dewar J, Macpherson E, et al. Activity of fulvestrant 500 mg versus anastrozole 1 mg as first-line treatment for advanced breast cancer: results from the FIRST study. J Clin Oncol. 2009;27:4530–5. doi: 10.1200/JCO.2008.21.1136. [DOI] [PubMed] [Google Scholar]
- 16.Robertson JF, Lindemann JP, Llombart-Cussac A, Rolski J, Feltl D, Dewar J, et al. Fulvestrant 500 mg versus anastrozole 1 mg for the first-line treatment of advanced breast cancer: follow-up analysis from the randomized ‘FIRST’ study. Breast Cancer Res Treat. 2012;136:503–11. doi: 10.1007/s10549-012-2192-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuter I, Gee JM, Hegg R, Singer CF, Badwe RA, Lowe ES, et al. Dose-dependent change in biomarkers during neoadjuvant endocrine therapy with fulvestrant: results from NEWEST, a randomized Phase II study. Breast Cancer Res Treat. 2012;133:237–46. doi: 10.1007/s10549-011-1947-7. [DOI] [PubMed] [Google Scholar]
- 18.Robertson JF, Nicholson RI, Bundred NJ, Anderson E, Rayter Z, Dowsett M, et al. Comparison of the short-term biological effects of 7alpha-[9-(4,4,5,5,5-pentafluoropentylsulfinyl)-nonyl]estra-1,3,5, (10)-triene-3,17beta-diol (Faslodex) versus tamoxifen in postmenopausal women with primary breast cancer. Cancer Res. 2001;61:6739–46. [PubMed] [Google Scholar]
- 19.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 20.Manning DL, Nicholson RI. Isolation of pMGT1: a gene that is repressed by oestrogen and increased by antioestrogens and antiprogestins. Eur J Cancer. 1993;29A:759–62. doi: 10.1016/s0959-8049(05)80362-4. [DOI] [PubMed] [Google Scholar]
- 21.Thomas P, Pang Y, Filardo EJ, Dong J. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology. 2005;146:624–32. doi: 10.1210/en.2004-1064. [DOI] [PubMed] [Google Scholar]
- 22.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–30. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 23.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–4. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 24.Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–24. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 25.Pietras RJ, Arboleda J, Reese DM, Wongvipat N, Pegram MD, Ramos L, et al. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10:2435–46. [PubMed] [Google Scholar]
- 26.Nicholson RIGJ, Francis AB, Manning DL, Wakeling AE, Katzenellenbogen BS. Observations arising from the use of pure antioestrogens on oestrogen-responsive (MCF-7) and oestrogen growth-independent (K3) human breast cancer cells. Endocr Relat Cancer. 1995:115–21. [Google Scholar]
- 27.Miller TW, Balko JM, Fox EM, Ghazoui Z, Dunbier AK, Anderson H, et al. ERα-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discovery. 2011;1:338–51. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith IE, Walsh G, Skene A, Llombart A, Mayordomo JI, Detre S, et al. A phase II placebo-controlled trial of neoadjuvant anastrozole alone or with gefitinib in early breast cancer. J Clin Oncol. 2007;25:3816–22. doi: 10.1200/JCO.2006.09.6578. [DOI] [PubMed] [Google Scholar]
- 29.Dunbier AK, Ghazoui Z, Anderson H, Salter J, Nerurkar A, Osin P, et al. Molecular profiling of aromatase inhibitor-treated post-menopausal breast tumors identifies immune-related correlates of resistance. Clin Cancer Res. 2013;19:2775–86. doi: 10.1158/1078-0432.CCR-12-1000. [DOI] [PubMed] [Google Scholar]
- 30. www.ebi.ac.uk/arrayexpress. (E-MTAB-887)
- 31. www.synapse.org. (Dataset:16243)
- 32. www.rock.icr.ac.uk/collaborations/Patani.
- 33.Cleator SJ, Powles TJ, Dexter T, Fulford L, Mackay A, Smith IE, et al. The effect of the stromal component of breast tumours on prediction of clinical outcome using gene expression microarray analysis. Breast Cancer Res. 2006;8:R32. doi: 10.1186/bcr1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maggiolini M, Picard D. The unfolding stories of GPR30, a new membrane-bound estrogen receptor. J Endocrinol. 2010;204:105–14. doi: 10.1677/JOE-09-0242. [DOI] [PubMed] [Google Scholar]
- 35.Mehta RS, Barlow WE, Albain KS, Vandenberg TA, Dakhil SR, Tirumali NR, et al. Combination anastrozole and fulvestrant in metastatic breast cancer. N Engl J Med. 2012;367:435–44. doi: 10.1056/NEJMoa1201622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Need EF, Selth LA, Harris TJ, Birrell SN, Tilley WD, Buchanan G. Research resource: interplay between the genomic and transcriptional networks of androgen receptor and estrogen receptor alpha in luminal breast cancer cells. Mol Endocrinol. 2012;26:1941–52. doi: 10.1210/me.2011-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 2012;26:1252–67. doi: 10.1210/me.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Research. 2009;69:6131–40. doi: 10.1158/0008-5472.CAN-09-0452. [DOI] [PubMed] [Google Scholar]
- 39.Bennett N, Hooper JD, Lee CS, Gobe GC. Androgen receptor and caveolin-1 in prostate cancer. IUBMB Life. 2009;61:961–70. doi: 10.1002/iub.244. [DOI] [PubMed] [Google Scholar]
- 40.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109:16654–9. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu K, Gore C, Yang L, Fazli L, Gleave M, Pong RC, et al. Slug, a unique androgen-regulated transcription factor, coordinates androgen receptor to facilitate castration resistance in prostate cancer. Mol Endocrinol. 2012;26:1496–507. doi: 10.1210/me.2011-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patani N, Martin LA, Reis-Filho JS, Dowsett M. The role of caveolin-1 in human breast cancer. Breast Cancer Res Treat. 2012;131:1–15. doi: 10.1007/s10549-011-1751-4. [DOI] [PubMed] [Google Scholar]
- 43.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007;282:22278–88. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- 44.Come C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF, Theillet C, et al. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- 45.Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, et al. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Res. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol. 2008;214:25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- 47.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glinka Y, Mohammed N, Subramaniam V, Jothy S, Prud’homme GJ. Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-kappaB activation and tumor sphere formation. Biochem Biophys Res Commun. 2012;425:775–80. doi: 10.1016/j.bbrc.2012.07.151. [DOI] [PubMed] [Google Scholar]
- 49.Xin Y, Li J, Wu J, Kinard R, Weekes CD, Patnaik A, et al. Pharmacokinetic and pharmacodynamic analysis of circulating biomarkers of anti-NRP1, a novel antiangiogenesis agent, in two phase I trials in patients with advanced solid tumors. Clin Cancer Res. 2012;18:6040–8. doi: 10.1158/1078-0432.CCR-12-1652. [DOI] [PubMed] [Google Scholar]
- 50.Robertson JF. Fulvestrant (Faslodex) -- how to make a good drug better. Oncologist. 2007;12:774–84. doi: 10.1634/theoncologist.12-7-774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.