Abstract
Paracetamol is the commonest cause of acute liver failure in the Western world and biomarkers are needed that report early hepatotoxicity. The liver enriched microRNA, miR-122, is a promising biomarker currently being qualified in humans. For biomarker development and drug toxicity screening the zebrafish has advantages over rodents, however, blood acquisition in this model remains technically challenging. We developed a method for collecting blood from the adult zebrafish by retro-orbital (RO) bleeding and compared it to the commonly used ‘lateral incision’ (LI) method. The RO technique was more reliable in terms of the blood yield and minimum amount per fish. This new RO technique was used in a zebrafish model of paracetamol toxicity. Paracetamol induced dose-dependent increases in liver cell necrosis, serum ALT activity and mortality. In situ hybridization localised expression of miR-122 to the cytoplasm of zebrafish hepatocytes. After collection by RO bleeding, serum miR-122 could be measured and this microRNA was substantially increased by paracetamol 24 hours after exposure, an increase that was prevented by delayed (3 hours post start of paracetamol exposure) treatment with acetylcysteine. In summary, collection of blood by RO bleeding facilitated measurement of miR-122 in a zebrafish model of paracetamol hepatotoxicity. The zebrafish represents a new species for measurement of circulating microRNA biomarkers that are translational and can bridge between fish and humans.
Introduction
At therapeutic doses, paracetamol (acetaminophen) is a safe analgesic drug used by millions of people worldwide. However, in overdose, paracetamol is hepatotoxic, being the most common cause of acute liver failure in the United States and Europe.1-3 The mechanism of paracetamol hepatotoxicity is well described; in overdose, the reactive metabolite N-acetyl-p-benzoquinone imine (NAPQI) is generated in excess and cellular glutathione (GSH) is depleted, NAPQI then binds covalently to cellular proteins resulting in oxidative stress and consequent hepatocyte necrosis.4 The current antidote, acetylcysteine (AC), replenishes cellular GSH and is highly effective if administered within 8 hours of overdose. However, this treatment is time-consuming and commonly results in adverse drug reactions. Therefore, there is a pressing need for new biomarkers that identify hepatotoxicity soon after overdose to stratify patients and improve the targeting of treatment to those at most risk of adverse outcomes.
MicroRNAs (miRNAs) are small (~22 nucleotides long) non-protein coding RNA species involved in post-transcriptional gene product regulation. In the blood, miRNA is protected from degradation by microvesicles5 and RNA binding protein complexes.6 As they are amplifiable and tissue restricted, miRNAs represent a new reservoir for biomarker discovery. The liver enriched miRNA, miR-122, is a circulating biomarker for paracetamol-induced liver toxicity in rodents and humans.7,8 It has greater specificity for liver injury than serum alanine transaminase (ALT) activity9 and in humans and rodents is elevated soon after overdose when ALT activity is still normal.10 MiRNAs are highly conserved across species and so represent bridging biomarkers that facilitate translation of toxicity signals from animal models to human clinical trials.
The zebrafish is used to study drug effects on organ systems such the heart, CNS and gastrointestinal tract.11,12 Recent studies have demonstrated that paracetamol exposure induces hepatotoxicity in zebrafish.13 The zebrafish has many mammalian homologs at multiple loci and several advantages as a vertebrate model compared to rodents and other higher order species. These include lower financial costs, convenient drug delivery, genetically tractable and visually accessible organs.11,14,15 However, the small size of the adult fish means that circulating blood volume is low which makes blood acquisition technically challenging. A number of blood collection techniques exist for zebrafish, such as a lateral incision (LI) in the dorsal aorta, yet significant limitations remain with regard to the volume of blood that can be collected. For measurement of circulating biomarkers such as miRNA there is a need for new approaches to maximise blood yield. With regard to the clinical translatability of circulating miRNA, it is important to note that the zebrafish liver expresses) miR-122 which has an identical nucleotide sequence (UGGAGUGUGACAAUGGUGUUUG) to rodent and human.16
To date, zebrafish miRNA expression has been studied only in tissues, and no data have been published on circulating miRNA. In this paper, we describe a new method for blood collection based on retro orbital (RO) sinus bleeding, a technique commonly used in rodents.17 This technique was used to collect blood for measurement of circulating miR-122 in a clinically relevant zebrafish model of paracetamol hepatotoxicity.
Materials and Methods
Ethical approval
All experiments were conducted in accordance with the Animals (Scientific Procedures) Act 1986 in a UK Home Office approved establishment.
Materials
Paracetamol, MS-222 (tricaine methanesulfonate) and EDTA (Ethylenediaminetetraacetic acid) were obtained from Sigma-Aldrich (Gillingham, UK). The MiRNeasy and the MinElute Cleanup kit were obtained from Qiagen Benelux B.V. (Venlo, the Netherlands). Specific RT and PCR primers were obtained from Applied Biosystems (Foster City, CA) (assay number 002245 for miR-122, 004468_mat for let-7d).
Zebrafish
Fish were 5-24 months old. In all studies the age of fish was the same across experimental groups. All fish were maintained at 28.50C, pH 7.2 and conductivity of ≈400μS. Fish were euthanized with 4mg/ml MS-222 before all blood and tissue collection procedures. All experimental procedures were performed at room temperature (23±0.5 °C).
Blood collection
To study the relationship between fish size and blood yield, after euthanasia each fish was weighed and the total body length was measured (from the tip of the snout to the origin of the caudal fin). Two methods of blood collection were compared; the established lateral incision (LI) technique and a new RO approach. LI was performed as previously described.18 In brief, after euthanasia, fish were dried then the tail was cut at the caudal peduncle with a surgical knife. Blood was collected, with the aid of a pipette, into an eppendorf tube containing heparin solution (0.5 μL of 25mg/ml). For RO bleeding, following euthanasia, fish were quickly dried then one eye was removed using a pair of fine forceps. Heparin (0.5μL of 25mg/ml stock) or EDTA (0.5 μL of 2% stock) solution was pipetted onto the eye socket to prevent the blood from clotting. Once the orbital space had filled with blood, it was pipetted off and collected in an eppendorf tube. After this step, pressure was applied to the fish from tail towards head to accumulate more blood into the orbit for collection. This last step was repeated until no more blood could be collected.
Paracetamol toxicity model
Zebrafish were exposed to paracetamol (20-40mM), dissolved in system water, for 3 hours (or system water alone for negative controls). This was followed by a change of system water for 2-21 hours with or without AC (3mM). This brief exposure to high concentration paracetamol followed by delayed AC treatment was used to replicate a human single, acute overdose. The drug concentrations were based on published data.13 Experiments were terminated 5-24 hours after the start of paracetamol exposure (5-24hpe).
All histology samples were placed in Dietrich’s fixative and left to fix for at least 24 hours before processing, cutting and haematoxylin staining using standard protocols. All fish were sectioned sagittally along the midline to allow examination of the hepatic tissue. Hepatic lesions were scored by a boarded pathologist (author JDP) who was blinded to the treatment groups. Zebrafish blood was obtained by the RO technique and pooled as described in the results. Serum was separated by centrifugation and ALT activity was measured as described by Bergmeyer et al,19 utilising a commercial kit (Alpha Laboratories Ltd., Eastleigh, UK) adapted for use on a Cobas Fara centrifugal analyser (Roche Diagnostics Ltd, Welwyn Garden City, UK). Paracetamol concentration was measured by an aryl acylamidase enzymatic assay in the Royal Infirmary of Edinburgh Clinical Biochemistry laboratory.
MiR-122 localisation by in situ hybridization
MiR-122 was localised using double-DIG labelled miRCURY LNA microRNA detection probes (Exiqon A/S, Vedbaek, Denmark) specific for miR-122. The in situ hybridisation procedure was performed on a fully-automated Ventana Discovery Ultra (Roche diagnostics AG, Rotkreuz, Switzerland). All reagents were provided by Roche Diagnostics. Briefly, whole zebrafish sections were deparaffinized at 62 °C for 12 minutes before proteinase K digestion step (Exiqon A/S, Vedbaek, Denmark) treatment (37 °C, 24 min). The anti-miR-122 probe (25 pmoles) was then incubated at 54 °C for 3 h with deparaffinized sections. Sections were then washed with 3 × 8 mins cycles of saline sodium citrate (SSC) at increasing stringency per cycle (2.0×, 1.0×, 0.5×). The secondary antibody (alkaline phosphatase-linked Sheep anti-DIG, dilution 1:500 in antibody diluent) was then incubated at 37 °C for 32 mins. Chromogenic detection was performed using the BlueMap® kit as per the manufacturer’s instructions. The substrate was allowed to develop for 6 h before counterstaining with Red Stain II for 4 mins. Slides were manually washed before mounting on laboratory-grade glycerol gelatin (Sigma-Aldrich, Buchs, Switzerland). Slides were scanned using a Nanozoomer 2.0-HT digital slide scanner (Hamamatsu photonics, Hamamatsu, Japan). Images were captured using NDP.view2 software (Hamamatsu photonics, Hamamatsu, Japan).
MiRNA measurement
For miRNA analysis, zebrafish blood was obtained by RO technique and pooled as described in results. Serum was separated by centrifugation and miRNA was extracted using a miRNeasy kit (Qiagen, Venlo, Netherlands) after which the small RNA fraction was purified using a MinElute kit (Qiagen, Venlo, Netherlands).8 MiRNAs were measured using Taqman-based quantitative PCR.
The small RNA elute was reverse transcribed using specific stem-loop reverse-transcription RT primers (Applied Biosystems, Foster City, CA, USA) for each target miRNA species, following the manufacturer’s instructions. In the reverse transcription reaction, 2 μl of RNA was used to produce the complementary DNA (cDNA) template in a total volume of 15 μl. Then, 1.33 μl of cDNA was used in the PCR mixture with specific PCR primers (Applied Biosystems, Foster City, CA) in a total volume of 20 μl. Levels of miRNA were measured using the Light Cycler 480 (Roche, Basel, Switzerland). All samples were assayed in duplicate. MiRNA levels were normalised to levels of let-7d which has been reported to be an appropriate internal normaliser in humans.20 At the time of writing the optimum normaliser for zebrafish was not described.
Statistical analysis
Statistical analysis was performed with GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). Differences in blood yield between the LI and RO techniques and biomarker differences between experimental groups were analysed with Student’s unpaired t test. For correlation analysis, a Pearson’s correlation test was used. Nominal statistical significance was set at P <0.05.
Results
Blood yield was higher with the retro orbital technique
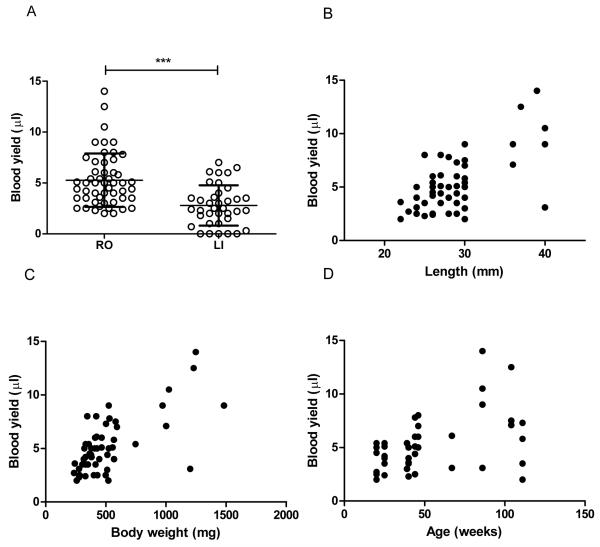
The volume of blood obtained by RO (N=52) and LI (N=37) techniques was compared (figure 1); the RO technique resulted in significantly higher blood yields than the LI technique (mean per fish±SD: 5.3±2.6μl vs. 2.8±2.0μl, P<0.0001). Furthermore, the LI technique did not result in successful blood collection from every fish, with 6 from 37 providing insufficient or no blood. In comparison, RO technique resulted in blood collection from every fish, with a minimum yield of 2μl.
Figure 1.
(A) Blood yield for the retro orbital (RO) technique (N=52) and for the lateral incision (LI) technique (N=37). Each point represents an individual fish. Horizontal bar represent the mean. Error bars represent standard deviation. ***P < 0.0001. Scatter graphs of RO blood yield in relation to body length (B), body weight (C) and age (D). Each point represents an individual fish. Pearson R values (statistical probabilities) are 0.63 (P<0.0001), 0.67 (P<0.0001) and 0.46 (P=0.001) for length, weight and age, respectively.
Individual fish age, body length (BL) and body weight (BW) were not significantly different between RO and LI groups (mean±SD: age 52.2±30.3 vs. 53.9±32.48 weeks, P=0.81, BL 28.5±4.5 vs. 27.8±3.6 mm, P=0.44, BW 518.6±284.1 vs. 492.7±181.9 mg, P=0.62, for RO vs. LI respectively). Within the RO group, blood yield correlated significantly with all three parameters. However, BL and BW were more strongly correlated with blood yield than age: BL(R=0.63, P<0.0001), BW(R=0.67, P<0.0001) and age (R=0.46, P=0.001) (figure 1).
Paracetamol induced liver toxicity in adult zebrafish
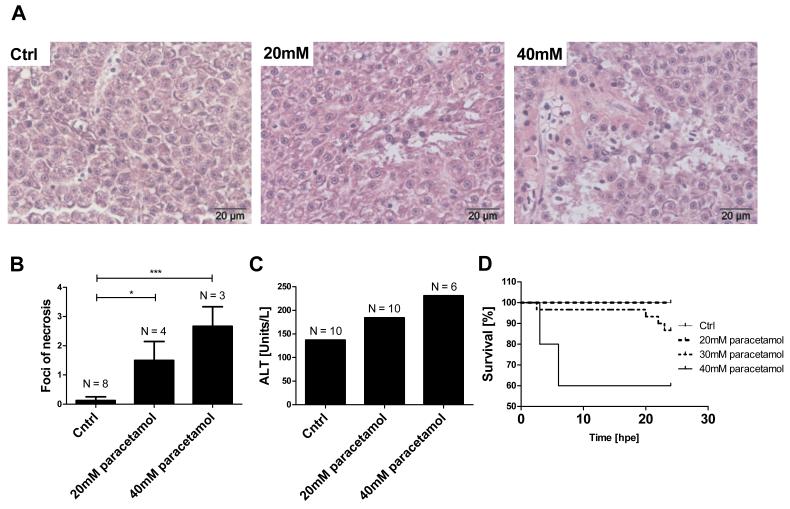
Next, the RO blood collection technique was applied to a zebrafish model of paracetamol hepatotoxicity. Paracetamol dissolved in the system water entered the zebrafish circulation. In a pooled serum sample from 10 fish, the serum paracetamol concentration was 962 mg/L immediately following 3 hours exposure to paracetamol (30mM). Exposure to 40mM paracetamol for 3 hours resulted in 4 out of 10 fish dying 3-6hpe. In the 30mM group, 1 out of 30 fish died at 2.5 hpe and 3 out of 30 fish died at 20-23 hpe (figure 2). No fish died after 20mM paracetamol exposure (P=0.02 for mortality following paracetamol exposure). Exposure to 20mM and 40mM paracetamol for 3 hours resulted in hepatocyte necrosis at 24 hpe (figure 2). There was a dose response relationship in the mean number of necrotic foci per liver section (mean±SEM: control 0.13±0.1, 20mM 1.5±0.7, 40mM 2.7±0.7), and both treatment groups induced a significant increase in necrosis compared to control (P=0.02 and P<0.001 for 20mM and 40mM paracetamol, respectively) (figure 2). Other histological features included pericapsular macrophage clustering, pigment-laden macrophages and cytoplasmic granularity of hepatocytes. The former two features were noted in both cases and controls, suggesting these are background findings. Cytoplasmic granularity of hepatocytes appeared to be associated with paracetamol exposure. At 24hpe, there was a paracetamol induced dose-dependent increase in serum ALT activity (figure 2).
Figure 2.
Effect of paracetamol exposure on adult zebrafish. Paracetamol treatment was for 3 hours, all measurements made 24 hpe. (A) Histological images of adult zebrafish liver at the indicated paracetamol concentrations; both treatment groups displayed liver necrosis. (B) Mean foci of necrosis per liver section at the indicated paracetamol concentrations. N= number of fish per treatment group. Error bars represent SEM, *p<0.05, ***p<0.001 (C) Serum ALT activity in pooled samples from each treatment group. N = the number of pooled fish for ALT measurement (D) Survival curves for experimental groups n = 10/group. There was a significant difference between treatment groups, P=0.02.
MiR-122 localised to zebrafish hepatocytes
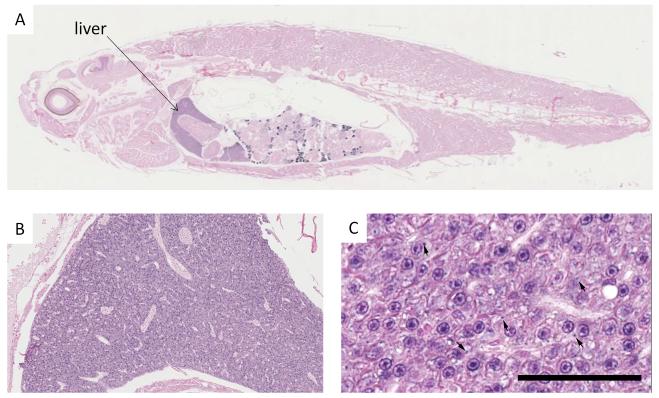
In situ-hybridisation with the miR-122 probe revealed positive intra-cytoplasmic staining in zebrafish. ISH positive staining was noted in variable numbers of hepatocytes scattered throughout the hepatic parenchyma, and was characterised by one or more focal deposits within the cytoplasm (figure 3).
Figure 3.
MiR-122 In-situ hybridization. A: whole zebrafish section demonstrates localization to the liver. B and C liver microphotographs: Widespread intracytoplasmic deposition of ISH substrate in the hepatocytes of a fish (arrows indicate focal deposits) Scale bar:50μm.
Paracetamol hepatotoxicity was associated with increased circulating miR-122
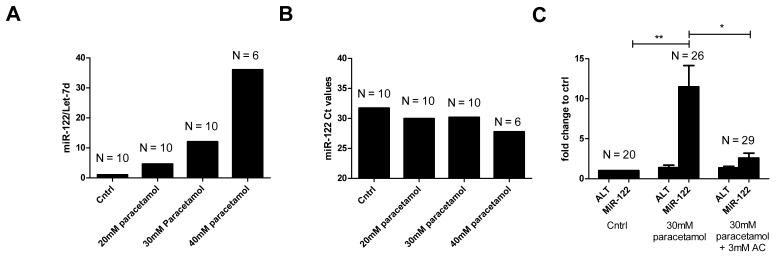
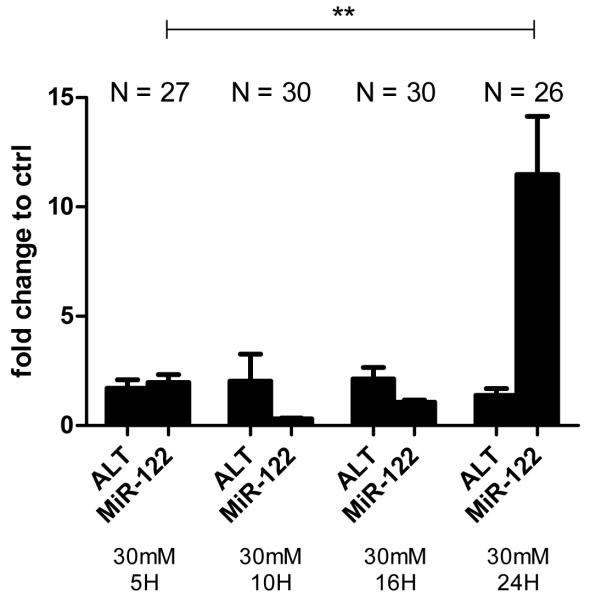
Measurement of miRNA in pooled serum from adult zebrafish was possible, and this was facilitated by the increased blood yields from RO bleeding. Exposure to paracetamol (20-40mM for 3 hours) resulted in a dose-dependent increase of miR-122 concentration, measured at 24hpe (figure 4). This increase was still apparent when raw ct values were displayed without normalisation to the miRNA Let-7d (figure 4). A paracetamol concentration of 30mM was chosen to study the effect of AC as this concentration induced a significant increase in circulating miR-122 at 24hpe (miR-122/Let-7d: mean±SEM; control 0.1±0.005, paracetamol 1.0±0.2. P=0.01) with lower mortality than 40mM. Zebrafish were divided across 3 replicate experiments giving total numbers per group as presented in figure 4. Blood was pooled within groups from each experiment. Expressed with reference to each experiment’s system water control, there was an 8 fold larger increase of miR-122 compared with ALT activity for the same paracetamol dose at 24hpe (fold change relative to control, 30mM paracetamol (mean±SEM): 1.4±0.3 ALT, 11.5±2.7 miR-122). (figure 4). After 3 hours exposure to paracetamol the fish were treated with AC (3mM) in the water or water alone for 21 hours. With AC 1 of 30 fish died (3.3%); where as 4 of 30 fish died (13.3%) in the absence of AC. Consistent with delayed AC treatment preventing liver injury, AC prevented the paracetamol-induced increase in circulating miR-122 (miR-122/Let-7d: mean±SEM; paracetamol 1.0±0.2, paracetamol & AC 0.2±0.05. P=0.009). Figure 5 presents the temporal profile of circulating miR-122 and ALT activity at 5–24hpe to 30mM paracetamol for 3h. This demonstrates that miR-122 was not significantly elevated until 24hpe. With this dose of paracetamol there was no significant increase in serum ALT activity.
Figure 4.
(A) Circulating miR-122 24hpe to 3 hours of paracetamol at the concentrations indicated. miR-122 was normalised to Let-7d. N=number of fish pooled for microRNA measurement. (B) Circulating miR-122 24hpe to 3 hours of paracetamol at the concentrations indicated. Raw ct values are presented without normalisation N=number of fish pooled for microRNA measurement. (C) Serum ALT activity and miR-122 24hpe to 3 hours of paracetamol (30mM) followed by 21hours of AC (3mM) or water. ALT and miR-122 are expressed as fold change compared to control. Values are mean, error bars are SEM. Zebrafish were equally divided across 3 replicate experiments giving the indicated total numbers per group (N). *P<0.05, **P<0.001 for comparison of miR-122/Let7d values between groups indicated.
Figure 5.
Serum ALT activity and miR-122 5-24hpe to 3 hours of paracetamol (30mM). ALT and miR-122 are expressed as fold change compared to controls run at each timepoint. Values are mean, error bars are SEM. Zebrafish were equally divided across 3 replicate experiments giving the indicated total numbers per group (N). **P<0.001 for comparison of miR-122/Let7d values between groups indicated.
Discussion
For zebrafish to be useful in circulating biomarker discovery and validation studies, it is vital to maximize the yield of blood from every fish. In this study, we demonstrated that a new RO technique was more efficient than the commonly used LI approach in terms of blood yield. We developed a zebrafish model for paracetamol-induced hepatotoxicity in which we used the new RO blood draw technique to, for the first time, measure circulating miRNA in zebrafish. We noted that circulating liver enriched miRNA (miR-122), was elevated in a dose dependent pattern as a result of paracetamol hepatotoxicity. This is consistent with findings in humans and rodents.
In the present study the LI blood collection technique yielded a mean of 2.8 μl of blood per fish but no blood or blood volumes of less than 1 μl, were obtained from around 19 % of fish. These results are consistent with previous studies.21,22 The new RO technique yielded a higher mean of 5.3 μl of blood per fish, and a maximum of 14μl. Additionally, the RO technique resulted in a minimum of 2 μl from all fish used in this study. When calculating the number of fish that need to be pooled (with a power of 80%) to ensure a sample of 40 μl of blood; for the RO technique 13 fish would be needed, whereas 36 fish would be needed if the LI technique is used. Therefore, fewer animals may be needed for a successful experiment if the RO technique is used, which is in compliance with the principle of reduction in number of experimental animals (http://www.nc3rs.org.uk/).
Fish age, length and weight correlated significantly with the amount of blood drawn by the RO technique (longer, heavier and older fish yielded higher volumes of blood). However, size correlated more strongly with blood yield than fish age, which makes fish size a more useful predictor of successful blood acquisition. In our study relatively small fish were mostly used; approximately 22-30 mm in length. However, a few larger fish, up to 40 mm, were included which is a size that is rarely exceeded in captivity.23 These large fish yielded up to 14 μl of blood. The largest reported blood yield was from the study of Pedroso et al.,18 who reported a yield of up to 20 μl however, fish size was not disclosed in their report and the anaesthetic agent used might have increased blood yield. Pedroso et al.18 used ice cold water which slows fish metabolism24,25, however, the UK Animals (Scientific Procedures) Act 1986, does not allow this approach. Jensen et al.,26 who, like our study, used MS-222 to anesthetise the fish before the LI technique, reported blood yields of 2-8 μl, which is in line with our LI results. Recently, a new technique has been described in which fish were centrifuged in a perforated eppendorf tube after the administration of a LI.27 They reported comparable blood yields (5.5–11 μL) to the RO approach. However, centrifugation may lead to blood contamination with other body fluids, where as the RO blood collection approach results in a reduced risk of contamination. Zang and colleagues28, described a recovery method that allows for serial blood sampling from adult zebrafish, but it takes one to two weeks for fish to recover normal haemoglobin values after taking a small blood sample of 2 μL. This delayed recovery may limit the value of serial blood sampling in toxicity studies.
Paracetamol was associated with hepatocyte necrosis in the liver of zebrafish. This is consistent with previous studies in zebrafish that have demonstrated paracetamolinduced formation of NAPQI through the enzymatic action of CYP3A65;29 hepatic glutathione depletion and necrotic cell death.13 In the present study the histological evidence of liver injury was accompanied by modestly increased serum ALT activity and clearly increased mortality. The zebrafish liver expressed miR-122 as detected by ISH. To the best of our knowledge, this is the first study to localise zebrafish miR-122 by in situ hybridisation and to measure miRNA in zebrafish serum, facilitated by the increased blood volumes collected by the RO technique. In paracetamol exposed fish, there was a clear, dose-response elevation in circulating miR-122. This is consistent with release from the injured liver as is described in rodents and humans.7,8 Furthermore, circulating miR-122 is elevated in humans and rodents soon after a paracetamol overdose at a time when ALT is still normal, therefore, miR-122 appears more sensitive than ALT for reporting hepatotoxicity.7,10 In the present study 24 hours after paracetamol exposure, when there was histologically proven hepatocyte necrosis, there was a 8-fold larger increase of miR-122 than ALT, suggesting greater sensitivity for reporting tissue injury. More work is needed to fully define the kinetics of circulating miRNA in fish using different dosing regimens for paracetamol and using other hepatotoxic drugs. However, this study suggests miRNA may have significant advantages over ALT measurement in zebrafish.
Delayed AC treatment (administered after paracetamol exposure for 3 hours) prevented the increase in circulating miR-122 at 24hpe. This is, to the authors’ knowledge, the first demonstration that effective antidote treatment is reported by a circulating miRNA biomarker. MiRNAs are very promising disease biomarkers because they are stable in blood, some display tissue-restricted expression, they rarely undergo post-processing modification, and they are readily amplifiable signals. Another major strength of miRNA is that they are translational across species, and this study adds zebrafish to the list of species (rodent and humans) that report hepatotoxicity by elevation in circulating miR-122.
The results of this study suggests that with further development, zebrafish may be valuable as a model to identify and explore drug-induced liver injury using miR-122 as a read-out of toxicity that is directly translatable to rodents and humans. The value of such a zebrafish model lies in the simplicity of drug administration, low financial cost and ease of genetic manipulation. In the present study we measured the serum paracetamol concentration which was 962 mg/L immediately following 3 hours exposure to paracetamol (30mM), clear evidence that drug in the water enters the fish circulation. This paracetamol concentration is within the range reported in humans and would be expected to produce hepatotoxicity if untreated in clinical practice.
There are limitations to the model that will be a focus of further work. Although RO bleeding increased the blood yield per fish, there was still a need to pool samples to measure biomarkers. Future work will focus on refining techniques to allow miRNA measurement on single fish to produce a significant reduction in numbers needed for experiments. Zebrafish could represent a high throughput screen but measurement of miRNA is time-consuming by PCR. There is a need for a rapid ‘point of care’ assay for miR-122 to speed up the time to results both in pre-clinical and clinical studies. In this study we normalised miR-122 using circulating Let-7d, as per our published human studies. This is a new field, and further work is needed to determine the optimal normalising approach for zebrafish blood miRNA.
In summary, collection of blood by RO bleeding facilitated measurement of circulating miR-122 in a model of paracetamol hepatotoxicity. The adult zebrafish represents a new species for measurement of circulating miRNA biomarkers that can bridge from fish to human.
Acknowledgments
All the authors acknowledge the contribution of the British Heart Foundation Centre of Research Excellence Award and author JWD the financial support of NHS Research Scotland (NRS), through NHS Lothian. Author ADBV was supported by UK NC3Rs Studentship. We thank Forbes Howie for analysis of serum ALT activity.
References
- 1.Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, et al. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42(6):1364–72. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 2.Møller LR, Nielsen GL, Olsen ML, Thulstrup AM, Mortensen JT, Sørensen HT. Hospital discharges and 30-day case fatality for drug poisoning: a Danish population-based study from 1979 to 2002 with special emphasis on paracetamol. European journal of clinical pharmacology. 2004;59(12):911–5. doi: 10.1007/s00228-003-0713-0. [DOI] [PubMed] [Google Scholar]
- 3.Gunnell D, Murray V, Hawton K. Use of paracetamol (acetaminophen) for suicide and nonfatal poisoning: worldwide patterns of use and misuse. Suicide and life-threatening behavior. 2000;30(4):313–26. [PubMed] [Google Scholar]
- 4.James LP, Mayeux PR, Hinson J a. Acetaminophen-induced hepatotoxicity. Drug metabolism and disposition. 2003;31(12):1499–506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 5.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PloS one. 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DGN, Antoine DJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–76. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clinical chemistry. 2010;56(12):1830–8. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 10.Antoine DJ, Dear JW, Starkey-Lewis P, Platt V, Coyle J, Masson M, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013 doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGrath P, Li C-Q. Zebrafish: a predictive model for assessing drug-induced toxicity. Drug discovery today. 2008;13(9-10):394–401. doi: 10.1016/j.drudis.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Barros TP, Alderton WK, Reynolds HM, Roach a G, Berghmans S. Zebrafish: an emerging technology for in vivo pharmacological assessment to identify potential safety liabilities in early drug discovery. British journal of pharmacology. 2008;154(7):1400–13. doi: 10.1038/bjp.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North TE, Babu IR, Vedder LM, Lord AM, Wishnok JS, Tannenbaum SR, et al. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(40):17315–20. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nature reviews. Genetics. 2007;8(5):353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 15.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nature reviews. Drug discovery. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 16.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 17.Hoff J. Methods of Blood Collection in the Mouse. Lab animal. 2000;29(10):47–53. [Google Scholar]
- 18.Pedroso GL, Hammes TO, Escobar TDC, Fracasso LB, Forgiarini LF, da Silveira TR. Blood collection for biochemical analysis in adult zebrafish. Journal of visualized experiments : JoVE. 2012;(63):e3865. doi: 10.3791/3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergmeyer HU, Scheibe P, Wahlefeld AW. Optimization of methods for aspartate aminotransferase and alanine aminotransferase. Clinical chemistry. 1978;24(1):58–73. [PubMed] [Google Scholar]
- 20.Starkey Lewis PJ, Merz M, Couttet P, Grenet O, Dear J, Antoine DJ, et al. Serum microRNA biomarkers for drug-induced liver injury. Clinical pharmacology and therapeutics. 2012;92(3):291–3. doi: 10.1038/clpt.2012.101. [DOI] [PubMed] [Google Scholar]
- 21.Eames SC, Philipson LH, Prince VE, Kinkel MD. Blood sugar measurement in zebrafish reveals dynamics of glucose homeostasis. Zebrafish. 2010;7(2):205–13. doi: 10.1089/zeb.2009.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murtha JM, Qi W, Keller ET. Hematologic and serum biochemical values for zebrafish (Danio rerio) Comparative medicine. 2003;53(1):37–41. [PubMed] [Google Scholar]
- 23.Spence R, Gerlach G, Lawrence C, Smith C. The behaviour and ecology of the zebrafish, Danio rerio. Biological reviews of the Cambridge Philosophical Society. 2008;83(1):13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 24.Johnston IA, Dunn J. Temperature acclimation and metabolism in ectotherms with particular reference to teleost fish. Symposia of the Society for Experimental Biology. 1987;41:67–93. [PubMed] [Google Scholar]
- 25.Denvir M a, Tucker CS, Mullins JJ. Systolic and diastolic ventricular function in zebrafish embryos: influence of norepenephrine, MS-222 and temperature. BMC biotechnology. 2008;8:21. doi: 10.1186/1472-6750-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen FB. Nitric oxide formation from nitrite in zebrafish. The Journal of experimental biology. 2007;210(Pt 19):3387–94. doi: 10.1242/jeb.008748. [DOI] [PubMed] [Google Scholar]
- 27.Babaei F, Ramalingam R, Tavendale A, Liang Y, Yan LSK, Ajuh P, et al. Novel Blood Collection Method Allows Plasma Proteome Analysis from Single Zebrafish. Journal of proteome research. 2013 doi: 10.1021/pr3009226. [DOI] [PubMed] [Google Scholar]
- 28.Zang L, Shimada Y, Nishimura Y, Tanaka T, Nishimura N. A novel, reliable method for repeated blood collection from aquarium fish. Zebrafish. 2013;10(3):425–32. doi: 10.1089/zeb.2012.0862. [DOI] [PubMed] [Google Scholar]
- 29.Chng HT, Ho HK, Yap CW, Lam SH, Chan ECY. An investigation of the bioactivation potential and metabolism profile of Zebrafish versus human. Journal of biomolecular screening. 2012;17(7):974–86. doi: 10.1177/1087057112447305. [DOI] [PubMed] [Google Scholar]