Abstract
Calcium homeostasis balances passive calcium leak and active calcium uptake. Human Bax inhibitor 1 (hBI-1) is an anti-apoptotic protein that mediates a calcium leak and is representative of highly conserved and widely distributed family, the transmembrane Bax inhibitor motif (TMBIM) proteins. Here we present crystal structures of a bacterial homolog and characterize its calcium leak activity. The structure has a seven-transmembrane-helix fold that features two triple-helix sandwiches wrapped around a central C-terminal helix. Structures obtained in closed and open conformations are reversibly inter-convertible by change of pH. A hydrogen-bonded, pKa-perturbed pair of conserved aspartate residues explains the pH dependence of this equilibrium, and biochemical studies show that pH regulates calcium influx in proteoliposomes. Homology models for hBI-1 provide insights into TMBIM-mediated calcium leak and cytoprotective activity.
Calcium (Ca2+) is a ubiquitous intracellular messenger that regulates cellular and physiological activities. Cytosolic Ca2+ is kept at a low level to assure responsiveness to Ca2+ signals, but subcellular organelles such as the endoplasmic reticulum (ER) and Golgi apparatus maintain calcium stores. Upon activation, Ca2+ is mobilized to cross membrane barriers through calcium-release channels and calcium uptake pumps (1). Under resting conditions, intracellular and subcellular calcium homeostasis is dynamically regulated to equilibrate between active calcium uptake and passive calcium leak. Calcium homeostasis is cytoprotective (2, 3). An overloaded ER calcium content promotes cell death (4); inversely, lowering of ER calcium content by anti-apoptotic proteins Bcl-2/Bcl-xL or Bax inhibitor-1 (BI-1) elicits a survival signal (5–7). Bcl-2/Bcl-xL and BI-1have been suggested to regulate ER calcium leak, either directly by forming a leaky pore or by modulating calcium-release channels such as inositol trisphosphate receptors (IP3Rs) (8–10).
Human BI-1 (hBI-1) was discovered as a human gene product that can block lethality of the pro-apoptotic Bax protein in yeast (8). BI-1 is localized to the ER membrane where, among other functions, it mediates a calcium leak downstream of Bcl-2/Bcl-xL (8, 11). By sequence similarity to hBI-1, a highly conserved TMBIM (Transmembrane Bax Inhibitor Motif) family was identified (12) and assigned the Pfam (13) name of Bax1-I (PF01027). TMBIM proteins are present in prokaryotes, fungi, plants, and metazoans, including invertebrates and mammals (12) (fig. S1). Humans have six identified TMBIM proteins (TMBIM1-6), each containing seven presumed transmembrane helices (14) and with variations mainly in their N-terminal extensions (fig. S2). Besides hBI-1 (TMBIM6) in the ER membrane, human Golgi anti-apoptotic protein (hGAAP/TMBIM4) is in the Golgi membrane where it mediates Golgi calcium leak, providing another identified connection to calcium and apoptosis (15). Other human TMBIM proteins are diversely localized and less well characterized (12).
Accumulating evidence has demonstrated the calcium-leak activity of the TMBIM proteins and their regulatory roles in apoptosis (11, 15); however, little is known about the structure or mechanism of action for these proteins beyond recent topological studies on hBI-1 and hGAAP (16, 17). Seeking structural clues into the mechanism of calcium flux activity, we undertook structural studies of TMBIM proteins. Here we present crystal structures of a bacterial homolog in inter-convertible conformational states dependent on pH; we demonstrate pH-sensitive calcium permeation by this protein consistent with the calcium-leak activity of hBI-1 and hGAAP; and we build a homology model of hBI-1 to provide structural insights into the calcium leak and anti-apoptotic functions of the TMBIM family.
Structural analyses
To address the structural challenge of the TMBIM family, we identified prokaryotic homologs of human BI-1 that might provide structural insights into function. After screening 51 bacterial relatives for expression in Escherichia coli, we identified YetJ from Bacillus subtilis (BsYetJ), a previously uncharacterized protein, as a family member with satisfactory biochemical properties. The detergent-extracted protein was purified and crystallized in two crystal forms.
Form-1 crystals grew at pH 8 in space group P6522 with one protein molecule per asymmetric unit. We solved this structure by multi-crystal native-SAD phasing (18) using relatively low energy x-rays (~ 6 keV) to enhance anomalous signal-to-noise ratios. The eight ordered sulfur atoms contributed a Bijvoet-diffraction ratio of ~1.4%. Diffraction data up to 2.8 Å spacings were measured from 12 crystals, and 10 of these met criteria for statistical equivalence (fig. S3A). Previously established analytical procedures (18) allowed both substructure determination and native-SAD phasing. The resulting electron-density map (fig. S3B) permitted automatic tracing of a nearly complete model, which was further refined at 1.95 Å resolution against a separate high-energy dataset (table S1 and fig. S3C).
Form-2 crystals grew at pH 6 in space group C2221 and also have one molecule per asymmetric unit. Native crystals diffracted x-rays only to ~4.5 Å with severe anisotropy. Attempts at structure solution by molecular replacement from the form-1 structure did not succeed, suggesting a different conformation. Fortunately, a platinum derivative diffracted better, and the structure was determined by Pt-SAD phasing at 3.6 Å resolution (fig. S3D), and a conformationally distinct model was built with reference to the form-1 structure, mainly by displacing one helix. We also found that BsYetJ can undergo an intra-crystalline transition when form-1 crystals, as grown, are soaked in medium at pH 6. The resulting low pH conformation is almost identical to that in the orthorhombic form-2 crystals (fig. S3E). The converted structure in the hexagonal form-1 lattice diffracted better and could be refined to 2.5 Å resolution (table S1), and this structure was used for further structural characterization.
Structural features
The structure of BsYetJ in each of its conformations comprises seven transmembrane helices, which by structure-based alignment compares with anticipated helix boundaries for the entire family of TMBIM proteins (fig. S2). The higher pH form has a compact, closed conformation (Fig. 1A and fig. S4); whereas the lower pH form has an opened conformation with helix TM2 displaced (Fig. 1B). Based on topology assays of the human homologs (16, 17), TMBIM proteins have their N-termini in the cytoplasm, which is consistent with the positive-inside rule (19) as applied to the electrostatic potential surfaces of the BsYetJ structures (fig. S5). The tightly packed helices of the closed form give this structure a barrel-like shape. The barrel is about 56 Å high by 34 Å in diameter, and judged by electrostatics its axis is tilted in the lipid bilayer by 9°, so that 31 Å is embedded in lipid bilayer. The open conformation produces a pore through the lipid bilayer (figs. S5E-H) that is wide open (11 Å) at the periplasmic side and narrows to a 5 Å-wide bottleneck near the cytoplasmic side.
Fig. 1. Structures of BsYetJ.
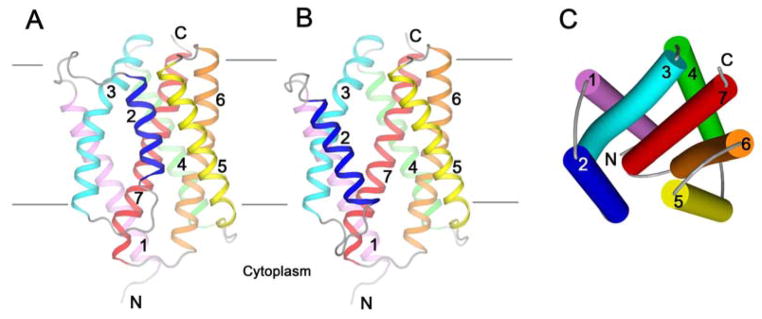
(A) Ribbon drawing of form-1 structure determined at pH 8. (B) Ribbon drawing of form-2 structure determined at pH 6. The views of (A) and (B) are from the membrane, and each structure has its seven TM helices color-coded. All connecting loops are colored in gray. (C) A cylinder diagram of the form-2 structure to show the structural features from a periplasmic view. The coloring is as for (B).
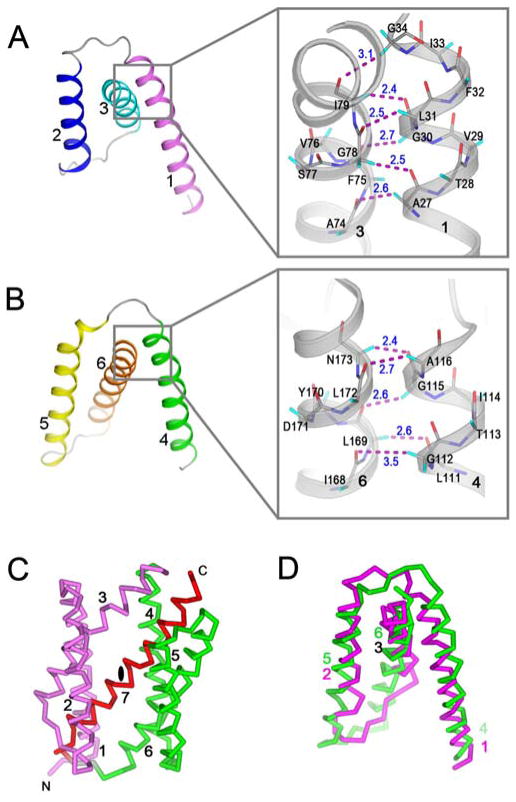
The polypeptide folding in BsYetJ is topologically different from that of any known structures, as revealed by a DALI search (20). As best seen in the open conformation, conceptually the overall structure has three components, TM1-3, TM4-6 and TM7 (Fig. 1C). TM1-3 and TM4-6 are similar, each forming a triple-helix-sandwich substructure: TM3 is clamped by helix-loop-helix TM1-2; TM6 is clamped by helix-loop-helix TM4-5 (Fig. 1C and Figs. 2A-B). In forming the triple-helix-sandwich, short side-chain hydrophobic residues on the transmembrane helices are crucial (21) as they provide sticky patches to allow close TM1/TM3 and TM4/TM6 contacts for clamping (Figs. 2A-B, insets). TM3 and TM6 are each bent at the sticky patch. Although there is no obvious sequence similarity between TM1-3 and TM4-6, the two components are superimposable when inverted by a pseudo two-fold symmetry viewed from within the membrane (Figs. 2C-D). C-terminal helix TM7 is in the center of the structure, parallel to TM3 and antiparallel to TM6; these three helices together form a central layer that is sandwiched by the four peripheral helices TM1-2 and TM4-5 (Fig. 1C).
Fig. 2. Structural features.
(A–B) Two triple-helix sandwich substructures consist of TM1-3 (A) and TM4-6 (B). The color scheme is as Fig. 1B. Insets in (A) and (B) are, respectively, the close α-helical contacts between TM1 and TM3 and between TM4 and TM6. Cα-H······O contacts between 2.3 Å - 3.5 Å were drawn as magenta dashes. (C) Overall pseudo-inverse symmetry with the triple-helix sandwiches in magenta for TM1-3 and green for TM4-6. The pseudo twofold axis is on the middle of the red TM7. (D) Superimposition of the two symmetric components.
As a seven transmembrane helix (7-TM) protein, BsYetJ compares with G-protein coupled receptors (GPCRs) (22), and recent CAAX metalloprotease structures (23, 24). The fold of BsYetJ is distinct from these (fig. S6). Notably, the CAAX protease helices encompass an intramembrane chamber and cross-sectional registrations of these helices are unrelated. The GPCR fold has TM3 somewhat central, which has been proposed to have functional importance in ligand binding and signal transduction (25). For BsYetJ, TM7 has the central position and it too has functional importance as suggested by sequence conservation and biochemical analyses on it human relative hBI-1 (17). By topology, BsYetJ somewhat resembles insect olfactory receptors, a special family of 7-TM proteins that also have their N-termini inside (26); however, sequence alignments did not reveal any homology between the two families.
Regulation of pore opening and closing by pH
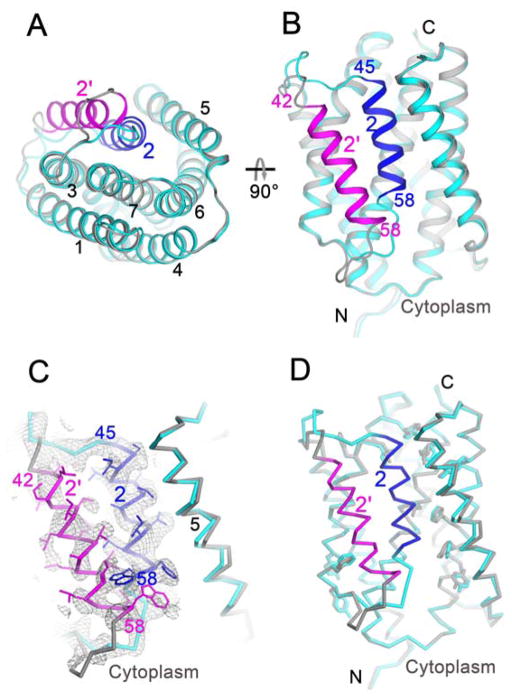
The fact that the pore-closed and pore-open conformations were obtained at pH 8 and pH 6 and that the pore can be opened by intra-crystalline transition (Figs. 3A–B) suggest a role for pH in regulation of conformational transition. Superimposition of the pore-open and pore-closed structures indicates substantial structural changes for TM2 and the two loops connecting it to TM1 and TM3 (Figs. 3A–B). Relative to the closed conformation, TM2 in the open conformation swings away by as far as 13.5 Å to form a transmembrane pore bordered by it, TM5-TM7 and the presumed lipid bilayer (Fig. 3A; fig. S5F). In the closed conformation, TM2 is rather short (14 residues); while in the open conformation, TM2 is extended by roughly one helical turn at its N-terminus. The transformation from the closed to open conformation (rotation χ =38.6°; translation tχ=3.4 Å) displaces TM2 away from contact with TM6 and also moves it along its axis toward the cytoplasm (Fig. 3B).
Fig. 3. Pore opening and closing regulated by pH.
(A–B) Superimposition of the two conformationally different structures at pH 8 (cyan and blue) and at pH 6 (gray and magenta). (A) is cytoplasmic view and (B) is membrane view. (C) Electron density of the pH 7 structure showing alternative conformations of TM2. The 2Fo-Fc electron densities were drawn as gray isomeshes at 0.8s. The side chains of TM2 in closed and open conformations were drawn respectively as blue and magenta sticks. (D) Overall pH 7 structure with two alternative conformations. Deviated side chains are shown as sticks: cyan for closed conformation and gray for open conformation.
During the pH 6 to pH 8 transition, the c-axis of the P6522 lattice shrinks from 289 Å to 276 Å, but the resulting open-conformation structure is essentially the same as that obtained in the C2221 lattice as grown at pH 6 (fig. S3E). To test for reversibility of this pH-driven conformational transition, we soaked the closed-form crystals at pH 6; took diffraction images to confirm unit-cell shrinkage, the hallmark of pore-opening upon soaking; and then back-soaked the exact crystals to pH 8. This re-established pH 8 structure, which has a diagnostic c-axis of 293 Å, is re-closed to be almost identical to the initial pore-closed conformation. Thus, pH can regulate the opening and closing of the pore.
To further test the pH-driven conformational changes, we moved the opened crystals from pH 6 to pH 7 and determined this pH 7 structure by molecular replacement, using the back-soaked pH 8 structure since their lattice parameters were nearly identical (table S1). Electron density was seen for TM2 in both closed and open conformations, consistent with alternate states in equilibrium (Fig. 3C). The ratio of closed:open component was determined as 60:40 by occupancy refinement with the program PHENIX (27). The pH 7 structure as refined with two alternative conformations shows differences propagated into neighboring helices with associated main-chain and side-chain shifts (Fig. 3D). Thus the conformation of BsYetJ is pH sensitive, and we presume that the ratio of TM2 occupancies reflects the conformational equilibrium. Based on the 60:40 closed:open ratio at pH 7, the apparent pKa of the protein in the crystal may be estimated to be slightly below pH 7.
Di-aspartyl pH sensor
When closed at pH 8, TM2 and cytoplasmic loop
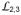 (TM2-TM3 connection) engage in several hydrogen bonds with residues from TM3, TM6 and TM7; and all of these are disrupted in the open conformation at pH 6 (Fig. 4A). A key interaction at the closed-conformation interface appears to be a latch of Arg60 with the TMBIM-conserved di-aspartyl unit Asp171/Asp195 (Fig. 4B). The guanidinium moiety from Arg60 forms a doubly hydrogen-bonded salt bridge with Asp171 when the protein is at pH 8 (Fig. 4B), but this is broken at pH 6 (Fig. 4C) and reestablished upon back-soaked to pH 8 (Fig. 4D). Most remarkably, in all cases the carboxylate groups of the two aspartates are hydrogen bonded to one another. Electron density that is continuous through the di-aspartate unit, even for the 1.95 Å-resolution structure, and refined O-O distances of 2.55 Å at pH 8 and 2.71 Å at pH 6, clearly demonstrate the hydrogen bonding and anomalous aspartate protonation.
(TM2-TM3 connection) engage in several hydrogen bonds with residues from TM3, TM6 and TM7; and all of these are disrupted in the open conformation at pH 6 (Fig. 4A). A key interaction at the closed-conformation interface appears to be a latch of Arg60 with the TMBIM-conserved di-aspartyl unit Asp171/Asp195 (Fig. 4B). The guanidinium moiety from Arg60 forms a doubly hydrogen-bonded salt bridge with Asp171 when the protein is at pH 8 (Fig. 4B), but this is broken at pH 6 (Fig. 4C) and reestablished upon back-soaked to pH 8 (Fig. 4D). Most remarkably, in all cases the carboxylate groups of the two aspartates are hydrogen bonded to one another. Electron density that is continuous through the di-aspartate unit, even for the 1.95 Å-resolution structure, and refined O-O distances of 2.55 Å at pH 8 and 2.71 Å at pH 6, clearly demonstrate the hydrogen bonding and anomalous aspartate protonation.
Fig. 4. Di-aspartyl pH sensor.
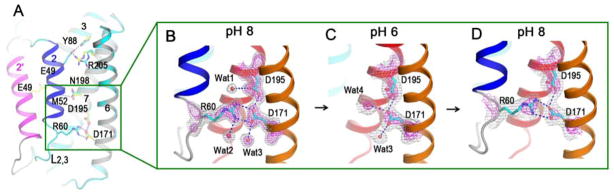
(A) H-bond interactions within the pore of the closed-conformation structure. (B–D) Successive structures from intra-crystalline transitions with superimposed 2Fo-Fc electron densities contoured at two levels, 1.2s (gray) and 3.0s (magenta). (B) Starting form-1 structure at pH 8. (C) Structure after soaking a form-1 crystal into a medium at pH 6, disrupting the interactions between Arg60 and Asp171. (D) Structure after reversal, from first soaking at pH 6 and then back-soaking to pH 8, thereby reclosing the pore and restoring the interactions between Arg60 and Asp171.
The structural presumption based on the pH 8 structure would have Asp195 as the protonated group, leaving Asp171 free to form its salt bridge with Arg60. This presumption is validated by pKa calculations from the program PROPKA (28). For the closed conformation, the pKa values of Asp171 and Asp195 are 3.1 and 11.2, respectively, one depressed and the other elevated due to a presumed coupling effect (28). For the open conformation, the respective values are 6.2 and 12.0. The increased pKa of Asp171 in the open conformation may account for the equilibrated conformational states seen in our pH 7 structure. Thus, in absence of the guanidinium interaction, Asp171 is more readily protonated; alternatively, Asp171 when protonated is incompatible with Arg60 engagement. We suggest that the pH control of Asp171 thereby regulates the conformational transition and pore opening/closing. Asp195 is protonated throughout pH 6 to 8. Aspartate residues at positions 171 and 195 are strictly conserved in the TMBIM family (fig. S2), suggesting that a di-aspartyl pH sensor may be a family trait. The latch partner Arg60 is also conserved in TMBIM1-4 but not in TMBIM5-6, opening a question of alternative latch partners.
Both Arg60-bearing loop
 and loop
and loop
 , which connects TM2 to TM1, are quite flexible as seen in B-factor plots (fig. S7). This mobility is consistent with ready displacement of TM2 when the latch to Asp171 is broken at lower pH. When TM2 is displaced from its van der Waals contacts with TM5, both it and TM5 have increased flexibility. Thus, we picture a pH-sensitive conformational equilibrium between a rather flexible pore-open state seen at lower pH and a pore-closed state seen at higher pH.
, which connects TM2 to TM1, are quite flexible as seen in B-factor plots (fig. S7). This mobility is consistent with ready displacement of TM2 when the latch to Asp171 is broken at lower pH. When TM2 is displaced from its van der Waals contacts with TM5, both it and TM5 have increased flexibility. Thus, we picture a pH-sensitive conformational equilibrium between a rather flexible pore-open state seen at lower pH and a pore-closed state seen at higher pH.
Calcium leak
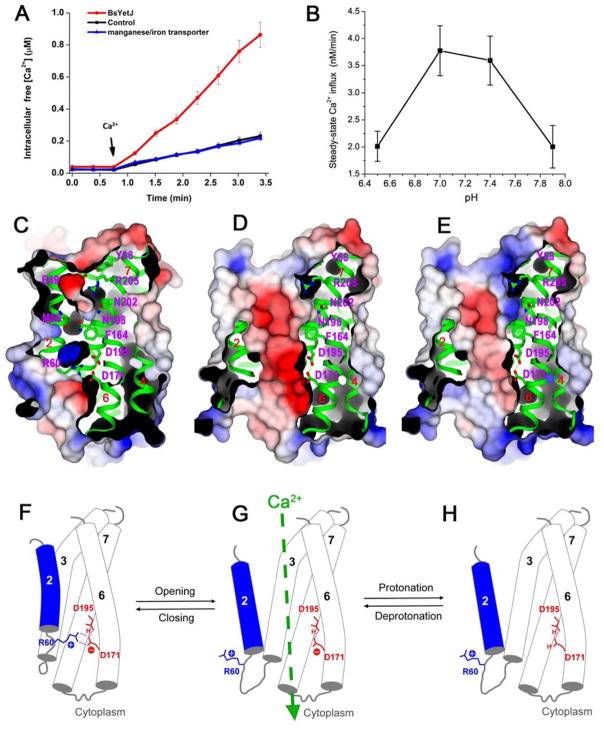
Both hBI-1 and hGAAP are able to mobilize calcium leak into the cytoplasm from stores in ER and Golgi compartments, respectively (7, 11, 15). To test whether our bacterial homolog has calcium-leak activity, we overexpressed BsYetJ in E. coli, added calcium extracellularly and measured the intracellular calcium concentration [Ca2+]cyto with the fluorescent calcium dye Fura-2/AM. Upon addition of external calcium, [Ca2+]cyto increased steadily in cells overexpressing BsYetJ, but not for controls of an empty plasmid or an unrelated membrane protein transporter (Fig. 5A). Thus BsYetJ is a bona fide functional bacterial TMBIM homolog with calcium-leak activity.
Fig. 5. Structural and functional characterization of calcium leak.
(A) Calcium influx into bacteria over-expressing BsYetJ. An empty plasmid and a manganese transporter were used as negative controls. Error bars indicate the standard error of the mean (±SEM, n=9). (B) Calcium influx into proteoliposomes. Error bars indicate the standard error of the mean (±SEM, n=3). (C) Electrostatic surface of the closed-conformation structure showing charged surface concavities and internal cavities, but a blocked pore. (D) Electrostatic surface of the open-conformation structure at pH 7.4 where the cleft is electronegative. (E) Electrostatic surface for the open-conformation structure at pH 6 where the cleft is more neutral. The contour level of the electrostatics surface is at ±5 kT/e. Red, negative potential; blue, positive potential. (F–H) A proposed model for pH-sensitive calcium leak. (F) At higher pH (e.g. 8), Asp195 is protonated and Asp171 is deprotonated. Asp171 forms two H-bonds with positively charged Arg60 and the Arg60/Asp171 latch closes the pore. (G) When in the open conformation at a more neutral pH (e.g. 7.4), Asp171 may equilibrate between protonated and deprotonated states. Calcium passage occurs only when Asp171 is transiently deprotonated. (H) At lower pH (e.g. 6), the equilibration will favor more complete protonation of Asp171, disfavoring calcium passage due to pore neutralization. Cartoons (F–H) correspond to structures (C–E) directly above.
Having shown that BsYetJ can produce a calcium leak in bacteria and that pH can control pore opening in BsYetJ, we explored the effect of pH on calcium influx. We constructed BsYetJ proteoliposomes at various pHs, preloaded them with the Fura-2 dye, and measured calcium influx when exposed to externally added calcium. We observed pH-dependent influxes of calcium, typically accumulating more rapidly at the outset of calcium application and then slowing somewhat to steady-state levels after ~10 min (fig. S8). The steady-state calcium influx was substantially lower when the pH was lower (pH 6.5) or higher (pH 7.9) than when under near-neutral conditions (pH 7.0 or 7.4) (Fig. 5B). We conclude that calcium-leak activity is intrinsic to BsYetJ because we prepared the proteoliposomes from pure components.
Multiple hydrophilic residues lie within the core of closed-conformation BsYetJ (Glu49, Arg205, Asp202, Asn198, Asp195, Asp171 and Arg60), lining-up from the periplasmic side to the cytoplasmic side (Fig. 5C). These residues border concave surfaces that invaginate from either side and form two charged internal cavities along with these residues. This incipient passage way for calcium ions is structurally blocked in the closed conformation by hydrophobic residue Phe164 (Fig. 5C). When TM2 is moved away from the closed conformation, the separated concave surfaces and internal cavities unify to form a transmembrane pore, largely electronegative at a physiological pH (7.4; Fig. 5D) and less electronegative at a lower pH (6.0; Fig. 5E).
The pore at a more neutral pH appears conducive to calcium passage; however, we did not detect calcium ions in the pore of any BsYetJ crystal structure even though calcium was included in all crystallization and soaking experiments. Evidently, the structure does not feature discrete calcium binding sites in the pore but does allow calcium passage with only transient interactions within the pore. Based on the alternative conformations in our pH 7 structure, we imagine BsYetJ in the lipid bilayer to be in a facile equilibrium between open and closed conformations.
Why is the flux rate higher at pH 7 where the pore is in equilibrium than at pH 6 where it is open? Fluctuations in the open/closed equilibrium may permit a calcium leak by opening a pore that is electronegative when at physiologically neutral pHs (Fig. 5D), as in our assays for uptake of calcium into bacteria and proteoliposomes (Figs. 5A–B), whereas either closure at higher pH or reduced electronegativity at lower pH will counteract calcium passage. A working model for this pH-sensitive calcium-leak activity is given in Figs. 5F-H. We propose that BsYetJ exists equilibrated among three states: closed, open deprotonated and open protonated. At higher pH, Asp171 is predominantly deprotonated and forms a doubly hydrogen-bonded salt bridge with Arg60 (Fig. 5F); with this Arg60/Asp171 latch in place, the structure is closed and the pore is sealed (Fig. 5C; 5F). At a more neutral pH, the open conformation becomes accessible, even if transiently (Fig. 5G); the opened pore is electronegative with Asp171 remaining deprotonated, and a calcium leak is then facilitated along the calcium gradient into the cytosol (Figs. 5D; 5G). At lower pH, the protonated state of Asp171 is favored since its pKa is raised in the open conformation, which precludes formation of the Arg60/Asp171 latch (Fig. 5H). In this state, electronegativity of the pore is reduced and calcium influx is impeded (Fig. 5E).
We conclude that BsYetJ is a pH sensitive calcium-leak channel. Its molecular architecture is distinct from that known for other calcium channels and exchangers (29–33), and its pH-dependent changes in conformation and electrostatics are compatible with observed calcium-flux activities. Human TMBIM homolog BI-1 also exhibits pH-sensitive calcium-leak activities, proposed to be mediated by aspartic acid residues on TM-7 (34) or by the C-terminal lysine-rich motif (35). Our results are consistent with the TMBIM-conserved di-aspartyl pH sensor, but not the C-terminal lysine-rich motif, as a shared mechanism for the pH-sensitive calcium leak.
Insights into the BI-1-mediated calcium leak and anti-apoptosis
To explore the structural linkage of BsYetJ to human homologs, we constructed homology models of hBI-1, the most studied of TMBIM proteins, using structure-based sequence alignment (fig. S2) and the program MODELLER (36).
Since BsYetJ is clearly homologous with its human TMBIM relatives (21% for TMBIM4 and 18% for TMBIM6), the hBI-1 models are highly similar to their BsYetJ templates while with instructive differences. The di-aspartyl pH sensor Asp188/Asp213 (respectively Asp171/Asp195 in BsYetJ) is intact in both states; however, the Arg60 latch of BsYetJ (and TMBIM1-4) is replaced by His78 in hBI-1. In the closed conformation of hBI-1, His78 forms an alternative latch by hydrogen bonding to Asp209 (Ser191 in BsYetJ) (Fig. 6A). In the open conformation, Asp209 is freed from its interaction with His78, and it will likely exacerbate the pKa elevations of Asp188/Asp213 expected by analogy with BsYetJ. At a suitably low pH, we expect that Asp209 will adopt an alternative conformation and form a hydrogen bond with protonated Asp188 (Fig. 6B). Analogous to residues in BsYetJ, side chains of Leu71, Thr74 and Met181 (Phe164 in BsYetJ) are expected to provide pore-sealing functions in the closed state, separating concave indentations from the opposing membrane surfaces. Although many residues that line the open-state pore differ in hBI-1 as compared to BsYetJ, the shape and electrostatic features of the two pores remain very similar; notably, the narrow opening on the cytoplasmic side of the hBI-1 pore is like that of BsYetJ.
Fig. 6. Homology models of human Bax inhibitor 1 (hBI-1).
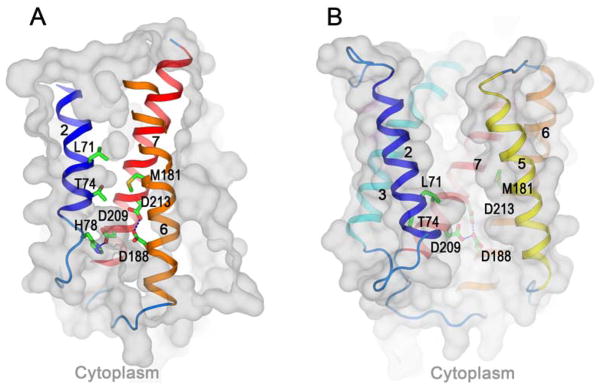
(A–B) Homology models of hBI-1 in its closed conformation (A) and open conformation (B). The conserved di-aspartyl Asp188/Asp213 unit is shown within the membrane in each case, and a third aspartate, Asp209, is shown interacting with His78 in (A) and with Asp188 in (B). In the closed-conformation (A), the indicated pore-sealing residues, Leu71, Thr74 and Met181, separate concave surfaces invaginating from opposite membrane face.
Based on topology studies, hBI-1 was proposed to have a 6-TM topology with both N-and C-termini in the cytoplasm, where the predicted TM7 segment is either disordered or hemi-penetrant back into the membrane (16, 17). On the other hand, TM7 is in the middle of the 7-TM structure, of BsYetJ, where it forms extensive interactions with other TM helices. These include the conserved di-aspartyl pH sensor between TM6 and TM7 and a contact between TM4 and TM7 (Thr104/121 and Phe200/218 in BsYetJ and hBI-1, respectively). The hydrophobicity analysis, secondary structure predictions, and conservation also suggest that hBI-1 has a 7-TM topology like that in BsYetJ (fig. S9).
Besides calcium-leak activity, which has been observed for TMBIM proteins wherever tested, TMBIM interactions with other proteins have also been identified. For example hBI-1 and hGAAP co-immunoprecipitate with IP3R and modulate IP3-induced Ca2+ release (9, 15), and hBI-1 interacts with Bcl-2 as shown by in-vivo cross-linking and co-immunoprecipitation (8, 11). Based on the BsYetJ structures and the homology models for hBI-1, a plausible mode of TMBIM-mediated protein-protein interactions would have a transmembrane helix from the partner protein taking the place of TM2 after its displacement in the open conformation of the TMBIM protein (figs. S10A-B).
It has been reported that Bax/Bak activation and mitochondria outer membrane permeabilization (MOMP) are enhanced by overloaded calcium stores (2). The protective role of hBI-1 in decreasing [Ca2+]ER is expected to reduce MOMP and thus suppress the activation of Bax/Bak toward the initiation of apoptosis. We propose that human TMBIM proteins function in maintaining a dynamic homeostasis of stored [Ca2+] and cytosolic [Ca2+] through the pH-sensitive calcium-leak mechanism. The overexpression of hBI-1 in various cancers, including prostate, breast, glioma, uterine, ovarian and lung, presumably reflects recruitment of its anti-apoptotic activity (37–40). Knockdown of hBI-1 expression by RNA interference has shown effectiveness in inducing spontaneous apoptosis of cancer cells in prostate and breast (37, 38). The structures of BsYetJ and its derivative hBI-1 models provide substantial insights into the functioning of TMBIM proteins and offer therapeutic prospects for intervention of anti-apoptotic functions in treatment of cancers.
Acknowledgments
We thank John Schwanof and Randy Abramowitz at National Synchrotron Light Source (NSLS) beamlines X4A and X4C for their assistance in data collection, James Love for help in initial high-throughput screening, Marco Punta for help in initial target selection, Min Su for help with the phylogenetic presentation, and Filippo Mancia and Lawrence Shapiro for discussions. This work was supported in part by NIH grant GM095315. Beamlines X4A and X4C of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory, a DOE facility, is supported by the New York Structural Biology Center. Atomic coordinates and structure factor files have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 4PGR for closed-form at pH 8, 4PGS for open-form at pH 6 by soaking, 4PGU for closed/open-from at pH 7 by back-soaking, 4PGV for closed-form at pH 8 by back-soaking, and 4PGW for open-form in C2221 lattice. Q.L., Y.C., and W.A.H. are inventors on a patent application filed by The New York Structural Biology Center on uses of the three-dimensional structures of BsYetJ and homolog models of human BI-1.
References and Notes
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Smaili SS, Pereira GJ, Costa MM, Rocha KK, Rodrigues L, et al. The role of calcium stores in apoptosis and autophagy. Curr Mol Med. 2013;13:252–265. doi: 10.2174/156652413804810772. [DOI] [PubMed] [Google Scholar]
- 3.Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- 4.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 5.Palmer AE, Jin C, Reed JC, Tsien RY. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci USA. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ihara-Ohori Y, Nagano M, Muto S, Uchimiya H, Kawai-Yamada M. Cell death suppressor Arabidopsis Bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant physiol. 2007;143:650–660. doi: 10.1104/pp.106.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westphalen BC, Wessig J, Leypoldt F, Arnold S, Methner A. BI-1 protects cells from oxygen glucose deprivation by reducing the calcium content of the endoplasmic reticulum. Cell Death Differ. 2005;12:304–306. doi: 10.1038/sj.cdd.4401547. [DOI] [PubMed] [Google Scholar]
- 8.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 9.Kiviluoto S, Schneider L, Luyten T, Vervliet T, Missiaen L, et al. Bax inhibitor-1 is a novel IP(3) receptor-interacting and -sensitizing protein. Cell Death Dis. 2012;3:e367. doi: 10.1038/cddis.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distelhorst CW, Bootman MD. Bcl-2 interaction with the inositol 1,4,5-trisphosphate receptor: role in Ca2+ signaling and disease. Cell Calcium. 2011;50:234–241. doi: 10.1016/j.ceca.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Xu W, Palmer AE, Reed JC. BI-1 regulates endoplasmic reticulum Ca2+ homeostasis downstream of Bcl-2 family proteins. J Biol Chem. 2008;283:11477–11484. doi: 10.1074/jbc.M708385200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henke N, Lisak DA, Schneider L, Habicht J, Pergande M, et al. The ancient cell death suppressor BAX inhibitor-1. Cell Calcium. 2011;50:251–260. doi: 10.1016/j.ceca.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Mattia F, Gubser C, van Dommelen MM, Visch HJ, Distelmaier F, et al. Human Golgi antiapoptotic protein modulates intracellular calcium fluxes. Mol Biol Cell. 2009;20:3638–3645. doi: 10.1091/mbc.E09-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrara G, Saraiva N, Gubser C, Johnson BF, Smith GL. Six-transmembrane topology for Golgi anti-apoptotic protein (GAAP) and Bax inhibitor 1 (BI-1) provides model for the transmembrane Bax inhibitor-containing motif (TMBIM) family. J Biol Chem. 2012;287:15896–15905. doi: 10.1074/jbc.M111.336149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bultynck G, Kiviluoto S, Henke N, Ivanova H, Schneider L, et al. The C terminus of Bax inhibitor-1 forms a Ca2+-permeable channel pore. J Biol Chem. 2012;287:2544–2557. doi: 10.1074/jbc.M111.275354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα ---H…O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 23.Quigley A, Dong YY, Pike AC, Dong L, Shrestha L, et al. The structural basis of ZMPSTE24-dependent laminopathies. Science. 2013;339:1604–1607. doi: 10.1126/science.1231513. [DOI] [PubMed] [Google Scholar]
- 24.Pryor EE, Jr, Horanyi PS, Clark KM, Fedoriw N, Connelly SM, et al. Structure of the integral membrane protein CAAX protease Ste24p. Science. 2013;339:1600–1604. doi: 10.1126/science.1232048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 26.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sondergaard CR, Olsson MHM, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pK(a) values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 29.Liao J, Li H, Zeng W, Sauer DB, Belmares R, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335:686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 30.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waight AB, Pedersen BP, Schlessinger A, Bonomi M, Chau BH, et al. Structural basis for alternating access of a eukaryotic calcium/proton exchanger. Nature. 2013;499:107–110. doi: 10.1038/nature12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M, Tong S, Waltersperger S, Diederichs K, Wang M, et al. Crystal structure of Ca2+/H+ antiporter protein YfkE reveals the mechanisms of Ca2+ efflux and its pH regulation. Proc Natl Acad Sci USA. 2013;110:11367–11372. doi: 10.1073/pnas.1302515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishizawa T, Kita S, Maturana AD, Furuya N, Hirata K, et al. Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger. Science. 2013;341:168–172. doi: 10.1126/science.1239002. [DOI] [PubMed] [Google Scholar]
- 34.Kiviluoto S, Luyten T, Schneider L, Lisak D, Rojas-Rivera D, et al. Bax inhibitor-1-mediated Ca2+ leak is decreased by cytosolic acidosis. Cell Calcium. 2013;54:186–192. doi: 10.1016/j.ceca.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Kim HR, Lee GH, Ha KC, Ahn T, Moon JY, et al. Bax Inhibitor-1 Is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J Biol Chem. 2008;283:15946–15955. doi: 10.1074/jbc.M800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 37.Grzmil M, Kaulfuss S, Thelen P, Hemmerlein B, Schweyer S, et al. Expression and functional analysis of Bax inhibitor-1 in human breast cancer cells. J Pathol. 2006;208:340–349. doi: 10.1002/path.1902. [DOI] [PubMed] [Google Scholar]
- 38.Grzmil M, Thelen P, Hemmerlein B, Schweyer S, Voigt S, et al. Bax inhibitor-1 is overexpressed in prostate cancer and its specific down-regulation by RNA interference leads to cell death in human prostate carcinoma cells. Am J Pathol. 2003;163:543–552. doi: 10.1016/S0002-9440(10)63682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka R, Ishiyama T, Uchihara T, Inadome Y, Iijima T, et al. Expression of the Bax inhibitor-1 gene in pulmonary adenocarcinoma. Cancer. 2006;106:648–653. doi: 10.1002/cncr.21639. [DOI] [PubMed] [Google Scholar]
- 40.Sano R, Hou YCC, Hedvat M, Correa RG, Shu CW, et al. Endoplasmic reticulum protein BI-1 regulates Ca2+-mediated bioenergetics to promote autophagy. Gene Dev. 2012;26:1041–1054. doi: 10.1101/gad.184325.111. [DOI] [PMC free article] [PubMed] [Google Scholar]