Abstract
Many studies have identified metabolic pathways that underlie cellular transformation, but the metabolic drivers of cancer progression remain less well understood. The Hippo transducer pathway has been shown to confer malignant traits on breast cancer cells. In this study, we used metabolic mapping platforms to identify biochemical drivers of cellular transformation and malignant progression driven through RAS and the Hippo pathway in breast cancer, and identified platelet activating factor acetylhydrolase 1B3 (PAFAH1B3) as a key metabolic driver of breast cancer pathogenicity that is upregulated in primary human breast tumors and correlated with poor prognosis. Metabolomic profiling suggests that PAFAH1B3 inactivation attenuates cancer pathogenicity through enhancing tumor-suppressing signaling lipids. Our studies provide a map of altered metabolism that underlies breast cancer progression and put forth PAFAH1B3 as a critical metabolic node in breast cancer.
Keywords: cancer metabolism, PAFAH1B3, TAZ, Hippo, activity-based protein profiling, metabolomics, lipids
Introduction
Cancer cells possess fundamentally dysregulated metabolism that underlies their pathogenic features (Benjamin et al., 2012; Vander Heiden et al., 2009). Targeting altered metabolism in cancer cells has historically served as a validated strategy for treating cancer patients. These therapies include chemotherapeutics such as 5-fluorouracil, methotrexate, and anastrozole that target nucleotide, folate, and estrogen metabolism, respectively (Nov. 2013). More recent discoveries have focused on targeting other aspects of dysregulated cancer metabolism including glycolysis, TCA cycle, and lipogenesis with pyruvate kinase M2 (PKM2) activators, isocitrate dehydrogenase (IDH) inhibitors, and fatty acid synthase (FASN) inhibitors, respectively (Benjamin et al., 2012; Menendez and Lupu, 2007; Vander Heiden et al., 2009). While targeting dysregulated metabolism is a promising strategy for cancer treatment, currently available chemotherapies, including those that target certain aspects of cancer metabolism, fail to eradicate the most aggressive cancers (Chambers et al., 2002).
Studies over the past decade have uncovered a subpopulation of cancer cells termed cancer stem/precursor cells (CSCs) that, like embryonic stem cells, can undergo a process of epithelial-to-mesenchymal transition (EMT), which is associated with self-renewing and tumor initiating capabilities, poor prognosis, and chemotherapy-resistance within breast tumors (Polyak and Weinberg, 2009). Recent studies have identified TAZ, a transducer of the Hippo pathway, as a critical factor that is elevated in poorly differentiated human breast tumors, is correlated with poor prognosis, promotes EMT and is stabilized by EMT, and is required to sustain self-renewal and tumor-initiation capacities of breast CSCs (Cordenonsi et al., 2011; Hong and Guan, 2012). Identifying altered metabolic pathways that underlie both cellular transformation and malignant progression through TAZ may thus yield novel therapeutic strategies for combatting malignant breast cancers.
While targeting dysregulated metabolism is a promising therapeutic strategy for cancer treatment, nearly all research in cancer metabolism has focused on well-understood metabolic pathways in central carbon metabolism and has largely ignored the majority of cellular metabolic pathways, despite genetic or metabolic evidence of their involvement in cancer. This is in large part because our incomplete understanding of metabolic networks in normal physiology, let alone in dysregulated or rewired biochemical networks in human cancer cells (Benjamin et al., 2012).
In this study, we used proteomic, functional proteomic, and metabolomic platforms to more broadly identify commonly altered metabolic pathways that underlie both mammary epithelial cell transformation by the oncogene HRAS as well as malignant progression, EMT, and cancer stem-cell like features through the constitutive activation of TAZ in the MCF10A non-transformed mammary epithelial cell line background. Through these profiling efforts, we have identified platelet activating factor acetylhydrolase 1B3 (PAFAH1B3) as a critical enzyme in maintaining breast cancer cell aggressiveness and tumor growth through regulating tumor-suppressing lipids.
Results and Discussion
Breast Cancer Progression Model
In this study, we used integrated metabolic mapping platforms to identify enzymes and metabolites that are dysregulated in a MCF10A breast cancer progression model, a series of isogenically derived human mammary cell lines consisting of non-transformed MCF10A cells, HRAS-transformed MCF10A-T1k cells (premalignant, also known as MII) and fully malignant MCF10A-CA1a (MIV) cells, derived in vivo from spontaneous progression of MII cells (Santner et al., 2001) (Fig. S1). With in vivo orthotopic models, MII cells generate low-grade tumors in approximately 25% of xenografts, while the MIV lines form high-grade tumors, resembling grade III human breast tumors, at a much higher frequency. This well-characterized progression model displays many important features of breast cancer progression found in highly aggressive metaplastic and claudin-low breast tumor subtypes including EMT, expansion of CSC population and the associated increase in expression of the stem cell-associated CD44+/CD24−/low antigenic profile, self-renewal capabilities, and resistance to conventional therapies (Chaffer and Weinberg, 2011; Gupta et al., 2009). In particular, Cordenonsi et al. recently reported that MIV cells show a significantly higher self-renewal ability, tumorigenic potential, and an increased CSC population than MII cells, resembling the difference between grade III and grade I human breast tumors (Cordenonsi et al., 2011). By analyzing a large human patient dataset, they identified TAZ as a key signature that is over-represented in poorly differentiated high-grade tumors and correlates with increased CSC, metastasis, and reduced survival. TAZ, a transducer of the Hippo signaling pathway that mediates cell-cell contact and polarity signals to control cell proliferation and organ size (Chan et al., 2011), is also expressed at higher levels in MIV cells than MII cells and is required to sustain self-renewal and tumor-initiation capacities in breast CSCs. Consistent with previous reports, we show that expression of a constitutively active TAZ, TAZ S89A, in MCF10A or MII cells results in increased EMT, colony formation in soft-agar, and cellular migration (Cordenonsi et al., 2011) (Fig. S1).
Identifying Dysregulated Metabolic Pathways Underlying Cellular Transformation and Malignant Progression
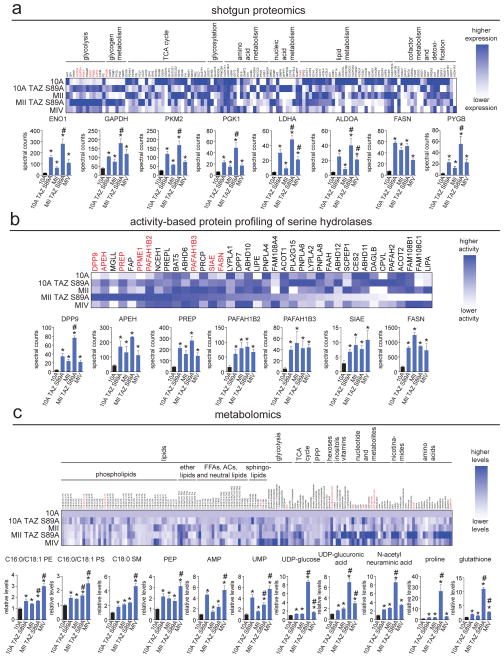
Our goal was to employ multiple metabolic mapping platforms to broadly identify dysregulated metabolic pathways that underlie cellular transformation and malignant progression using the aforementioned breast cancer model. We performed shotgun proteomic analysis, activity-based protein profiling (ABPP) using the serine hydrolase-directed activity based probe, and targeted single reaction monitoring (SRM) liquid chromatography/mass spectrometry (LC/MS)-based metabolomic analyses to identify commonly altered changes in protein expression of metabolic enzymes, activities of serine hydrolases, and metabolite levels, respectively, that may underlie cellular transformation and TAZ-mediated malignant progression. While shotgun proteomic profiling provides broad coverage of alterations in protein expression, ABPP uses active-site directed chemical probes to identify dysregulated activities of large numbers of enzymes (Nomura et al., 2010a). We chose to profile the serine hydrolase superfamily for this study since this enzyme class is one of the largest metabolic enzyme classes in the human genome with a broad range of functions including esterase, lipase, hydrolase, deacetylase, thioesterase, protease, and peptidase activities and many serine hydrolases have been shown to be important in cancer (Long and Cravatt, 2011). Through these profiling efforts, we identified several enzymes and lipids that were either specifically upregulated by constitutive activation of TAZ or commonly upregulated in 10A TAZ S89A, MII, MII TAZ S89A, and MIV cells (Fig. 1a–c; Fig. S1; Table S1). The dysregulated enzymes identified through shotgun proteomics include glycolytic enzymes (enolase 1 (ENO1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), pyruvate kinase MII (PKM2), phosphoglycerate kinase (PGK1), lactate dehydrogenase A (LDHA), and aldolase A (ALDOA)), the de novo lipogenesis enzyme fatty acid synthase (FASN), and the glycogen metabolizing enzyme glycogen phosphorylase B (PYGB) (Fig. 1a; Fig. S1; Table S1). ABPP of serine hydrolases also revealed FASN upregulation, in addition to peptidases (dipeptidylpeptidase 9 (DPP9), acylpeptide hydrolase (APEH), prolyl endopeptidase (PREP)), lipases (platelet activating factor acetylhydrolase 1B2 (PAFAH1B2) and PAFAH1B3), and sialic acid acetylesterase (SIAE) (Fig. 1b; Table S1). Metabolomic analysis yielded several metabolites that were commonly heightened across the four cell lines, including lipids (phosphatidyl ethanolamine (PE), phosphatidyl serine (PS), sphingomyelin (SM)), the glycolytic intermediate phosphoenolpyruvate (PEP), nucleotides (adenosine monophosphate (AMP), uridine monophosphate (UMP)), uridine diphosphate -conjugated sugars (UDP-glucose, UDP-glucuronic acid), the sialic acid N-acetylneuraminic acid, the amino acid proline, and the antioxidant glutathione (Fig. 1c; Table S1). Certain enzymes such as monoacylglycerol lipase (MGLL), the serine protease fibroblast activation protein (FAP), and protein methyl esterase 1 (PPME1) were upregulated specifically by constitutive activation of TAZ (Fig. S1; Table S1). Anti-oncogenic enzymes such as succinate dehydrogenases (SDHA), as well as several other citric acid cycle enzymes were specifically down-regulated upon induction with TAZ S89A (Fig. S1; Table S1).
Figure 1. Identifying dysregulated metabolic enzymes in breast cancer progression model.
(a) Shotgun proteomic profiling of metabolic enzyme expression in the breast cancer progression model by MudPIT. Upper heat map shows relative protein expression of each protein, normalized to the highest expression of each protein across the 5 cell lines. Dark blue corresponds to high expression and white or light blue corresponds to lower expression. Lower bar graphs show the metabolic enzymes that were significantly upregulated across 10A TAZ S89A, MII, MII TAZ S89A, and MIV cells. (b) ABPP of serine hydrolase activities in the breast cancer progression model by ABPP-MudPIT using the fluorophosphonate-biotin serine hydrolase activity-based probe. Upper heat map shows relative serine hydrolase activities of each protein, normalized to the highest expression of each protein across the 5 cell lines. Dark blue corresponds to high activity and white or light blue corresponds to lower activity. Lower bar graphs show the serine hydrolase activities that were significantly upregulated across 10A TAZ S89A, MII, MII TAZ S89A, and MIV cells. (c) Metabolomic profiling of the breast cancer progression model using SRM-based targeted LC/MS/MS. Upper heat map shows relative metabolite activities normalized to MCF10A control cells. Dark blue corresponds to high levels and white corresponds to lower levels. Lower bar graphs show the metabolites that were significantly elevated in levels across 10A TAZ S89A, MII, MII TAZ S89A, and MIV cells. Data in bar graphs are presented as mean ± sem; n = 4–6/group. Significance is presented as *p < 0.05 compared to MCF10A control cells, #p<0.05 compared to MII cells. Detailed data is shown in Table S1. Characterization of breast cancer progression model, TAZ S89A-specific changes in metabolic enzymes, additional data showing regulation of these metabolic enzyme targets by the Hippo pathway, and additional data showing regulation of glycolytic metabolism by RAS and TAZ S89A are shown in Figure S1.
Overall, our data indicate that cellular transformation and malignant progression heighten glycolysis and glycogen metabolism, de novo lipogenesis, nucleic acid metabolism, sialic acid metabolism, oxidative stress, and specific branches of lipid metabolism. Many of these pathways, including glycolytic and glycogen metabolism, nucleic acid metabolism, sialic acid metabolism, oxidative stress, and de novo lipogenesis have been previously shown to be critical pathways that underlie cancer metabolism and pathogenicity. Nonetheless, we show that both HRAS transformation as well as constitutive activation of TAZ induces glycolytic, glycogen, and sialic acid metabolism and that TAZ and HRAS act synergistically in many cases (e.g. ENO1, GAPDH, PKM2, LDHA, ALDOA, PGK1, PYGB, UDP-glucose, N-acetylneuraminic acid) to enhance these metabolic pathways (Fig. S1). Additionally, we confirmed heightened glycolytic metabolism in these lines by measuring glucose consumption and lactic acid secretion and found that both are heightened upon transduction with TAZ S89A or HRAS and further heightened upon transduction with both TAZ S89A and HRAS. The highly malignant MIV cells also show significantly heightened glycolytic metabolism beyond that of MII cells (Fig. S1). Interestingly, the enzymes specifically upregulated by TAZ, such as MGLL, FAP, and PPME1, or downregulated by TAZ, such as SDHA, have all been implicated as critical drivers of cancer progression (Bachovchin et al., 2011; Kelly, 2005; Nomura et al., 2011; Nomura et al., 2010b; Puustinen et al., 2009; Yang et al., 2013).
While our data indicate that many of the aforementioned metabolic enzymes are upregulated upon induction with TAZ S89A, we wanted to determine whether these enzymes are regulated by TAZ in aggressive MIV cells. Upon knocking down TAZ in MIV cells, we observed only modest reductions in the expression of the presumably TAZ-regulated targets, including PAFAH1B2, ENO1, ALDOA, DPP9, and NANS (Fig. S1). Since both YAP and TAZ may regulate overlapping transcriptional targets, we proceeded to knockdown both YAP and TAZ in MIV cells. Interestingly, YAP and TAZ dual knockdown led to more robust reductions in the expression of most TAZ-regulated targets, including PAFAH1B2, ENO1, ALDOA, PGK1, LDHA, PJM2, DPP9, MGLL, and NANS (Fig. S1). PAFAH1B3 was not altered, even upon TAZ and YAP knockdown (Fig. S1). We interpret this lack of change to perhaps indicate that other oncogenic pathways, such as HRAS, may regulate this enzyme or that PAFAH1B3 may be regulated by post-translational mechanisms.
Screening for Metabolic Enzymes that are required for Breast Cancer Pathogenicity
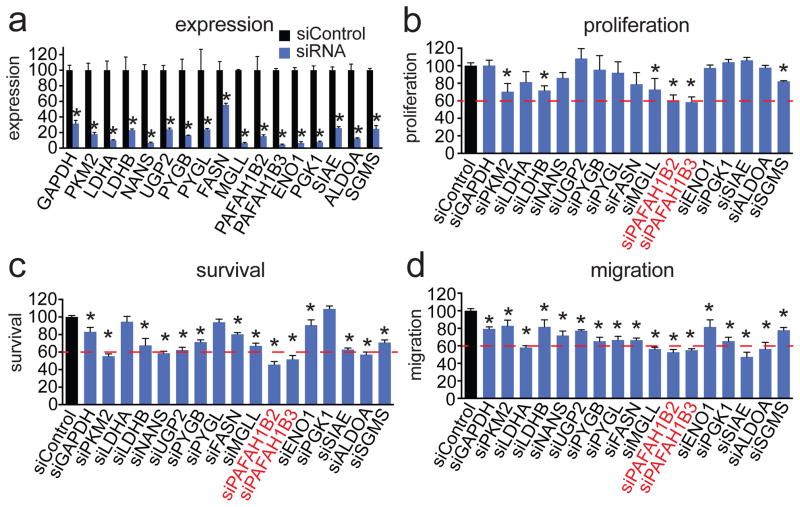
We next sought to identify metabolic enzymes that, when inactivated would impair cancer pathogenicity. We used small interfering RNA (siRNA) to transiently knockdown the expression of the dysregulated metabolic enzymes that we found through our profiling studies and identified those enzymes that impaired cellular proliferation, survival, or motility upon siRNA inactivation in MII TAZ S89A cells (Fig. 2a–d). To filter a large list of dysregulated metabolic enzyme targets, we narrowed our focus on those enzymes found by shotgun proteomics and ABPP and enzymes that control metabolites found by metabolomics that were altered across 10A TAZ S89A, MII, MII TAZS89A, and MIV cells. While inactivation of glycolytic, glycogen, and sialic acid metabolizing enzymes modestly impaired proliferative, survival, or migratory capacity, we were surprised to find that knockdown of PAFAH1B2 and PAFAH1B3 caused the most significant and dramatic impairments in the proliferative, survival, and migratory phenotypes in MII TAZ S89A cells (Fig. 2a–d). Thus, we decided to further investigate the role of PAFAH1B2 and PAFAH1B3 in breast cancer pathogenicity.
Figure 2. Screening for nodal metabolic enzymes in breast cancer.
(a) We transiently knocked down the expression of representative metabolic enzymes in the pathways that we identified as consistently dysregulated in the breast cancer progression model with siRNA in MII TAZ S89A cells. Knockdowns were confirmed by qPCR 48 h after siRNA transfection (b–d). We screened for enzymes that, when inactivated, impaired various aspects of cancer pathogenicity including cellular proliferation (b), serum-free cell survival (c), and cell migration (d) in MII TAZ S89A cells. Cells were transfected with siRNA for 48 h before seeding into phenotypic experiments. Proliferation and cell survival were assessed 48 h after seeding cells by the WST1 cell viability assay. Migration was assessed by counting the number of cells migrated through Transwell chambers over 24 h. Data in bar graphs are presented as mean ± sem; n = 3–5/group normalized to siControl. Significance is presented as *p < 0.05 compared to siControl MII TAZ S89A control cells.
Characterizing the pathological role of PAFAH1B2 and PAFAH1B3 in Breast Cancer
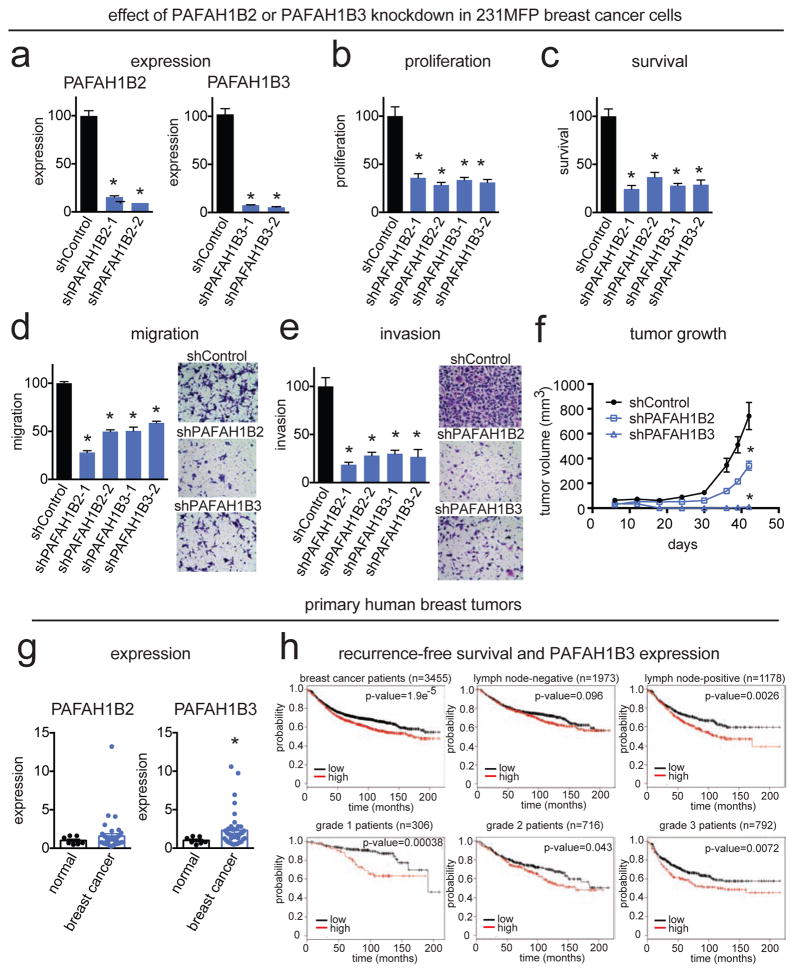
Intrigued by our initial findings in our screen in the MII TAZ S89A cells, we next wanted to ascertain whether PAFAH1B2 and PAFAH1B3 inactivation also impaired breast cancer pathogenicity in a more realistic patient-derived breast cancer cell line. We thus stably inactivated the expression of PAFAH1B2 and PAFAH1B3 in the highly aggressive estrogen receptor/progesterone receptor/HER2 receptor-negative (i.e. triple negative) human breast cells, 231MFP, a line derived from a breast cancer patient adenocarcinoma MDA-MB-231 that has been in vivo passaged in mice to derive a malignant and more aggressive variant (Jessani et al., 2004). Using two independent short hairpin RNA oligonucleotides (shPAFAH1B2-1, shPAFAH1B2-2, shPAFAH1B3-1, shPAFAH1B3-2), PAFAH1B2 and PAFAH1B3 were knocked down by >80 % as measured by both qPCR and ABPP-MudPIT (Fig. 3a; Fig. S2). Knockdown of PAFAH1B2 or PAFAH1B3 significantly impaired 231MFP cell proliferation, survival, migration, and invasiveness in culture, and slowed or eliminated in vivo tumor growth in immune-deficient mice (Fig. 3b–f; Fig. S2). Thus, our results suggest that PAFAH1B enzymes are important in maintaining aggressive features of cancer.
Figure 3. PAFAH1B2 and PAFAH1B3 are critical metabolic enzymes in maintaining breast cancer pathogenicity.
(a) We stably knocked down the expression of PAFAH1B2 and PAFAH1B3 in the triple negative aggressive 231MFP breast cancer cells with two independent shRNA oligonucleotides by >80 % as confirmed by qPCR. (b–e) PAFAH1B2 and PAFAH1B3 knockdown impairs proliferation (b), serum-free cell survival (c), migration (d), and invasion (e). (f) PAFAH1B2 and PAFAH1B3 knockdown also impairs tumor xenograft growth in immune-deficient SCID mice. (g) Expression of PAFAH1B2 and PAFAH1B3 in normal mammary tissue and primary human breast tumors assessed by qPCR. (h) Recurrence-free survival stratified by high versus low PAFAH1B3 expression in breast cancer patients overall and by lymph-node-negative/positive patients and grade 1–3 breast cancer patients. This data is derived from the online resource known as the Kaplan-Meier Plotter (www.kmplotter.com) (Gyorffy et al., 2010). Data is presented as mean ± sem; n = 4–8/group for (a–f), n-7-41 patients/group for (g), and 306–3455 patients for (h) as indicated. Significance is presented as *p < 0.05 compared to normal tissue (in (a)), and compared to shControl 231MFP cells for (b–g). p-values for (h) are indicated on the figure itself. Protein expression data of PAFAH1B2 and PAFAH1B3 knockdown cells, a recapitulation of tumor growth deficits in shPAFAH1B3 cells, and PAFAH1B3 overexpression studies are shown in Fig. S2.
Upon profiling the gene expression of PAFAH1B2 and PAFAH1B3 in primary breast tumors, we found that PAFAH1B3, but not PAFAH1B2, was significantly heightened in human breast tumors compared to normal mammary tissue (Fig. 3g). Based on the online Kaplan-Meier Plotter resource for breast cancer recurrence-free survival, PAFAH1B3 expression is correlated with low recurrence-free survival in breast cancer patients overall, in lymph-node-positive tumors, and in grade 1–3 breast cancer patients (Fig. 3h) (Gyorffy et al., 2010).
From our phenotypic studies, we found that PAFAH1B3 inactivation produced more striking anti-tumorigenic effects compared with that of PAFAH1B2. Furthermore, PAFAH1B3 expression, but not PAFAH1B2 expression, was significantly higher in primary human breast tumors compared to normal tissue. Thus, we focused our attention on further investigating the pathophysiological and metabolic roles of PAFAH1B3 in driving breast cancer pathogenicity. While we do not understand the mechanistic basis for the differences in tumor growth deficits conferred by PAFAH1B2 or PAFAH1B3 knockdown, it will be of future interest to analyze the tumors that grow out in shPAFAH1B2 xenograft tumors.
We next overexpressed both the catalytically active and the inactive serine to alanine mutant form of PAFAH1B3 (Ser47Ala, S47A) in MCF10A cells to determine whether this enzyme was sufficient to confer oncogenic properties (Fig. S2). While overexpression of the wild-type form of PAFAH1B3 did not alter proliferation or survival, wild-type PAFAH1B3, but not the catalytically inactive mutant, significantly enhanced cell migration indicating that PAFAH1B3 exerts at least some of its pathogenic effects through its catalytic activity (Fig. S2). Our studies also indicate that while PAFAH1B3 overexpression may be not sufficient to confer heightened proliferation or survival in the nontransformed MCF10A cells, its activity is sufficient to heighten mammary epithelial cell migration. Since PAFAH1B3 has been shown to interact with PAFAH1B2, PAFAH1B1, as well as ubiquitin ligase LNX1, it will be of future interest to co-express these other binding partners to determine whether this coupled expression is sufficient to drive other malignant features of cancer (Rual et al., 2005; Sweeney et al., 2000).
Metabolomic Characterization of PAFAH1B3-Regulated Pathways in Breast Cancer
The putative function of PAFAH1B3 is to act as a deacetylase for the signaling lipid platelet activating factor (PAF) (Hattori et al., 1995; Manya et al., 1999). Although recombinant PAFAH1B3 can cleave PAF in vitro (Fig. S3), we found that extracellular and intracellular PAF levels are unchanged in shPAFAH1B2 and shPAFAH1B3 231MFP cells and PAFAH hydrolytic activity was not reduced upon PAFAH1B2 and PAFAH1B3 knockdown, indicating that PAFAH1B is not the dominant enzyme controlling PAF levels or PAF hydrolytic activity in 231MFP cells. Thus, PAFAH1B3 is likely not controlling cancer pathogenicity through controlling PAF signaling pathways (Fig. S3).
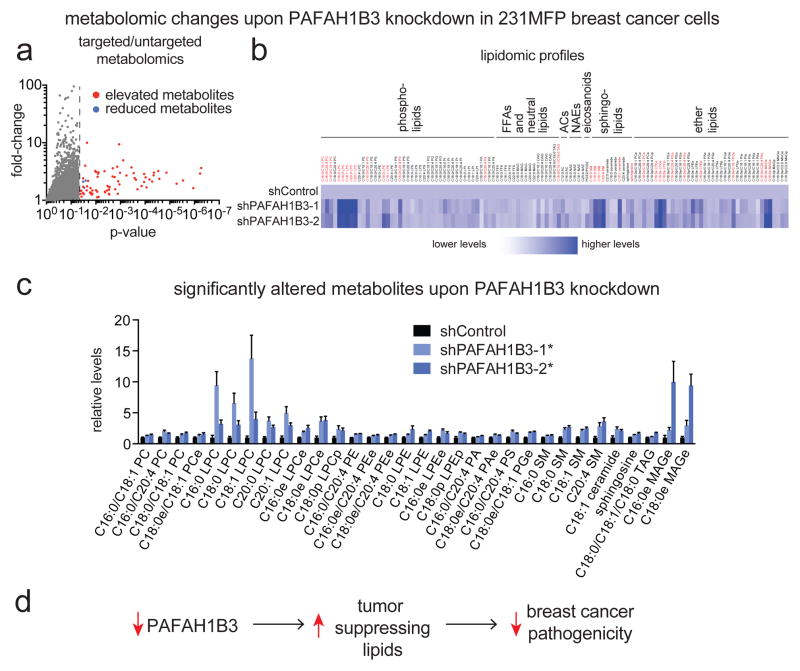
We next performed targeted and untargeted liquid chromatography/mass spectrometry (LC/MS)-based metabolomic profiling to identify metabolic changes in the lipidome conferred by PAFAH1B3 knockdown in 231MFP breast cancer cells (Fig. 4a, 4b; Table S2). We subsequently interpreted only those changes in lipid levels that were significant between the two sh-oligonucleotides and in more than one experiment to reduce false positives or artifacts. We found that PAFAH1B3 knockdown raises the levels of several lipid species including phosphatidylcholines (PCs), lysophosphatidyl choline (LPCs), phosphatidyl ethanolamines (PEs), lysophosphatidyl ethanolamines (LPEs), phosphatidic acids (PAs), phosphatidyl serines (PSs), phosphatidyl glycerol (PGs), sphingomyelins (SMs), ceramide, sphingosine, triacylglycerols, and monoalkylglycerols, as well as ether lipid counterparts (designated as “e” after lipid nomenclature) (Fig. 4c; Table S2). We screened the lipids that contain potential hydrolyzable bonds against purified PAFAH1B3, but none of these lipids are direct substrates of PAFAH1B3 and were not turned over (Fig. S3). Thus, we interpret these changes to be indirect alterations in lipid metabolism downstream of accumulation of a so-far unknown substrate of PAFAH1B3.
Figure 4. Metabolomic profiling of PAFAH1B3 knockdown cells reveals elevations in anti-tumorigenic lipids.
(a) Volcano plot showing all significantly altered metabolites from untargeted and targeted lipidomic profiling in shPAFAH1B3 knockdown 231MFP cancer cells. The red and blue points correspond to elevated and reduced metabolites, respectively. The points to the left of the dotted line are ions detected but not changing upon PAFAH1B3 knockdown, while points to the right of the line are metabolites that are significantly changing (p<0.05 compared to shControl cells). (b) Heat map showing representative metabolites identified through targeted SRM-based lipidomic analysis. Dark blue corresponds to highest levels and white corresponds to lowest levels. Metabolites highlighted in red are significantly changed (p<0.05) in both shPAFAH1B3 lines and repeated in at least two independent experiments. (c) Bar graph showing relative levels of metabolites that are significantly altered in PAFAH1B3 knockdown cells normalized to shControl cells. (d) Our results show that PAFAH1B3 inactivation leads to a metabolomic signature enriched in heightened levels of tumor-suppressing lipids such as ceramides and PPARα ligands (e.g. PC, PS, and LPC). We postulate that this lipidomic signature contributes to reduced cancer pathogenicity upon PAFAH1B3 knockdown in breast cancer cells. Data is presented as mean ± sem; n = 5–6/group. Significance is presented as *p < 0.05 compared to shControl 231MFP breast cancer cells. Detailed metabolomic data is shown in Table S2. PAF levels and PAFAH hydrolytic activity, lipid hydrolytic activities against PAFAH1B3, and studies testing the role of PPARα in PAFAH1B3 effects upon cancer are shown in Fig. S3.
Interestingly, several of the lipids that were elevated upon PAFAH1B3 knockdown have been shown to possess anti-tumorigenic or pro-apoptotic effects or impair cancer pathogenicity. For example, ceramide levels are elevated upon PAFAH1B3 knockdown and ceramide is a well-established pro-apoptotic lipid (Morad and Cabot, 2013). The metabolites involved in ceramide biosynthesis, including phosphatidylcholine (PC) and sphingomyelin, also show heightened levels upon PAFAH1B3 knockdown. Phosphatidylserine (PS) has been shown to be elevated in cancer cells undergoing apoptosis after chemotherapy or radiation treatment and is involved in immune-recognition and tumor clearance (Chao et al., 2012). C16:0/C18:1 Phoshphatidylcholine (PC) has been shown to act as a PPARα agonist and PPARα agonists have been shown to have antitumorigenic and antiangiogenic properties (Chakravarthy et al., 2009; Huang et al., 2013; Liang et al., 2013; Pozzi and Capdevila, 2008). Consistent with this premise, we found that the PPARα agonist fenofibrate impaired cancer cell survival and proliferation in 231MFP cells and the PPARα antagonist GW6471 rescued the proliferative and survival defects of PAFAH1B3 knockdown (Fig. S3). Interestingly, we also found that the elevated C16:0 LPC and C16:0/C18:1 PS also stimulate PPARα activity (Fig. S3).
While our metabolomic data still did not reveal the direct substrate of PAFAH1B3, our results indicate that PAFAH1B3 inactivation leads to a metabolomic signature that is enriched in tumor-suppressing signaling lipids that may be in-part responsible for the observed impairments in breast cancer pathogenicity and tumorigenicity.
Conclusions
Here, we have used multiple platforms, including protein expression profiling, ABPP, and metabolomics to broadly profile dysregulated metabolism in a breast cancer progression model that incorporates cellular transformation through HRAS and EMT, CSC-like features, and malignant progression through the constitutive activation of TAZ. In addition to identifying several canonical metabolic pathways, such as glycolysis, glycogen, and lipogenic metabolism, that have already been shown to metabolic drivers of cancer, we have discovered PAFAH1B3 as a novel metabolic enzyme that is important in driving aggressive and tumorigenic features of breast cancer. Metabolomic profiling reveals that PAFAH1B3 controls cancer cell pathogenicity through regulating an optimal landscape of signaling lipids that may impair cancer aggressiveness.
While PAFAH1B3 has been implicated as a PAF acetylhydrolase, our data indicate that PAF is likely not the endogenous substrate of PAFAH1B3 in cancer cells. While we still do not know the identification of this endogenous substrate, the answer may lie in the altered metabolites that still remain unannotated from our metabolomic analyses of PAFAH1B3 inactivation. Identifying these unknown metabolites may further yield mechanisms through which PAFAH1B3 exerts its pathogenic effects in breast cancer cells. Alternatively, the endogenous substrate may also not be a lipid, or even a metabolite, and may operate on peptides, proteins, or protein post-translational modifications. Thus, it would be of future interest to further investigate the direct substrates of PAFAH1B3 and how these substrates lead to the lipidomic changes that we observe in this study.
While we show that PAFAH1B2 and 1B3 knockdown lead to impairments in cellular proliferation, survival, migration, and invasiveness, the motility and invasiveness deficits may in-part be due to the proliferative and survival deficits. Thus, it may be interesting to pursue those metabolic enzyme targets, such as SIAE or NANS that do not show proliferative deficits but confer migratory impairments, since these targets may regulate invasiveness of the cancer cells. It will also be of future interest to investigate the roles of the other pathways and enzymes that are under the control of HRAS, TAZ, or HRAS and TAZ.
Overall, our results show many metabolic enzymes and biochemical pathways that are dysregulated during cellular transformation and also under malignant progression. While we focused on PAFAH1B3 in this study due to the striking phenotypes observed, many of the other targets may represent unique metabolic nodes that influence various aspects of cancer pathogenicity. In this study, we have also identified multiple metabolic genes, activities, metabolites, and pathways that are regulated by the Hippo signaling pathway. Previous studies have shown that the Hippo transducer pathway controls a diverse transcriptional network that drives EMT, cancer stem-like properties, proliferation, survival, motility, invasiveness, metastasis and transformation (Cordenonsi et al., 2011; Lamar et al., 2012). Here, we show that the Hippo transducers YAP and TAZ collectively regulate the expression and activities of critical metabolic drivers of cancer pathogenicity. We show that the Hippo transducer pathway strongly regulates glycolytic enzymes and glycolytic metabolism alone or in conjunction with HRAS, thus likely contributing to the glycolytic addiction or the “Warburg effect” that subserves cancer pathogenicity. We also show here that the Hippo transducer pathway regulates many other metabolic enzymes that have been linked to controlling pathogenic features of cancer, including SDHA, FASN, MGLL, FAP, PPME1, and neutral cholesteryl ester hydrolase 1 (NCEH1, also known as KIAA1363). SDHA loss-of-function mutations, through accumulation of succinate and inhibition of alpha-ketoglutarate-dependent oxygenases, have been shown to enhance glycolysis, angiogenesis, and epigenetic alterations that fuel cancer (Yang et al., 2013). Both FASN and MGLL have been previously shown to be critical in cancer pathogenicity through controlling lipogenic and lipolytic pathways in cancer cells, respectively (Menendez and Lupu, 2007; Nomura et al., 2011; Nomura et al., 2010b). FAP, a serine protease selectively expressed on tumor-associated fibroblasts and pericytes, has been shown to drive tumor stromagenesis and tumor growth (Santos et al., 2009). PPME1, a demethylase and inhibitor of protein phosphatase 2A, that helps to promote pro-tumorigenic phosphorylation signaling cascades (Bachovchin et al., 2011; Puustinen et al., 2009). Interestingly, NCEH1 is upregulated only upon induction of HRAS and TAZ S89A, indicating that both oncogenic stimuli are required for elevating KIAA1363 activity. This enzyme has been shown to drive cancer migration and tumorigenesis through controlling ether lipid signaling pathways (Chang et al., 2011; Chiang et al., 2006). Thus, we show that the Hippo transducer is a critical factor in controlling the metabolic programming that drives the pathogenic properties of cancer. Here, we show that PAFAH1B2 and PAFAH1B3 are both regulated by a combination of Hippo transducer pathway and HRAS and are also critical metabolic drivers of cancer.
Our studies underscore the utility of multidimensional metabolic profiling efforts in uncovering unique metabolic nodes in cancer and we put forth PAFAH1B3 as a novel potential therapeutic target for the treatment of aggressive breast cancers.
Significance
While many recent studies have revealed key metabolic pathways that underlie cellular transformation, the metabolic drivers of cancer malignancy are less well understood. Here, we have performed massively parallel metabolic profiling study to identify metabolic drivers underlying a breast cancer progression model that incorporates both cellular transformation by RAS and malignant progression by the Hippo transducer TAZ. We have discovered metabolic pathways including glycolytic, lipogenic, and lipolytic processes that have been previously linked to cancer pathogenicity that are coordinately regulated through both the RAS and Hippo pathways. Most notably, we have uncovered a novel metabolic enzyme PAFAH1B3 as a critical driver of breast cancer pathogenicity through controlling tumor-suppressing signaling lipids. Overall, we show the utility of multidimensional metabolic profiling strategies in assembling a metabolic map of breast cancer progression and put forth PAFAH1B3 as a potential metabolic enzyme target for breast cancer therapy.
Methods
Cell Lines
The MCF10A, MII, MII TAZ S89A, and MIV lines were provided by Professor Stefano Piccolo at the University of Padua (Cordenonsi et al., 2011). The 231MFP cells were generated from explanted xenograft tumors of MDA-MB-231 cells, as described previously (Jessani et al., 2004).
Cell Culture Conditions
HEK293T cells were cultured in DMEM media containing 10% FBS and maintained at 37°C with 5% CO2. 231MFP cells were cultured in L15 media containing 10% FBS and glutamine and were maintained at 37°C in 0% CO2. MCF10A and MCF10A-derived lines were cultured in DMEM/F12K media containing 5% (vol/vol) horse serum, 1% (vol/vol) penicillin-streptomycin-glutamine, 20 ng/mL EGF, 100 ng/mL cholera toxin, 10 ng/mL insulin, and 500 ng/mL hydrocortisone and maintained at 37 °C at 5% (wt/vol) CO2 as described previously (Cordenonsi et al., 2011).
qPCR
qPCR was performed using the manufacturer’s protocol for Fischer Maxima SYBRgreen, with 10 μM primer concentrations. Primer sequences were derived from Primer Bank (Spandidos et al., 2010).
Cancer phenotype studies
Migration, invasion, cell proliferation, and survival studies were performed as described previously (Nomura et al., 2010b).
Shotgun Proteomic Profiling of cancer cells
Proteins (100 μg) were TCA precipitated in 20% TCA at −80°C overnight then centrifuged at 10,000 × g at 4°C for 10 min to pellet protein. The protein pellets were washed three times with 8 M urea in PBS. After solubilization, 30μL of 0.2% Protease Max Surfactant (Promega) was added and the resulting mixture was vortexed followed by the addition of 40 μL of 100mM ammonium bicarbonate and 10 mM TCEP. After 30 minutes, 12.5 mM iodoacetamide was added and allowed to react for 30 min in the dark before adding 120 μL PBS and 1.2 μL 1% Protease Max Surfactant (Promega). The protein solution was vortexed and 0.5 μg/μL Sequencing Grade Trypsin (Promega) was added and allowed to react overnight at 37°C. The peptide solution was subjected to a 30 min 10,000 × g spin before the supernatant was transferred to a new tube and loaded onto a biphasic (strong cation exchange/reverse phase) capillary column and analyzed by two-dimensional liquid chromatography in combination with tandem mass spectrometry on a nanospray Thermo LTQ-XL MS/MS. The data was analyzed using the Integrated Proteomics Pipeline (IP2) as described previously (Nomura et al., 2010b).
Activity-based protein profiling of cancer cells
ABPP-MudPIT analysis was performed using previously established procedures (Nomura et al., 2010b). Briefly, 1 mg protein was labeled with 5 μM fluorophosphonate-biotin in 1 mL PBS for 1 h at room temperature, solubilized in 1% Triton X-100 for 1 h, denatured, and labeled enzymes were enriched using avidin beads, reduced and alkylated, and trypsinized. Tryptic peptides were analyzed on a Thermo LTQ-XL MS/MS as described previously (Nomura et al., 2010b).
Metabolomic profiling of cancer cells
Metabolite measurements were conducted using modified versions of previous procedures (Kopp et al., 2010; Nomura et al., 2010b). Details are described in Supplementary Materials.
RNA interference of Enzymes
For small interfering RNA (siRNA) knockdown, cells were treated with ON-TARGETplus SMARTpool siRNA (Thermo) per the manufacturer’s protocol. 200,000 cells were treated with 50 nM siRNA using DharmaFECT 1 Transfection Reagent. Assays were performed after 48 h of treatment. We confirmed knockdown by qPCR.
We used short-hairpin RNA (shRNA) using two independent silencing oligonucleotides to knockdown the expression of PAFAH1B2 and PAFAH1B3 using previously described procedures (Nomura et al., 2010b). Details are described in Supplementary Materials.
Overexpression of PAFAH1B2 or PAFAH1B3 in MCF10A cells
Stable PAFAH1B3 overexpression was achieved by subcloning the PAFAH1B3 gene from MGC fully sequenced human cDNA (Thermo Scientific) into the pLenti CMV puro DEST vector (addgene 17452) using the gateway cloning system (Life Technologies). The PAFAH1B3 Ser 47 Ala mutant was made using Quick-Change Mutagenesis to mutate base 139 (Strategene).
Breast Tumor array qPCR
Breast Cancer tumor array 1 was purchased from Origene and qPCR was performed on the array using the protocol described above.
Tumor Xenograft Studies
Human tumor xenografts were established by transplanting cancer cells ectopically into the flank of C.B17 SCID mice (Taconic Farms) as described previously (Nomura et al., 2010b). Briefly, cells were washed two times with PBS, trypsinized, and harvested in serum-containing medium. Next, the harvested cells were washed two times with serum-free medium and resuspended at a concentration of 2.0 × 104 cells/μl and 100 μl was injected. Growth of the tumors was measured every 3–6 days with calipers.
Supplementary Material
Highlights.
We have performed a metabolic profiling study to identify drivers of breast cancer
Cancer metabolism is regulated by RAS and Hippo pathways
PAFAH1B3 is a driver of breast cancer
PAFAH1B3 may be a novel metabolic target for breast cancer therapy
Acknowledgments
We thank the members of the Nomura Research Group and the Luo laboratory for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (R21CA170317, R01CA172667, R00DA030908 for MMM, DIB, SML, and DKN and R01CA101891 for KL) and the Searle Scholar Foundation (DKN).
Footnotes
Author Contributions: MMM designed research, performed research, analyzed data, and wrote the paper. DIB performed research and analyzed data. AS, MG, TN, and PJM performed research. PJM contributed reagents. ELS and XJ performed research, contributed new reagents, and analyzed data. KL designed research, analyzed data, contributed new reagents, wrote the paper. DKN designed research, performed research, analyzed data, and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Drugs Approved for Breast Cancer. National Cancer Institute; Nov, 2013. http://www.cancer.gov/cancertopics/druginfo/breastcancer. [Google Scholar]
- Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schurer SC, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Cravatt BF, Nomura DK. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell metabolism. 2012;16:565–577. doi: 10.1016/j.cmet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, Hong W. The Hippo pathway in biological control and cancer development. Journal of cellular physiology. 2011;226:928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chemistry & biology. 2011;18:476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2012;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chemistry & biology. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang GZ, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li QY, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Tr. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hattori K, Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Purification and characterization of platelet-activating factor acetylhydrolase II from bovine liver cytosol. The Journal of biological chemistry. 1995;270:22308–22313. doi: 10.1074/jbc.270.38.22308. [DOI] [PubMed] [Google Scholar]
- Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Seminars in cell & developmental biology. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Das SK, Jha P, Al Zoughbi W, Schauer S, Claudel T, Sexl V, Vesely P, Birner-Gruenberger R, Kratky D, et al. The PPARalpha agonist fenofibrate suppresses B-cell lymphoma in mice by modulating lipid metabolism. Biochimica et biophysica acta. 2013;1831:1555–1565. doi: 10.1016/j.bbalip.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. Fibroblast activation protein-alpha and dipeptidyl peptidase IV (CD26): Cell-surface proteases that activate cell signaling and are potential targets for cancer therapy. Drug Resist Update. 2005;8:51–58. doi: 10.1016/j.drup.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Kopp F, Komatsu T, Nomura DK, Trauger SA, Thomas JR, Siuzdak G, Simon GM, Cravatt BF. The glycerophospho metabolome and its influence on amino acid homeostasis revealed by brain metabolomics of GDE1(−/−) mice. Chemistry & biology. 2010;17:831–840. doi: 10.1016/j.chembiol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Kowalczyk P, Junco JJ, Klug-De Santiago HL, Malik G, Wei SJ, Slaga TJ. Differential Effects on Lung Cancer Cell Proliferation by Agonists of Glucocorticoid and PPARalpha Receptors. Molecular carcinogenesis. 2013 doi: 10.1002/mc.22029. [DOI] [PubMed] [Google Scholar]
- Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chemical reviews. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manya H, Aoki J, Kato H, Ishii J, Hino S, Arai H, Inoue K. Biochemical characterization of various catalytic complexes of the brain platelet-activating factor acetylhydrolase. Journal of Biological Chemistry. 1999;274:31827–31832. doi: 10.1074/jbc.274.45.31827. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Morad SA, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer. 2013;13:51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer. 2010a;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chemistry & biology. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010b;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Capdevila JH. PPAR alpha Ligands as Antitumorigenic and Antiangiogenic Agents. Ppar Res. 2008 doi: 10.1155/2008/906542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen P, Junttila MR, Vanhatupa S, Sablina AA, Hector ME, Teittinen K, Raheem O, Ketola K, Lin S, Kast J, et al. PME-1 protects extracellular signal-regulated kinase pathway activity from protein phosphatase 2A-mediated inactivation in human malignant glioma. Cancer research. 2009;69:2870–2877. doi: 10.1158/0008-5472.CAN-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast cancer research and treatment. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Santos AM, Jung J, Aziz N, Kissil JL, Pure E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. The Journal of clinical investigation. 2009;119:3613–3625. doi: 10.1172/JCI38988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic acids research. 2010;38:D792–799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney KJ, Clark GD, Prokscha A, Dobyns WB, Eichele G. Lissencephaly associated mutations suggest a requirement for the PAFAH1B heterotrimeric complex in brain development. Mechanisms of development. 2000;92:263–271. doi: 10.1016/s0925-4773(00)00242-2. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. Journal of Clinical Investigation. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.