Summary
Southern China is proposed as an influenza epicentre. At least two of the three pandemics in the last century, including 1957 and 1968 influenza pandemics, originated from this area. In 1996, A/goose/Guangdong/1/1996 (H5N1), the precursor of currently circulating highly pathogenic H5N1 avian influenza viruses (HPAIVs) was identified in farmed geese in southern China. These H5N1 HPAIVs have been spread across Asia, Europe and Africa and poses a continuous threat to both animal and human health. However, how and where this H5N1 HPAIV emerged are not fully understood. In the past decade, many influenza surveillance efforts have been carried out in southern China, and our understanding of the genetic diversity of non-human influenza A viruses in this area has been much better than ever. Here, the historical and first-hand experimental data on A/goose/Guangdong/1/1996(H5N1)-like HPAIVs are reviewed within the context of the findings from recent surveillance efforts on H5N1 HPAIVs and other non-human influenza A viruses. Such a retrospective recapitulation suggests that long-term and systematic surveillance programmes should continue to be implemented in southern China that the wet markets on the animal–human interface shall be the priority area and that the surveillance on the animal species bridging the interface between wildlife and domestic animal populations and the interface between the aquatics and territories shall be the strengthened.
Keywords: Influenza A virus, ecology, molecular evolution, southern China, H5N1 highly pathogenic avian influenza virus, A/goose/Guangdong/1/1996(H5N1)
Introduction
Southern China has a unique ecological system with many bodies of water, intensive farming systems, a highly dense human population and dynamic industrial settings. This area is designated as a putative influenza epicentre (Shortridge and Stuart-Harris, 1982). Both the 1957 and 1968 influenza pandemic originated from this area (Chang, 1969; Cox and Subbarao, 2000), and the emergence of 1918 influenza pandemic was also proposed to emerge from southern China (Shortridge, 1999). About 15 years ago, A/goose/Guangdong/1/1996(H5N1) (briefly Gs/Gd/96), the precursor of currently circulating H5N1 highly pathogenic avian influenza viruses (HPAIVs), was identified in farmed geese at Sanshui, Foshan, a rural area in southern China (Wan, 1998). Since then, this emerging H5N1 HPAIV has spread across Asia, Europe and Africa and caused more than 600 human cases and millions of deaths in birds (WHO, 2012, Peiris et al., 2007), and these numbers are still increasing. Thus, these H5N1 HPAIVs pose a continuing threat to both animal and human health. Currently, how and where Gs/Gd/96-like H5N1 viruses emerged are not understood fully.
In the past decade, many influenza surveillance efforts, especially from the surveillance programme led by the University of Hong Kong, have been conducted in southern China. Knowledge about influenza prevalence and evolution in this area is becoming much clearer. The genomic dynamics of H5N1 HPAIVs (Duan et al., 2008), H9N2 low-pathogenic avian influenza viruses (LPAIVs) (Xu et al., 2007), H6 LPAIVs (Huang et al., 2010; Cheung et al., 2007) and recently H1N1 swine influenza virus (Vijaykrishna et al., 2010) have been illustrated in wet markets. Sporadic cases of H9N2 avian influenza infections in humans were identified (Peiris et al., 1999). A number of novel non-human influenza viruses, such as H3N2 canine influenza virus (Li et al., 2010) and H6N6 swine influenza viruses (Zhang et al., 2011), were detected recently. Findings from these surveillance efforts helped provide a baseline to define an influenza situation as normal or abnormal.
This review article recapitulates the history and firsthand experimental data on A/goose/Guangdong/1/1996(H5N1), emergence and evolution of H5N1 HPAIVs and other non-human influenza A viruses, and recent influenza surveillance efforts. The insights on the influenza surveillance programme are then discussed.
Ecosystem and Animal Agricultural System in Southern China
Geographically, China is classified into two regions, northern China and southern China, whose common boundary is defined broadly by the Huai River-Qinling Mountains line and narrowly by the Dayuling-Qitianling-Dupangling-Mengzhuling-Yuechengling Mountains line. In the context of this article, southern China is referred to the one by the narrow definition.
Southern China includes Guangdong, Guangxi, Hainan, Hong Kong and Macau SAR, Taiwan, and southern parts of Fujian, Jiangxi and Hunan Provinces (Fig. 1). Southern China has a unique ecosystem with a complicated agricultural system and interwoven with numerous lakes, rivers, creeks and ponds. The major river systems in this area include the Pearl River, the third largest river in China. The Pearl River basin drains the majority of Guangdong and Guangxi Provinces and four other provinces, Yunan, Guizhou, Hunan, and Jiangxi Provinces.
Fig. 1.
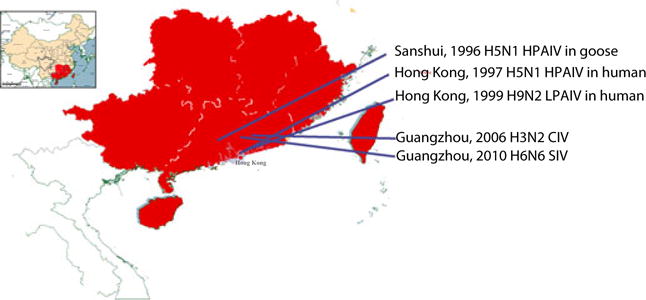
Geographic map of southern China. Southern China and northern China are narrowly defined by the boundary of the Dayuling-Qitian-ling-Dupangling-Mengzhuling-Yuechengling Mountains line. Southern China includes Guangdong, Guangxi, Hainan, Hong Kong and Macau SAR, Taiwan, and southern parts of Fujian, Jiangxi and Hunan Provinces. The locations of recently emerging influenza A viruses are marked.
Southern China is an important wintering area for migratory birds, for example the Haifeng wetland in eastern Guandong, the Futian and Mai Po wetlands in southeastern Guangdong, and coastal wetlands in the Leizhou Peninsula in southern Guangdong (Wang et al., 1993; Hu et al., 2011; Zou et al., 2006). Southern China is one of the major animal agricultural systems in China. In addition to chickens, waterbirds such as geese and ducks are major poultry species in this area. In the past three decades, the domestic poultry and animal population sizes have increased dramatically. For instance, in Guangdong Province, the chicken meat production in 1995 doubled from that in 1990, and the pork production in 2004 increased 70% over that in 1990 and 300% over that in 1980 (Guangdong Statistical Bureau, 2005). These large industrialized chicken and swine production units have becoming an important part of the influenza ecosystem (Fig. 2). This surge of domestic animal and bird population increases the likelihood of contacts among domestic, commercial and wild animals.
Fig. 2.
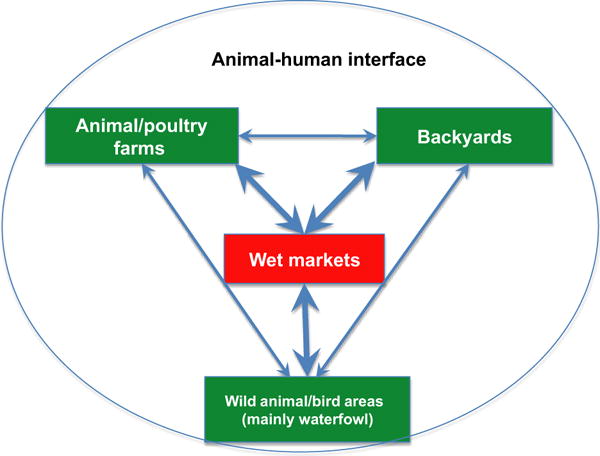
A model for the influenza ecosystem. The major bird species in poultry farms, backyards and wet markets in southern China include chickens, ducks, geese and quails; the major animal species in farms and wet markets in southern China is swine.
Circulation of Avian Influenza Viruses in Southern China Before 1996
Since the 1970s, many avian influenza virus types have been isolated from domestic duck and goose in southern China (Shortridge, 1992). From 1975 to 1979, 480 avian influenza viruses with at least 43 combinations of haemagglutinin (HA) and neuraminidase (NA) subtypes were identified from domestic ducks, geese or chickens imported to Hong Kong from mainland China or from those raised in Hong Kong (Shortridge et al., 1977a, 1979; Shortridge, 1979, 1982). A total of 10 HA subtypes (H1–H7, H9–H11) and 9 NA subtypes (N1-N9) were recovered. The HA/NA subtypes recovered from chickens includes H1N1, H3N2, H3N6, H3N9 and H6N4; and those from ducks include H1N1, H1N2, H1N3, H2N2, H2N3, H2N9, H3N2, H3N3, H3N4, H3N6, H3N8, H4N1, H4N2, H4N3, H4N4, H4N5, H4N6, H4N7, H4N8, H5N2, H5N3, H6N1, H6N2, H6N3, H6N4, H6N5, H6N6, H6N8, H6N9, H7N2, H9N2, H9N6, H10N1, H10N2, H10N3, H10N4, H10N5, H10N8, H10N9, H11N2, H11N3, H11N9; those from goose include H1N1, H3N2, H3N8, H4N2, H4N5, H4N6, H5N3, H6N1, H6N2, H6N4 and H6N9 (Shortridge, 1982). Most of the HA–NA combinations in contemporary non-human influenza A viruses in southern China were among those identified in the 1970s.
With such prevalence and diversity of avian influenza viruses in domestic poultry raised in close proximity to humans, southern China was designated by Shortridge and Stuart-Harris in 1982 as a hypothetical epicentre for the emergence of pandemic influenza viruses (Shortridge and Stuart-Harris, 1982).
Emergence of A/goose/Guangdong/1/1996 (H5N1) in Southern China
In early spring of 1996, several outbreaks occurred in farmed geese at Sanshui, a small town about 50 miles west of Guangzhou, the capital of Guangdong Province (Fig. 1). Sanshui occupies the section of Pearl River where three river sub-branches interact, one branch from Guangxi Province, one from Guangdong, and one from Jiangxi.
In 1996, the mortality of H5N1 in farmed geese in Sanshui was more than 40%, and typical symptoms included bleeding and neurological dysfunction. Two viruses, A/goose/Guangdong/1/1996 (Gs/Gd/96-1) and A/goose/Guangdong/2/1996 (Gs/Gd/96-2), were isolated from these sick geese and were subtyped by the Chinese National Influenza Center as H5N4 initially but retyped as H5N1 in 1997 (Xu et al., 1999; Guo et al., 1998, 1999a,b). These results and associated characterization were reported first on 6 October 1996, at the 8th symposium on avian medicine for the Chinese Society of Animal Husbandry and Veterinary Medicine, Qingdao, China. Both Gs/Gd/96-1 and Gs/Gd/96-2 were shown to be HPAIVs. The genomic sequencing showed that both Gs/Gd/96-1 and -2 have the connecting peptide of R-E-R-R-R-K-K-R between HA1 and HA2, which is similar to those connecting peptides of the H5N1 isolates isolated from humans and chickens at Hong Kong in 1997 (Guo et al., 1999b).
The pathogenesis of Gs/Gd/96-2 was tested in 4-week-old Qingyuan ducks, which were grouped 8 in an experimental group and 8 in a control group. Each duck was infected with 1.2 × 106 EID50 of Gs/Gd/96-2. None of the infected ducks showed any symptoms within 2 weeks post-infection (Wan, 1998).
Two breeds of chickens, Shiqiza broilers (locally grown species) and AA broilers (commercially imported species), were housed in pens off the floor. These birds were infected with Gs/Gd/96-2 virus through five different routes: intravenous injection, intramuscular injection, ocular and nasal instillation, drinking water, and penmate contact (Wan, 1998). The results showed that this H5N1 virus was more pathogenic to AA broilers than to Shiqiza broilers. For instance, Gs/Gd/96-2 (about 1.2 EID50 as infection dose) did not cause any death in birds through intramuscular injection, whereas the same amount of Gs/Gd/96-2 viruses given by intramuscular injection caused six deaths among the eight infected AA broilers. Similar observations were seen in experiments using both ocular and nasal instillation routes. Both chicken species did not show any clinical symptoms when infected via drinking water or penmate contact, which indicated that this precursor virus was not transmitted efficiently among chickens.
From January 1996 to December 1997, influenza surveillance of sick chickens on poultry farms was performed across Guangdong Province, and none of the viral isolates obtained from the chicken population was H5N1 HPAIV (Wan, 1998). The failure to identifying this Gs/Gd/96-like virus (we called PR1 (Zhao et al., 2008)) in the Guangdong chicken population suggested that this virus was not likely to be transmitted efficiently among chickens (in addition, no large outbreaks were reported) during this time period.
However, later findings showed that Gs/Gd/96-like virus mutated and became more easily transmitted by faecal-oral ingestion from waterfowl to land-based (territory) poultry, such as chickens and quails. A Gs/Gd/96-like virus, A/goose/Hong Kong/437-4/99, was detected and shown to be transmitted from infected geese to chickens by faecal contact only but to quail by either aerosol or faecal routes (Webster, 2002).
The emergence of Gs/Gd/96-like virus in the Sanshui area was associated with an avian influenza virus transmitted possibly by birds migrating across the Eurasian land mass. Our results showed that both HA and NA genes of Gs/Gd/96-like viruses most likely evolved from A/turkey/England/50-92/91(H5N1) (E91-like viruses), whereas polymerases (PB2, PB1, PA), nucleoprotein (NP) and non-structural (NS) genes originated from A/duck/Nanchang/1681/92(H3N8) (NC92-like viruses), which were isolated in southern China about 3 years earlier than the detection of Gs/Gd/96-like viruses. The matrix (MP) genes of Gs/Gd/96-like viruses were from an unknown lineage. This reassortant Gs/Gd/96-like virus (named also as PR1) (Zhao et al., 2008) was reported last in 2001 in the waterfowl in Hong Kong (Webster et al., 2002).
Emergence of Human H5N1 HPAIVs in Hong Kong
In late April and May of 1997, two outbreaks with high avian mortality were reported on chicken farms located in a northwest area of Hong Kong (Sims et al., 2003). This outbreak occurred about 1 year later than did the avian influenza outbreaks in farmed geese in Guangdong Province. The first human case of H5N1 avian influenza was confirmed in May 1997, and 17 more occurred in November and December 1997. This outbreak led to a depopulation of about 1.5 million birds in Hong Kong.
In the literature and reports, investigations have always reported that the H5N1 HPAIVs in Hong Kong were brought in along with the poultry imported from Guangdong Province and were linked directly to Gs/Gd/96-like isolates. We attempted to identify the progenitor genes for these H5N1 viruses isolated from the birds and patients in Hong Kong in 1997 (so called HK/97-like virus)(Zhao et al., 2008). Our results show that the Gs/Gd/96-like virus has a very different genomic constellation from that in HK/97-like virus, and only HA, PB2, PB1 and NP segments of Gs/Gd/96-like virus and those of HK/97-like virus share similar progenitors. In HK/97-like virus, NA evolved possibly from A/duck/Nanchang/04/92(H7N1)-like virus; PA from A/turkey/England/N28/73(H5N2)-like virus; and matrix (MP) and NS from A/chicken/Hong Kong/739/94(H9N2)-like virus. This HK/97-like virus (genotype PR2) disappeared after slaughtering 1.5 million chickens in Hong Kong and stopping the live poultry trade for 7 weeks (Sims et al., 2003).
Re-Emergence of H5N1 HPAIVs
The H5N1 HPAIV did not disappear from Hong Kong, although HK/97-like virus was not detected any more. In 1999, H5N1 HPAIV was detected again in waterfowl in Hong Kong (Webster et al., 2002). Moreover, in February 2003, two human cases were confirmed and caused by a novel H5N1 genetic variant in Hong Kong. Since the end of 2003, H5N1 outbreaks in chickens have spread over eastern and southeastern Asia (Li et al., 2004). In May 2005, another novel H5N1 genetic variant was identified in Qinghai Lake (Chen et al., 2005) and that variant later spread further to the Mideast, Europe and Africa.
Genotypic analysis indicated at least 20 reassortants emerged since the first detection of Gs/Gd/96-like virus (Zhao et al., 2008) and new reassortants continue to emerge. Among these reassortants, the genotype of PR7 (Zhao et al., 2008) has become predominant and that genotype was responsible for all human infections since 2003 as well as for the major outbreaks in poultry and wild birds. The genomic constellation of PR7 is different from those of both Gs/Gd/96-like (PR1) and HK/97-like (PR2) viruses. However, PR7 is more similar to Gs/Gd/96-like virus because between PR7 and PR1 only the progenitor of NS genes was different. In vitro experiments showed that this NS gene enhanced virus replication in mammalian cells (Twu et al., 2007). Different from PR1, the viruses in PR7 were identified in land-based bird, such as chickens.
During 2003/2004 H5N1 outbreaks in Southern Asia, the viruses evolved to be transmitted efficiently in and among various hosts, especially between waterfowl (e.g., ducks and geese) and land-based birds (e.g., chickens). The host range for H5N1 HPAIVs has been broadened, and these H5N1 viruses have been isolated from a broad range of species, such as crane, chicken, crow, duck, egret, grey heron, gull, partridge, pheasant, quail, sparrow, swan, swine, teal, tiger, and turkey. These H5N1 viruses can be transmitted among ducks even through feather dust (Yamamoto et al., 2007).
Waterfowl are accepted generally as the major natural reservoir for avian influenza viruses. Generally, these viruses do not cause any disease in waterfowl. Before 2002, the H5N1 HPAIVs isolated from birds in China did not cause any symptoms or deaths in ducks (Chen et al., 2004). However, in 2002, H5N1 HPAIVs with a high virulence to ducks were isolated (Pantin-Jackwood and Swayne, 2007). During the 2003/2004 outbreaks, the virulence for ducks of H5N1 isolates varied: some of these H5N1 strains were of low pathogenicity to mallards, whereas other strains were highly pathogenic to mallards (Sturm-Ramirez et al., 2005). However, experiments demonstrated further that the pathogenesis of these H5N1 viruses varied according to duck species. For example, in five tested duck species in Northern America, only the wood duck is sensitive to A/duck meat/Anyang/01 (H5N1) and A/W.swan/Mongolia/244/05 (H5N1) (Hurt et al., 2007). Besides duck species, the age of the duck hosts may affect results in pathogenesis experiments (Steensels et al., 2007; Pantin-Jackwood et al., 2007). In general, the pathogenesis of these H5N1 viruses in waterfowl has increased since 1996. Experiments showed that H5N1 HPAIVs can revert from high to low pathogenicity after infecting ducks (Hulse-Post et al., 2005). This suggests the important role of ducks in virus evolution and transmission.
Emergence and Re-Emergence of Low-Pathogenic Avian Influenza Viruses, Canine Influenza Viruses and Swine Influenza Viruses in Southern China
H5N1 incidences, especially the outbreak of H5N1 human infections in Hong Kong in 1997, re-emergence of H5N1 human cases since 2002, and the 2003–2004 H5N1 outbreaks in Southeast Asia raised public concern for emergence of another influenza pandemic. More funds and efforts were made available to conduct influenza surveillance in southern China. It is worth mentioning that, especially after 2000, a systematic surveillance programme has been established in this area by Guan and other colleagues from Hong Kong University. These surveillance efforts have greatly enhanced our understanding about influenza genetic diversity in both domestic chicken and ducks in southern China. Besides the discovery of genomic dynamics of H5N1 HPAIVs described above, a number of other subtypes of non-human influenza A viruses have been detected in domestic animals and birds. This review does not intend to review all findings derived from influenza surveillance carried out in southern China area but rather to highlight a few subtypes of avian influenza viruses, especially those reported in the animal species (e.g., swine and canine), other than avian species.
The serotype of H9 avian influenza virus was among those identified in the surveillance performed by Shortridge and other colleagues in 1970s (Shortridge, 1982). One H9N6 and sixteen H9N2 isolates were recovered from domestic ducks in southern China. In the earlier 1990s, the H9N2 LPAIV was endemic in southern China, and consequently inactivated H9N2 vaccine has used widely in the poultry industry in this area (and across most areas in China). Genomic analysis showed that H9N2 LPAIV evolved rapidly in the past two decades, and many genotypes of H9N2 have emerged (Choi et al., 2004; Chu et al., 2011; Xu et al., 2007; Li et al., 2003). The wide prevalence of H9N2 virus in domestic chicken and duck populations presents a large impact on the evolution of other subtypes of influenza A viruses. The genetic pool of H9N2 viruses was shown to supply donor genes in the emergence of novel genotypes of H5N1 HPAIVs (Guan et al., 1999), and reassortants between H9N2 and other subtypes of avian influenza viruses, such as H6, are very common also. In addition, the subtype of H9N2 influenza A virus was circulating in the swine population in southern China (Zhang et al., 2011). The H9N2 LPAIV was confirmed to cause at least four human infections (Peiris et al., 1999; Butt et al., 2005), and up to 15.5% of poultry retailers had a seroconversion against H9N2 LPAIV (Li et al., 2003).
Through surveillance in the live bird market since 2002, H6 subtypes of LPAIVs (such as H6N1, H6N2 and H6N6) were shown to be endemic, primarily in ducks (Huang et al., 2010; Cheung et al., 2007). Frequent reassortments between H6 and H5N1 HPAIVs were observed also (Chin et al., 2002). Recently, an avian-origin H6N6 influenza A virus was detected in the swine population, and serological assay demonstrated there was 1.9% prevalence of H6 subtypes in swine population southern China (Zhang et al., 2011).
Compared with H9 and H6 AIVs, H3 subtypes of AIVs were isolated less frequently from wet market poultry, although this subtype had been reported earlier in the domestic duck population (Shortridge et al., 1979). The H3 subtypes from live bird market surveillance included H3N2, H3N3, H3N6 and H3N8. However, serological surveillance from 2006 to 2007 demonstrated about 2.83% of farmed poultry in China was positive for H3 subtype of avian influenza virus (Pu et al., 2009). In the winter of 2006, H3N2 avian influenza virus was identified in sick dogs (Li et al., 2010). Interestingly, this virus was similar to the H3N2 canine influenza virus reported in Korea during approximately the similar time period (Song et al., 2009). This H3N2 canine influenza virus under experimental conditions caused both dog and cat infections (Song et al., 2009, 2011). It is not known whether this avian-origin H3N2 canine influenza virus emerged from southern China or was transmitted to this area from Korea.
In swine, besides the newly emerging H9N2 and H6N6 swine influenza viruses, classic H1N1, H1N2 reassortant and H3N2 subtypes of influenza A viruses were identified within this swine population (Yu et al., 2011; Zhang et al., 2011; Shortridge and Webster, 1979; Shortridge et al., 1977b). Recently, 2009 pandemic H1N1 was identified in the swine slaughter house, and a number of reassortants were observed (Vijaykrishna et al., 2010).
Lessons from Emergence of Gs/Gd/96-Like H5N1 HPAIVs and Recent Surveillance Efforts
Emergence of Gs/Gd/96-like H5N1 virus, especially HK97-like virus, reshaped the central dogma of influenza ecology, which used to indicate that influenza A virus must first adapt to swine (as a mixing vessel) before infecting human. From 2003 to 2009, a global framework for influenza surveillance and reporting system has been established towards pandemic influenza preparedness: ‘minimize the risk of further spread in animal populations’, ‘reduce the risk of human infections’ and ‘further support pandemic planning and preparedness’ (http://www.usaid.gov/our_work/global_health/home/News/news_items/ai_activities.html). Such a framework provided foundations for making policies and initiating rapid actions in 2009 swine-origin H1N1 influenza pandemic prevention and control.
The lessons derived from influenza surveillance carried out in Hong Kong, and other over the past four decades, especially the recent 10 years, set the scene for what was ‘normal’ and ‘abnormal’ influenza activity, and, also importantly, the development of a culture of awareness across government, university, medical personnel, veterinarian, the media and other groups. These surveillance efforts provide a foundation for review of the history of emergence of Gs/Gd/96-like viruses and HK/97-like viruses. This retrospective capitulation reflects the challenges in the current surveillance programme and contributes to designing a more effective influenza surveillance programme. More surveillance data are being generated will be helpful for the scientists and influenza workers on either side of the Pacific (as well as other regions) to detect introduction of Gs/Gd/96-like H5N1 HPAIVs or emergence of novel influenza viruses associated with Gs/Gd/96-like H5N1 HPAIVs thus to effectively implement tough decisions in influenza prevention and control.
Earlier genetic analyses have suggested waterfowl are the major sources of the progenitors of the Gs/Gd/96-like, HK/97-like viruses, and other genotypes of H5N1 HPAIVs. However, how and where Gs/Gd/96-like viruses emerged still remain a puzzle because many of the progenitors were from those avian influenza viruses identified in 1970s and from the areas other than southern China (e.g., England). These progenitor genes have large genetic divergences from those in Gs/Gd/96-like, HK/97-like, and other H5N1 HPAIVs (Zhao et al., 2008). This suggested that there was still a large gap in the knowledge of influenza virus genetic diversity in both domestic and wild bird populations in southern China, mostly due to the lack of systematic surveillance before and even after the detection of Gs/Gd/96-like viruses in southern China. This confirms further that influenza surveillance is the key to identifying novel influenza viruses and potential threats and to monitoring the dynamics of influenza A viruses.
The progenitor genes for Gs/Gs/96-like viruses were shown to generate reassortants as LPAIVs, such as A/swan/Hokkaido/51/96(H5N3), in Japan. Recent surveillance findings, especially the discovery of H5N1 HPAIVs in Qinghai Lake (Chen et al., 2005; Liu et al., 2005) and the links of H5N1 HPAIVs from Qinghai Lake and Poyang Lake (Li et al., 2004), have confirmed the roles of migratory birds in the emergence of H5N1 HPAIVs. The detection of genetically diverse H5N1 HPAIVs in wild birds, especially duck and goose, highlights the potential roles of these viruses in emergence of Gs/Gd/96-like and HK/97-like viruses. Five of the gene progenitors of Gs/Gd/96-like viruses were associated genetically with those in A/duck/Nanchang/1681/92(H3N8) (NC92-like viruses), which were identified in Nanchang, about 100 miles southwest Poyang Lake. Recent surveillance in the Poyang Lake area suggested there was a large genetic diversity of avian influenza viruses in migratory duck and sentical ducks in this area (Duan et al., 2011). As mentioned in the section of Re-emergence of H5N1 HPAIV, pathogenesis of H5N1 HPAIVs may vary according to duck species (Hurt et al., 2007) or age (Steensels et al., 2007; Pantin-Jackwood et al., 2007), and this finding suggested great challenges in influenza surveillance in migratory waterfowl: which bird species should be included?
In 1996, all the domestic geese on the farms with Gs/Gd/96 H5N1 outbreaks were culled soon after the disease outbreaks were reported. However, Gs/Gd/96-like viruses did not disappear and were detected again in domestic geese in Hong Kong in 1999 (Webster et al., 2002). It is very likely that these Gs/Gd/96-like H5N1 viruses were present asymptotically in some avian species (e.g., duck). Other surveillance suggested there were a large genetic diversity of H5N1 HPAIVs in domestic and wild ducks in eastern China before the 2002–2003 H5N1 outbreaks (Chen et al., 2004). Whether Gs/Gd/96-like viruses emerged first in other areas (e.g., Poyang Lake or even eastern China) and transmitted to southern China is still a mystery.
The transmission of H5N1 and H9N2 avian influenza viruses to humans strengthened the concept of ‘one health’ in influenza surveillance. The exchange of genomic segments among influenza A viruses, especially among avian influenza viruses, swine influenza viruses and human seasonal influenza strains, can facilitate the emergence of a novel pandemic influenza A virus. The prevalence of H3, H4, H5, H6, H9 in avian, emergence of H4N8, H6N6, H5N1 (Shi et al., 2008) and H9N2 in swine, and H3 in canine (antigenically different from currently circulating human H3N2 seasonal influenza viruses) has posed threats to human health. The genomic characterization and across species transmission of H5N1 and other low-pathogenic avian influenza viruses, such as H3, H4, H6 and H9, strengthened the concept of ‘one flu’ in influenza surveillance. Thus, interface surveillance of the animal–human interface shall be a top priority, and genomic constellation of the influenza viruses recovered from animal–human interface will need to be characterized.
The lessons through the emergence of Gs/Gd/96-like viruses and the findings from recent surveillance in southern China provide us knowledge about an ecological setting as an influenza epicentre, which can help identify potential influenza epicentres in other areas. For instance, a novel swine-origin H1N1 pandemic virus emerged in North America in 2009 (Dawood et al., 2009). The experience in southern China could be useful in identifying the influenza epicentre in North America. Mississippi Bird Migration Flyway spans the Northern, Midwestern and Southeastern United States, and these areas form a unique ecological system: (i) the 8 of top 10 swine producing states are located along this flyway; (ii) Arkansas, Mississippi and Missouri produce more than 25% of poultry in the United States; and (iii) Large aquatic bodies and fish farming are located in the area, for example the Great Lakes and Mississippi Delta. Would these areas across Mississippi Bird Migration Flyways form another influenza epicentre?
In summary, continuous and systematic influenza surveillance shall be implemented in southern China, and the animal–human interface will be a top priority. Besides southern China, other neighbouring areas, such as central China and eastern China, should be included in the surveillance system, if possible. Findings from influenza surveillance in other countries neighbouring southern China, for example Vietnam, should be integrated into influenza data analyses.
The Priority of Influenza Surveillance in Southern China
In general, the influenza ecosystem can be summarized into four major subsystems: farms, backyards, wildlife and wet markets (Fig. 2). The wet markets here included both the live animal market and the animal slaughtering houses. The farms and wildlife subsystems, with the largest domestic and wild animal populations, respectively, serve as two major natural reservoirs for influenza viruses. The wet markets facilitate influenza A viruses to overcome geographic barriers by connecting farms, backyards and wildlife subsystems. Instead, backyard exposures play an important role in facilitating the contacts between wild and domestic animals. Because influenza A viruses have a wide range of natural hosts, it is not possible to design a surveillance protocol to cover all the species in these four subsystems, but what should be the priority in an effective surveillance programme in southern China?
Because influenza A viruses from farms, backyards and wildlife are able, potentially, to mix at wet markets, the genetic pool from wet markets could mimic those from influenza ecosystems. On the other hand, at the wet markets, viruses from farms, backyards and wild areas may interact with pre-existing influenza viruses present in the wet markets, including those from animals and humans, and such interactions facilitate emergence of novel influenza viruses. Thus, wet markets serve sources for the emergence of novel influenza A viruses and are major sources for human H5N1 infections in China (Wan et al., 2011). Compared with the surveillance in the other three subsystems, surveillance in wet markets, such as live bird markets and swine slaughter houses, is relatively easier and more economic to be conducted. Thus, long-term and systematic surveillance in wet markets should be continued, and the species covered should be as wide as possible, and at least include swine and the majority poultry species, chicken, goose and duck, which are the major components among the animal–human interface.
Compared with wet markets, such surveillance of farmed animals (e.g., swine, chicken, duck and goose) in southern China is relatively limited. Because the ecology of the wet market subsystem is different from that of the farm subsystem (e.g. the number and species diversity of hosts), the genomic constellation and prevalence of viral population recovered from the wet markets do not necessarily reassemble those on farms. In the 2003–2004 H5N1 outbreaks, the H5N1 genotypes identified from farms were not the same as those reported elsewhere from live bird markets (Wan et al., 2005; Li et al., 2004). Thus, given the phenomenal increases in chicken and swine production, disease-based surveillance will be critical not only in detecting the emerging and re-emerging influenza strains but also in assessing the epidemic situation. A sound disease-based virus surveillance system will need to consider a number of factors, such as animal species, animal population size, and animal population distribution. Seroconversion rate is commonly used as an important parameter in disease-based influenza surveillance.
Migratory waterfowl serves as natural reservoirs of influenza A viruses. Lessons from emergence of Gs/Gd/96-like H5N1 HPAIVs suggested the important role of migratory birds in influenza transmission and epidemics: failure of Eastern Asia to react to the detection of Gs/Gd/96-like viruses in geese in Hong Kong in 1999 and the development of H5N1 genotypes in China thereafter until the H5N1 situation could no longer be contained in 2003; failure to prevent the spread of H5N1 viruses from eastern China to Qinghai Lake in 2005 and subsequent spread across the Eurasian landmass. Compared with those in wild bird populations, the genotypic diversity of influenza viruses exhibited in domestic ducks could represent only as ‘a tip of an iceberg’ because migratory waterfowl serve as natural reservoirs of influenza A viruses. Surveillance in wild bird population has been very challenging and many times not achievable without banding or even hunting those birds.
Among all the natural hosts of influenza A viruses, geese are likely to play a unique role in influenza ecology in this area. Geese act as one of the major farmed and backyard poultry species. Geese belong to the genus of Anser under the family of Anatidae. The goose has an avian-like α2,3 linked sialic acid receptor through the respiratory track and human-like α2,6 sialic acid in the colon (Kimble et al., 2010). Although ducks are members of Anatidae, they have different habitats from geese. Ducks spend most of their time in wetlands, whereas geese are free-range birds and spend much time on the dry land. Thus, geese have opportunities to serve as an intermediate host among waterfowl (domestic and wild), land-based poultry (e.g., chicken and quail), swine and other domestic animals, such as dogs (Fig. 2). A variety of influenza viruses were identified in domestic geese in southern China (Guan et al., 2002; Wan, 1998; Shortridge et al., 1977a). In summary, geese could play an important role in the ecology of influenza, connecting wild animal populations to domestic poultry and waterfowl to land-based birds and animals. Thus, geese must be one major species in influenza surveillance and be strengthened in southern China.
In summary, with the importance of all four subsystems covered in influenza surveillance, wet markets represent one of the major animal–human interfaces and thus shall be continued as a priority in influenza surveillance. On the other hand, the animal species, for example geese, bridging the interface between wild and domestic animals and the interface between the water and land should be strengthened.
Conclusions
Southern China provides a unique ecological system optimal for influenza viral emergence. The lessons from the emergence of A/goose/Guangdong/1/1996 (H5N1) and recent virus surveillance efforts suggested these emerging influenza A viruses in southern China as a continuous threat to human health. A long-term and systematic surveillance programme should continue to be implemented in this area. Genomic characterization of influenza viruses helps us understand their genomic constellation and evolutionary pathway and detect a novel influenza virus, thus genomic characterization shall be an essential component of influenza surveillance programme.
Impacts.
Southern China provides a unique ecological system optimal for influenza viral emergence, and emergence of A/goose/Guangdong/1/1996 (H5N1) (Gs/Gd/96) presents a continuous threat to both animal and human health.
Gs/Gd/96-like viruses, HK/97-like viruses, and currently circulating H5N1 HPAIVs have different genomic constellations, and where and how these viruses emerged are not fully understood.
Long-term and systematic surveillance programmes shall continue to be implemented in southern China, the wet markets on the animal–human interface should be priority study area, and the surveillance on the animal species, such as the geese, bridging the interface between wild and domestic animals and the interface between the aquatics and territories shall be the strengthened.
Acknowledgments
This project was supported by NIH NIAID RC1AI086830. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH. We thank three anonymous reviewers for critical comments and Dr. John Harkness for editorial support.
Footnotes
Conflicts of interest
The authors have not declared any potential conflicts.
References
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WK. National influenza experience in Hong Kong, 1968. Bull World Health Organ. 1969;41:349–351. [PMC free article] [PubMed] [Google Scholar]
- Chen H, Deng G, Li Z, Tian G, Li Y, Jiao P, Zhang L, Liu Z, Webster RG, Yu K. The evolution of H5N1 influenza viruses in ducks in southern China. Proc Natl Acad Sci USA. 2004;101:10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJ, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JS, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Vijaykrishna D, Smith GJ, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, Poon LL, Peiris JS, Chen H, Guan Y. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin PS, Hoffmann E, Webby R, Webster RG, Guan Y, Peiris M, Shortridge KF. Molecular evolution of H6 influenza viruses from poultry in Southeastern China: prevalence of H6N1 influenza viruses possessing seven A/Hong Kong/156/97 (H5N1)-like genes in poultry. J Virol. 2002;76:507–516. doi: 10.1128/JVI.76.2.507-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, Butt C, Leung YH, Guan Y. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol. 2004;78:8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YC, Cheung CL, Hung Leung CY, Man Poon LL, Chen H, Peiris JS, Guan Y. Continuing evolution of H9N2 influenza viruses endemic in poultry in southern China. Influenza Other Respi Viruses. 2011;5(Suppl. 1):68–71. [PubMed] [Google Scholar]
- Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Duan L, Bahl J, Smith GJ, Wang J, Vijaykrishna D, Zhang LJ, Zhang JX, Li KS, Fan XH, Cheung CL, Huang K, Poon LL, Shortridge KF, Webster RG, Peiris JS, Chen H, Guan Y. The development and genetic diversity of H5N1 influenza virus in China, 1996–2006. Virology. 2008;380:243–254. doi: 10.1016/j.virol.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Zhu H, Wang J, Huang K, Cheung CL, Peiris JS, Chen H, Guan Y. Influenza virus surveillance in migratory ducks and sentinel ducks at Poyang Lake, China. Influenza Other Respi Viruses. 2011;5(Suppl. 1):65–68. [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Peiris M, Kong KF, Dyrting KC, Ellis TM, Sit T, Zhang LJ, Shortridge KF. H5N1 influenza viruses isolated from geese in Southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology. 2002;292:16–23. doi: 10.1006/viro.2001.1207. [DOI] [PubMed] [Google Scholar]
- Guangdong Statistical Bureau. Guangdong Statistical Yearbook. China Statistics Press; Beijing, China: 2005. [Google Scholar]
- Guo Y, Xu X, Wan X. Genetic characterization of an avian influenza A (H5N1) virus isolated from a sick goose in China. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1998;12:322–325. [PubMed] [Google Scholar]
- Guo Y, Wang M, Guo J. The complete nucleotide sequences of A/Goose/Guangdong/2/96(H5N1) virus RNA segment 1–3 and 5. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999a;13:205–208. [PubMed] [Google Scholar]
- Guo Y, Wang M, Xu X. Nucleotide sequence of A/Goose/Guangdong/2/96 (H5N1) virus M and NS RNA. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999b;13:305–308. [PubMed] [Google Scholar]
- Hu J, Jiang Z, Zhang C, Xiao F, Hu H. Bird diversity and the conservation value of a new Ramsar site: Guangdong Haifeng Wetlands, China. Integr Zool. 2011;6:266–278. doi: 10.1111/j.1749-4877.2011.00252.x. [DOI] [PubMed] [Google Scholar]
- Huang K, Bahl J, Fan XH, Vijaykrishna D, Cheung CL, Webby RJ, Webster RG, Chen H, Smith GJ, Peiris JS, Guan Y. Establishment of an H6N2 influenza virus lineage in domestic ducks in southern China. J Virol. 2010;84:6978–6986. doi: 10.1128/JVI.00256-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse-Post DJ, Sturm-Ramirez KM, Humberd J, Seiler P, Govorkova EA, Krauss S, Scholtissek C, Puthavathana P, Buranathai C, Nguyen TD, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci USA. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Selleck P, Komadina N, Shaw R, Brown L, Barr IG. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antiviral Res. 2007;73:228–231. doi: 10.1016/j.antiviral.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kimble B, Nieto GR, Perez DR. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol J. 2010;7:365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KS, Xu KM, Peiris JS, Poon LL, Yu KZ, Yuen KY, Shortridge KF, Webster RG, Guan Y. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J Virol. 2003;77:6988–6994. doi: 10.1128/JVI.77.12.6988-6994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long LP, Cai Z, Zhu X, Liao M, Wan XF. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol. 2010;10:1286–1288. doi: 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, Zhang XL, Zhao D, Wang G, Feng Y, Ma J, Liu W, Wang J, Gao GF. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Swayne DE. Pathobiology of Asian highly pathogenic avian influenza H5N1 virus infections in ducks. Avian Dis. 2007;51:250–259. doi: 10.1637/7710-090606R.1. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Suarez DL, Spackman E, Swayne DE. Age at infection affects the pathogenicity of Asian highly pathogenic avian influenza H5N1 viruses in ducks. Virus Res. 2007;130:151–161. doi: 10.1016/j.virusres.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Zhang GZ, Ma JH, Xia YJ, Liu QF, Jiang ZL, Wang Z, Brown EG, Tian FL, Liu JH. Serologic evidence of prevalent avian H3 subtype influenza virus infection in chickens. Avian Dis. 2009;53:198–204. doi: 10.1637/8410-071708-Reg.1. [DOI] [PubMed] [Google Scholar]
- Shi WF, Gibbs MJ, Zhang YZ, Zhang Z, Zhao XM, Jin X, Zhu CD, Yang MF, Yang NN, Cu YJ, Ji L. Genetic analysis of four porcine avian influenza viruses isolated from Shandong, China. Arch Virol. 2008;153:211–217. doi: 10.1007/s00705-007-1083-1. [DOI] [PubMed] [Google Scholar]
- Shortridge KF. H2N2 influenza viruses in domestic ducks. Lancet. 1979;1:439. doi: 10.1016/s0140-6736(79)90911-5. [DOI] [PubMed] [Google Scholar]
- Shortridge KF. Avian influenza A viruses of southern China and Hong Kong: ecological aspects and implications for man. Bull World Health Organ. 1982;60:129–135. [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- Shortridge KF. The 1918‘Spanish’ flu: pearls from swine? Nat Med. 1999;5:384–385. doi: 10.1038/7383. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Stuart-Harris CH. An influenza epicentre? Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Webster RG. Geographical distribution of swine (Hsw1N1) and Hong Kong (H3N2) influenza virus variants in pigs in Southeast Asia. Intervirology. 1979;11:9–15. doi: 10.1159/000149006. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Butterfield WK, Webster RG, Campbell CH. Isolation and characterization of influenza A viruses from avian species in Hong Kong. Bull World Health Organ. 1977a;55:15–20. [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977b;196:1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Butterfield WK, Webster RG, Campbell CH. Diversity of influenza A virus subtypes isolated from domestic poultry in Hong Kong. Bull World Health Organ. 1979;57:465–469. [PMC free article] [PubMed] [Google Scholar]
- Sims LD, Ellis TM, Liu KK, Dyrting K, Wong H, Peiris M, Guan Y, Shortridge KF. Avian influenza in Hong Kong 1997–2002. Avian Dis. 2003;47:832–838. doi: 10.1637/0005-2086-47.s3.832. [DOI] [PubMed] [Google Scholar]
- Song D, Lee C, Kang B, Jung K, Oh T, Kim H, Park B, Oh J. Experimental infection of dogs with avian-origin canine influenza A virus (H3N2) Emerg Infect Dis. 2009;15:56–58. doi: 10.3201/eid1501.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song DS, An DJ, Moon HJ, Yeom MJ, Jeong HY, Jeong WS, Park SJ, Kim HK, Han SY, Oh JS, Park BK, Kim JK, Poo H, Webster RG, Jung K, Kang BK. Interspecies transmission of the canine influenza H3N2 virus to domestic cats in South Korea, 2010. J Gen Virol. 2011;92:2350–2355. doi: 10.1099/vir.0.033522-0. [DOI] [PubMed] [Google Scholar]
- Steensels M, Van Borm S, Boschmans M, van den Berg T. Lethality and molecular characterization of an HPAI H5N1 virus isolated from eagles smuggled from Thailand into Europe. Avian Dis. 2007;51:401–407. doi: 10.1637/7554-033106R.1. [DOI] [PubMed] [Google Scholar]
- Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, Buranathai C, Nguyen TD, Chaisingh A, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu KY, Kuo RL, Marklund J, Krug RM. The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells. J Virol. 2007;81:8112–8121. doi: 10.1128/JVI.00006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS, Guan Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XF. Isolation and Characterization of Avian Influenza Viruses in China. College of Veterinary Medicine, South China Agricultural University; Guangzhou: 1998. [Google Scholar]
- Wan XF, Ren T, Luo KJ, Liao M, Zhang GH, Chen JD, Cao WS, Li Y, Jin NY, Xu D, Xin CA. Genetic characterization of H5N1 avian influenza viruses isolated in southern China during the 2003–04 avian influenza outbreaks. Arch Virol. 2005;150:1257–1266. doi: 10.1007/s00705-004-0474-9. [DOI] [PubMed] [Google Scholar]
- Wan XF, Dong L, Lan Y, Long LP, Xu C, Zou S, Li Z, Wen L, Cai Z, Wang W, Li X, Yuan F, Sui H, Zhang Y, Dong J, Sun S, Gao Y, Wang M, Bai T, Yang L, Li D, Yang W, Yu H, Wang S, Feng Z, Wang Y, Guo Y, Webby RJ, Shu Y. Indications that live poultry markets are a major source of human H5N1 Influenza Virus Infection in China. J Virol. 2011;85:13432–13438. doi: 10.1128/JVI.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu Z, Chen X. A survey on winter birds in shenzhen Futian Mangrove. Ecol Sci. 1993;2:10. [Google Scholar]
- Webster RG. The importance of animal influenza for human disease. Vaccine. 2002;20(Suppl. 2):S16–S20. doi: 10.1016/s0264-410x(02)00123-8. [DOI] [PubMed] [Google Scholar]
- Webster RG, Guan Y, Peiris M, Walker D, Krauss S, Zhou NN, Govorkova EA, Ellis TM, Dyrting KC, Sit T, Perez DR, Shortridge KF. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J Virol. 2002;76:118–126. doi: 10.1128/JVI.76.1.118-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. 2010 Available at: http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/index.html (accessed on 13 April 2012)
- Xu X, Subbarao, Cox NJ, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- Xu KM, Li KS, Smith GJ, Li JW, Tai H, Zhang JX, Webster RG, Peiris JS, Chen H, Guan Y. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol. 2007;81:2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Nakamura K, Kitagawa K, Ikenaga N, Yamada M, Mase M, Narita M. Pathogenesis in call ducks inoculated intranasally with H5N1 highly pathogenic avian influenza virus and transmission by oral inoculation of infective feathers from an infected call duck. Avian Dis. 2007;51:744–749. doi: 10.1637/0005-2086(2007)51[744:PICDII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kong W, Qi W, Long LP, Cao Z, Huang L, Qi H, Cao N, Wang W, Zhang F, Ning Z, Liao M, Wan XF. Identification of a H6N6 swine influenza virus in southern China. Infect Genet Evol. 2011 doi: 10.1016/j.meegid.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZM, Shortridge KF, Garcia M, Guan Y, Wan XF. Genotypic diversity of H5N1 highly pathogenic avian influenza viruses. J Gen Virol. 2008;89:2182–2193. doi: 10.1099/vir.0.2008/001875-0. [DOI] [PubMed] [Google Scholar]
- Zou F, Yang Q, Dahmer T, Cai J, Zhang W. Habitat use of waterbirds in Coastal Wetland on Leizhou Peninsula, China. Waterbirds. 2006;29:459–464. [Google Scholar]


