Abstract
A significant portion of the world’s population is infected with herpes simplex virus type 1 and/or type 2 (HSV-1 and/or HSV-2), that cause a wide range of diseases including genital herpes, oro-facial herpes, and the potentially blinding ocular herpes. While the global prevalence and distribution of HSV-1 and HSV-2 infections cannot be exactly established, the general trends indicate that: (i) HSV-1 infections are much more prevalent globally than HSV-2; (ii) Over half billion people worldwide are infected with HSV-2; (iii) the sub-Saharan African populations account for a disproportionate burden of genital herpes infections and diseases; (iv) the dramatic differences in the prevalence of herpes infections between regions of the world appear to be associated with differences in the frequencies of human leukocyte antigen (HLA) alleles. The present report: (i) analyzes the prevalence of HSV-1 and HSV-2 infections across various regions of the world; (ii) analyzes potential associations of common HLA-A, HLA-B and HLA-C alleles with the prevalence of HSV-1 and HSV-2 infections in the Caucasoid, Oriental, Hispanic and Black major populations; and (iii) discusses how our recently developed HLA-A, HLA-B, and HLA-C transgenic/H-2 class I null mice will help validate HLA/herpes prevalence associations. Overall, high prevalence of herpes infection and disease appears to be associated with high frequency of HLA-A*24, HLA-B*27, HLA-B*53 and HLA-B*58 alleles. In contrast, low prevalence of herpes infection and disease appears to be associated with high frequency of HLA-B*44 allele. The finding will aid in developing a T-cell epitope-based universal herpes vaccine and immunotherapy.
INTRODUCTION
A significant portion of the world’s population is infected with herpes simplex virus type 1 and/or type 2 (HSV-1 and/or HSV-2) [1-3]. Herpes is epidemic in some regions and ethnicities of the world but is at its lowest prevalence in other regions and ethnicities (Fig. 1 and Table 1). For instance, a high HSV-2 prevalence of 39.2% is recorded among Non-Hispanic Blacks while a low prevalence of 12.3% is recorded among Non-Hispanic Whites. While the exact estimates of HSV-1 and HSV-2 prevalence in the world are limited by poor availability of sero-epidemiological data, general trends indicate that HSV-2 infections continue to spread globally: (i) HSV-2 now affects over 540 million people, aged 14-49 years, worldwide, with an estimate of 23.6 million people newly infected per year [1-4]; (ii) In the United States, about one in six individuals from the sexually active population (i.e. between 14 and 49 years of age) are infected with HSV-2, which is equivalent to approximately 60 million people, with the highest prevalence among non-Hispanic black individuals and the lowest among those of Asian descent; (iii) globally, more women are infected by HSV-2 than men [4, 5]; (iv) the number of infected men and women increases with age [4]; (v) although the prevalence of herpes infection varies substantially by region, it is mostly higher in developing than in developed regions; (vi) HSV-2 and HSV-1 prevalence varies markedly by country, by region within country, and by population subgroup [6]; (vii) globally, genital herpes caused by HSV-2 is particularly frequent in sub-Saharan Africa, countries form Latin America, and South East Asia, with greater than 20% of the populations in the sub-regions suffering from genital herpes infection [6]. In contrast, the lowest HSV-2 prevalence is recorded in Western and Southern Europe, Middle East, North Africa, Japan, Australia and New Zealand, with less than 10% of the populations in the sub-regions exhibiting HSV-2 infection [6, 7]. Countries from central Europe and North America have an intermediate 11-20% HSV-2 prevalence [4] (Fig. 1 and Table 1); (viii) HSV-1 infections, acquired during childhood and adolescence [6], are globally, much more prevalent than HSV-2 [7], causing significant morbidity especially among young adults in western societies, where up to 80% are sero-positive; (ix) HSV-2, but not HSV-1, leads to two- to three-fold increase in risk of HIV-1 infection, supported by an increase in prevalence of both HSV-2 and HIV-1 in the same regions of sub-Saharan African regions [8-12]; (x) HSV-1 and HSV-2 infections are a large and worldwide public health problem [11, 13-17]. In addition to causing genital herpes and oro-facial herpes (i.e. cold sores), HSV-1 and HSV-2 also cause encephalitis and death in newborns due to vertical transmission, while HSV-1 causes blinding ocular herpes [18, 19]. In the United States, over 400,000 individuals have a history of ocular herpes disease caused by HSV-1; and finally; (xi) there is no vaccine available today to protect against HSV-1 or HSV-2 infection and disease, and patients must rely on sustained or intermittent antiviral drugs (Acyclovir and derivatives) [20, 21]; (xii) the association of the prevalence of HSV-1 & HSV-2 infections and diseases with the frequency of human leukocyte antigens (HLA) alleles is not fully elucidated. While a handful of studies point to association trends between the frequency of some common HLA class I alleles and the high/low prevalence of genital HSV-2 infection/disease, few studies have reported association of HLA alleles with infection and severity of ocular HSV-1 infection and disease, such as recurrent herpetic stromal keratitis (rHSK), the most common cause of cornea-derived blindness in developed nations [13, 17, 22-28].
Figure 1.
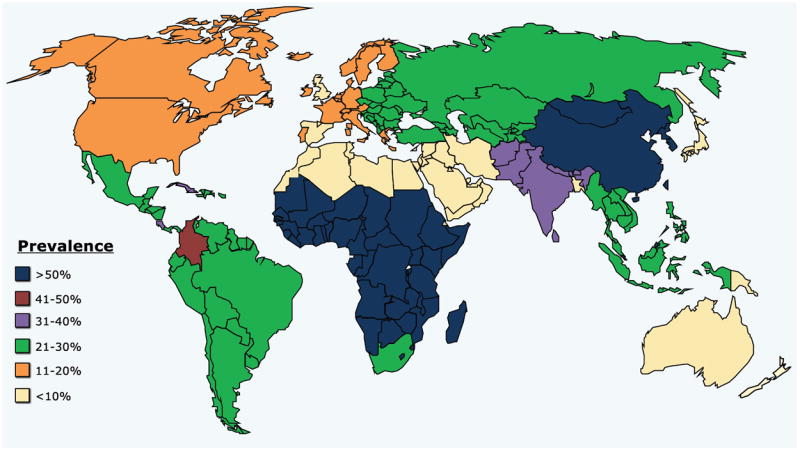
The HSV-2 prevalence is generalized by world major regions, including Asia, Australia, Europe, Middle East, North Africa, North America, South/Central America and sub-Saharan Africa. HSV-2 prevalence per region was predicted based on representative populations within that region.
Table 1.
Prevalence of HSV-2 infection.
| Region | Sub-region 1 | Prevalence of HSV-2 Infection |
|---|---|---|
| Asia | Bangladesh, Brunei Darussalam, China, Japan, Mongolia, North Korea, Russia, Singapore, South Korea | Variable by Sub-regions |
| Australia | - | Low |
| Europe | Basque, Belgium, Corsica, Denmark, France, Germany, Northern Ireland, Norway, Poland, Republic of Ireland, Romania, Scotland, Spain, Sweden, United Kingdom, | Low |
| Middle East | Arab Nations, Iran, Israel | Low |
| North Africa | Algeria, Morocco, Tunisia | Low |
| North America | United States | Variable by Ethnic groups |
| South/Central America | Colombia, Costa Rica, Cuba | High |
| Sub-Saharan Africa | Botswana, Burkina Faso, Cameroon, Central African Republic, Ethiopia, Guinea, Guinea Bissau, Kenya, Mali, Natal Zulu, Rwanda, Senegal, South Africa, Uganda, Zambia, Zimbabwe | High |
Sub-regions listed are specifically referenced in the text
Six co-dominantly expressed HLA class I alleles are encoded by three loci (HLA-A, -B, and -C). The frequency of HLA class I alleles differ significantly among the four major world’s populations (i.e. Caucasoids, Orientals, Hispanics and Blacks) as the HLA system is the most polymorphic of all human genetic systems [29]. Many DNA-defined alleles with identical serotypes may have variable frequencies in different populations [29]. About 7000 HLA-I polymorphisms have been reported to date (http://hla.alleles.org/). Each HLA class I molecule has a distinct peptide binding specificity. As a result, HLA-I/peptide complexes that drive the expansion of pathogen-specific CD8+ T cells are extremely diverse in each individual. However, it is possible to account for the predominance of all known HLA class I with only eleven main functional binding specificities.
This report will: (i) analyze the existing literature depicting prevalence of HSV-1 and HSV-2 infections across various human populations (Fig. 1); (ii) provide a comparative review of potential associations of the most common HLA-A, HLA-B and HLA-C alleles with prevalence of HSV-1 and HSV-2 infections and diseases in the four major world’s populations (i.e. Caucasoids, Orientals, Hispanics and Blacks) (Fig. 2 and Table 2 to 4); and (iii) discuss how our recently developed HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02 and HLA-C*07:01 monochain transgenic/H-2 class I null mice, will help validate these associations and aid in developing a universal T-cell epitope-based herpes vaccine that would provide a broad coverage of human population, regardless of race and ethnicity.
Figure 2.
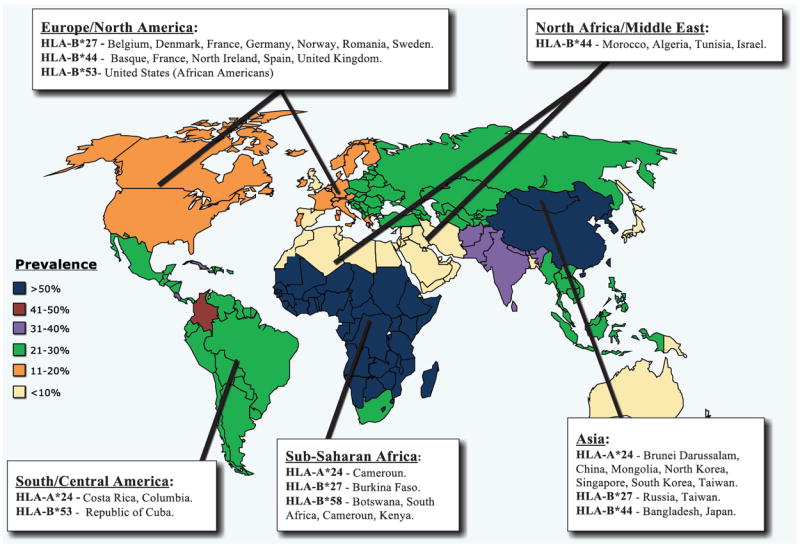
High frequency of HLA-B*44 allele is reported in regions and ethnicities with low to moderate HSV-2 prevalence. High frequency of HLA-B*27 allele frequency is reported in regions and ethnicities with moderate to high HSV-2 prevalence. High frequencies of HLA-A*24, HLA-B*27, HLA-B*53 and HLA-B*58 alleles are only reported in regions and ethnicities with high prevalences.
Table 2.
| A: Association of high frequency of HLA-B*44 allele with low/moderate prevalence of HSV-2 infection in various regions of the world. | |||
|---|---|---|---|
| Sub regions | Prevalence of HSV-2 infection | HLA-B*44 allele frequency 1 | |
| Asia | Bangladesh | Low | High |
| Japan | Low | High | |
| Europe | Basque | Low | Most frequent HLA-B (Caucasoid) |
| France | Low | Most frequent HLA-B | |
| North Ireland | Low | 35.7% | |
| Spain | Low (3.9% men genital herpes) | Most frequent HLA-B | |
| United Kingdom | Low (4% England) | 32% | |
| Middle East | Israel | Low | High |
| North Africa | Algeria | Low | High |
| Morocco | Low | High | |
| Tunisia | Low | High | |
| Sub-Saharan Africa | Cameroon | 55% women, 36% men | Low |
| Kenya | 55% women, 36% men | Low | |
| South Africa | High | Low | |
| B: Association of high frequency of HLA-A*24, HLA-B*27, HLA-B*53, and HLA-B*58 alleles with moderate/high prevalence of HSV-2 infection in various regions of the world. | ||||||
|---|---|---|---|---|---|---|
| Sub regions | HSV-2 infection prevalence | HLA-A*24 allele frequency 1 | HLA-B*27 allele frequency 1 | HLA-B*53 allele frequency 1 | HLA-B*58 allele frequency 1 | |
| Asia | Oriental Asia | Low | - | - | <3% | - |
| Bangladesh | Low (7% women genital herpes) | - | - | - | Low | |
| Japan | Low | - | 0.1-0.5% | - | Low | |
| Russia | 21-30% | - | 10.2% | - | - | |
| Taiwan | High | 48.9% | 15% | - | - | |
| Brunei Darussalam | 61.8% women and 47.8% men aged 15-49 | Most frequent HLA-A | - | - | - | |
| China | 2.9% | - | - | |||
| Mongolia | <2% | - | - | |||
| North Korea | <2% | - | - | |||
| Singapore | - | - | - | |||
| South Korea | <2% | - | - | |||
| Europe | Caucasoid Europe | Low | - | - | <3% | - |
| Belgium | Low | - | 14.3% | - | - | |
| Denmark | Low | - | 24% | - | - | |
| France | Low | - | 12% | - | - | |
| Germany | Low | - | 13% | - | - | |
| Norway | Low | - | 24% | - | - | |
| Romania | Low | - | 11.2% | - | - | |
| Spain (men) | 3.9% genital herpes | - | - | - | Low | |
| Sweden | Low | - | 16% | - | - | |
| United Kingdom | Low | Low | ≤ 8% (*27:05) | - | - | |
| Middle East | Arab Nations | Low | - | low | - | Low |
| Israel | Low | - | low | - | Low | |
| North Africa | Morocco | Low | - | 4% | <3% (Caucasoid) | - |
| Algeria | Low | - | 4% | <3% (Caucasoid) | - | |
| Tunisia | Low | - | 4% | <3% (Caucasoid) | - | |
| Lybia | Low | - | 4% | <3% (Caucasoid) | - | |
| Egypt | Low | - | 4% | <3% (Caucasoid) | - | |
| North America | United States (African Americans) | High | - | ≤ 3% | 25.5%, most frequent HLA-B | - |
| United States (Caucasoid) | Low | - | - | <3% | - | |
| South/Central America | South/Central America | Moderate | - | < 2% | - | - |
| Costa Rica | 39% genital herpes women | 2nd most frequent HLA-A | - | - | - | |
| Columbia (Black, Mestizo) | 50% women | Most frequent HLA-A | - | - | - | |
| Republic of Cuba | High | - | - | Most frequent HLA | - | |
| Sub-Saharan Africa | sub-Saharan Africa (Blacks) | High | - | ≤ 3% | - | 2nd most frequent HLA |
| Botswana | High | - | - | - | 17.9% | |
| South Africa | High | - | - | - | Most frequent HLA-B, 39% | |
| Burkina Faso | High | - | Most frequent HLA | - | - | |
| Cameroun | 55% women, 36% men | 3rd most frequent HLA-A | - | - | Most frequent HLA-B | |
| Kenya | - | - | - | |||
Table 4.
Summary of all HLA class I allele associations with prevalence of HSV-2 infection.
| HLA allele | Association with HSV-2 infections and diseases | Regions associated with HIGH allele frequency |
|---|---|---|
| HLA-A*01 | Unclear | Australia, Europe, Middle East, North Africa, sub-Saharan Africa |
| HLA-A*02 | Unclear | Australia, Europe, North Africa, South/Central America, sub-Saharan Africa |
| HLA-A*24 | Positive 1 | Asia, sub-Saharan Africa, South/Central America |
| HLA-B*07 | Unclear | Europe, sub-Saharan Africa |
| HLA-B*08 | Unclear | Sub-Saharan Africa |
| HLA-B*15 | Unclear | Asia, North America, South/Central America, sub-Saharan Africa |
| HLA- B*27 | Positive 1 | Asia, Europe, sub-Saharan Africa |
| HLA-B*35 | Unclear | Asia, Europe, Middle East, North Africa, South/Central America |
| HLA-B*44 | Negative 2 | Asia, Europe, Middle East, North Africa |
| HLA-B*53 | Positive 1 | South/Central America |
| HLA-B*58 | Positive 1 | Sub-Saharan Africa |
| HLA-C*02 | Unclear | Europe, sub-Saharan Africa |
| HLA-C*07 | Unclear | Europe, Middle East, North Africa, North America, South/Central America |
HLA allele association with HIGH prevalence of HSV-2 infection, i.e. “susceptibility”
HLA allele association with LOW prevalence of HSV-2 infection, i.e. “resistance”
MATERIALS AND METHODS
1. Analyses of potential association of the HLA with high vs. low prevalence of herpes
HLA genes are involved in inducing either susceptibility or resistance to many infectious diseases [30-34]. In order to disentangle the respective contribution of the HLA-A, HLA-B and HLA-C alleles in HSV-1 and HSV-2 infections: (1) We performed a systematic haplotypic analysis in the four major world’s populations (Caucasoids, Orientals, Hispanics and Blacks), which display various prevalence of HSV-1 and HSV-2 infection; (2) PubMed® (1966-present) was used to identify cross-sectional studies with HSV-1 and HSV-2 sero-prevalence data and to identify the highest HLA class I allele that are specific to distinct populations with low vs. high prevalence of HSV-1 and HSV-2 infections; (3) The potential association of the HLA class I alleles [35-37] with high vs. low prevalence of herpes infections (which can be translated into “herpes susceptibility or resistance”) was then determined. For the HLA frequency in ethnically and geographically distinct populations; the databases from the following websites were consulted: http://www.ncbi.nlm.nih.gov/projects/gv/mhc/ [38, 39], http://www.allelefrequencies.net/default.asp [38, 39], and http://hla-net.eu [40-42].
2. Statistical analyses
Data for each assay were compared by analysis of variance (ANOVA) and Student’s t-test using Graph Pad Prism 5 software (San Diego, CA). Differences between the groups were identified by ANOVA, multiple comparison procedures, as we previously described [14, 24, 25, 27, 43-54]. Statistical analysis also included Fisher’s exact test and multivariate analyses, using logistic and linear regression models. Results are considered statistically significant if p < 0.05.
RESULTS & DISCUSSIONS
1. A comparative review of HLA-A, HLA-B, and HLA-C associations with prevalence of HSV-1 and HSV-2 infections and diseases across various human populations (Fig. 2 and Table 2 to 4)
The clinical spectrum of HSV-1 & HSV-2 infections, ranging from asymptomatic to frequently distressing symptomatic outbreaks appear to be associated with HLA class I molecules [55-57]. These associations suggest that CD8+ T cell-mediated immune mechanism(s), influenced by HLA class I genes at the A, B, and C main loci, may influence the outcome of recurrent herpes infections and diseases [14, 58]. The HLA-B locus is the most polymorphic, with 1795 known proteins, followed by HLA-A with 1290, and HLA-C with 946 [58]. Genetic variation within the highly polymorphic HLA class I region contributes to the diversity of viral pathogens recognized by CD8+ T cells [59, 60]. The dramatic differences in the prevalence of HSV-1 and HSV-2 between regions of the world prompted us to explore potential associations between frequencies of most common HLA-A, HLA-B, and HLA-C alleles and prevalence of HSV-1 and HSV-2 infections and diseases (i.e., “resistance vs. susceptibility” to infections and diseases) [56].
α) HLA-A
HLA-A*01: According to the Allele Frequency website [39, 61], HLA-A*01 is highly represented in the sub-Saharan populations. For instance, the HLA-A*01:03 allele is one of the most frequent alleles in Ethiopia. Thus, the HLA-A*01 allele appears to be linked with high prevalence to genital herpes disease. However, HLA-A*01 is also the most frequent allele in North Africans where genital herpes is not that endemic. For instance, 25.6% of Moroccans, who have one of the lowest rate of genital herpes, express the HLA-A*01 allele [62-65]. Spain and Israeli populations, which have one of the lowest rates of herpes infection in the world, also express a high frequency of the HLA-A*01 allele [66]. About 33.6% of Australians, which also have one of the lowest rates of herpes, express the HLA-A*01 allele [67, 68]. Thus, HLA-A*01 allele may not be considered as a factor in resistance/susceptibility to herpes.
HLA-A*02 allele has become an important target for T cell-based herpes immunotherapy reflecting its high prevalence in most populations worldwide, regardless of race and ethnicity [69-72]. HLA-A*0201 subtype is highly represented in human population regardless of race and ethnicity [39, 61]. HLA-A*0201 is also highly represented in sub-Saharan populations with up to 50% of the population in Cameroun expressing this haplotype [73]. In Kenya, where 20% of individuals are affected by genital herpes disease, 51% of individuals express the HLA-A*0201 subtype [38, 39]. In Senegal, where both HSV-1 and HSV-2 are endemic, up to 68% of the population express HLA-A*0201. However, the Australian population, which has one of the lowest rates of herpes, 46.5% of individuals express the HLA-A*0201 allele [67, 68]. Similarly, 50% of Belgians and 32.4% of Moroccans, both exhibit low rates of herpes infection, express the HLA-A*0201 allele [62-65, 74-77]. Thus, it is unclear whether HLA-A*0201 is a factor in resistance/susceptibility to herpes.
Of those tested positive for HSV-1 and HSV-2 infection, up to 90% said a health-care professional had never told them that they had genital herpes, ocular or oro-facial herpes (Asymptomatic individuals, ASYMP). The remaining 10% are symptomatic individuals (SYMP) often-express herpetic disease, some of them multiple times a year. In a recent study [78], we performed HLA-A*02 sub-typing of HSV-1 seropositive symptomatic (SYMP) vs. asymptomatic (ASYMP) individuals using SSP genotyping [79]. The results showed that all, but one of the HLA-A*02 individuals in our cohort were of the HLA-A*02:01 subtype, with the remaining (SYMP) individual of the HLA-A*02:06 subtype. Although each HLA-A*02 subtype molecule can be expected to have a unique specificity, large overlaps in the repertoires of HLA-A*02 molecules have been reported [72]. Indeed, over 70% of the peptides that bind HLA-A*02:01, the most common HLA-A*02 subtype, with high affinity were found to also bind at least two other of the 4 next most common HLA-A*02-subtypes, including the HLA-A*02:06 subtype [72]. Thus, HLA-A*02:01-restricted epitopes may be useful in vaccine studies with individuals expressing alleles other than HLA-A*02:01. At the same time, however, while cross-reactivity at the binding level appears to be particularly high in the case of HLA-A*02, it must also be noted that differences between the molecules may encompass residues at positions involved in TCR recognition, and thus may be associated with differences that lead to divergence in T cell repertoires.
HLA-A*24 is the most frequently represented allele of the HLA-A locus in sub-Saharan African countries, where the highest prevalence of herpes infection and disease is recorded. HLA-A*24 is frequent in Cameroun and Guinea populations with the highest worldwide prevalence of both HSV-1 and HSV-2 infections [39]. In Cameroun, where the HSV-2 infection rate is at 55% for women and 36% for men, HLA-A*24 is the third most frequent allele of the HLA-A locus (after A*33 and A*30) [73]. The population of Costa Rica, which has one the highest rates of genital herpes in the world (39% in women) also has HLA-A*24 as the second most frequent allele, after HLA-A*02 [80]. HLA-A*24 is also frequent in some Asian populations where a high prevalence of HSV-2 infection is also recorded. For instance, HLA-A*24 is highly represented in individuals from China [81, 82] and represented in up to 48.9% of individuals from Taiwan [83]. In populations from Eastern Asia (Brunei Darussalam, China, Democratic People’s Republic of Korea, Mongolia, Republic of Korea, Singapore), where 61.8% of females aged 15-49 and 47.8% of males aged 15-49 are seropositive for HSV-2, HLA-A*24 is the most frequent HLA-A allele [4]. HLA-A*24 is also the most frequent HLA-A allele in South East Asian populations with HLA-A*02 and HLA-A*11 as the 2nd and 3rd most frequent alleles. HLA-A*24:02 is one of the most common HLA subtypes in Southeast Asian populations (29.9%) [84]. HLA-A*24 is the most frequent allele of the HLA-A locus in Colombian black and Mestizos ethnic groups of the population where one of the highest prevalence of genital HSV-2 infection is recorded (50% in women) [85, 86]. In contrast, this haplotype is not well represented in North African countries with the lowest rate of herpes infection. HLA-A*24 is not frequent in the United Kingdom where one of the lowest rate of genital herpes is recorded [87] (i.e. the sixth most frequent after HLA-A*01, HLA-A*02, HLA-A*03, HLA-A*11 and HLA-A*23). Thus, overall, it appears that the high frequency of HLA-A*24 allele positively correlates with high prevalence of HSV-2 infection in the world populations (Table 2B).
β) HLA-B
HLA-B*07 is the most frequent allele of HLA-B locus in Spain [87, 88], Germany [87, 89] and the United Kingdom, where some of lowest rates of genital herpes are recorded. However, HLA-B*07 is also a frequent allele in the sub-Saharan population where herpes infection is endemic [87]. For instance, between 9 and 19% of individuals from South Africa, depending on ethnicity, and 13% of Kenyans express the HLA-B*07:02 subtype [38, 39]. South African and Kenyan populations have one of the highest rates of herpes in the world. This suggests that HLA-B*07 might not be a factor in resistance/susceptibility to herpes infection.
The HLA-B*08 allele is more common in many populations of African ancestry who continue to account for a disproportionate burden of the HSV-1 and HSV-2 epidemic. For instance, up to 29% of Senegalese express HLA-B*08, and 14% of individuals from South Africa Natal Zulu express the HLA-B*08:01 subtype. However, HLA-B*08 is also frequent in many western nations where the frequency of herpes in lowest. For instance HLA-B*08:01 subtype is represented in up to 35% of some Caucasoid western European populations [90-92]. This suggests that HLA-B*08 might not be a factor in resistance/susceptibility to herpes infection.
The HLA-B*15 allele is also highly represented in sub-Saharan populations that have the highest rates of HSV-1 and HSV-2 infections in the world. For instance, HLA-B*15:03 subtype is the highest alleles of the HLA-B locus expressed by 30% South Africans [93]. Up to 20% of Cameroonians express HLA-B*15 [73, 94]. HLA-B*15:03 is also the third highest alleles of the HLA-B locus (after HLA-B*42:01 and HLA-53:01) in Zambia [95]. 15.5% of Costa Ricans, which have one the highest rates of genital herpes in the world (39% in women), express the HLA-B*15 allele [80]. HLA-B*15 is also the most frequent allele of the HLA-B locus in the Colombian black ethnic group with one of the highest prevalence of genital herpes (50% in women) [85, 86, 96]. HLA-B*15 allele is also the second most frequent allele of the HLA-B locus (after HLA-B*53) in non-Hispanic African Americans (22.3%), a population with high rate of HSV-2 infection [97-100]. HSV-2 sero-prevalence is approximately three times greater among non-Hispanic blacks in the US (~39.2%) compared to non-Hispanic whites (~12.3%) [101]. However, HLA-B*15 is also the most frequent allele of HLA-B locus in the Japanese population, known for the lowest rates of genital herpes [102]. HLA-B*15 is also the most frequent allele of HLA-B locus in Bangladesh, also know for one of the lowest rates of herpes in the world [103]. Thus, the frequency of HLA-B*15 allele does not appear to correlate with prevalence of HSV-1 and HSV-2 epidemic, suggesting that this allele may not be associated to susceptibility to herpes infection and disease.
The HLA B*27 appears to promote recurrence of herpetic eye disease caused by HSV-1 infection [57]. Recurrent herpetic stromal keratitis (rHSK), caused by spontaneous reactivation of HSV-1 from latently infected neurons of the trigeminal ganglia, is the most common cause of cornea-derived blindness in developed Western nations [13, 17, 22-28]. Interestingly, in developed Western nations, where rHSK is more prevalent, the HLA-B*27 allele is more frequent in Caucasoid populations than in North Africans (4%), Chinese (2.9%), and Japanese (0.1-0.5%) descent. For instance, 24% of northern Scandinavian [104, 105], 16% of Swedish [106-109], 14.3% of Belgium [74], 13% of German [110, 111], 12% of French [112-115], 11.2% of Romanian [116-121], 10.2% of Russian [122] populations express HLA-B*27 allele. HLA-B*27 allele is, overall, not frequent in Oriental populations, except for Taiwanese where this allele is highly frequent (15%). For instance, HLA-B*27 allele is < 3% in Chinese Tibet, Southwest and North regions and Yannan Province [93, 123-125]. Only ≤ 3% of HLA-B*27 allele is detected in Black Sub-Saharan population [126-129] and African-American populations [130, 131]. The frequency of HLA-B*27 allele is also low in Arab (<3%) [132-139] and Israel Jewish populations (3.4%) [140]. One of the lowest frequencies of HLA-B*27 allele is detected in Hispanic (<2%) [100, 141-144], Mongolian (<2%) [145], Japanese (<2%) [146], Singaporean Chinese (<2%) [93, 123, 124, 147-149], and in Korean (<2%) populations [150-155]. A genital herpes study comparing HLA frequencies among HSV-seronegative individuals, HSV-2 seropositive “asymptomatic” individuals and HSV-2 seropositive symptomatic individuals reported an association between HLA-B*27 high frequency and an increase in symptomatic genital herpes infection [56]. HLA-B*27 is the second most frequent allele of the HLA-B locus in Burkina Faso where herpes is endemic [156]. HLA-B*27:05 is represented less than 8% of the Caucasian populations where herpes not endemic [97, 157]. The HLA-B*27 allele is associated with exacerbation of herpes disease. While HLA-B*27 allele is ubiquitous in the world population, it is significantly more common in herpes endemic regions than in non-endemic regions, suggesting that the HLA-B*27 allele is associated with herpes infection and disease. This suggests that HLA-B*27 might be a factor of susceptibility to genital (Table 2B) and ocular herpes infection and disease (not shown). Since HLA-B*27:05 monochain transgenic/H-2 class I null mice are now available [158], it would be interesting to see whether these mice will be more susceptible to genital and ocular herpes infection and disease.
HLA-B*35 association with prevalence of herpes infection and disease is rather controversial. HLA-B*35 expression has been previously linked with resistance to recurrent herpes labialis in HSV-1 seropositive individuals (cold sores) [159, 160]. Inversely, a low expression of HLA-B*35 was associated with a significant increase in symptoms from recurrent herpes [159, 160]. However, high frequency of the HLA-B*35 in herpes endemic populations appears to be associated with high prevalence of HSV-2 infection. Particularly, HLA-B*35 is the most frequent allele of HLA-B locus in population from Costa Rica (31.5% of Costa Ricans express this allele), which have one the highest rate of genital herpes in the world (39% in women) [80]. HLA-B*35 is also a frequent allele in South America, where a moderate prevalence of genital HSV-2 infection is recorded [6, 7]. However, HLA-B*35 is also well represented in the Middle East populations where the rates of genital herpes are one of the lowest in the world. High frequency of HLA-B*35 is also detected in Bangladesh [103], North Africa [64], Corsica [161, 162] and Israel [140] all of these regions have the low rate of genital herpes. HLA-B*35 is the most frequent allele of the HLA-B locus (28.8 %) in the Iranian population which has one of the lowest rate of herpes [163]. Inversely, low frequency of HLA-B*35 haplotype was detected in Australians adults who also have one of the lowest prevalence of HSV-2 infection (12%) [7]. We should emphasize that: (1) HLA-B*35 is associated with increased susceptibility to HIV-1 infection; (2) a rapid progression of HIV-1 [164, 165] and HIV-2 [166] infections to AIDS; and (3) genital herpes is associated with a two- to three-fold increase in risk of HIV-1 infection in sub-Saharan African population. Thus, it is likely that a negative immuno-synergy occurs between the two sexually transmitted viruses (HIV-1 and HSV-2) in HLA-B*35 positive individuals from the same sub-Saharan region.
HLA-B*44 is the most frequent allele of HLA-B locus in Caucasoid European population. Thus, HLA-B*44 expression may positively correlate with low prevalence of HSV-2 infection known in European populations. For instance, 35.7% of North Ireland [93, 124, 147] and 32% of English [167] populations that have low frequency of herpes infection express the HLA-B*44 allele. HLA-B*44 is the most frequent allele of the HLA-B locus in French [112-115] Spanish [168] and Basques Caucasoid populations [169, 170], which have some of the lowest rates of herpes infection and disease. HLA-B*44 is also frequent in North African [64], Bangladeshi [103], Japanese [171] and Israeli [140] populations, which have some of the lowest rates of genital herpes. In contrast, the HLA-B*44 is not frequent in sub-Saharan Africans, where the prevalence of HSV-2 infection is relatively high. Thus, the high frequency of HLA-B*44 appears to positively correlate with low prevalence of HSV-2 infection and disease. This suggests that the HLA-B*44 allele may constitute an excellent marker, or a factor, of genetic resistance to herpes infection and disease (Table 2A).
The HLA-B*53 allele is the most frequent allele of the HLA-B locus in African Americans (25.5%), a population with a high rate of HSV-2 infection [97-100]. HLA-B*53 allele is also the most frequent allele in Cuban population with a highest prevalence of herpes [93, 124, 147-149]. In contrast, HLA-B*53 allele is not frequent in Caucasian and oriental populations (<3%) with a lowest rate of herpes [93, 124, 132, 172]. Thus, the high frequency of HLA-B*53 appears to positively correlate with high prevalence of HSV-2 infection and disease (Table 2B).
The HLA-B*58 allele is the most frequent HLA-B allele in South Africa (39%), Cameroun and Kenya, where HSV-2 infection rates are at 55% for women and 36% for men [73, 93, 124, 147]. HLA-B*58 is the second most frequent haplotype of the HLA-B locus (after HLA-B*15 allele), in many other sub-Saharan African countries where herpes is endemic. For instance, HLA-B*58 is the most frequent generic HLA-B type in Botswana (17.9%) [164]. HLA-B*58 is also the second most frequent allele, after HLA-B*15, in South African regions where herpes in endemic [93]. This association suggests that immunological factors linked to HLA-B*58, such as CD8+ T cell responses may actually increase the risk of HSV-2 infection and disease. In contrast, the HLA-B*58 allele is not frequent in many South East Asia where the rate of genital herpes is low (compared to HLA-B*13, HLA-B*15 and HLA-B*40, the three most represented alleles) [39]. For instance, HLA-B*58 allele is at its lowest rate in individuals from Japan [173], which also have one of the lowest rate of genital herpes in the world. HLA-B*58 allele is not a frequent allele in the population of Bangladesh, where a low genital herpes infection rate of 7% was recorded in women. In western European countries that also have a low prevalence of genital herpes, such as Spain with a 3.9 prevalence of genital herpes in men, HLA-B*58 is not well represented [87, 88]. The HLA-B*58 allele is also not frequent in the Middle East countries where the rates of genital herpes is one of the lowest in the world. Thus, overall, a high frequency of HLA-B*58 allele appears to be associated with increased frequency of HSV-2 infections and diseases (Table 2B).
One should also emphasize that, among the three HLA-A, HLA-B and HLA-C alleles, the HLA-B alleles are the most associated with resistance/susceptibility to herpes. This suggests that the HLA-B locus, but not HLA-A and HLA-C, might encode for one or several immunopathological factors that would rather lead to herpes infection and diseases.
γ) HLA-C
HLA-C*02 is the most frequent allele of HLA-C locus in sub-Saharan countries known for a high rate of genital herpes, such as Burkina Faso [156, 174], Cameroun [73], Central African Republic (where 77% of the population express this allele) [94, 175]. A recent study showed an association between HLA-C*02 high frequency with an increase in herpes infection and disease [56]. However, HLA-C*02 is the second most frequent allele of the HLA-C locus in the Spanish population (7.2%, after HLA-C*07), which has one of the lowest prevalence of genital herpes in men (3.9% [87, 88]) as well as in overall Caucasoid Western Europe [110, 111]. Thus, HLA-C*02 association with prevalence of herpes infection and disease is rather controversial.
HLA-C*07 is one of the most frequent alleles of the HLA-C locus in sub-Saharan population. For instance, over 49.5% of individuals in Central African Republic express this allele and have a high prevalence of herpes. In Zambia, where the rate of genital herpes is at 55% in women and 36% in men, HLA-C*07 haplotype is the third most frequent HLA-C allele. In Cameron where HSV-2 infection rate is at 55% for women and 36% for men, HLA-C*07 is the second frequent allele of the HLA-C locus [73]. HLA-C*07 is the most highly frequent allele of HLA-C locus in population from Costa Rica, which have one the highest rate of genital herpes in the world (39% women have genital herpes) [80]. HLA-C*07 is also the most frequent allele of the HLA-C locus in Columbian black and Mestizos ethnic group of the population with one of the highest prevalence of genital herpes (50% in women) [85, 86]. However, HLA-C*07 is also a frequent allele in the European and oriental populations with ~41 % of Caucasians and 28% of Orientals expressing HLA-C*07, both known for low rates of genital herpes [176, 177]. The HLA-C*07 expression is at 60.3% in England [178-180], 66% in Poland [181], 64% in Scotland [182] and 63% in Ireland [124, 125]. The 60.3 % of England population expresses HLA-C*07 have only 4% prevalence of genital herpes HSV-2 infection. 45 % of Spanish population do express HLA-C*07 and have one of the lowest prevalence of genital herpes HSV-2 infection (4%) [87, 183-188]. HLA-C*07:01 is also the most frequent alleles of HLA-C locus in North American Caucasians (i.e. USA), European Caucasians (i.e. France) and Middle Eastern (i.e. Saudi Arabia) and North African (i.e. Morocco, Tunisia and Algeria) populations with low prevalence of genital HSV-2 infections. HLA-C*07 is also the second most represented alleles (after HLA-C*04) for the HLA-C locus in Middle-East countries, such as Syria [189], where the lowest rates of HSV-2 infection in the word are recorded. HLA-C*07 also appears to be the most frequent HLA-C allele in Latin America where a high prevalence of HSV-2 infection is recorded [4]. In addition, although HLA-C*07 is frequent in some regions of sub-Saharan Africa where genital herpes caused by HSV-2, is endemic, its frequency appears lower compared to European and north American populations [64, 95, 190-194]. For instance, in South Africa and Cameron (Swa, Pygmy and Bamileke ethnic groups) where genital herpes is endemic only 18% of the population express HLA-C*07 [195-197]. In Cameron HLA-C*07:01 is the second most frequent HLA-C allele, after HLA-C*02:01 [39]. HLA-C*07:01 is the second or third most frequent allele in Kenya after HLA-C*06:02 and HLA-C*0401 [39]. In Kenya and Zimbabwe, where the prevalence of HSV-2 infection in women is at 68% and 67%, respectively, the HLA-C*07 haplotype is not frequent in the adult population [64, 95, 190-194]. Depending on the ethnicity, HLA-C*07:01 is the second or third frequent allele in Kenya after HLA-C*06:02 and HLA-C*0401 [39]. In Cameron, Zimbabwe and Zambia, HLA-C*07:01 is the second frequent allele after HLA-C*04:01 [39]. HLA-C*07 appears to be poorly represented in Mali [39]. HLA-C*07:01 is not well represented in Burkina Faso nor in Senegal ethnic groups [39]. However, in Equatorial Guinea, Ughanda, Rowanda and Kenya HLA-C*07:01 appears to be the most frequent allele for HLA-C locus [39]. About 49.5% of the population in Central African Republic express HLA-C*07:01 [94, 175]. In Burkina Faso (Fulani ethnic group), HLA-C*07:01 is the third most frequent allele for HLA-C locus, after HLA-C*02 and HLA-C*16:02 [39]. In North-East Asia, which constitutes a region with high rates of herpes infection, the HLA-C*07 is not a frequently represented allele. Nevertheless, in Asia HLA-C*07 is the second most frequently represented allele for HLA-C locus (after HLA-C*03), and this region constitutes one of the lowest rates of herpes infection in the world. While the HLA-C*07 is highly expressed in the four major population groups (Caucasoid, Hispanics, Orientals and Blacks), its association with resistance vs. susceptibility to herpes in unclear.
In this review, we have elucidated the association of the prevalence of HSV-2 infection with HLA class I allele frequency for thirteen different HLA-A/B/C alleles (Table 4). In summary, the subset of “risk” HLA class I alleles that appears more strongly and consistently associated with high prevalence of herpes infections and diseases (i.e. susceptibility to herpes infections and diseases) included HLA-A*24, HLA-B*27, HLA-B*53 and HLA-B*58. In contrast, high frequency of HLA-B*44 allele appears to be associated with low prevalence of herpes infections and diseases (i.e. resistance to herpes infections and diseases) (Fig. 2 and Table 3). This HLA association should not be interpreted as the sole factor driving the prevalence of herpes nor should be considered as the sole cause of the susceptibility vs. resistance to herpes.
Table 3.
HLA allele association with low/moderate or moderate/high prevalence of HSV-2 infection organized by region: HLA-A*24, HLA-B*27, HLA-B*44, HLA-B*53, HLA-B*58.
| Region associated with HIGH allele frequency | Association of high frequency of HLA alleles with moderate/high HSV-2 prevalence | Association of high frequency of HLA alleles with low/moderate HSV-2 prevalence | HSV-2 Infection Prevalence |
|---|---|---|---|
| Asia | HLA-A*24 | HLA-B*44 | Variable by sub-region |
| HLA-B*27 | |||
|
| |||
| Europe | HLA-B*27 | HLA-B*44 | Low/Moderate |
|
| |||
| Middle East | - | HLA-B*44 | Low |
|
| |||
| North Africa | - | HLA-B*44 | Low |
|
| |||
| South/Central America | HLA-A*24, HLA-B*53 | - | High |
|
| |||
| Sub-Saharan Africa | HLA-A*24, HLA-B*27, HLA-B*58 | - | High |
2. Associations of HLA class I haplotypes with herpetic eye disease
While a handful of studies point to an emergence of association trends between the frequency of some HLA class I alleles and the high/low prevalence of genital HSV-2 infection/disease, few studies have reported association of HLA with the prevalence of ocular HSV-1 infection or with the severity of recurrent herpetic stromal keratitis (rHSK) [13, 17, 22-28]. In an early 1970s study a significant association of HLA-B5 locus with recurrent herpes was reported [198]. However, in that study it is unclear which allele of the HLA-B5 locus is most associated with recurrent herpes. A later 1980 study showed that HLA-A*30 allele occurred three times more frequently in patients with rHSK than in those who lack the HLA-A*30 allele [199]. Note that HLA-A*30 is the second highest frequent allele of HLA-A locus in many sub-Saharan countries (where both HSV-1 and HSV-2 is endemic). For example in Cameroun, 18% of the population express HLA-A*30 [73, 94], whereas the HLA-A*30 is not frequent in the Caucasoid population where HSV-1 infections are more frequent [40-42, 200, 201]. A more recent study reported that HLA B*27 alleles promotes recurrence of HSV-1 herpetic eye disease [57]. Thus far, HLA-B5, HLA B*27 and HLA-A*30 are the only alleles reported as associated to exacerbation of recurrences of ocular herpetic eye disease [57]. However, much work is needed to investigate whether ocular herpes infection and disease are affected by HLA-A, HLA-B and HLA-C alleles. We are in the process of evaluation whether HLA-B*27:05 monochain transgenic/H-2 class I null mice, recently developed, will be more susceptible to ocular herpes infection and disease.
3. How would the knowledge of association of HLA frequency with herpes prevalence help develop a universal T cell epitope-based herpes vaccine?
T cell responses to peptide epitope-based vaccines are dependent on HLA molecules, which play a pivotal role in the selection of the T cell repertoire in the thymus and presentation of “protective” and “pathogenic” HSV T cell epitopes in the periphery. Although many HLA-A*0201-restricted epitopes have been identified from HSV-1 and HSV-2 protein antigens, a question of practical importance is the translation of these pre-clinical findings based solely on HLA-0201-restricted epitopes, for the development of a universal vaccine for a genetically heterogeneous human population [17, 27, 202-205]. In an attempt to develop a universal T cell epitope-based vaccine, and since CD8+ T cells appear to be crucial effectors of protective immunity against HSV-1 and HSV-2 infections [14, 24, 25, 27, 43-54], past-decade research from our laboratory has focused on two strategies: (1) identifying HLA-restricted epitopes on herpes protein antigens that are strongly recognized by human CD8+ T cell from seropositive “asymptomatic” individuals; and (2) immunization of “humanized” HLA transgenic animal models (mice and rabbits) with antigenic epitopes to determine their immunogenicity and protective efficacy against ocular and genital herpes infection and disease. In our ongoing prophylactic and therapeutic herpes vaccine program, we have already successfully identified several “symptomatic” and “asymptomatic” CD8+ T cell epitopes and have demonstrated pathogenic vs. protective efficacy in “humanized” HLA-A*0201 transgenic animal models [24, 43, 44, 158]. However, although the HLA-A*02 allele is frequent in three major population groups (i.e. in over 50% of Caucasoid, Hispanic and Oriental individuals), it is represented in the African ethnic groups in only 30% of individuals. Therefore, a universal herpes vaccine that provides a perfect coverage of human population, regardless of race and ethnicity, should include multiple T-cell epitopes restricted by common HLA-A, HLA-B and HLA-C alleles worldwide, not just the HLA-A*0201 allele. A combination of nine HLA supertypes of cumulative high frequencies has been defined to provide an almost perfect coverage (greater than 99%) of the entire repertoire of HLA molecules [206]. Since a mixture of multiple epitopes is likely to stimulate polyclonal T-cell lines (one T cell clone for each epitope) and more effector T cells than a single epitope [26], we used multiple CD8+ T cell epitopes to induce a broader T cell response [26, 27]. Therefore, for a better coverage of all ethnic groups it is imperative to extend the discovery process and identify epitopes that are restricted to other HLA-A, HLA-B and HLA-C alleles, by selecting the HLA molecules with high cumulative frequencies in human populations around the world.
A universal herpes vaccine that is based on T cell epitopes presented by most common HLA-A, HLA-B, and HLA-C alleles of cumulative high frequencies in the four major world’s populations (i.e. Caucasoids, Orientals, Hispanics and Blacks) would allow full coverage of the world population and thus a superior clinical response. Additionally, a therapeutic vaccine that would control the disease (in contrast to a prophylactic vaccine that prevent transmission) would be relatively more feasible and the results would be more testable, requiring a much smaller sample size. This is especially true in sub-Saharan regions where the prevalence of genital herpes disease remains high (e.g., in Kenya were greater than 50% are infected by genital herpes).
Towards this goal, our first step in driving a protective cellular immunity is the discovery of “protective” HLA-A, HLA-B, and HLA-C-restricted epitopes that are: (1) presented by common and highly frequent alleles that are cumulatively distributed throughout the 5 continents, regardless of race and ethnicity, making those T cell epitopes “universal” candidates to be included in a universal vaccine. For instance, because of the high frequency of the HLA-C*07, HLA-A*24, HLA-B*35 alleles of the five continents, epitopes restricted to those three alleles would be a good candidates to be included in an universal vaccine for a genetically heterogeneous human population [17, 27, 202-205]; (2) include “asymptomatic” presented by HLA-A, HLA-B, and HLA-C alleles that are mostly represented in non-endemic areas, because those alleles, and associated epitopes, might be involved in resistance to HSV infection and/or to a slow progression to herpetic disease. For instance, HLA-B*44 high expression and frequency that appears to positively correlate with low prevalence of HSV-2 infection known in Caucasoid European population (Fig. 2), makes HLA-B*44-restricted epitopes “good” candidates to be included in future herpes vaccine, and finally; (3) to exclude the “symptomatic” epitopes that are associated with HLA-A, HLA-B, and HLA-C alleles most represented in endemic areas, because those HLA-A, HLA-B, and HLA-C alleles and associated epitopes might actually be involved in susceptibility to HSV infection and may facilitate progression to herpetic disease. For instance, because HLA-A*24 is the most frequent allele of the HLA-A locus in sub-Saharan populations, where one of the highest prevalence of genital HSV-2 infection is recorded (Fig. 2), it makes HLA-A*24-restricted epitopes undesirable candidates to be included in future herpes vaccine candidates because these epitopes may actually be involved in the mechanisms that facilitate herpes infection and/or in the immuno-pathological mechanisms that exacerbate herpes disease.
The repertoire of epitope peptides presented by HLA class I molecules is largely determined by the affinity that is dictated by the structure of the peptide itself and of the HLA binding groove. It is expected that the molecules having similar grooves (i.e., belonging to the same supertype) might present similar/overlapping peptides. While T cell responses are detected against epitopes presented by alternative molecules across HLA class I supertypes and loci, peptide elution studies report minute overlaps between the peptide repertoires of even related HLA molecules. However, we should emphasize that different HLA class I molecules can nevertheless present largely overlapping epitope peptide sets, and that “functional” HLA polymorphism on the individual and population level is probably much lower than previously anticipated [207]. Thus, a recent study showed that an unexpectedly large fraction of epitopes (> 50%) appeared bind two or more HLA molecules, often across supertype or even loci [207]. It also appears from a most recent study that HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and Immunogenicity [208]. An in silico analysis of potential CD8+ T cell epitopes of consensus 84+ ORF sequences of HSV-1 and HSV-2 strains revealed relatively higher number of promiscuous epitopes in contrast to limited numbers of binders restricted by one haplotype at a time. This suggests that the extent of promiscuity of T cell epitopes of HSV restricted by HLA alleles exerting opposing effects might differ.
4. HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02 and HLA-C*07:01 transgenic mice: novel versatile pre-clinical models to determine the role of HLA class I in susceptibility/resistance to herpes infection and disease
There a lack of “humanized” small animal models to study the role of HLA alleles and their restricted epitopes in protection vs. susceptibility to HSV-1/HSV-2 infection and disease. HLA transgenic mice represent a suitable versatile small animal model system to quickly evaluate T cell responses specific to human epitopes and complex pathogens, such HSV-1 and HSV-2. To facilitate the design of an effective HLA-restricted epitope-based herpes vaccine, several HLA-A, HLA-B, and HLA-C monochain transgenic/H-2 class I null mice have been recently developed [158].
The second step in driving cellular protective immunity against herpes is the in vivo immunogenicity study and protective efficacy of naturally presented human CD8+ T cell epitopes in reliable HLA transgenic (Tg) animal models [14]. In our ongoing HSV therapeutic vaccine program, we have successfully used HLA-A*0201 Tg mice to determine the immunogenicity and protective efficacy of many human HSV-1 and HSV-2 epitopes [24, 43, 44]. However, for a full coverage of the world’s European, African, Asian and Hispanic populations it is imperative to determine the immunogenicity of human epitopes that are restricted to other HLA class I (A, B and C) haplotypes that are highly represented in the populations where herpes is endemic [61]. We are currently mapping herpes epitopes restricted to other HLA class I and, in particular we are studying the immunogenicity protective efficacy of herpes epitopes in recently developed HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02 and HLA-C*07:01 mono-chain transgenic/H-2 class I null mice. The above set of HLA class I mono-chain transgenic mice combined with the already available HLA-A*02:01 and HLA-B*07:02 transgenic, H-2 KO mice [209, 210] would represent a large coverage of the four major world’s populations (European, African, Asian and Hispanic) as detailed below:
α) HLA-A
As discussed above, the HLA-A*01 appears to be frequent in regions with high endemic herpes, such as sub-Saharan Africa. HLA-A*01:03 monochain transgenic/H-2 class I null mice, now available, will be used to study susceptibility/resistance to herpes infection and disease [158]. In addition, the immunogenicity and protective efficacy of herpes epitopes restricted to the HLA-A*01:03 molecule can be studied in HLA-A*01:03 transgenic mice [158]. While HLA-A*24 is the most frequent allele of the HLA-A locus in population with the highest prevalence of genital HSV-2 infection [85, 86], the question remains whether HLA-A*24 allele constitutes a resistance or susceptible factor for HSV-2 infection. Thus, the HLA*A24:02 monochain transgenic/H-2 class I null mice which we recently developed could also be used to determine whether they are more susceptible to HSV-1 and HSV-2 infection and disease as compared to their age- and sex-matched wild type mice counterparts. HLA-A*24:02 transgenic mice can also be used to determine whether HLA class I-mediated susceptibility to HSV-2 is associated with HLA-A*24 and to study the immunogenicity of potential “asymptomatic” herpes epitopes restricted to HLA-A*24:02 [158].
β) HLA-B
The HLA-B*08 allele is also highly represented in sub-Saharan populations which have the highest rates of HSV-1 and HSV-2 infections. HLA-B*08:01 monochain transgenic/H-2 class I null mice will be used to study whether these mice are more susceptible to HSV-1 and HSV-2 infection and disease [158]. In addition, the patho-immunogenicity of potential “symptomatic” herpes epitopes restricted to HLA-B*08:01 molecule can be studies in these new mice [158]. The question also remains whether this HLA-B*35 allele constitutes a resistance or a susceptible factor to HSV-1 and HSV-2 infection. HLA-B*35:01 monochain transgenic/H-2 class I null mice are also now available to study the immunogenicity of herpes epitopes restricted to HLA-B*35 haplotype [158]. It will be of interest to determine whether HLA-B*35:01 transgenic mice will resist herpes infection and disease compared to HLA-A*24:02 monochain transgenic/H-2 class I null mice. We expect HLA-B*35:01 transgenic mice to be resistant to infection while HLA-A*24:02 transgenic mice to be susceptible to infection.
A study comparing HLA frequencies among seronegative, HSV-2 seropositive asymptomatic individuals to HSV-2 seropositive symptomatic individuals appears to show association of HLA-B*27 with an increase in symptomatic herpes disease [56]. HLA B*27 is the only allele reported as associated to exacerbation of recurrence of ocular herpetic eye disease [57]. Since HLA-B*27:05 monochain transgenic/H-2 class I null mice are now available, it would be interesting to see whether these mice will be more susceptible or more resistant to genital and ocular herpes infection and disease.
The HLA-B*44 is so far the most frequent allele of the HLA-B locus in Caucasoid European population but not in sub-Saharan African nations where the prevalence of HSV-2 infection in highest (Fig. 2). Thus, HLA-B*44 allele may constitute a direct or indirect factor in resistance to HSV-2 infection. HLA-B*44:02 monochain transgenic/H-2 class I null mice, now available, will be used to determine whether they will be more resistant to herpes compared to their wild type. It will also be of interest to determine whether HLA-B*44 transgenic mice will resists herpes infection compared to HLA-A*24:02 monochain transgenic/H-2 class I null mice. We expect HLA-B*44 transgenic mice to be resistant to infection while HLA-A*24:02 transgenic mice to be susceptible to infection. HLA-B*44:02 mice will also be used to study the immunogenicity and protective efficacy of herpes epitopes restricted to HLA-B*44 molecule [158].
γ) HLA-C
Although HLA-C*07 is the most frequent allele in the four major world’s populations (Caucasoid, Hispanics, Orientals and Blacks), its association with resistance vs. susceptibility to herpes is unclear. The recently available HLA-C*07:01 monochain transgenic/H-2 class I null will be used to determine whether HLA-C*07 allele will lead to resistance or susceptibility to herpes infection and disease compared to wild type mice and to HLA-A*24:02 mice.
5. Concluding Remarks
Several points could be made from the above analysis:
While the present study is focused on determining the association of HLA alleles with the prevalence of HSV-2 infections and disease; we do not ignore that other genetic, environmental, and socio-economic factors might also be involved in shaping up the prevalence of HSV-2 infections and diseases. For instance, while the high prevalence of herpes recorded in Sub-Saharan African populations is associated with high frequency of certain HLA alleles, it might also be affected by other co-infections, such as HIV, malaria and tuberculosis, that are also endemic in those regions. Nevertheless, HIV is far too recent to have exerted a pressing on the frequency of HLA alleles and on the prevalence of herpes. Moreover, malaria is geographically limited in its scope while tuberculosis is also typically limited to urban settings.
The subset of “risk” HLA class I alleles that appears more strongly and consistently associated with high prevalence of herpes infections and diseases, i.e. “susceptibility” included HLA-A*24, HLA-B*27, HLA-B*53 and HLA-B*58 (Fig. 2 and Table 2B). We are evaluating whether HLA-B*27:05 monochain transgenic/H-2 class I null mice, recently developed, will be more susceptible to ocular herpes infection and disease
High frequency of HLA-B*44 “protective” allele appears to be more strongly and consistently associated with low prevalence of herpes infections and diseases. (Fig. 2 and Table 2A). This makes HLA-B*44-restricted epitopes “good” candidates to be included in future herpes vaccine. We are also evaluating whether HLA-B*44 monochain transgenic/H-2 class I null mice, recently developed, will be more resistant to ocular herpes infection and disease.
HLA-B5, HLA B*27 and HLA-A*30 are the only alleles reported as associated to exacerbation of recurrences of ocular herpetic eye disease [57]. We are in the process of evaluation whether HLA-B*27:05 monochain transgenic/H-2 class I null mice, recently developed, will be more susceptible to ocular herpes infection and disease.
The HLA B*27 allele is so far the only allele that is positively associated with exacerbation of ocular herpes disease and with high prevalence of genital herpes infection.
Because of the high frequency of the HLA-C*07, HLA-A*24, HLA-B*35 alleles of the five continents, epitopes restricted to those three alleles would be a good candidates to be included in an universal vaccine for a genetically heterogeneous human population.
Because HLA-A*24 is the most frequent allele of the HLA-A locus in populations where one of the highest prevalence of genital HSV-2 infection is recorded, it makes HLA-A*24 -restricted epitopes undesirable candidates to be included in a future herpes vaccine because these epitopes may be involved in an immune mechanism that increase herpes infection and/or exacerbates herpes disease.
The findings in this report are a pure HLA-herpes-infection-disease associations and do not exclude other implications and immunogenetic factors that can affect the prevalence of HSV-2 infections and the resistance/susceptibility to herpes. Therefore, it would be of interest to further explore the influence of the genetic make-up of HSV-infected symptomatic vs. asymptomatic individuals. Comparing the immunogenetic profile of symptomatic vs. asymptomatic individuals might give clues for the individual course of the HSV infection and disease and will possibly lead to tailored therapeutic strategies in HSV-infected persons. Furthermore, sociologic factors such as sexual promiscuity and other high-risk sexual behaviors are likely to be associated with susceptibility to herpes infection but their role is beyond the scope of this review.
The associations of certain HLA alleles with susceptibility or resistance to HSV-1 and HSV-2 infection may help develop effective epitope-based vaccine against herpes infection and disease. However, the above analysis on the association of the frequency of HLA class I alleles with the prevalence of herpes infection does not imply to exclude a linkage between HLA class II (HLA-DR, HLA-DP and HLA-DQ) alleles and resistance/susceptibility to herpes infection and disease.
The present data constitute an association (rather than a cause or effect) and there may or may not be a direct relationship between an HLA haplotype and resistance/susceptibility to herpes. To directly assess whether HLA class I haplotypes play a role in susceptibility/resistance to herpes simplex virus infection and disease we have recently generated HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02 and HLA-C*07:01 monochain transgenic / H-2 Class I null mice. These novel, versatile pre-clinical mouse models of human T cell responses will also help us identify human epitopes restricted to specific HLA haplotype from the 84+ herpes proteins and assess the role of these epitopes in protection against genital and ocular herpes. The development of an effective vaccine against herpes is badly needed and would present an unparalleled economical alternative for herpes control, as it provide the most cost-effective and safe means to reduce the estimated yearly toll of 400 000 ocular herpes and 200,000 to 500,000 cases of genital herpes reported so far in the US [101, 211-219]. These numbers should be viewed cautiously, because many herpes infections are not reported, many are not easily diagnosed, and many are asymptomatic or inapparent.
Acknowledgments
This work is supported by Public Health Service research grants NIH-EY14900 and NIH-EY019896 to LBM, by The Discovery Eye Foundation, by The Henry L. Guenther Foundation, and by an unrestricted Research to Prevent Blindness Challenge grant.
Footnotes
Conflict of Interest: The authors have declared that no conflict of interest exists
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chentoufi AA, Kritzer E, Yu DM, Nesburn AB, BenMohamed L. Towards a rational design of an asymptomatic clinical herpes vaccine: the old, the new, and the unknown. Clin Dev Immunol. 2012;2012:187585. doi: 10.1155/2012/187585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awasthi S, Friedman HM. Status of prophylactic and therapeutic genital herpes vaccines. Curr Opin Virol. 2014;6C:6. doi: 10.1016/j.coviro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Knipe DM, Corey L, Cohen JI, Deal CD. Summary and recommendations from a National Institute of Allergy and Infectious Diseases (NIAID) workshop on “Next Generation Herpes Simplex Virus Vaccines”. Vaccine. 2014;32:1561. doi: 10.1016/j.vaccine.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Looker KJ, Garnett GP, Schmid GP. An estimate of the global prevalence and incidence of herpes simplex virus type 2 infection. Bull World Health Organ. 2008;86:805. doi: 10.2471/BLT.07.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haddow LJ, Sullivan EA, Taylor J, Abel M, Cunningham AL, Tabrizi S, et al. Herpes simplex virus type 2 (HSV-2) infection in women attending an antenatal clinic in the South Pacific island nation of Vanuatu. Sex Transm Dis. 2007;34:258. doi: 10.1097/01.olq.0000237774.29010.30. [DOI] [PubMed] [Google Scholar]
- 6.Smith JS, Robinson NJ. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J Infect Dis. 2002;186(Suppl 1):S3. doi: 10.1086/343739. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham AL, Taylor R, Taylor J, Marks C, Shaw J, Mindel A. Prevalence of infection with herpes simplex virus types 1 and 2 in Australia: a nationwide population based survey. Sex Transm Infect. 2006;82:164. doi: 10.1136/sti.2005.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mbopi-Keou FX, Gresenguet G, Mayaud P, Weiss HA, Gopal R, Matta M, et al. Interactions between herpes simplex virus type 2 and human immunodeficiency virus type 1 infection in African women: opportunities for intervention. J Infect Dis. 2000;182:1090. doi: 10.1086/315836. [DOI] [PubMed] [Google Scholar]
- 9.Ouedraogo A, Nagot N, Vergne L, Konate I, Weiss HA, Defer MC, et al. Impact of suppressive herpes therapy on genital HIV-1 RNA among women taking antiretroviral therapy: a randomized controlled trial. Aids. 2006;20:2305. doi: 10.1097/QAD.0b013e328010238d. [DOI] [PubMed] [Google Scholar]
- 10.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;20:73. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 11.Chentoufi AA, Dervillez X, Rubbo PA, Kuo T, Zhang X, Nagot N, et al. Current trends in negative immuno-synergy between two sexually transmitted infectious viruses: HIV-1 and HSV-1/2. Curr Trends Immunol. 2012;13:51. [PMC free article] [PubMed] [Google Scholar]
- 12.Rubbo PA, Tuaillon E, Nagot N, Chentoufi AA, Bollore K, Reynes J, et al. HIV-1 infection impairs HSV-specific CD4(+) and CD8(+) T-cell response by reducing Th1 cytokines and CCR5 ligand secretion. Journal of acquired immune deficiency syndromes. 2011;58:9. doi: 10.1097/QAI.0b013e318224d0ad. [DOI] [PubMed] [Google Scholar]
- 13.Chentoufi AA, BenMohamed L. Mucosal herpes immunity and immunopathology to ocular and genital herpes simplex virus infections. Clin Dev Immunol. 2012;2012:149135. doi: 10.1155/2012/149135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta G, BenMohamed L. Of mice and not humans: how reliable are animal models for evaluation of herpes CD8(+)-T cell-epitopes-based immunotherapeutic vaccine candidates? Vaccine. 2011;29:5824. doi: 10.1016/j.vaccine.2011.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta G, Chentoufi AA, Nesburn AB, Wechsler SL, BenMohamed L. New concepts in herpes simplex virus vaccine development: notes from the battlefield. Expert Rev Vaccines. 2009;8:1023. doi: 10.1586/erv.09.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasgupta G, Chentoufi AA, Nesburn AB, Wechsler SL, BenMohamed L. New concepts in herpes simplex virus vaccine development: notes from the battlefield. Expert review of vaccines. 2009;8:1023. doi: 10.1586/erv.09.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nesburn AB, Bettahi I, Zhang X, Zhu X, Chamberlain W, Afifi RE, et al. Topical/mucosal delivery of sub-unit vaccines that stimulate the ocular mucosal immune system. Ocul Surf. 2006;4:178. doi: 10.1016/s1542-0124(12)70164-7. [DOI] [PubMed] [Google Scholar]
- 18.Whitley RJ. Herpes simplex virus infections of women and their offspring: implications for a developed society. Proc Natl Acad Sci U S A. 1994;91:2441. doi: 10.1073/pnas.91.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westhoff GL, Little SE, Caughey AB. Herpes simplex virus and pregnancy: a review of the management of antenatal and peripartum herpes infections. Obstet Gynecol Surv. 2011;66:629. doi: 10.1097/OGX.0b013e31823983ec. [DOI] [PubMed] [Google Scholar]
- 20.Corey L, Wald A, Patel R, Sacks SL, Tyring SK, Warren T, et al. Once-daily valacyclovir to reduce the risk of transmission of genital herpes. N Engl J Med. 2004;350:11. doi: 10.1056/NEJMoa035144. [DOI] [PubMed] [Google Scholar]
- 21.Conant MA, Spicer DW, Smith CD. Herpes simplex virus transmission: condom studies. Sex Transm Dis. 1984;11:94. doi: 10.1097/00007435-198404000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Chentoufi AA, BenMohamed L, Van De Perre P, Ashkar AA. Immunity to ocular and genital herpes simplex viruses infections. Clin Dev Immunol. 2012;2012:732546. doi: 10.1155/2012/732546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chentoufi AA, Dasgupta G, Nesburn AB, Bettahi I, Binder NR, Choudhury ZS, et al. Nasolacrimal duct closure modulates ocular mucosal and systemic CD4(+) T-cell responses induced following topical ocular or intranasal immunization. Clin Vaccine Immunol. 2010;17:342. doi: 10.1128/CVI.00347-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chentoufi AA, Dasgupta G, Christensen ND, Hu J, Choudhury ZS, Azeem A, et al. A novel HLA (HLA-A*0201) transgenic rabbit model for preclinical evaluation of human CD8+ T cell epitope-based vaccines against ocular herpes. J Immunol. 2010;184:2561. doi: 10.4049/jimmunol.0902322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesburn AB, Bettahi I, Dasgupta G, Chentoufi AA, Zhang X, You S, et al. Functional Foxp3+ CD4+ CD25(Bright+) “natural” regulatory T cells are abundant in rabbit conjunctiva and suppress virus-specific CD4+ and CD8+ effector T cells during ocular herpes infection. J Virol. 2007;81:7647. doi: 10.1128/JVI.00294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettahi I, Nesburn AB, Yoon S, Zhang X, Mohebbi A, Sue V, et al. Protective immunity against ocular herpes infection and disease induced by highly immunogenic self-adjuvanting glycoprotein D lipopeptide vaccines. Invest Ophthalmol Vis Sci. 2007;48:4643. doi: 10.1167/iovs.07-0356. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Issagholian A, Berg EA, Fishman JB, Nesburn AB, BenMohamed L. Th-cytotoxic T-lymphocyte chimeric epitopes extended by Nepsilon-palmitoyl lysines induce herpes simplex virus type 1-specific effector CD8+ Tc1 responses and protect against ocular infection. J Virol. 2005;79:15289. doi: 10.1128/JVI.79.24.15289-15301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesburn AB, Ramos TV, Zhu X, Asgarzadeh H, Nguyen V, BenMohamed L. Local and systemic B cell and Th1 responses induced following ocular mucosal delivery of multiple epitopes of herpes simplex virus type 1 glycoprotein D together with cytosine-phosphate-guanine adjuvant. Vaccine. 2005;23:873. doi: 10.1016/j.vaccine.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Hollenbach JA, Thomson G, Cao K, Fernandez-Vina M, Erlich HA, Bugawan TL, et al. HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives. Hum Immunol. 2001;62:378. doi: 10.1016/s0198-8859(01)00212-9. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N, Tanabe K, Sugamori T, Sato M, Kitamura J, Sato H, et al. Association between Takayasu arteritis and ulcerative colitis - case report and review of serological HLA analysis. Med Sci Monit. 2011;17:CS81. doi: 10.12659/MSM.881837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somkrua R, Eickman EE, Saokaew S, Lohitnavy M, Chaiyakunapruk N. Association of HLA-B*5801 allele and allopurinol-induced Stevens Johnson syndrome and toxic epidermal necrolysis: a systematic review and meta-analysis. BMC Med Genet. 2011;12:118. doi: 10.1186/1471-2350-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huy NT, Hamada M, Kikuchi M, Lan NT, Yasunami M, Zamora J, et al. Association of HLA and post-schistosomal hepatic disorder: a systematic review and meta-analysis. Parasitol Int. 2011;60:347. doi: 10.1016/j.parint.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 33.de Menthon M, Lavalley MP, Maldini C, Guillevin L, Mahr A. HLA-B51/B5 and the risk of Behcet’s disease: a systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 2009;61:1287. doi: 10.1002/art.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awata T. Association of HLA genes with IDDM: a review. Nihon Rinsho. 1997;55(Suppl):370. [PubMed] [Google Scholar]
- 35.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. An update to HLA nomenclature, 2010. Bone Marrow Transplant. 2010;45:846. doi: 10.1038/bmt.2010.79. [DOI] [PubMed] [Google Scholar]
- 36.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holdsworth R, Hurley CK, Marsh SG, Lau M, Noreen HJ, Kempenich JH, et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens. 2009;73:95. doi: 10.1111/j.1399-0039.2008.01183.x. [DOI] [PubMed] [Google Scholar]
- 38.NIH Allele Frequency. 2013 http://www.ncbi.nlm.nih.gov/projects/gv/mhc/
- 39.Allele Frequency. 2013 http://www.allelefrequencies.net/default.asp.
- 40.Sulcebe G, Cuenod M, Sanchez-Mazas A, Tiercy JM, Zhubi B, Shyti E, et al. Human leukocyte antigen-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in an Albanian population from Kosovo. Int J Immunogenet. 2013;40:104. doi: 10.1111/j.1744-313X.2012.01136.x. [DOI] [PubMed] [Google Scholar]
- 41.Riccio ME, Buhler S, Nunes JM, Vangenot C, Cuenod M, Currat M, et al. 16(th) IHIW: analysis of HLA population data, with updated results for 1996 to 2012 workshop data (AHPD project report) Int J Immunogenet. 2013;40:21. doi: 10.1111/iji.12033. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Mazas A, Vidan-Jeras B, Nunes JM, Fischer G, Little AM, Bekmane U, et al. Strategies to work with HLA data in human populations for histocompatibility, clinical transplantation, epidemiology and population genetics: HLA-NET methodological recommendations. Int J Immunogenet. 2012;39:459. doi: 10.1111/j.1744-313X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Castelli FA, Zhu X, Wu M, Maillere B, BenMohamed L. Gender-dependent HLA-DR-restricted epitopes identified from herpes simplex virus type 1 glycoprotein D. Clin Vaccine Immunol. 2008;15:1436. doi: 10.1128/CVI.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chentoufi AA, Zhang X, Lamberth K, Dasgupta G, Bettahi I, Nguyen A, et al. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glycoprotein D. J Immunol. 2008;180:426. doi: 10.4049/jimmunol.180.1.426. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Dervillez X, Chentoufi AA, Badakhshan T, Bettahi I, BenMohamed L. Targeting the genital tract mucosa with a lipopeptide/recombinant adenovirus prime/boost vaccine induces potent and long-lasting CD8+ T cell immunity against herpes: importance of MyD88. J Immunol. 2012;189:4496. doi: 10.4049/jimmunol.1201121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X, Chentoufi AA, Hsiang C, Carpenter D, Osorio N, BenMohamed L, et al. The herpes simplex virus type 1 latency-associated transcript can protect neuron-derived C1300 and Neuro2A cells from granzyme B-induced apoptosis and CD8 T-cell killing. J Virol. 2011;85:2325. doi: 10.1128/JVI.01791-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chentoufi AA, Kritzer E, Tran MV, Dasgupta G, Lim CH, Yu DC, et al. The herpes simplex virus 1 latency-associated transcript promotes functional exhaustion of virus-specific CD8+ T cells in latently infected trigeminal ganglia: a novel immune evasion mechanism. J Virol. 2011;85:9127. doi: 10.1128/JVI.00587-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chentoufi AA, Kritzer E, Tran MV, Dasgupta G, Lim CH, Yu DC, et al. The herpes simplex virus 1 latency-associated transcript promotes functional exhaustion of virus-specific CD8+ T cells in latently infected trigeminal ganglia: a novel immune evasion mechanism. Journal of Virology. 2011;85:9127. doi: 10.1128/JVI.00587-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen SJ, Hamrah P, Gate D, Mott KR, Mantopoulos D, Zheng L, et al. The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. J Virol. 2011;85:4184. doi: 10.1128/JVI.02290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen SJ, Hamrah P, Gate D, Mott KR, Mantopoulos D, Zheng L, et al. The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. Journal of virology. 2011;85:4184. doi: 10.1128/JVI.02290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang X, Chentoufi AA, Hsiang C, Carpenter D, Osorio N, Benmohamed L, et al. The herpes simplex virus type 1 latency associated transcript (LAT) can protect neuronal derived C1300 and Neuro2A cells from Granzyme B induced apoptosis and CD8 T-cell killing. J Virol. 2010 doi: 10.1128/JVI.01791-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Chentoufi AA, Dasgupta G, Nesburn AB, Wu M, Zhu X, et al. A genital tract peptide epitope vaccine targeting TLR-2 efficiently induces local and systemic CD8+ T cells and protects against herpes simplex virus type 2 challenge. Mucosal Immunol. 2009;2:129. doi: 10.1038/mi.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mott KR, Chentoufi AA, Carpenter D, BenMohamed L, Wechsler SL, Ghiasi H. The role of a glycoprotein K (gK) CD8+ T-cell epitope of herpes simplex virus on virus replication and pathogenicity. Invest Ophthalmol Vis Sci. 2009;50:2903. doi: 10.1167/iovs.08-2957. [DOI] [PubMed] [Google Scholar]
- 54.Mott KR, Bresee CJ, Allen SJ, BenMohamed L, Wechsler SL, Ghiasi H. Level of herpes simplex virus type 1 latency correlates with severity of corneal scarring and exhaustion of CD8+ T cells in trigeminal ganglia of latently infected mice. J Virol. 2009;83:2246. doi: 10.1128/JVI.02234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moraru M, Cisneros E, Gomez-Lozano N, de Pablo R, Portero F, Canizares M, et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J Immunol. 2012;188:4412. doi: 10.4049/jimmunol.1103434. [DOI] [PubMed] [Google Scholar]
- 56.Lekstrom-Himes JA, Hohman P, Warren T, Wald A, Nam JM, Simonis T, et al. Association of major histocompatibility complex determinants with the development of symptomatic and asymptomatic genital herpes simplex virus type 2 infections. J Infect Dis. 1999;179:1077. doi: 10.1086/314729. [DOI] [PubMed] [Google Scholar]
- 57.Bohringer D, Sundmacher R, Reinhard T. HLA B27 seems to promote graft failure following penetrating keratoplasties for herpetic corneal scars. Ophthalmologe. 2007;104:705. doi: 10.1007/s00347-007-1550-9. [DOI] [PubMed] [Google Scholar]
- 58.Groothuis TA, Griekspoor AC, Neijssen JJ, Herberts CA, Neefjes JJ. MHC class I alleles and their exploration of the antigen-processing machinery. Immunol Rev. 2005;207:60. doi: 10.1111/j.0105-2896.2005.00305.x. [DOI] [PubMed] [Google Scholar]
- 59.Klein J, Sato A. The HLA system. First of two parts. N Engl J Med. 2000;343:702. doi: 10.1056/NEJM200009073431006. [DOI] [PubMed] [Google Scholar]
- 60.Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000;343:782. doi: 10.1056/NEJM200009143431106. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones A. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acid Research. 2011;39:D913. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canossi A, Piancatelli D, Aureli A, Oumhani K, Ozzella G, Del Beato T, et al. Correlation between genetic HLA class I and II polymorphisms and anthropological aspects in the Chaouya population from Morocco (Arabic speaking) Tissue Antigens. 2010;76:177. doi: 10.1111/j.1399-0039.2010.01498.x. [DOI] [PubMed] [Google Scholar]
- 63.Canossi A, Liberatore G, Del Beato T, Oumhani K, Adorno D. Sequence-based typing characterization of the novel HLA-Cw*1609 allele in the Moroccan Chaouya group. Tissue Antigens. 2007;69:367. doi: 10.1111/j.1399-0039.2006.00799.x. [DOI] [PubMed] [Google Scholar]
- 64.Piancatelli D, Canossi A, Aureli A, Oumhani K, Del Beato T, Di Rocco M, et al. Human leukocyte antigen-A, -B, and -Cw polymorphism in a Berber population from North Morocco using sequence-based typing. Tissue Antigens. 2004;63:158. doi: 10.1111/j.1399-0039.2004.00161.x. [DOI] [PubMed] [Google Scholar]
- 65.Oumhani K, Canossi A, Piancatelli D, Di Rocco M, Del Beato T, Liberatore G, et al. Sequence-Based analysis of the HLA-DRB1 polymorphism in Metalsa Berber and Chaouya Arabic-speaking groups from Morocco. Hum Immunol. 2002;63:129. doi: 10.1016/s0198-8859(01)00370-6. [DOI] [PubMed] [Google Scholar]
- 66.Lopez-Nevot MA, Garcia E, Romero C, Oliva MR, Serrano S, Garrido F. Phenotypic and genetic analysis of HLA class I and HLA-DR antigen expression on human melanomas. Exp Clin Immunogenet. 1988;5:203. [PubMed] [Google Scholar]
- 67.Ozawa M, Terasaki PI, Lee JH, Castro R, Alberu J, Alonso C, et al. 14th International HLA and Immunogenetics Workshop: report on the Prospective Chronic Rejection Project. Tissue Antigens. 2007;69(Suppl 1):174. doi: 10.1111/j.1399-0039.2006.00765.x. [DOI] [PubMed] [Google Scholar]
- 68.Ozawa M, Terasaki PI, Castro R, Alberu J, Morales-Buenrostro L, Alvarez I, et al. 14th International HLA and Immunogenetics Workshop Prospective Chronic Rejection Project: a three-year follow-up analysis. Clin Transpl. 2007:255. [PubMed] [Google Scholar]
- 69.El-Awar N, Lee JH, Tarsitani C, Terasaki PI. HLA class I epitopes: recognition of binding sites by mAbs or eluted alloantibody confirmed with single recombinant antigens. Hum Immunol. 2007;68:170. doi: 10.1016/j.humimm.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Pekiner FN, Aytugar E, Demirel GY, Borahan MO. HLA-A, B (Class I) and HLA-DR, DQ (Class II) Antigens in Turkish Patients with Recurrent Aphthous Ulceration and Behcet’s Disease. Med Princ Pract. 2013 doi: 10.1159/000348366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castelli EC, Gil DS, Veiga LC, de Camargo JL. Typing class I HLA-A gene using a nested PCR-RFLP procedure. Braz J Med Biol Res. 2005;38:837. doi: 10.1590/s0100-879x2005000600004. [DOI] [PubMed] [Google Scholar]
- 72.Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum Immunol. 2001;62:1200. doi: 10.1016/s0198-8859(01)00319-6. [DOI] [PubMed] [Google Scholar]
- 73.Torimiro JN, Carr JK, Wolfe ND, Karacki P, Martin MP, Gao X, et al. HLA class I diversity among rural rainforest inhabitants in Cameroon: identification of A*2612-B*4407 haplotype. Tissue Antigens. 2006;67:30. doi: 10.1111/j.1399-0039.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 74.Dupont E, Toungouz M, Andrien M, Vereerstraeten P. Compressive HLA matching. Nephrol Dial Transplant. 1997;12:2048. doi: 10.1093/ndt/12.10.2048. [DOI] [PubMed] [Google Scholar]
- 75.Toungouz M, Denys C, Andrien M, De Groote D, Dupont E. Interleukin 12 unmasks HLA class I differences during mixed lymphocyte reaction induced interferon gamma production. Hum Immunol. 1995;44:145. doi: 10.1016/0198-8859(95)00096-8. [DOI] [PubMed] [Google Scholar]
- 76.Andrien M, Tiercy JM, Defleur V, Bouillenne C, Toungouz M, Jeannet M, et al. HLA-B locus DNA typing: detection of B*7801 and seven additional alleles by BW6-specific exon 2 amplification. Tissue Antigens. 1993;42:480. doi: 10.1111/j.1399-0039.1993.tb02192.x. [DOI] [PubMed] [Google Scholar]
- 77.Andrien M, Stordeur P, Vamos E, Mascart Lemone F, Toungouz M, Monfils G, et al. Differential inducibility of HLA class I and II antigens by r-IFN gamma in type III bare lymphocyte syndrome. Transplant Proc. 1991;23:441. [PubMed] [Google Scholar]
- 78.Dervillez X, Qureshi H, Chentoufi AA, Khan AA, Kritzer K, Yu DC, et al. “Asymptomatic” HLA-A*02:01-Restricted Epitopes from Herpes Simplex Virus Glycoprotein B Preferentially Recall Polyfunctional CD8+ T Cells from Seropositive Asymptomatic Individuals and Protect HLA Transgenic Mice Against Ocular Herpes. J Immunol. 2013 doi: 10.4049/jimmunol.1301415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bunce M. PCR-sequence-specific primer typing of HLA class I and class II alleles. Methods Mol Biol. 2003;210:143. doi: 10.1385/1-59259-291-0:143. [DOI] [PubMed] [Google Scholar]
- 80.Arrieta-Bolanos E, Maldonado-Torres H, Dimitriu O, Hoddinott MA, Fowles F, Shah A, et al. HLA-A, -B, -C, -DQB1, and -DRB1,3,4,5 allele and haplotype frequencies in the Costa Rica Central Valley Population and its relationship to worldwide populations. Hum Immunol. 2011;72:80. doi: 10.1016/j.humimm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Miao KR, Pan QQ, Tang RC, Zhou XP, Fan S, Wang XY, et al. The polymorphism and haplotype analysis of HLA-A, -B and -DRB1 genes of population in Jiangsu province of China. Int J Immunogenet. 2007;34:419. doi: 10.1111/j.1744-313X.2007.00713.x. [DOI] [PubMed] [Google Scholar]
- 82.Miao KR, Pan QQ, Xue M, Zhou XY, Fei XM, Tang YH, et al. A novel HLA allele, DRB1*1609, identified in the Chinese Han population*. Tissue Antigens. 2005;66:248. doi: 10.1111/j.1399-0039.2005.00443.x. [DOI] [PubMed] [Google Scholar]
- 83.Yang KL, Lee SK, Kao RH, Lin CL, Lin PY. Recognition of HLA-A*24: 137 allele in a Taiwanese unrelated bone marrow stem cell donor and the plausible HLA haplotype associated with A*24:137. Int J Immunogenet. 2012;39:530. doi: 10.1111/j.1744-313X.2012.01134.x. [DOI] [PubMed] [Google Scholar]
- 84.Rivino L, Tan AT, Chia A, Kumaran EA, Grotenbreg GM, Macary PA, et al. Defining CD8+ T Cell Determinants during Human Viral Infection in Populations of Asian Ethnicity. J Immunol. 2013 doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
- 85.Fleischhauer K, Agostino A, Zino E, Mazzi B, Benazzi E, Arevalo-Herrera M, et al. Molecular characterization of HLA class I in Colombians carrying HLA-A2: high allelic diversity and frequency of heterozygotes at the HLA-B locus. Tissue Antigens. 1999;53:519. doi: 10.1034/j.1399-0039.1999.530601.x. [DOI] [PubMed] [Google Scholar]
- 86.Fleischhauer K, Zino E, Arevalo-Herrera M, Herrera S, Valmori D, Cerottini JC, et al. Differential expression of HLA-A*02 subtypes in Colombian Blacks and Mestizos. Tissue Antigens. 1998;51:204. doi: 10.1111/j.1399-0039.1998.tb02966.x. [DOI] [PubMed] [Google Scholar]
- 87.Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G, Schmidt AH. High-resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches? Hum Immunol. 2013;74:330. doi: 10.1016/j.humimm.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 88.Pingel S, Weyer S, Galle PR, Lohr HF. Evolution of viral quasispecies in four dominant HlA-A2 restricted T cell epitopes is not a major reason for viral persistence in interferon-treated patients with chronic hepatitis C. J Med Virol. 2002;66:472. doi: 10.1002/jmv.2168. [DOI] [PubMed] [Google Scholar]
- 89.Schmidt J, Neumann-Haefelin C, Altay T, Gostick E, Price DA, Lohmann V, et al. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J Virol. 2011;85:5232. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunne C, Crowley J, Hagan R, Rooney G, Lawlor E. HLA-A, B, Cw, DRB1, DQB1 and DPB1 alleles and haplotypes in the genetically homogenous Irish population. Int J Immunogenet. 2008;35:295. doi: 10.1111/j.1744-313X.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 91.Crowley J, Dunne C, Cox ST, Little AM. An unlikely rare result of HLA-A*0236, *3601 masking the presence of a novel allele A*0114. Tissue Antigens. 2007;69:200. doi: 10.1111/j.1399-0039.2006.00748.x. [DOI] [PubMed] [Google Scholar]
- 92.McGuigan C, Dunne C, Crowley J, Hagan R, Rooney G, Lawlor E, et al. Population frequency of HLA haplotypes contributes to the prevalence difference of multiple sclerosis in Ireland. J Neurol. 2005;252:1245. doi: 10.1007/s00415-005-0850-8. [DOI] [PubMed] [Google Scholar]
- 93.Middleton D, Williams F, Meenagh A, Daar AS, Gorodezky C, Hammond M, et al. Analysis of the distribution of HLA-A alleles in populations from five continents. Hum Immunol. 2000;61:1048. doi: 10.1016/s0198-8859(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 94.Bruges Armas J, Destro-Bisol G, Lopez-Vazquez A, Couto AR, Spedini G, Gonzalez S, et al. HLA class I variation in the West African Pygmies and their genetic relationship with other African populations. Tissue Antigens. 2003;62:233. doi: 10.1034/j.1399-0039.2003.00100.x. [DOI] [PubMed] [Google Scholar]
- 95.Cao K, Moormann AM, Lyke KE, Masaberg C, Sumba OP, Doumbo OK, et al. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens. 2004;63:293. doi: 10.1111/j.0001-2815.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 96.Fleischhauer K, Tanzarella S, Wallny HJ, Bordignon C, Traversari C. Multiple HLA-A alleles can present an immunodominant peptide of the human melanoma antigen Melan-A/MART-1 to a peptide-specific HLA-A*0201+ cytotoxic T cell line. J Immunol. 1996;157:787. [PubMed] [Google Scholar]
- 97.O’Hanlon TP, Rider LG, Mamyrova G, Targoff IN, Arnett FC, Reveille JD, et al. HLA polymorphisms in African Americans with idiopathic inflammatory myopathy: allelic profiles distinguish patients with different clinical phenotypes and myositis autoantibodies. Arthritis Rheum. 2006;54:3670. doi: 10.1002/art.22205. [DOI] [PubMed] [Google Scholar]
- 98.Gao X, Single RM, Karacki P, Marti D, O’Brien SJ, Carrington M. Diversity of MICA and linkage disequilibrium with HLA-B in two North American populations. Hum Immunol. 2006;67:152. doi: 10.1016/j.humimm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 99.Klitz W, Gragert L, Maiers M, Tu B, Lazaro A, Yang R, et al. Four-locus high-resolution HLA typing in a sample of Mexican Americans. Tissue Antigens. 2009;74:508. doi: 10.1111/j.1399-0039.2009.01374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68:779. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 101.Xu F, Sternberg MR, Markowitz LE. Women who have sex with women in the United States: prevalence, sexual behavior and prevalence of herpes simplex virus type 2 infection-results from national health and nutrition examination survey 2001-2006. Sex Transm Dis. 2010;37:407. doi: 10.1097/OLQ.0b013e3181db2e18. [DOI] [PubMed] [Google Scholar]
- 102.Morishima Y, Sasazuki T, Inoko H, Juji T, Akaza T, Yamamoto K, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99:4200. doi: 10.1182/blood.v99.11.4200. [DOI] [PubMed] [Google Scholar]
- 103.Ali ME, Ahmed MU, Alam S, Rahman MH. HLA-A, -B and -DRB1 allele frequencies in the Bangladeshi population. Tissue Antigens. 2008;72:115. doi: 10.1111/j.1399-0039.2008.01079.x. [DOI] [PubMed] [Google Scholar]
- 104.Olsson KS, Ritter B, Raha-Chowdhury R. HLA-A3-B14 and the origin of the haemochromatosis C282Y mutation: founder effects and recombination events during 12 generations in a Scandinavian family with major iron overload. Eur J Haematol. 2010;84:145. doi: 10.1111/j.1600-0609.2009.01376.x. [DOI] [PubMed] [Google Scholar]
- 105.Smestad C, Brynedal B, Jonasdottir G, Lorentzen AR, Masterman T, Akesson E, et al. The impact of HLA-A and -DRB1 on age at onset, disease course and severity in Scandinavian multiple sclerosis patients. Eur J Neurol. 2007;14:835. doi: 10.1111/j.1468-1331.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 106.Hammond L, Bengtsson M, Street J, Pepperall J, Darke C. HLA-A*2312--a novel allele found during testing for the UK NEQAS for H&I DNA HLA-typing scheme. Tissue Antigens. 2006;68:175. doi: 10.1111/j.1399-0039.2006.00639.x. [DOI] [PubMed] [Google Scholar]
- 107.Bengtsson M, Danielsson F, Jansson IE, Senkas A. Identification of a new HLA-Cw*05 allele, HLA-Cw*0505*. Tissue Antigens. 2004;63:600. doi: 10.1111/j.0001-2815.2004.00206.x. [DOI] [PubMed] [Google Scholar]
- 108.Bengtsson M, Danielsson F, Jansson IE, Jidell E, Johansson U. Two new HLA Cw* alleles, Cw*0105 and Cw*1405, detected by sequence based typing. Tissue Antigens. 2002;59:226. doi: 10.1034/j.1399-0039.2002.590309.x. [DOI] [PubMed] [Google Scholar]
- 109.Kippner L, Klint C, Sturfelt G, Bengtsson AA, Eriksson H, Truedsson L. Increased level of soluble HLA class I antigens in systemic lupus erythematosus: correlation with anti-DNA antibodies and leukopenia. J Autoimmun. 2001;16:471. doi: 10.1006/jaut.2001.0509. [DOI] [PubMed] [Google Scholar]
- 110.Ottinger HD, Ferencik S, Beelen DW, Lindemann M, Peceny R, Elmaagacli AH, et al. Impact of HLA-A,B,C allele mismatches on outcome after unrelated blood stem cell transplantation in whites. Transplantation. 2004;78:1077. doi: 10.1097/01.tp.0000137791.28140.93. [DOI] [PubMed] [Google Scholar]
- 111.Kubens BS, Ferencik S, Grosse-Wilde H. An HLA-A/C,B recombination detected by one-dimensional isoelectric focusing gel electrophoresis. Tissue Antigens. 1991;38:92. doi: 10.1111/j.1399-0039.1991.tb01886.x. [DOI] [PubMed] [Google Scholar]
- 112.Svejgaard A, Hauge M, Jersild C, Platz P, Ryder LP, Nielsen LS, et al. The HLA system. An introductory survey. Monogr Hum Genet. 1975;7:1. [PubMed] [Google Scholar]
- 113.Staub Nielsen L, Jersild C, Ryder LP, Svejgaard A. HL-A antigen, gene, and haplotype frequencies in Denmark. Tissue Antigens. 1975;6:70. doi: 10.1111/j.1399-0039.1975.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 114.Fleischhauer K, Kernan NA, O’Reilly RJ, Dupont B, Yang SY. Bone marrow-allograft rejection by T lymphocytes recognizing a single amino acid difference in HLA-B44. N Engl J Med. 1990;323:1818. doi: 10.1056/NEJM199012273232607. [DOI] [PubMed] [Google Scholar]
- 115.Bera O, Cesaire R, Quelvennec E, Quillivic F, de Chavigny V, Ribal C, et al. HLA class I and class II allele and haplotype diversity in Martinicans. Tissue Antigens. 2001;57:200. doi: 10.1034/j.1399-0039.2001.057003200.x. [DOI] [PubMed] [Google Scholar]
- 116.Schulman LL, Weinberg AD, McGregor C, Galantowicz ME, Suciu-Foca NM, Itescu S. Mismatches at the HLA-DR and HLA-B loci are risk factors for acute rejection after lung transplantation. Am J Respir Crit Care Med. 1998;157:1833. doi: 10.1164/ajrccm.157.6.9707007. [DOI] [PubMed] [Google Scholar]
- 117.Liu Z, Colovai AI, Tugulea S, Reed EF, Fisher PE, Mancini D, et al. Indirect recognition of donor HLA-DR peptides in organ allograft rejection. J Clin Invest. 1996;98:1150. doi: 10.1172/JCI118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harris PE, Liu Z, Suciu-Foca N. MHC class II binding of peptides derived from HLA-DR 1. J Immunol. 1992;148:2169. [PubMed] [Google Scholar]
- 119.Igarashi M, Suciu-Foca N, Ferrone S. Differential expression of HLA-DR allospecificities on antigen specific T cell clones. Tissue Antigens. 1987;29:160. doi: 10.1111/j.1399-0039.1987.tb01569.x. [DOI] [PubMed] [Google Scholar]
- 120.Yunis EJ, Awdeh Z, Johnson A, Suciu-Foca N, Robinson MA, Hartzman R, et al. Complotype genetic loci segregate more frequently with HLA-DR than with HLA-B. Immunogenetics. 1985;21:25. doi: 10.1007/BF00372238. [DOI] [PubMed] [Google Scholar]
- 121.Suciu-Foca N, Reed E, Rohowsky C, Lewison A, King DW. Influence of race on the predictability of mixed lymphocyte culture identity by HLA-DR matching. Transplantation. 1983;35:35. doi: 10.1097/00007890-198301000-00008. [DOI] [PubMed] [Google Scholar]
- 122.Bubnova MG, Oganov RG. Treatment of patients with arterial hypertension and other stroke risk factors in clinical practice. The PROGNOS program. Ter Arkh. 2009;81:19. [PubMed] [Google Scholar]
- 123.Williams F, Meenagh A, Darke C, Acosta A, Daar AS, Gorodezky C, et al. Analysis of the distribution of HLA-B alleles in populations from five continents. Hum Immunol. 2001;62:645. doi: 10.1016/s0198-8859(01)00247-6. [DOI] [PubMed] [Google Scholar]
- 124.Middleton D, Williams F, Hamill MA, Meenagh A. Frequency of HLA-B alleles in a Caucasoid population determined by a two-stage PCR-SSOP typing strategy. Hum Immunol. 2000;61:1285. doi: 10.1016/s0198-8859(00)00186-5. [DOI] [PubMed] [Google Scholar]
- 125.Williams F, Meenagh A, Maxwell AP, Middleton D. Allele resolution of HLA-A using oligonucleotide probes in a two-stage typing strategy. Tissue Antigens. 1999;54:59. doi: 10.1034/j.1399-0039.1999.540107.x. [DOI] [PubMed] [Google Scholar]
- 126.Pinto C, Smith AG, Larsen CE, Fernandez-Vina M, Husain Z, Clavijo OP, et al. HLA-Cw*0409N is associated with HLA-A*2301 and HLA-B*4403-carrying haplotypes. Hum Immunol. 2004;65:181. doi: 10.1016/j.humimm.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 127.Moraes ME, Romero M, Salomao I, Correa M, Gouvea N, Hue MI, et al. Strong linkage disequilibrium between HLA-B*3913 and DRB1*0807 in Brazilians. Transplant Proc. 2004;36:823. doi: 10.1016/j.transproceed.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 128.Henry JB, Hubbell CA, Davis MC, Fernandez-Vina MA, Yunis EJ, Shrimpton AE. A new HLA-A1 mutation: a novel, null variant allele. Am J Clin Pathol. 2004;122:185. doi: 10.1309/8DE1-2YU6-5BKH-HQW6. [DOI] [PubMed] [Google Scholar]
- 129.Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 130.Mack SJ, Tu B, Lazaro A, Yang R, Lancaster AK, Cao K, et al. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens. 2009;73:17. doi: 10.1111/j.1399-0039.2008.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tu B, Mack SJ, Lazaro A, Lancaster A, Thomson G, Cao K, et al. HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens. 2007;69:73. doi: 10.1111/j.1399-0039.2006.00728.x. [DOI] [PubMed] [Google Scholar]
- 132.Elbjeirami WM, Abdel-Rahman F, Hussein AA. Probability of finding an HLA-matched donor in immediate and extended families: the Jordanian experience. Biol Blood Marrow Transplant. 2013;19:221. doi: 10.1016/j.bbmt.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 133.Yabuki K, Ota M, Goto K, Kimura T, Nomura E, Ohno S, et al. Triplet repeat polymorphism in the MICA gene in HLA-B27 positive and negative caucasian patients with ankylosing spondylitis. Hum Immunol. 1999;60:83. doi: 10.1016/s0198-8859(98)00092-5. [DOI] [PubMed] [Google Scholar]
- 134.Yabuki K, Ohno S, Mizuki N, Ando H, Tabbara KF, Goto K, et al. HLA class I and II typing of the patients with Behcet’s disease in Saudi Arabia. Tissue Antigens. 1999;54:273. doi: 10.1034/j.1399-0039.1999.540308.x. [DOI] [PubMed] [Google Scholar]
- 135.Yabuki K, Mizuki N, Ota M, Katsuyama Y, Palimeris G, Stavropoulos C, et al. Association of MICA gene and HLA-B*5101 with Behcet’s disease in Greece. Invest Ophthalmol Vis Sci. 1999;40:1921. [PubMed] [Google Scholar]
- 136.Wallace GR, Verity DH, Delamaine LJ, Ohno S, Inoko H, Ota M, et al. MIC-A allele profiles and HLA class I associations in Behcet’s disease. Immunogenetics. 1999;49:613. doi: 10.1007/s002510050656. [DOI] [PubMed] [Google Scholar]
- 137.Mizuki N, Ota M, Katsuyama Y, Yabuki K, Ando H, Goto K, et al. Association analysis between the MIC-A and HLA-B alleles in Japanese patients with Behcet’s disease. Arthritis Rheum. 1999;42:1961. doi: 10.1002/1529-0131(199909)42:9<1961::AID-ANR23>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 138.Jawdat DM, Al Saleh S, Sutton P, Al Anazi H, Shubaili A, Tamim H, et al. Chances of finding an HLA-matched sibling: The Saudi experience. Biol Blood Marrow Transplant. 2009;15:1342. doi: 10.1016/j.bbmt.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 139.Hajeer AH, Sawidan FA, Bohlega S, Saleh S, Sutton P, Shubaili A, et al. HLA class I and class II polymorphisms in Saudi patients with myasthenia gravis. Int J Immunogenet. 2009;36:169. doi: 10.1111/j.1744-313X.2009.00843.x. [DOI] [PubMed] [Google Scholar]
- 140.Roitberg-Tambur A, Witt CS, Friedmann A, Safirman C, Sherman L, Battat S, et al. Comparative analysis of HLA polymorphism at the serologic and molecular level in Moroccan and Ashkenazi Jews. Tissue Antigens. 1995;46:104. doi: 10.1111/j.1399-0039.1995.tb02485.x. [DOI] [PubMed] [Google Scholar]
- 141.Maiers M. A community standard XML message format for sequencing-based typing data. Tissue Antigens. 2007;69(Suppl 1):69. doi: 10.1111/j.1399-0039.2006.76061.x. [DOI] [PubMed] [Google Scholar]
- 142.Kollman C, Maiers M, Gragert L, Muller C, Setterholm M, Oudshoorn M, et al. Estimation of HLA-A, -B, -DRB1 haplotype frequencies using mixed resolution data from a National Registry with selective retyping of volunteers. Hum Immunol. 2007;68:950. doi: 10.1016/j.humimm.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 143.Hurley CK, Maiers M, Marsh SG, Oudshoorn M. Overview of registries, HLA typing and diversity, and search algorithms. Tissue Antigens. 2007;69(Suppl 1):3. doi: 10.1111/j.1399-0039.2006.758_2.x. [DOI] [PubMed] [Google Scholar]
- 144.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68:392. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 145.Machulla HK, Batnasan D, Steinborn F, Uyar FA, Saruhan-Direskeneli G, Oguz FS, et al. Genetic affinities among Mongol ethnic groups and their relationship to Turks. Tissue Antigens. 2003;61:292. doi: 10.1034/j.1399-0039.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 146.Aizawa H, Kinouchi Y, Negoro K, Nomura E, Imai G, Takahashi S, et al. HLA-B is the best candidate of susceptibility genes in HLA for Japanese ulcerative colitis. Tissue Antigens. 2009;73:569. doi: 10.1111/j.1399-0039.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- 147.Williams F, Curran MD, Leheny WA, Daar AS, Middleton D. Characterization of HLA-B*3921 and confirmation of HLA-B*4415, two variant HLA-B alleles identified in the Omani population. Tissue Antigens. 2000;56:376. doi: 10.1034/j.1399-0039.2000.560411.x. [DOI] [PubMed] [Google Scholar]
- 148.Williams F, Curran MD, Padadoa Perez M, Acosta A, Middleton D. Characterisation of a new HLA-B allele, HLA-B*0720, identified in the Cuban population. Tissue Antigens. 2001;57:80. doi: 10.1034/j.1399-0039.2001.057001080.x. [DOI] [PubMed] [Google Scholar]
- 149.Middleton D, Curran MD, Anholts JD, Reilly ER, Schreuder GM. Characterisation of a new HLA-B allele, HLA-B*0724. Tissue Antigens. 2001;57:471. doi: 10.1034/j.1399-0039.2001.057005471.x. [DOI] [PubMed] [Google Scholar]
- 150.Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens. 2005;65:437. doi: 10.1111/j.1399-0039.2005.00386.x. [DOI] [PubMed] [Google Scholar]
- 151.Kwon OJ, Hwang SH, Lee MN, Oh HB. A novel HLA-A*2601 variant, A*260102, identified by sequence-based typing. Tissue Antigens. 2005;66:704. doi: 10.1111/j.1399-0039.2005.00487.x. [DOI] [PubMed] [Google Scholar]
- 152.Kwon OJ, Hwang SH, Hur SS, Lee MN, Oh HB. Novel HLA-Cw*01 allele, Cw*010203, identified by sequence-based typing*. Tissue Antigens. 2005;66:240. doi: 10.1111/j.1399-0039.2005.00455.x. [DOI] [PubMed] [Google Scholar]
- 153.Kwon OJ, Hwang SH, Heo YS, Hur SS, Lee MN, Oh HB. Novel HLA-A*11 allele, A*1120, identified by sequence-based typing. Tissue Antigens. 2005;66:141. doi: 10.1111/j.1399-0039.2005.00447.x. [DOI] [PubMed] [Google Scholar]
- 154.Hwang SH, Oh HB, Hur SS, Lee MN, Hong SA, Kwon OJ. Novel HLA-A*31 allele, A*3111 identified by sequence-based typing. Tissue Antigens. 2005;66:145. doi: 10.1111/j.1399-0039.2005.00439.x. [DOI] [PubMed] [Google Scholar]
- 155.Hwang SH, Cha CH, Kwon OJ, Heo YS, Hur SS, Lee MN, et al. Novel HLA-Cw*06 allele, Cw*0612, identified by sequence-based typing. Tissue Antigens. 2005;66:330. doi: 10.1111/j.1399-0039.2005.00460.x. [DOI] [PubMed] [Google Scholar]
- 156.Modiano D, Luoni G, Petrarca V, Sodiomon Sirima B, De Luca M, Simpore J, et al. HLA class I in three West African ethnic groups: genetic distances from sub-Saharan and Caucasoid populations. Tissue Antigens. 2001;57:128. doi: 10.1034/j.1399-0039.2001.057002128.x. [DOI] [PubMed] [Google Scholar]
- 157.O’Hanlon TP, Carrick DM, Arnett FC, Reveille JD, Carrington M, Gao X, et al. Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: distinct HLA-A, -B, -Cw, -DRB1 and -DQA1 allelic profiles and motifs define clinicopathologic groups in caucasians. Medicine (Baltimore) 2005;84:338. doi: 10.1097/01.md.0000189818.63141.8c. [DOI] [PubMed] [Google Scholar]
- 158.Boucherma R, Kridane-Miledi H, Bouziat R, Rasmussen M, Gatard T, Langa-Vives F, et al. HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02, and HLA-C*07:01 Monochain Transgenic/H-2 Class I Null Mice: Novel Versatile Preclinical Models of Human T Cell Responses. J Immunol. 2013 doi: 10.4049/jimmunol.1300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gallina G, Cumbo V, Messina P, Caprera V, Lio D, Caruso C. Lack of correlation between HLA-B35 resistance against herpes labialis and antibody titers to HSV-1. Oral Surg Oral Med Oral Pathol. 1989;68:167. doi: 10.1016/0030-4220(89)90187-4. [DOI] [PubMed] [Google Scholar]
- 160.Gallina G, Cumbo V, Messina P, Modica MA, Caruso C. MHC-linked genetic factors (HLA-B35) influencing recurrent circumoral herpetic lesions. Dis Markers. 1987;5:191. [PubMed] [Google Scholar]
- 161.Grimaldi MC, Crouau-Roy B, Contu L, Amoros JP. Molecular variation of HLA class I genes in the Corsican population: approach to its origin. Eur J Immunogenet. 2002;29:101. doi: 10.1046/j.1365-2370.2001.00287.x. [DOI] [PubMed] [Google Scholar]
- 162.Grimaldi MC, Crouau-Roy B, Amoros JP, Cambon-Thomsen A, Carcassi C, Orru S, et al. West Mediterranean islands (Corsica, Balearic islands, Sardinia) and the Basque population: contribution of HLA class I molecular markers to their evolutionary history. Tissue Antigens. 2001;58:281. doi: 10.1034/j.1399-0039.2001.580501.x. [DOI] [PubMed] [Google Scholar]
- 163.Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H. Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of Pakistan. Tissue Antigens. 2004;64:581. doi: 10.1111/j.1399-0039.2004.00302.x. [DOI] [PubMed] [Google Scholar]
- 164.Novitsky V, Flores-Villanueva PO, Chigwedere P, Gaolekwe S, Bussman H, Sebetso G, et al. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum Immunol. 2001;62:146. doi: 10.1016/s0198-8859(00)00236-6. [DOI] [PubMed] [Google Scholar]
- 165.Flores-Villanueva PO, Hendel H, Caillat-Zucman S, Rappaport J, Burgos-Tiburcio A, Bertin-Maghit S, et al. Associations of MHC ancestral haplotypes with resistance/susceptibility to AIDS disease development. J Immunol. 2003;170:1925. doi: 10.4049/jimmunol.170.4.1925. [DOI] [PubMed] [Google Scholar]
- 166.Diouf K, Sarr AD, Eisen G, Popper S, Mboup S, Kanki P. Associations between MHC class I and susceptibility to HIV-2 disease progression. J Hum Virol. 2002;5:1. [PubMed] [Google Scholar]
- 167.Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy JM, Marsh SG, Aubrey M, et al. A bioinformatics approach to ascertaining the rarity of HLA alleles. Tissue Antigens. 2009;74:480. doi: 10.1111/j.1399-0039.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 168.Sanchez-Velasco P, Gomez-Casado E, Martinez-Laso J, Moscoso J, Zamora J, Lowy E, et al. HLA alleles in isolated populations from North Spain: origin of the Basques and the ancient Iberians. Tissue Antigens. 2003;61:384. doi: 10.1034/j.1399-0039.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 169.Comas D, Mateu E, Calafell F, Perez-Lezaun A, Bosch E, Martinez-Arias R, et al. HLA class I and class II DNA typing and the origin of Basques. Tissue Antigens. 1998;51:30. doi: 10.1111/j.1399-0039.1998.tb02944.x. [DOI] [PubMed] [Google Scholar]
- 170.Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bertranpetit J. HLA evidence for the lack of genetic heterogeneity in Basques. Ann Hum Genet. 1998;62:123. doi: 10.1046/j.1469-1809.1998.6220123.x. [DOI] [PubMed] [Google Scholar]
- 171.Wakimoto A, Ohashi K, Koyama M, Kato M, Tsutsui T, Saji F, et al. Regulation of classical HLA class I genes in human choriocarcinoma cells by nuclear proteins binding to MHC class I regulatory elements. Am J Reprod Immunol. 1995;34:323. doi: 10.1111/j.1600-0897.1995.tb00959.x. [DOI] [PubMed] [Google Scholar]
- 172.Rendine S, Borelli I, Barbanti M, Sacchi N, Roggero S, Curtoni ES. HLA polymorphisms in Italian bone marrow donors: a regional analysis. Tissue Antigens. 1998;52:135. doi: 10.1111/j.1399-0039.1998.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 173.Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, et al. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 174.Lulli P, Mangano VD, Onori A, Batini C, Luoni G, Sirima BS, et al. HLA-DRB1 and -DQB1 loci in three west African ethnic groups: genetic relationship with sub-Saharan African and European populations. Hum Immunol. 2009;70:903. doi: 10.1016/j.humimm.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 175.Spinola H, Couto AR, Peixoto MJ, Anagnostou P, Destro-Bisol G, Spedini G, et al. HLA class-I diversity in Cameroon: evidence for a north-south structure of genetic variation and relationships with African populations. Ann Hum Genet. 2011;75:665. doi: 10.1111/j.1469-1809.2011.00672.x. [DOI] [PubMed] [Google Scholar]
- 176.Chaux P, Luiten R, Demotte N, Vantomme V, Stroobant V, Traversari C, et al. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J Immunol. 1999;163:2928. [PubMed] [Google Scholar]
- 177.Breckpot K, Heirman C, De Greef C, van der Bruggen P, Thielemans K. Identification of new antigenic peptide presented by HLA-Cw7 and encoded by several MAGE genes using dendritic cells transduced with lentiviruses. J Immunol. 2004;172:2232. doi: 10.4049/jimmunol.172.4.2232. [DOI] [PubMed] [Google Scholar]
- 178.Velickovic ZM, Delahunt B, Carter JM. HLA-DRB1 and HLA-DQB1 polymorphisms in Pacific Islands populations. Tissue Antigens. 2002;59:397. doi: 10.1034/j.1399-0039.2002.590506.x. [DOI] [PubMed] [Google Scholar]
- 179.Velickovic ZM, Carter JM. HLA-DPA1 and DPB1 polymorphism in four Pacific Islands populations determined by sequencing based typing. Tissue Antigens. 2001;57:493. doi: 10.1034/j.1399-0039.2001.057006493.x. [DOI] [PubMed] [Google Scholar]
- 180.Velickovic ZM, Carter JM. Nucleotide sequence of exons 2 and 3 of the novel HLA-Cw*12032 allele. Tissue Antigens. 2001;58:97. doi: 10.1034/j.1399-0039.2001.580206.x. [DOI] [PubMed] [Google Scholar]
- 181.Szczerkowska Dobosz A, Rebala K, Szczerkowska Z, Nedoszytko B. HLA-C locus alleles distribution in patients from northern Poland with psoriatic arthritis--preliminary report. Int J Immunogenet. 2005;32:389. doi: 10.1111/j.1744-313X.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- 182.Browning MJ, Madrigal JA, Krausa P, Kowalski H, Allsopp CE, Little AM, et al. The HLA-A,B,C genotype of the class I negative cell line Daudi reveals novel HLA-A and -B alleles. Tissue Antigens. 1995;45:177. doi: 10.1111/j.1399-0039.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 183.Lopez-Alvarez MR, Moya-Quiles MR, Minguela A, Gil J, Miras M, Campillo JA, et al. HLA-C matching and liver transplants: donor-recipient genotypes influence early outcome and CD8+KIR2D+ T-cells recuperation. Transplantation. 2009;88:S54. doi: 10.1097/TP.0b013e3181af7d84. [DOI] [PubMed] [Google Scholar]
- 184.Moya-Quiles MR, Alvarez R, Miras M, Gomez-Mateo J, Lopez-Alvarez MR, Marin-Moreno I, et al. Impact of recipient HLA-C in liver transplant: a protective effect of HLA-Cw*07 on acute rejection. Hum Immunol. 2007;68:51. doi: 10.1016/j.humimm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 185.Campillo JA, Martinez-Escribano JA, Muro M, Moya-Quiles R, Marin LA, Montes-Ares O, et al. HLA class I and class II frequencies in patients with cutaneous malignant melanoma from southeastern Spain: the role of HLA-C in disease prognosis. Immunogenetics. 2006;57:926. doi: 10.1007/s00251-005-0065-2. [DOI] [PubMed] [Google Scholar]
- 186.Moya-Quiles MR, Torio A, Muro M, Montes-Ares O, Marin L, Minguela A, et al. Impact of HLA-C on acute rejection in liver transplantation. Transplant Proc. 2003;35:1892. doi: 10.1016/s0041-1345(03)00636-5. [DOI] [PubMed] [Google Scholar]
- 187.Muro M, Marin L, Torio A, Moya-Quiles MR, Minguela A, Rosique-Roman J, et al. HLA polymorphism in the Murcia population (Spain): in the cradle of the archaeologic Iberians. Hum Immunol. 2001;62:910. doi: 10.1016/s0198-8859(01)00290-7. [DOI] [PubMed] [Google Scholar]
- 188.Torio A, Sanchez-Guerrero I, Muro M, Herrero N, Pagan J, Minguela A, et al. Analysis of the phenotypic distribution of HLA class I and class II in atopic and non-atopic asthma patients. Eur J Immunogenet. 2000;27:81. doi: 10.1046/j.1365-2370.2000.00205.x. [DOI] [PubMed] [Google Scholar]
- 189.Ibrahim AI, Kouwatli KM, Obeid MT. Frequency of herpes simplex virus in Syria based on type-specific serological assay. Saudi Med J. 2000;21:355. [PubMed] [Google Scholar]
- 190.Kloverpris HN, Stryhn A, Harndahl M, Carlson JM, Leslie AJ, Chen F, et al. HLA-A*68:02-restricted Gag-specific CTL responses can drive selection pressure on HIV but are subdominant and ineffective. AIDS. 2013 doi: 10.1097/QAD.0b013e32836146cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Paximadis M, Minevich G, Winchester R, Schramm DB, Gray GE, Sherman GG, et al. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PLoS One. 2011;6:e16541. doi: 10.1371/journal.pone.0016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang S, Cao J. Theoretical analysis and computer simulation of fluorescence lifetime measurements. II. Contour length dependence of single polymers. J Chem Phys. 2004;121:572. doi: 10.1063/1.1756578. [DOI] [PubMed] [Google Scholar]
- 193.Clavijo OP, Delgado JC, Yu N, Fraser PA, Yunis EJ. HLA-Cw*1701 is associated with two sub-Saharan African-derived HLA haplotypes: HLA-B*4201, DRB1*03 and HLA-B*4202 without DRB1*03. Tissue Antigens. 1999;54:303. doi: 10.1034/j.1399-0039.1999.540316.x. [DOI] [PubMed] [Google Scholar]
- 194.Whitley RJ, Lentnek A, McCracken GH, Sande MA, Scheld WM. Evaluation of new anti-infective drugs for the treatment of viral encephalitis. Infectious Diseases Society of America and the Food and Drug Administration. Clin Infect Dis. 1992;15(Suppl 1):S195. doi: 10.1093/clind/15.supplement_1.s195. [DOI] [PubMed] [Google Scholar]
- 195.Queiro R, Gonzalez S, Lopez-Larrea C, Alperi M, Sarasqueta C, Riestra JL, et al. HLA-C locus alleles may modulate the clinical expression of psoriatic arthritis. Arthritis Res Ther. 2006;8:R185. doi: 10.1186/ar2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Martinez-Borra J, Brautbar C, Gonzalez S, Enk CD, Lopez-Vazquez A, Lopez-Larrea C. The region of 150 kb telometic to HLA-C is associated with psoriasis in the Jewish population. J Invest Dermatol. 2005;125:928. doi: 10.1111/j.0022-202X.2005.23892.x. [DOI] [PubMed] [Google Scholar]
- 197.Martinez-Borra J, Gonzalez S, Santos-Juanes J, Sanchez del Rio J, Torre-Alonso JC, Lopez-Vazquez A, et al. Psoriasis vulgaris and psoriatic arthritis share a 100 kb susceptibility region telomeric to HLA-C. Rheumatology (Oxford) 2003;42:1089. doi: 10.1093/rheumatology/keg304. [DOI] [PubMed] [Google Scholar]
- 198.Colin J, Chastel C, Saleun JP, Morin JF, Renard G. Recurrent ocular herpes simplex infection and HLA antigens; preliminary results (author’s transl) J Fr Ophtalmol. 1979;2:263. [PubMed] [Google Scholar]
- 199.Meyers-Elliott RH, Elliott JH, Maxwell WA, Pettit TH, O’Day DM, Terasaki PI, et al. HLA antigens in recurrent stromal herpes simplex virus keratitis. Am J Ophthalmol. 1980;89:54. doi: 10.1016/0002-9394(80)90228-7. [DOI] [PubMed] [Google Scholar]
- 200.Nunes JM, Riccio ME, Buhler S, Di D, Currat M, Ries F, et al. Analysis of the HLA population data (AHPD) submitted to the 15th International Histocompatibility/Immunogenetics Workshop by using the Gene[rate] computer tools accommodating ambiguous data (AHPD project report) Tissue Antigens. 2010;76:18. doi: 10.1111/j.1399-0039.2010.01469.x. [DOI] [PubMed] [Google Scholar]
- 201.Sulcebe G, Sanchez-Mazas A, Tiercy JM, Shyti E, Mone I, Ylli Z, et al. HLA allele and haplotype frequencies in the Albanian population and their relationship with the other European populations. Int J Immunogenet. 2009;36:337. doi: 10.1111/j.1744-313X.2009.00868.x. [DOI] [PubMed] [Google Scholar]
- 202.BenMohamed L, Belkaid Y, Loing E, Brahimi K, Gras-Masse H, Druilhe P. Systemic immune responses induced by mucosal administration of lipopeptides without adjuvant. Eur J Immunol. 2002;32:2274. doi: 10.1002/1521-4141(200208)32:8<2274::AID-IMMU2274>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 203.BenMohamed L, Krishnan R, Auge C, Primus JF, Diamond DJ. Intranasal administration of a synthetic lipopeptide without adjuvant induces systemic immune responses. Immunology. 2002;106:113. doi: 10.1046/j.1365-2567.2002.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.BenMohamed L, Wechsler SL, Nesburn AB. Lipopeptide vaccines--yesterday, today, and tomorrow. Lancet Infect Dis. 2002;2:425. doi: 10.1016/s1473-3099(02)00318-3. [DOI] [PubMed] [Google Scholar]
- 205.Zhu X, Ramos TV, Gras-Masse H, Kaplan BE, BenMohamed L. Lipopeptide Epitopes Extended by Ne-Palmitoyl Lysine Moiety Increases Uptake and Maturation of Dendritic Cell Through a Toll-Like Receptor 2 Pathway and Triggers a Th1- Dependent Protective Immunity. Eur J Immunol. 2004;34:1142. doi: 10.1002/eji.200425166. [DOI] [PubMed] [Google Scholar]
- 206.Sette A, Newman M, Livingston B, McKinney D, Sidney J, Ishioka G, et al. Optimizing vaccine design for cellular processing, MHC binding and TCR recognition. Tissue Antigens. 2002;59:443. doi: 10.1034/j.1399-0039.2002.590601.x. [DOI] [PubMed] [Google Scholar]
- 207.Rao X, Hoof I, Costa AI, van Baarle D, Kesmir C. HLA class I allele promiscuity revisited. Immunogenetics. 2011;63:691. doi: 10.1007/s00251-011-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. HLA Class I Alleles Are Associated with Peptide-Binding Repertoires of Different Size, Affinity, and Immunogenicity. J Immunol. 2013 doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rohrlich PS, Cardinaud S, Firat H, Lamari M, Briand P, Escriou N, et al. HLA-B*0702 transgenic, H-2KbDb double-knockout mice: phenotypical and functional characterization in response to influenza virus. Int Immunol. 2003;15:765. doi: 10.1093/intimm/dxg073. [DOI] [PubMed] [Google Scholar]
- 210.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sudenga SL, Kempf MC, McGwin G, Jr, Wilson CM, Hook EW, 3rd, Shrestha S. Incidence, prevalence, and epidemiology of herpes simplex virus-2 in HIV-1-positive and HIV-1-negative adolescents. Sex Transm Dis. 2012;39:300. doi: 10.1097/OLQ.0b013e318244a90f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Patel P, Bush T, Mayer KH, Desai S, Henry K, Overton ET, et al. Prevalence and risk factors associated with herpes simplex virus-2 infection in a contemporary cohort of HIV-infected persons in the United States. Sex Transm Dis. 2012;39:154. doi: 10.1097/OLQ.0b013e318239d7fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang CC, Yepes LC, Danaher RJ, Berger JR, Mootoor Y, Kryscio RJ, et al. Low prevalence of varicella zoster virus and herpes simplex virus type 2 in saliva from human immunodeficiency virus-infected persons in the era of highly active antiretroviral therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109:232. doi: 10.1016/j.tripleo.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tobian AA, Charvat B, Ssempijja V, Kigozi G, Serwadda D, Makumbi F, et al. Factors associated with the prevalence and incidence of herpes simplex virus type 2 infection among men in Rakai, Uganda. J Infect Dis. 2009;199:945. doi: 10.1086/597074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hill JM, Ball MJ, Neumann DM, Azcuy AM, Bhattacharjee PS, Bouhanik S, et al. The high prevalence of herpes simplex virus type 1 DNA in human trigeminal ganglia is not a function of age or gender. J Virol. 2008;82:8230. doi: 10.1128/JVI.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Clark JL, Konda KA, Munayco CV, Pun M, Lescano AG, Leon SR, et al. Prevalence of HIV, herpes simplex virus-2, and syphilis in male sex partners of pregnant women in Peru. BMC Public Health. 2008;8:65. doi: 10.1186/1471-2458-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Abu-Raddad LJ, Magaret AS, Celum C, Wald A, Longini IM, Jr, Self SG, et al. Genital herpes has played a more important role than any other sexually transmitted infection in driving HIV prevalence in Africa. PLoS One. 2008;3:e2230. doi: 10.1371/journal.pone.0002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xu F, Markowitz LE, Sternberg MR, Aral SO. Prevalence of circumcision and herpes simplex virus type 2 infection in men in the United States: the National Health and Nutrition Examination Survey (NHANES), 1999-2004. Sex Transm Dis. 2007;34:479. doi: 10.1097/01.olq.0000253335.41841.04. [DOI] [PubMed] [Google Scholar]
- 219.Tassiopoulos KK, Seage G, 3rd, Sam N, Kiwelu I, Shao J, Ao TT, et al. Predictors of herpes simplex virus type 2 prevalence and incidence among bar and hotel workers in Moshi, Tanzania. J Infect Dis. 2007;195:493. doi: 10.1086/510537. [DOI] [PubMed] [Google Scholar]


