Abstract
Background and Aims
MicroRNAs (miRNAs) are small non-coding RNAs, which regulate gene expression and are thus of interest as diagnostic markers, and as clues to etiology and targets of intervention. This pilot study examined whether circulating miRNAs are differentially expressed in patients with IBS.
Methods
miRNA microarrays (Nanostring) were run on the whole blood of 43 participants.
Results
hsa-miR-150 and hsa-miR-342-3p were found to be significantly elevated (FDR adjusted p ≤ 0.05, ≥1.6 fold change) in IBS patients compared to healthy controls. Neither of these miRNAs showed any relationship to race or sex. hsa-miR-150 is associated with inflammatory bowel disorders and pain, and interacts with a protein kinase (AKT2) through which it may affect inflammatory pathways. hsa-miR-342-3p is predicted to interact with mRNAs involved in pain signaling, colonic motility, and smooth muscle function.
Conclusions
This preliminary study reports the association of two miRNAs, detected in whole blood, with IBS. These miRNAs link to pain and inflammatory pathways both of which are thought to be dysregulated in IBS. Larger samples sizes are needed to confirm their importance and potential as biomarkers.
Keywords: irritable bowel syndrome, microRNA, hsa-miRNA-150, hsa-miR-342-3p
Introduction
Irritable Bowel Syndrome (IBS) is a common gastrointestinal (GI) disorder characterized by chronic abdominal pain (CAP) and changes in bowel habits, including the frequent occurrence of diarrhea (IBS-D), constipation (IBS-C) or a combination of both (IBS-M) (Drossman, 2007). The disorder affects an estimated 10–15% of the global population and is estimated to have a significant economic cost in terms of healthcare and lost productivity in the USA alone (Ashburn and Gupta, 2006). A better understanding of IBS pathogenesis and symptom etiology is important to improve patient outcomes.
The etiology and pathogenesis underlying IBS is currently unclear. Much of recent research, including ours, suggests dysregulated inflammatory and immune pathways and changes to epithelial barrier function in the etiology IBS (Henderson et al., 2012; Chadwick et al., 2002; Vivinus-Nebot et al., 2013). These results derive from studies based on the morphology (Demaude et al., 2006), functional performance (Piche et al., 2009), or protein (Miwa et al., 2001) and/or gene (Kerchoffs et al., 2012) expression of intestinal tissues. Recently, the expression of microRNAs (miRNAs), which are small RNAs capable of regulating protein levels, has piqued interests as diagnostic biomarkers, as indicators of etiology and as potential therapeutic targets. Although miRNAs are routinely examined in the study of a wide array of diseases relatively few studies of IBS have focused on miRNA expression.
Some notable studies have demonstrated the diagnostic and functional importance of miRNAs in IBS. Zhou et al. (2010) identified hsa-miR-29a as up-regulated in blood and colon tissue and described its role in modulating epithelial barrier function in patients with IBS-D and increased intestinal permeability. Kappelar et al. (2008) demonstrated the expression regulation of a serotonin receptor gene, associated with IBS-D, by hsa-miR-510. These discoveries have led to miRNA based therapeutic interventions in IBS being proposed (Zhou et al., 2011).
Most studies of gene expression in IBS focus on intestinal gene expression; these require invasive sampling of the intestinal mucosa. The purpose of this preliminary investigation was to examine circulating miRNA expression as a less invasive measure of molecular dysregulation in patients with IBS.
Materials and Methods
Sample
Patients who met Rome II criteria (i.e., chronic or recurrent abdominal pain, and altered stooling habits) for IBS (IBS-D = 5, IBS-C = 5, or IBS-M = 2) and healthy controls (i.e., no known organic disease or GI symptoms, n = 31) were recruited (clinicaltrial.gov #NCT00824921; see Table 1 for demographic and clinical indicators), at the National Institutes of Health (NIH) Hatfield Clinical Research Center. Participants signed a written consent to have their samples stored and used for research purposes. Whole blood samples were collected via venipuncture in PAXGene tubes (PreAnalytiX, Qiagen, Valencia, CA, USA) as per the manufacturer’s instructions, and stored at −80°C until RNA purification.
Table 1.
Sample characteristics and clinical results
| Variable | Healthy Controls (n=31) | IBSa patients (n=12) | P value |
|---|---|---|---|
| Gender (n) | |||
| Male | 15 | 3 | - |
| Female | 16 | 9 | - |
| Race (n) | |||
| Asian | 7 | 2 | - |
| African American/Black | 13 | 8 | - |
| Caucasian | 7 | 1 | - |
| Other | 4 | 1 | - |
| Age | 27.9±8.5 | 29.2±7 | p>0.05 |
| Range (years) | 13–45 | 16–45 | |
| IBS-Diarrhea | - | 5 | |
| IBS-Constipation | - | 5 | |
| IBS-Mixed | - | 2 | |
| BMIb | 24.5±4.7 | 26.6±5.8 | p>0.05 |
| Range | 28.5–48.2 | 20.5–41.3 | |
| Hematocrit | 40.4±5.0 | 39.4±4.6 | p>0.05 |
| Range (%) | 28.5–48.2 | 30.2–47.9 | |
| CRPc | 1.8±1.8 | 3.1±4.8 | p>0.05 |
| Range (mg L−1) | 0.2–7.3 | 0.2–17.5 | |
| ESRd | 9.6±8.4 | 12.5±7.7 | p>0.05 |
| Range (mm/hr) | 2.0–30.0 | 3.0–25.0 | |
| Alk. Phos.e | 75.2±56.6 | 66.0±19.6 | p>0.05 |
| Range (U L−1) | 36.0–367.0 | 40.0–110.0 | |
| ALTf | 27.5±10.6 | 28.6±13.7 | p>0.05 |
| Range (U L−1) | 15.0–66.0 | 17.0–55.0 | |
| ASTg | 17.8±8.9 | 20.5±15.0 | p>0.05 |
| Range (U L−1) | 6.0–53.0 | 6.0–61.0 | |
| GGTPh | 25.0±12.6 | 34.2±28.3 | p>0.05 |
| Range (U L−1) | 9.0–57.0 | 11.0–113.0 | |
| Amylase | 64.5±20.8 | 71.4±20.9 | p>0.05 |
| Range (U L−1) | 29.0–117.0 | 46.0–109.0 | |
| Lipase | 127.6±46.5 | 168.5±72.3 | p=0.034 |
| Range (U L−1) | 63.0–255.0 | 85.0–357.0 |
All values expressed as mean ±standard deviation.
IBS, Irritable Bowel Syndrome.
BMI, Body Mass Index.
CRP, C-reactive protein.
ESR, Erythrocyte sedimentation rate.
Alk. Phos., Alkaline phosphatase.
ALT, Alanine transaminase.
AST, Aspartate Aminotransferase.
GGTP, Gamma Glutamyl Transpeptidase. Normal ranges – Hematocrit: 34.1–44.9%, CRP: <3.0mg L−1, ESR: <42mm hr−1, Alk Phos.: 37–116 U L−1, ALT: 6–41 U L−1, AST: 9–34 U L−1, GGTP: 5–85 U L−1, Amylase: 25–115 U L−1, Lipase: 76–491 U L−1.
microRNA Expression
Total RNA, including miRNA, was extracted using the PAXGene Blood miRNA extraction kit (PreAnalytiX, Qiagen, Valencia, CA, USA). The nCounter Human miRNA Expression Assay Kit V1.4 (NanoString Technologies, Seattle, WA, USA) was used to anneal miRNAs to target specific barcode probes. No amplification was required. The sample was then immobilized on an nCounter Cartridge using the nCounter Prep Station. Barcode probes were counted and recorded for each miRNA target (735 human miRNAs) using the nCounter Digital Analyzer. The data were normalized to the top 100 expressed miRNAs, and the background was corrected using the nSolver software package. A 25 count threshold was set. miRNA species with more than 75% of cases at this threshold were excluded as such low counts cannot be accurately reproduced. Using these criteria 138 miRNAs were included in subsequent analyses. Repeated samples and internal controls show a high degree of consistency between assays.
Statistical Analysis
Statistical analysis of the demographic and clinical data was performed using SPSS v15.0 with a priori statistical significance set at p ≤ 0.05. Comparisons of each parameter were conducted using the parametric independent sample Student’s t-test.
Statistical analysis of the genetic data was performed using BRB-Array Tools (Biometric Research Branch, National Cancer Institute) with the a priori statistical significance set at p ≤ 0.05. A priori p-values were adjusted for False Discovery Rate (FDR). miRNA data from IBS patients and healthy controls were compared while assessing the presence of race and gender effects. Integrated Pathway Analysis (IPA, Ingenuity Systems, Inc.) was used to explore the functional associations of miRNA species of interest.
Results
Demographic and Clinical Data
The IBS group was not significantly different in terms of age, BMI, hematocrit, CRP, ESR, ALT, AST, or amylase compared to healthy controls (Table 1). Lipase was significantly higher in IBS patients compared to healthy controls (Table 1). Although a difference in lipase was observed among groups, none of the values fell outside of the normal clinical range for these indices.
miRNA Expression
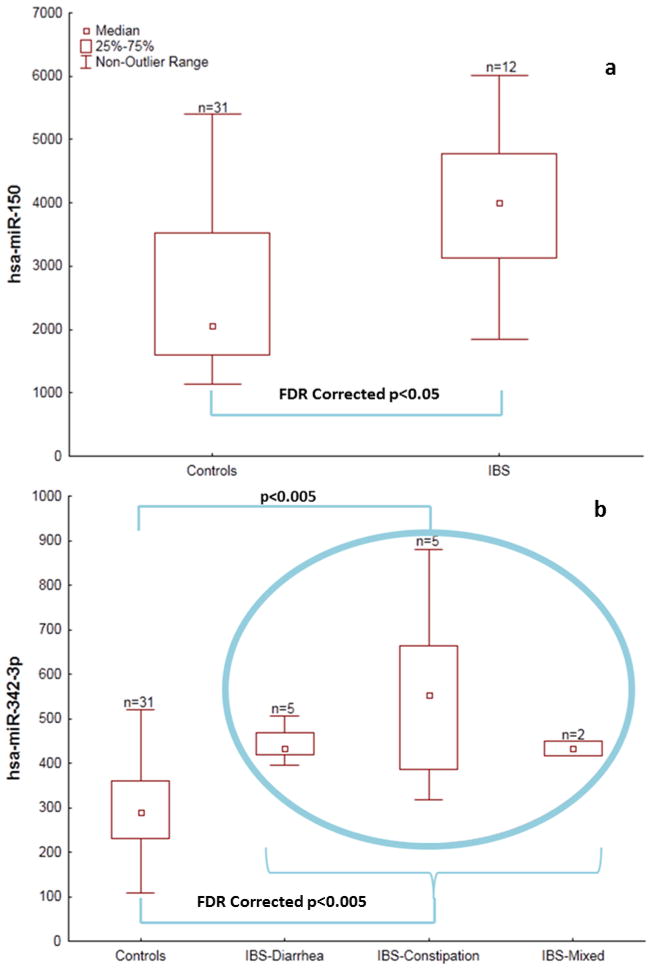
Both hsa-miR-150 and hsa-miR-342-3p were found to be elevated (FDR adjusted p ≤ 0.05) in patients with IBS compared to healthy controls (Figures 1a and b). The expression of hsa-miR-150 and hsa-miR-342-3p were 1.6 and 1.7 fold higher, respectively, in IBS patients (mean ± 1 standard deviation: hsa-miR-150 = 3988 ± 1195; hsa-miR-342-3p = 491 ± 152) than in healthy controls (hsa-miR-150 = 2509 ± 1135; hsa-miR-342-3p = 292 ± 87). The IBS-constipation (hsa-miR-342-3p = 560 ± 225) cohort but not the IBS-diarrhea (hsa-miR-342-3p = 444 ± 44) cohort had elevated (p<0.005) hsa-miR-342-3p expression compared to healthy controls (Figure 1b). Expression of these miRNAs was not found to have any relationship to race or gender.
Figure 1.
Figure 1a and 1b: Box plots showing the differential expression of (a) hsa-miR-150 and (b) hsa-miR-342-3p in healthy controls and IBS patients.
Integrated Pathway Analysis of IBS-Associated miRNAs
To explore possible interactions of these miRNAs with other genes and proteins, and their role in different molecular pathways, they were entered into an Ingenuity Pathway Analysis (IPA; Ingenuity Systems Inc., Redwood City, CA). The analysis merged networks associated with the following diseases: cancer, hematological disease, organismal injury and abnormalities, endocrine system disorders, and reproductive system disease. In addition, functional networks related to cellular development, growth, proliferation, movement, death, survival, and cell-to-cell signaling were merged. Several genes with both direct and indirect bindings to the two miRNA species were revealed, including the calcium channel voltage-dependent L type alpha 1C subunit (CACNA 1C), caspase-3, and the serine-threonine protein kinase AKT2 (Figure 2).
Figure 2.
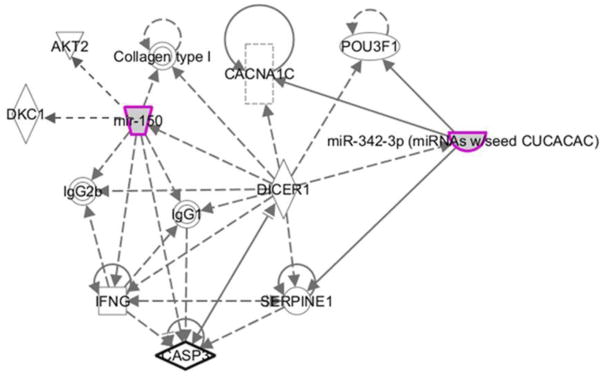
Integrated Pathway Analysis (IPA) network generated outlining proable relationships between hsa-miR-342-3p and hsa-miR-150 and related mRNAs and proteins. Direct and indirect relationships are shown by solid and dashed lines, respectively. The arrow indicates specific directionality of interactions.
Discussion
In this preliminary study of circulating miRNA expression in IBS, hsa-miR-150 and hsa-miR-342-3p were identified as up-regulated in IBS patients compared to healthy controls. Despite the small sample size, a consistent differential expression of hsa-miR-342-3p between the different IBS-subtypes (diarrhea, constipation, or mixed) compared to healthy controls is evident. Neither race nor gender was found to explain variation in these miRNAs. We have previously identified hsa-miR-342-3p as part of a dysregulated miRNA profile in a weight- and sex-matched Caucasian cohort of IBS patients and healthy controls (Peace et al., in review). This dysregulation of miR-342-3p in independent cohorts provides additional evidence for the consistency of an association between this miRNA species and IBS.
The hsa-miR-342-3p has been found to be up-regulated in Bladder Pain Syndrome (BPS) (Gheinani et al., 2013). This finding is intriguing, because similar to the symptom of abdominal pain with unknown cause presenting in our IBS patients, BPS patients present with pelvic pain in the absence of a specific cause. The IPA provides a potential molecular link between this miRNA and pain signaling pathways by revealing an association with the voltage-dependent L type calcium channel alpha 1C protein (CACNA 1C). Regulated processing of the CACNA1 mRNA has been functionally associated with nociceptive pain transmission (Lipscombe et al., 2013). Dysregulated expression of L-type calcium channels is also associated with altered colonic motility and smooth muscle contraction in a rat model of IBS 9Zhang et al., 2010). In addition to chronic pain, inflammation also underlies the pathogenesis of BPS. We have previously observed that inflammatory markers are disproportionately represented in colonic tissues from IBS patients, which suggests a sub-clinical inflammatory component in IBS (Henderson et al., 2012). Taken together, it is likely that the functional role of the hsa-miR-342-3p involves inflammatory, pain signaling, smooth muscle contractility, and GI tract motility pathways.
The hsa-miR-150 also has a postulated role in inflammation and has been previously associated with Crohn’s disease (CD), ulcerative colitis (UC), and colorectal cancer (Pekow et al., 2012; Ma et al., 2012). This miRNA was found to be over expressed in the colon epithelium of patients with CD and UC (Pekow et al., 2012) and has also been associated with pain (Orlova et al., 2011). Targets of the hsa-miR-150 include the telomerase-related proteins Dyskerin (dyskeratosis congenita 1, DKC1), as well as AKT2, a pro-survival protein kinase activated via the PI3K pathway, and is expressed in many human malignancies. AKT, which is an important factor in inflammatory pathways, has also been linked to inflammatory bowel diseases (Anderson and Wong, 2010).
In summary, our findings identify two miRNA species, hsa-miR-150 and hsa-miR-342-3p, that are differentially expressed in the peripheral circulation of patients suffering from IBS. Both miRNAs are implicated in inflammation and have previously been associated with pain. Further research will reveal the specific roles of hsa-miR-150 and hsa-miR-342-3p in these pathways and whether they may be developed as biomarkers and/or therapeutic targets in IBS.
Highlights.
Potential circulating miRNA biomarkers of IBS suggests non-invasive methods to understand, diagnose, and monitor the condition.
The primary aim of this study was to examine circulating miRNA expressions in patients with ROME II IBS and compare them to healthy controls.
Whole genome miRNA profiling was used to characterize peripheral circulating miRNA expression in patients with IBS and healthy controls.
miR-150 and miR-342-3p were significantly elevated in patients with IBS compared to healthy controls.
Acknowledgments
Funding
The authors acknowledge funding from the US Department of Health and Human Services, National Institutes of Health, National Institute of Nursing Research, Division of Intramural Research (to Wendy A. Henderson, 1ZIANR000018-01-04; Intramural Research Training Awards to Nicolaas H. Fourie, Ralph M. Peace, Sarah K. Abey, Leeanne B. Sherwin, and Paul A. Smyser; Howard Hughes-NIH Research Scholar RMP; Special support from the Burroughs Wellcome Fund to Ralph M. Peace.
Abbreviations
- AKT2
serine-threonine protein kinase
- Alk. Phos
alkaline phosphatase
- ALT
alanine transaminase
- AST
aspartate aminotransferase
- BMI
body mass index
- BPS
bladder pain syndrome
- CACNA 1C
calcium channel voltage-dependent L type alpha 1C subunit
- CAP
chronic abdominal pain
- CD
Crohn’s Disease
- CRP
C-reactive protein
- DKC1
dyskerin
- ESR
erythrocyte sedimentation rate
- FDR
false discovery rate
- GGTP
gamma glutamyl transpeptidase
- GI
gastrointestinal
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- IBS-C
irritable bowel syndrome constipation
- IBS-D
irritable bowel syndrome diarrhea
- IBS-M
irritable bowel syndrome mixed
- IPA
Ingenuity Pathway Analysis
- miRNA
microRNA
- NIH
National Institutes of Health
- UC
Ulcerative Colitis
Footnotes
Disclosure
None of the authors has potential conflicts of interest to disclose.
Author Contributions
Sarah K. Abey, Leeanne B. Sherwin and Wendy A. Henderson performed the patient enrollment and administered the patient protocols, patient data collection and sample collection; they also curate and formatted clinical and patient data for analysis. Ralph M. Peace, Nicolaas H Fourie and Paul A. Smyser performed sample preparation and curation. Ralph M. Peace performed the RNA purification and prepared RNA for miRNA analysis. Nicolaas H. Fourie conducted the data analysis, interpreted the data, and was responsible for manuscript generation. Sarah K. Abey, John W. Wiley and Bridgett Rahim-Williams contributed to the data interpretation, functional network analyses. Wendy A. Henderson was responsible for protocol generation. All co-authors were required to contribute to the writing and editing of the manuscript.
The opinions expressed herein and the interpretation and reporting of these data are the responsibility of the author(s) and in no way should be seen as an official recommendation, interpretation, or policy of the National Institutes of Health or the United States Government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson EC, Wong MH. Caught in the Akt: regulation of Wnt signaling in the intestine. Gastroenterology. 2010;139:718–722. doi: 10.1053/j.gastro.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn TT, Gupta MS. The IBS market. Nat Rev Drug Discov. 2006;5:99–100. doi: 10.1038/nrd1961. [DOI] [PubMed] [Google Scholar]
- Chadwick VS, et al. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–83. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- Demaude J, et al. Phenotypic changes in colonocytes following acute stress or activation of mast cells in mice: implications for delayed epithelial barrier dysfunction. Gut. 2006;55:655–661. doi: 10.1136/gut.2005.078675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drossman DA. Introduction. The Rome Foundation and Rome III. Neurogastroenterol Motil. 2007;19:783–786. doi: 10.1111/j.1365-2982.2007.01001.x. [DOI] [PubMed] [Google Scholar]
- Gheinani AH, et al. Deciphering microRNA code in pain and inflammation: lessons from bladder pain syndrome. Cell Mol Life Sci. 2013;70:3773–3789. doi: 10.1007/s00018-013-1275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson WA, et al. Inverse relationship of interleukin-6 and mast cells in children with inflammatory and non-inflammatory abdominal pain phenotypes. World J. Gastrointest. Pathophysiol. 2012;3:102–108. doi: 10.4291/wjgp.v3.i6.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller J, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs AP, et al. SERT and TPH-1 mRNA expression are reduced in irritable bowel syndrome patients regardless of visceral sensitivity state in large intestine. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1053–1060. doi: 10.1152/ajpgi.00153.2011. [DOI] [PubMed] [Google Scholar]
- Lipscombe D, et al. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochimica et Biophysica Acta. 2013;1828:1522–1529. doi: 10.1016/j.bbamem.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–53. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- Miwa J, et al. Patients with constipation-predominant irritable bowel syndrome (IBS) may have elevated serotonin concentrations in colonic mucosa as compared with diarrhea-predominant patients and subjects with normal bowel habits. Digestion. 2001;63:188–194. doi: 10.1159/000051888. [DOI] [PubMed] [Google Scholar]
- Orlova IA, et al. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9:195–205. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace RM, et al. In Review. Dysregulation of inflammatory microRNAs associated with gastrointestinal. Am J Physiol-Gastr L [Google Scholar]
- Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:187–193. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piche T, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: involvement of soluble mediators. Gut. 2009;58:196–201. doi: 10.1136/gut.2007.140806. [DOI] [PubMed] [Google Scholar]
- Vivinus-Nébot M, et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: role of epithelial barrier disruption and low-grade inflammation. Gut. 2013 doi: 10.1136/gutjnl-2012-304066. in press. [DOI] [PubMed] [Google Scholar]
- Zhang M, et al. Increased colonic motility in a rat model of irritable bowel syndrome is associated with up-regulation of L-type calcium channels in colonic smooth muscle cells. Neurogastroenterol Motil. 2010;22:e162–170. doi: 10.1111/j.1365-2982.2009.01467.x. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Verne GN. miRNA-based therapies for the irritable bowel syndrome. Expert Opin Biol Ther. 2011;11:991–995. doi: 10.1517/14712598.2011.577060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775–784. doi: 10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]