Abstract
IL-23, a heterodimer of IL-12 p40 and IL-23 p19, is critical for an effective immune response to many infections and has been implicated in several autoimmune diseases, however, little is known about the regulation of IL-23 gene expression in monocytes. We found that poly I:C, LPS, flagellin, and zymogen activated significant IL-23 production in primary human monocytes. Using chromatin immunoprecipitation, we found that a distal upstream region of the IL-23 p19 promoter at −601 to −521 underwent extensive histone modifications in response to stimuli. This distal region of the promoter is not highly conserved between species and has not been previously implicated in the regulation of IL-23 expression. Knockdown of CBP markedly decreased IL-23 p19 responses to poly I:C but had a less dramatic effect on LPS responses, confirming different chromatin responses to these two stimuli. Our data suggest that one of the mechanisms regulating IL-23 expression is the regulation of histone modifications at this distal upstream region of the promoter.
Keywords: human, monocyte/macrophages, cytokines, histones, chromatin, gene regulation, molecular biology, CBP
INTRODUCTION
The discovery of IL-23, a heterodimer of the IL-12 p40 chain with the novel IL-23 p19 chain, clarified the conflicting data for the IL-12 p40 and IL-12 p35 knockout mice. IL-12 p35 knockout mice have increased susceptibility to infection, but IL-12 p40 knockout mice are even more susceptible to infections [14, 17, 23, 32, 36]. We now know that the key difference between the two is that IL-12 p40 mice lack both IL-12 and IL-23.
The role of IL-23 in the defense against intracellular pathogens in humans has been suggested by the identification of IL-12 p40 and IL-12Rβ1 (the common IL-12/IL-23 receptor subunit) mutations in individuals with an inherited predisposition to Salmonella and mycobacterial infections. No patients with mutations in either the IL-12 p35 or IL-12Rβ2 have been identified, suggesting that the IL-23 effect is biologically important [3, 13, 46]. In infection models, IL-23 p19 knockout mice have shown that IL-23, and the IL-17 immune response it directs, are critical for the maintenance of the immune response in the face of persistent infections as well as granuloma formation, although anti-fungal responses may be compromised in conditions of high IL-23 [18, 26, 27, 44, 48, 65]. IL-23 acts to facilitate expression of certain cytokines and chemokines and it increases antigen presentation by dendritic cells and microglia [5, 12, 27, 28].
IL-23 is produced primarily by antigen presenting cells, including macrophages and dendritic cells (DCs), in response to a variety of toll-like receptor (TLR) molecules. The IL-23 receptor is expressed on activated/memory T cells, NK cells, and, at a low level, on monocytes, macrophages, and DCs [25]. IL-23 production, in combination with TGFβ and IL-6, directs the development of Th17 T cells [54, 59]. Th17 cells appear to be important in tissue inflammatory responses and these cells have been implicated in some of the effects of IL-23 in both host defense and autoimmunity [16, 26, 30, 39, 43]. In addition, IL-23 can directly mediate inflammation [37].
In spite of its importance, the regulation of IL-23 expression has not been thoroughly examined. Bacteria and bacterial products tend to preferentially induce IL-12 expression in lieu of IL-23 [51]. Certain viruses induce IL-23 along with type I interferons in monocytes and recently fungal stimuli have been examined [47]. Stimulation via dectin-1 with purified fungal extracts results in increased IL-23 production compared to zymosan, which preferentially triggers IL-12 production [34].
The signaling pathways regulating IL-23 share much with those of IL-12 in spite of many examples of discordant expression [4, 9, 47]. Like IL-12 p35 and IL-12 p40, TLR signaling induces expression of IL-23 p19 [4]. Cytokine priming can dramatically alter IL-23 responses, with IFN-γ increasing responses and IL-4 decreasing responses to TLR ligands [47, 50]. NF-κB, PI3 kinase, and p38 activation all lead to increased IL-23, but mTOR/S6K1 and Rac1 both negatively regulate IL-23 expression [9, 10, 37, 57, 64].
Despite these recent advances, little is known about the transcriptional regulation of IL-23 compared to IL-12 p40 and IL-12 p35. Following cellular activation, the IL-12 p40 promoter binds NF-κB, C/EBP, AP-1, and NFAT. Transcription of IL-12 p40 also requires remodeling by SWI/SNF of the nucleosome positioned at the transcription start site [63, 66] and a recently characterized inducible enhancer has also been found to contribute to transcriptional regulation [66]. Similarly, IL-12 p35 transcription is controlled by c-Rel and p65, and Sp1 binding is necessary for nucleosomal remodeling to allow for transcription [20, 21]. Limited data on IL-23 p19 expression suggests a different mode of regulation from IL-12. ERK, AP-1 and cRel signaling appear to be required for IL-23 p19 expression [10, 19, 37, 41]. Little else is known about the transcriptional regulation of IL-23 p19 expression in either mouse or human. Contributing to the difficulties in understanding the regulation of IL-23 is the finding that IL-23 expression differs in murine and human systems.
This study focuses on histone modifications at the IL-23 p19 promoter. The histone tails of nucleosomes undergo post-translational modifications that impact chromatin conformation and promoter accessibility [29, 35]. Modifications singly or in combination serve as docking sites for proteins involved in transcriptional regulation, DNA repair, or mitotic machinery. Patterns of modifications are distinct for repressed, competent, and actively transcribing promoters [22, 45]. Here, we report the chromatin modifications associated with induction of transcription of IL-23 p19 in human monocytes and identify a novel upstream regulatory region.
METHODS
Tissue Culture
All culture dishes and plasticware used in these experiments were certified pyrogen free. Human peripheral blood monocytes were obtained from the Human Immunology Core at the University of Pennsylvania and were isolated by countercurrent elutriation [61]. Monocytes were cultured in RPMI 1640 supplemented with 10% cosmic calf serum, 2 mM glutamine, 200 U/ml of penicillin, and 200 µg/ml of streptomycin. Monocytes were rested overnight at 4°C on a nutator before being transferred to culture flasks and the incubator. Cells were warmed in the incubator for at least 2 h prior to stimulation. Flow cytometry analysis was used to confirm that the cells were >90% CD14 positive.
Reagents
LPS (Escherichia coli 011:B4) and the bacterial muramyl dipeptide N-acetylmuramyl-L-alanyl-D-isoglutamine hydrate (MDP), a ligand for the intracellular nuclear oligomerization domain receptor family (NOD2), were obtained from Sigma Chemical Co (St. Louis, MO) and LPS was used at a concentration of 1 µg/ml in all experiments. TLR ligands were obtained from InvivoGen (San Diego, CA) and used as previously described [15]: Pam3CSK4, a synthetic tripalmitoylated lipopeptide that activates TLR 1 and 2; zymosan, a derivative of Saccharomyces cerevisiae, that activates TLRs 2,6; poly(I):poly (C) (poly I:C), a synthetic analog of double stranded RNA for TLR3 (and/or MDA5); ultrapure LPS (Salmonella enterica serovar Minnesota) for TLR4; flagellin (Salmonella enterica serovar typhimurium) for TLR5; and loxoribine for TLR7. All ligands were prepared in ultra-pure endotoxin free water according to manufacturer’s recommendations.
RT-PCR
RNA was isolated from 2–3 million monocytes that had been cultured with 1µg/ml LPS or 25 µg/ml of poly I: C for the indicated time points using Qiagen RNeasy kit and on-column DNase digestion using Qiagen’s RNase free DNase set (Qiagen, Valencia, CA). Reverse transcription utilized BD Bioscience’s Advantage RT-for-PCR kit (San Jose, CA). The final reaction was diluted to 100 µl with DEPC treated water and the product was used for quantitative real time Taqman PCR using ABI’s IL-23 human mRNA primers and the 18S internal control (Foster City, CA). IL-23 signal was normalized to the 18S signal and calibrated to the matched unstimulated sample. CBP knockdown in primary monocytes was accomplished using 3 µg of a pool of siRNAs (Dharmacon, Lafayette, CO) per 20 million cells, transfected via Amaxa. Knockdown efficiency was typically ≈80% as assessed by Western blot. GFP siRNA was used as a negative control [42].
TLR Assay
The TLR assay was performed as previously described using TLR ligand concentrations optimized for this system [15]. Briefly, 2×105 monocytes were plated in a 96 well flat bottom plate. Triplicates were stimulated with the following: 25 (µg/ml poly I:C, 100 µM Loxoribine, 10 µg/ml zymosan, 40 µg/ml Pam3CSK4, 10 µg/ml flagellin, 1 µg/ml LPS (Salmonella enterica or E. coli), and 0.1µg/ml MDP or media alone. Cell-free supernatants were collected at 6 or 24 h for the ELISA which was performed using e-Biosciences IL-23 (p19/p40) Ready-Set-Go ELISA kit according to the manufacturer’s protocol. The plate was read at 450 nm. IL-12p70, Interferon α, and TNFα were detected by Searchlight multiplex (Pierce, Rockford, IL).
Chromatin Immunoprecipitation (ChIP)
The ChIP assays were performed as described previously according to a modified protocol based on Upstate Biotechnology (Lake Placid, NY) [53]. 5 µg of the following antibodies were used: acetylated histone 3, acetylated histone 4, pan-histone 4, dimethylated H3 lysine 4 (Upstate Biotechnology), trimethylated H3 lysine 4 (Abcam, Cambridge, MA), and rabbit anti-GST (Zymed, South San Francisco, CA).
Immunoprecipitated DNA was quantitated by realtime Taqman PCR from specific regions of the IL-23 p19 promoter. Each DNA sample was subjected to three PCR reactions to three regions of the IL-23 p19 promoter: IL-23 A (+93/+33): IL-23A Forward—5’-ACAACTGAG GGAACCAAACCA-3’, IL-23 A probe—5’-6FAM-AGACGCGCTGAACAG-MGBNFQ-3’, IL-23A Reverse—5’-CACTTGCTTTGAGCCTGATTCTC-3’; IL-23 B (−213/−130): IL-23B Forward—5’-TCGATTCCT GGAGAGCATTACAC-3’, IL-23B Probe—5’-6FAM-ATGTGTCCCATCCCAG-MGBNFQ-3’, IL-23B Reverse— 5’-TCAGTTCCAGGGGAAATGAGTA-3’; and IL-23 C(−601/−521): IL-23C Forward—5’-TTCCCAGT TCTCCAAGTTCCTTC-3’, IL-23C Probe—5’-6FAM-ATGCGCATTCTCT-MGBNFQ-3’, IL-23C Reverse— 5’-GTTGGAAACAACATTATTTCAA-3’. All three FAM-MGB primer/probe sets were designed using ABI Primer Express and optimized according to ABI’s recommendations. Duplicates of samples were run and averaged separately prior to analysis. Ct values were normalized to 10% input of each sample according to the formula 2^(10% input CT-Sample CT).
Standard curves using thymus genomic DNA were used as positive controls. In all experiments, rabbit GST antibody was used as a control antibody, but is only shown in Fig. 3 for simplicity. Normalized CT values for GST were similar in all experiments and always less than 0.02. ChIP results reflect duplicate PCRs of multiple experiments.
Fig. 3.
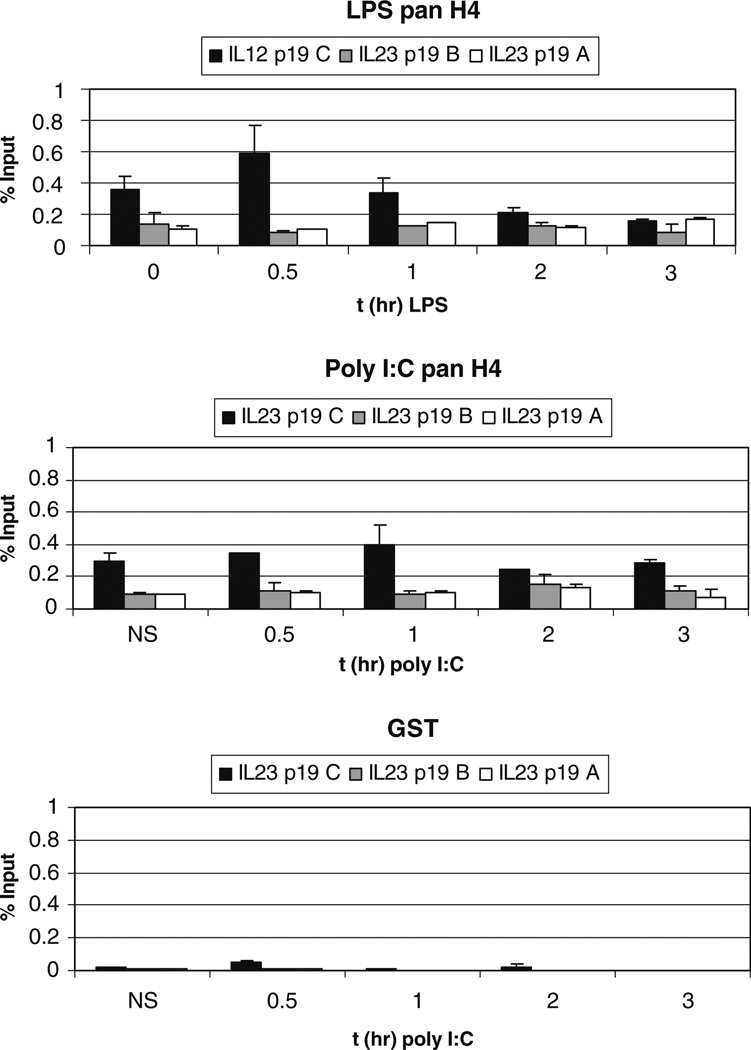
Nucleosome locations within the IL-23 p19 promoter. Ten million primary human monocytes were cultured and stimulated with 1 or 25 µg/ml LPS for the indicated timepoints. ChIP analyses were performed with antibodies to total H3 and H4. Anti-GST was used as a negative control and always gave a negligible signal. Graphs are the average of duplicates from two experiments.
Statistical Analysis
Significance calculations were performed using Student’s t tests. For CHiP data, Bonferroni’s correction was used. Only corrected p values are given. Error bars indicate standard deviation.
RESULTS
TLR Responses of Primary Human Monocytes
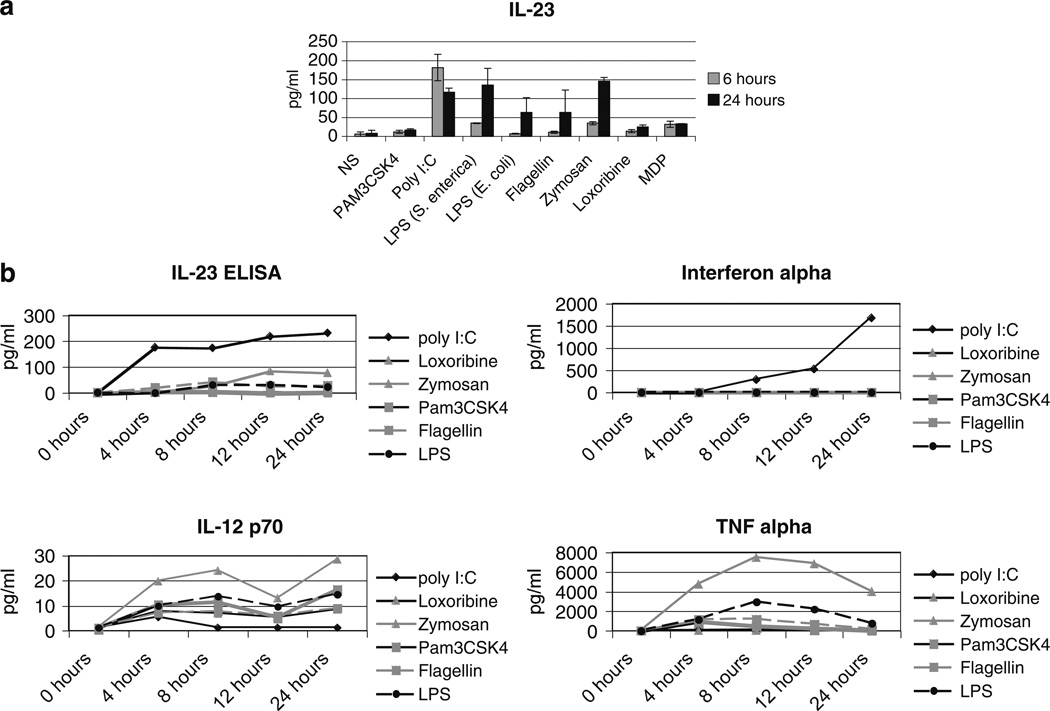
Most studies of IL-23 responses to TLR ligands have involved murine cells or human dendritic cells [10, 33, 58, 62]. There are limited data on human monocytes. We therefore defined the production of IL-23 p19/p40 in primary human peripheral blood monocytes by ELISA in response to specific pathogen ligands at both 6 and 24 h post stimulation (Fig. 1a). Of the six ligands tested, only two did not cause any significant IL-23 production at 6 or 24 h: PAM3CSK (TLR2), and synthetic muramyl dipeptide (MDP), a ligand for the intracellular protein, NOD2. After detecting very low levels of IL-23 in response to ultrapure Salmonella enterica derived LPS, we also tested the response to LPS from E. coli. E. coli LPS resulted in even lower levels of IL-23 production. Both S. enterica and E. coli are gram negative bacteria and LPS derived from each have previously been reported to induce IL-23 [40]. S. enterica LPS did result in significant levels of IL-23 after 24h of stimulation (6 h p=0.002; 24 h p=0.01).
Fig. 1.
a IL-23 response to TLR stimulation. 2×105 primary human monocytes were stimulated with the indicated ligands as described in “Methods” before assaying for IL-23 protein by ELISA. The grey bar indicates 6h of stimulation, the black bar indicates 24h. Results are representative of three experiments. Error bars represent the standard deviation between the triplicate samples. b IL-23 compared with other cytokine responses. Cells were stimulated as above and supernatants harvested at the indicated timepoints. IL-23 was measured by ELISA and the other cytokines were measured by Searchlight. Each ligand induced at least one cytokine response.
There was a distinct difference in the kinetics of IL-23 protein production after poly I:C, a synthetic double stranded RNA analog (which is a ligand for TLR3 and/or MDA5), with an early peak of protein production by 6 h. Poly I:C also induced the highest amount of IL-23 of the tested stimuli. The other stimulatory ligands by contrast, exhibited peak protein production at 24 h. Several other cytokines are also commonly induced by TLR ligands [55]. We compared TNFα, IL-12p70 and IFNα induction along with IL-23 (Fig. 1b). The pattern of expression for each of these cytokines is completely different. These data simply confirm the well known finding that although a given TLR induces the same signaling pathways, the response is distinct for each responding gene [55].
IL-23 p19 Promoter
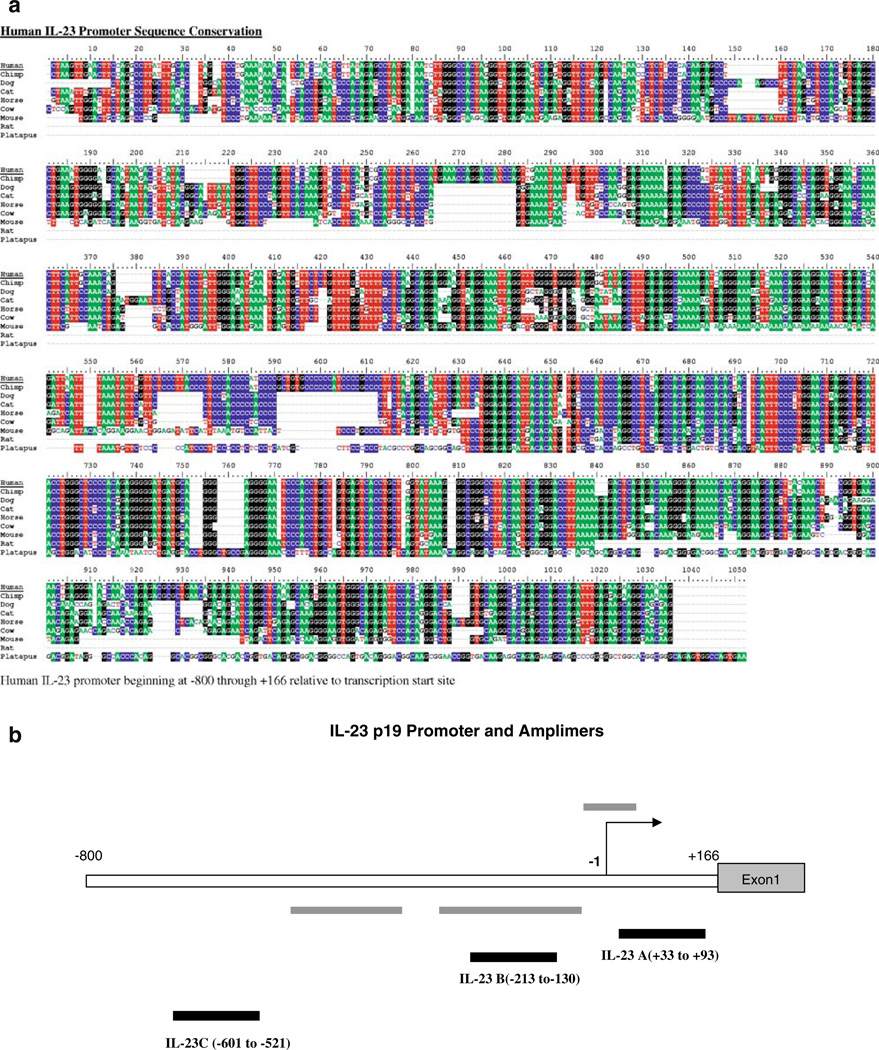
We examined the evolutionary conservation of DNA sequences between mouse and human in the IL-23 p19 gene (Fig. 2a) [11]. The promoter is generally well-conserved but there are three regions of high homology. Additionally, a portion of the first intron is also highly homologous between species. These areas of homology extend to multiple species.
Fig. 2.
a Sequence conservation is high in the IL-23 p19 promoter. Shading indicates >50% homology across species. Eight species are compared. b Schematic diagram of the human IL-23 promoter. Grey bars denote areas of conservation among species. Black bars show the three amplimers used in this study.
To study chromatin modifications within the promoter, we designed three Taqman MGB probes to the IL-23 p19 promoter using ABI Primer Express software (Fig. 2b). IL-23 A and B, respectively, correspond to the two proximal regions with high interspecies homology. Due to the large number of repeats and GC content of the IL-23 p19 promoter, our primer design had to exclude the most distal region of high homology within the promoter, and the IL-23C primer/probe set is located immediately upstream. This region still exhibits significant homology but has no recognized role in regulation.
We used an antibody to total histone H4 to determine if any nucleosomes were located in the three amplimer regions. Primary human monocytes were also stimulated with either LPS or poly I:C for different time points to determine if nucleosome depletion occurred during transcription. The IL-23 p19 C probe detected significant histone content, suggesting that the distal promoter region is characterized by the presence of nucleosomes at rest in monocytes (Fig. 3), while the proximal promoter appeared relatively depleted of nucleosomes. There was a significant increase in the level of total H4 at the distal promoter after LPS stimulation at 30 min ( p=0.03), followed by a decrease at 3 h. This increased nucleosome content after stimulation is currently without explanation but may reflect the increased turnover associated with transcription. During poly I:C stimulation, total H4 levels were constant during the 3 h time course.
Chromatin Modifications of the Il-23 p19 Promoter Following LPS Stimulation
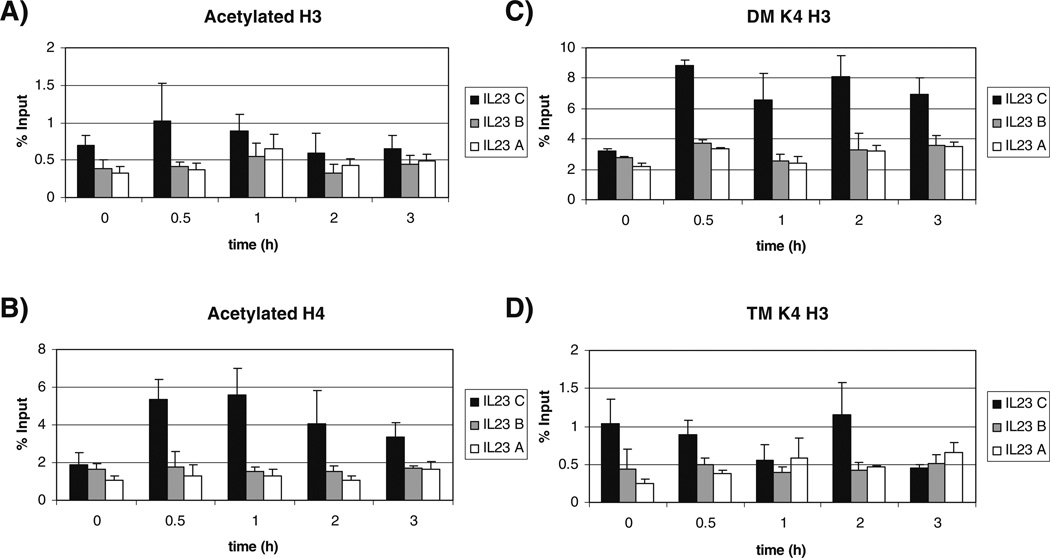
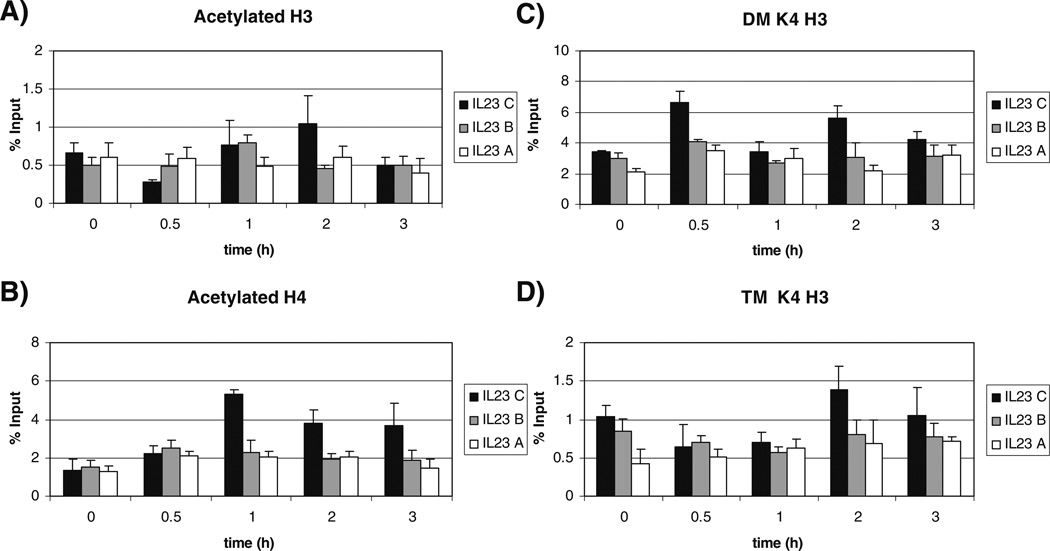
The IL-23 p19 promoter showed increased acetylation of H3 at the upstream IL-23 C region after 30 min of LPS stimulation compared to unstimulated cells. This diminished over time and roughly paralleled the nucleosome content (Figs. 3 and 4). Acetylation of H4 for the IL-23 C region was significantly increased after 30 min of stimulation ( p=0.001). Acetylation of H4 was maintained over time until 3 h after stimulation. Little change was seen at the IL-23 A and B regions because of the limited nucleosome content.
Fig. 4.
Histone modifications of the IL-23 p19 promoter following LPS stimulation. Ten million primary monocytes were cultured and stimulated with 1 µg/ml LPS for the indicated timepoints. a Acetylated H3, b acetylated H4, c dimethylated H3 lysine 4 (DM K4 H3), d trimethylated H3 lysine 4 (TM K4 H3). Graphs represent the average of three experiments with duplicates. (Black bar) IL-23 C, (grey bar) IL-23 B, and (white bar) IL-23 A. The error bars indicate standard deviation.
Methylation modifications of H3 lysine 4 have been linked to transcriptional activity [6, 49]. Dimethylation of H3 lysine 4 (DM K4 H3) is indicative of transcriptionally competent regions of DNA and trimethylation of H3 lysine 4 (TM K4 H3) at promoters has been linked to transcriptionally active genes [7]. After 30min of LPS stimulation, DM K4 H3 was significantly increased (IL-23 C p=2×10−6, IL-23 B p=0.0001, IL-23A p=0.0009). This increase in dimethylation was maintained throughout the entire 3h time course and the increases in IL-23 C were significant the entire time relative to unstimulated monocytes (1h p=2×10−6, 2 h p=0.001, 3h p=0.03).
For TM K4 H3, the IL-23 A region of the promoter showed significantly increased trimethylation after 30 min of LPS stimulation ( p=0.04) and this increase was maintained through the 3 h stimulation. Although the nucleosome content of the proximal promoter was limited, RNA pol II can deposit the trimethyl mark locally, perhaps explaining the finding. The IL-23 C region behaved differently in that TM K4 H3 decreased at 3 h.
Chromatin Modifications of the IL-23 p19 Promoter Following Poly I:C Stimulation
The different kinetics of IL-23 p19 protein production seen with LPS and Poly I:C stimulation suggested distinct mechanisms of regulation. In addition, these two ligands are known to activate different signaling pathways [55]. We therefore examined the kinetics of histone modifications of the IL-23 p19 promoter following stimulation with poly I:C (Fig. 5). The pattern of acetylation of H3 for poly I:C was significantly different than that seen after LPS. For poly I:C, acetylation of H3 in the IL-23 C region decreased significantly after 30min ( p=0.033). At 1 h, acetylation increased significantly compared to the levels at 0.5h ( p=0.01). These high levels of acetylation were maintained through 2 h of stimulation before decreasing at 3 h. The levels seen were not significantly different from their matching LPS time point, but the pattern was different.
Fig. 5.
Histone modifications of the IL-23 p19 promoter following Poly I:C stimulation. Ten million primary monocytes were cultured and stimulated with 25 µg/ml Poly I:C for the indicated timepoints. a Acetylated H3, b acetylated H4, c Dimethylated H3 lysine 4 (DM K4 H3), d Trimethylated H3 lysine 4 (TM K4 H3). Graphs represent the average of three experiments with duplicates. (Black bar) IL-23 C, (grey bar) IL-23 B, and (white bar) IL-23 A. The error bars indicate standard deviation.
Acetylation of H4 at the IL-23 C region was also slower following poly I:C stimulation compared to LPS stimulation, and the levels were maintained through 3 h of stimulation (1 h p=0.07, 2 h p=0.019, 3 h p=0.020). The level of acetylation of H4 by LPS at 0.5 h was significantly higher than that induced by poly I:C ( p=0.022), but the levels were not significantly different between the two stimuli at the other time points.
The pattern of dimethylation and trimethylation of H3 lysinea 4 induced by poly I:C was similar to that induced by LPS, although poly I:C induced significantly lower levels of dimethylation of IL-23 C region from 0.5 to 3 h of stimulation compared to LPS (0.5 h p=0.009, 1 h p=0.036, 2 h p=0.035, 3 h p=0.006).
In summary, H3 lysine 4 methylation was roughly comparable after LPS and poly I:C stimulation. H3 and H4 acetylation occurred more slowly after poly I:C stimulation compared to LPS stimulation which did not correlate with the pattern of protein production.
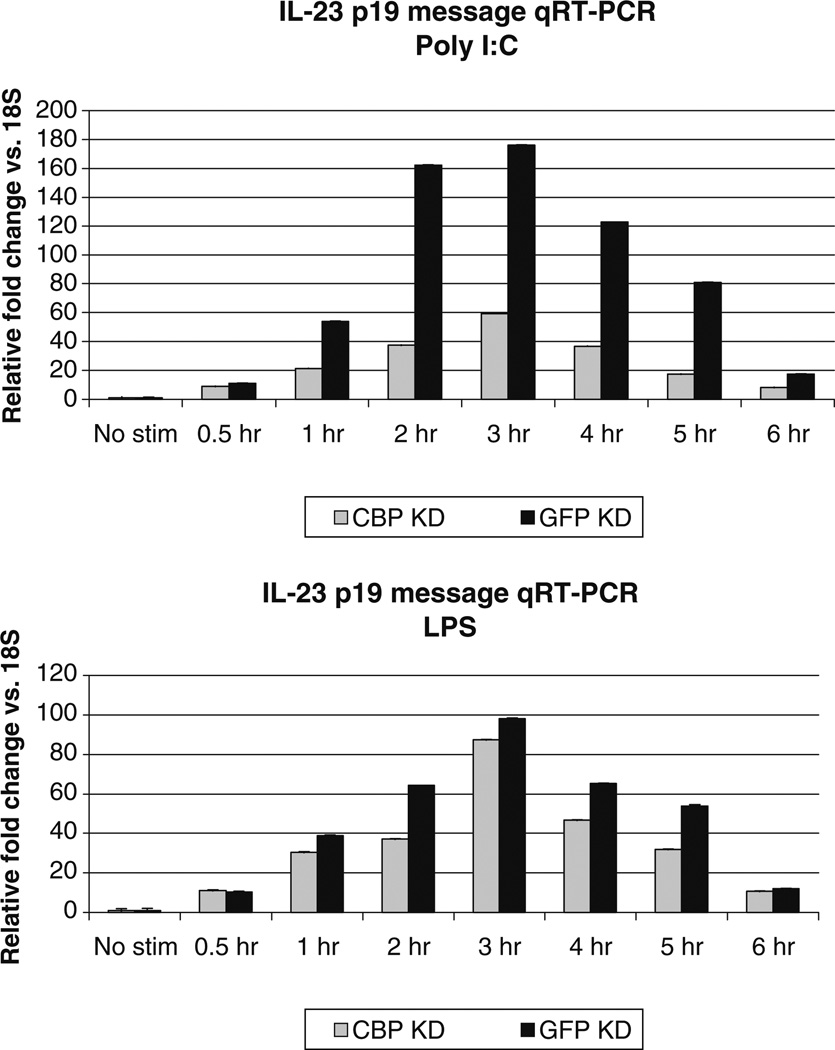
It was also surprising that the early protein production after poly I:C did not correlate with more rapid histone modifications. In fact, histone acetylation appeared to be slower in the poly I:C stimulated cells. To address both of these issues, we examined the distal region of the promoter in more detail. Just 40bp upstream of the IL-23 C region, a potential CBP binding site was found. CBP is activated after LPS though a PI3 kinase-glycogen synthase kinase 3 pathway [38]. In contrast, poly I:C activates CBP via IRF3 [52]. To determine whether CBP differentially affected IL-23 expression, we knocked down CBP in primary monocytes using a pool of siRNAs and stimulated with either LPS or poly I:C. IL-23 p19 message was detected by qRT-PCR. Message levels appeared to increase slightly more rapidly in poly I:C stimulated cells compared to LPS stimulated cells, but the difference was not significant. CBP knockdown significantly compromised responses to poly I:C but had little effect on responses to LPS (Fig. 6). These data confirm that the chromatin responses seen at the distal IL-23 p19 promoter are unique to different stimuli.
Fig. 6.
CBP knockdown analysis of IL-23 p19 message. Primary monocytes were transfected using Amaxa with either pooled siRNAs directed to CBP or an siRNA directed to GFP, as a negative control. Cells were stimulated 72 h after transfection. RNA was isolated from 2 million monocytes following stimulation for the indicated times with 1 µg/ml LPS or 25 µg/ml poly I:C. Although not obvious, error bars represent standard deviations. CBP plays an important role in IL-23 p19 responses to poly I:C.
DISCUSSION
In primary human peripheral blood monocytes, LPS, poly I:C, flagellin, and zymogen were found to be the primary activators of IL-23 p19/p40 production and secretion. Cytokine priming or examination of distinct human monocyte subsets may show a different pattern of IL-23 responses [24, 31, 47, 56]. Poly I:C, a synthetic double stranded RNA analog led to the highest IL-23 protein production as well as the earliest. The role of IL-23 in viral infections is not well established but these results suggest that innate responses to viruses could be a major stimulus for IL-23 production in vivo. This has important implications for models of both infection and autoimmunity.
Very little is known about the transcriptional regulation of IL-23 p19. This is the first study to examine histone modifications at this gene. Using an antibody for total H4, we found that the distal region of the IL-23 p19 promoter had a higher nucleosome content than the proximal promoter. This, combined with the generally negligible levels of histone modifications at the proximal promoter following stimulation, led us to believe that unlike IL-12 p40 and p35 gene promoters, IL-23 p19 expression is not controlled by a nucleosome proximal to the site of gene transcription [2, 20].
We were surprised to see that LPS induced IL-23 p19 transcription as early as 30min after stimulation when significant IL-23 p19/p40 protein was only found after 24 h of stimulation. These data are consistent with several nonexclusive explanations: (1) IL-23 p19 is strongly post-transcriptionally and translationally regulated, (2) IL-12 p40 kinetics control the rate of IL-23 p19/p40 protein production, and (3) primary human monocytes require another signal in addition to LPS to secrete high levels of IL-23 p19/p40.
Stimulation with either LPS or poly I:C resulted in significant histone modifications of the nucleosome located in the upstream IL-23 C region compared to unstimulated cells. For LPS-stimulated monocytes, H3 and H4 acetylation of the IL-23 C region was detected after only 30 min of stimulation before decreasing at 2 and 3 h post stimulation. In contrast, poly I:C did not induce increased acetylation until after 1h of stimulation.
Although the differences in the patterns of histone acetylation after LPS and poly I:C were not dramatic, it is clear that CBP plays a much more significant role in the response to poly I:C compared to LPS. This implies that chromatin effects are significant and mold the individual cell’s response to stimulation.
The pattern of dimethylation and trimethylation of H3 lysine 4 (DM K4 H3, TM K4 H3) following LPS and poly I:C treatment was very similar although LPS induced significantly higher levels of dimethylation.
It is worth noting that the pattern of histone modifications after LPS at the IL-23 p19 promoter is completely different than that seen at the TNFα promoter after LPS stimulation of human monocytes [53]. This suggests that the signaling molecules themselves are incompletely responsible for the modifications. Pre-existing chromatin marks and recruited transcription factors must mold the histone responses to stimuli. Additional studies of transcription factors could provide insight into the regulation of chromatin at the IL-23 p19 promoter.
In summary, we identified a novel upstream promoter region in IL-23 p19 which appears to be involved in the epigenetic regulation of expression. This IL-23 p19 C amplimer region is somewhat conserved between species and has no recognized function. Nevertheless, just 40bp upstream of the distal edge of the amplimer is a CBP recognition motif which could regulate histone acetylation within the −601–521 IL-23 C upstream region. Knockdown of CBP diminished responses to poly I:C but had little effect on LPS responses. Additional motifs which could be important in regulating histone modifications are a GCN4 site approximately 50bp downstream and several potential ATF-2 sites 75–90bp upstream of the IL-23 C region. These may be important in the LPS response. Taken together, our data describe a distal upstream promoter region which appears to be involved in chromatin regulation. Although transcription factor binding is typically limited to the 200bp surrounding the initiation site, it has recently become apparent that many regulatory motifs are located distantly from the transcribed region [1]. These data help to formulate a growing model for the transcription of IL-23.
A better understanding of the transcriptional regulation of IL-23 p19, as well as the epigenetic control of its promoter, may be helpful to design better therapies and adjuvants for autoimmunity and immunizations. Therapeutic interventions directed at IL-23 are already in development [8, 60]. Understanding the regulation of IL-23 p19 is critical for the rational development of interventions directed at this complex cytokine network.
ACKNOWLEDGEMENTS
This work was supported by NIH grants AI051323 and AI44127. The authors wish to acknowledge the Human Immunology Core at the University of Pennsylvania, which provided the primary monocytes for this study. Stacey Garrett was supported in part by The University of Pennsylvania School of Medicine Cancer Research Institute Predoctoral Emphasis on Pathways in Tumor Immunology Training Grant.
Footnotes
Authors’ Contributions SG carried out the ChIP studies and drafted the manuscript. MF performed the knockdown and the RT-PCR experiments. KS provided oversight of the study design and final wording of the text. All authors read and approved the final manuscript.
REFERENCES
- 1.Birney E. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht I, Tapmeier T, Zimmermann S, Frey M, Heeg K, Dalpke A. Toll-like receptors differentially induce nucleosome remodelling at the IL-12p40 promoter. EMBO Reports. 2004;5:172–177. doi: 10.1038/sj.embor.7400078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altare F, Lammas D, Revy P, Jouanguy E, Doffinger R, Lamhamedi S, Drysdale P, Scheel-Toellner D, Girdlestone J, Darbyshire P, Wadhwa M, Dockrell H, Salmon M, Fischer A, Durandy A, Casanova JL, Kumararatne DS. Inherited interleukin 12 deficiency in a child with bacille Calmette-Guerin and Salmonella enteritidis disseminated infection. J. Clin Invest. 1998;102:2035–2040. doi: 10.1172/JCI4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson A, Kokkola R, Wefer J, Erlandsson-Harris H, Harris RA. Differential macrophage expression of IL-12 and IL-23 upon innate immune activation defines rat autoimmune susceptibility. J. Leukoc. Biol. 2004;76:1118–1124. doi: 10.1189/jlb.0704385. [DOI] [PubMed] [Google Scholar]
- 5.Belladonna ML, Renauld JC, Bianchi R, Vacca C, Fallarino F, Orabona C, Fioretti MC, Grohmann U, Puccetti P. IL-23 and IL-12 have overlapping, but distinct, effects on murine dendritic cells. J. Immunol. 2002;168:5448–5454. doi: 10.4049/jimmunol.168.11.5448. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Billich A. Drug evaluation: Apilimod, an oral IL-12/IL-23 inhibitor for the treatment of autoimmune diseases and common variable immunodeficiency. IDrugs. 2007;10:53–59. [PubMed] [Google Scholar]
- 9.Butchar JP, Rajaram MV, Ganesan LP, Parsa KV, Clay CD, Schlesinger LS, Tridandapani S. Francisella tularensis induces IL-23 production in human monocytes. J. Immunol. 2007;178:4445–4454. doi: 10.4049/jimmunol.178.7.4445. [DOI] [PubMed] [Google Scholar]
- 10.Carmody RJ, Ruan Q, Liou HC, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J. Immunol. 2007;178:186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- 11.Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome. Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, Sedgwick JD. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 13.de Jong R, Altare F, Haagen IA, Elferink DG, Boer T, Vriesman PJ, van Breda Kabel PJ, Draaisma JM, Dissel JTvan, Kroon FP, Casanova JL, Ottenhoff TH. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 14.Decken K, Kohler G, Palmer-Lehmann K, Wunderlin A, Mattner F, Magram J, Gately MK, Alber G. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect. Immun. 1998;66:4994–5000. doi: 10.1128/iai.66.10.4994-5000.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deering RP, Orange JS. Development of a clinical assay to evaluate toll-like receptor function. Clin. Vaccine Immunol. 2006;13:68–76. doi: 10.1128/CVI.13.1.68-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins KL, Cooper A, Colombini SM, Cowley SC, Kieffer TL. In vivo clearance of an intracellular bacterium, Francisella tularensis LVS, is dependent on the p40 subunit of interleukin-12 (IL-12) but not on IL-12 p70. Infect. Immun. 2002;70:1936–1948. doi: 10.1128/IAI.70.4.1936-1948.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gafa V, Lande R, Gagliardi MC, Severa M, Giacomini E, Remoli ME, Nisini R, Ramoni C, Francesco PDi, Aldebert D, Grillot R, Coccia EM. Human dendritic cells following Aspergillus fumigatus infection express the CCR7 receptor and a differential pattern of interleukin-12 (IL-12), IL-23, and IL-27 cytokines, which lead to a Th1 response. Infect. Immun. 2006;74:1480–1489. doi: 10.1128/IAI.74.3.1480-1489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodridge HS, Harnett W, Liew FY, Harnett MM. Differential regulation of interleukin-12 p40 and p35 induction via Erk mitogen-activated protein kinase-dependent and -independent mechanisms and the implications for bioactive IL-12 and IL-23 responses. Immunology. 2003;109:415–425. doi: 10.1046/j.1365-2567.2003.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goriely S, Demonte D, Nizet S, Wit DDe, Willems F, Goldman M, Lint CVan. Human IL-12(p35) gene activation involves selective remodeling of a single nucleosome within a region of the promoter containing critical Sp1-binding sites. Blood. 2003;101:4894–4902. doi: 10.1182/blood-2002-09-2851. [DOI] [PubMed] [Google Scholar]
- 21.Grumont R, Hochrein H, O’Keeffe M, Gugasyan R, White C, Caminschi I, Cook W, Gerondakis S. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J. Exp. Med. 2001;194:1021–1032. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Calcar SVan, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 23.Holscher C, Atkinson RA, Arendse B, Brown N, Myburgh E, Alber G, Brombacher F. A protective and agonistic function of IL-12p40 in mycobacterial infection. J. Immunol. 2001;167:6957–6966. doi: 10.4049/jimmunol.167.12.6957. [DOI] [PubMed] [Google Scholar]
- 24.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: Related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 26.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. IL-23 and IL-17 in the establishment of protective pulmonary CD4+T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 27.Kleinschek MA, Muller U, Brodie SJ, Stenzel W, Kohler G, Blumenschein WM, Straubinger RK, McClanahan T, Kastelein RA, Alber G. IL-23 enhances the inflammatory cell response in Cryptococcus neoformans infection and induces a cytokine pattern distinct from IL-12. J. Immunol. 2006;176:1098–1106. doi: 10.4049/jimmunol.176.2.1098. [DOI] [PubMed] [Google Scholar]
- 28.Kopp T, Lenz P, Bello-Fernandez C, Kastelein RA, Kupper TS, Stingl G. IL-23 production by cosecretion of endogenous p19 and transgenic p40 in keratin 14/p40 transgenic mice: Evidence for enhanced cutaneous immunity. J. Immunol. 2003;170:5438–5444. doi: 10.4049/jimmunol.170.11.5438. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapenta C, Santini SM, Spada M, Donati S, Urbani F, Accapezzato D, Franceschini D, Andreotti M, Barnaba V, Belardelli F. IFN-alpha-conditioned dendritic cells are highly efficient in inducing cross-priming CD8(+) T cells against exogenous viral antigens. Eur. J. Immunol. 2006;36:2046–2060. doi: 10.1002/eji.200535579. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann J, Bellmann S, Werner C, Schroder R, Schutze N, Alber G. IL-12p40-dependent agonistic effects on the development of protective innate and adaptive immunity against Salmonella enteritidis. J. Immunol. 2001;167:5304–5315. doi: 10.4049/jimmunol.167.9.5304. [DOI] [PubMed] [Google Scholar]
- 33.Lehner M, Morhart P, Stilper A, Petermann D, Weller P, Stachel D, Holter W. Efficient chemokine-dependent migration and primary and secondary IL-12 secretion by human dendritic cells stimulated through Toll-like receptors. J. Immun-other(1997) 2007;30:312–322. doi: 10.1097/01.cji.0000211345.11707.46. [DOI] [PubMed] [Google Scholar]
- 34.Leibundgut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis ESC. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman LA, Cardillo F, Owyang AM, Rennick DM, Cua DJ, Kastelein RA, Hunter CA. IL-23 provides a limited mechanism of resistance to acute toxoplasmosis in the absence of IL-12. J. Immunol. 2004;173:1887–1893. doi: 10.4049/jimmunol.173.3.1887. [DOI] [PubMed] [Google Scholar]
- 37.Liu FL, Chen CH, Chu SJ, Chen JH, Lai JH, Sytwu HK, Chang DM. Interleukin (IL)-23 p19 expression induced by IL-1{beta} in human fibroblast-like synoviocytes with rheumatoid arthritis via active nuclear factor-{kappa}B and AP-1 dependent pathway. Rheumatology. 2007;46(8):1266–1273. doi: 10.1093/rheumatology/kem055. [DOI] [PubMed] [Google Scholar]
- 38.Martin M, Rehani K, Jope RS, Michalek SM. Tolllike receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333–1336. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23-IL-17 immune pathway. Trends. Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Mise-Omata S, Kuroda E, Niikura J, Yamashita U, Obata Y, Doi TS. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J. Immunol. 2007;179:6596–6603. doi: 10.4049/jimmunol.179.10.6596. [DOI] [PubMed] [Google Scholar]
- 42.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 43.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakano N, Nishiyama C, Kanada S, Niwa Y, Shimokawa N, Ushio H, Nishiyama M, Okumura K, Ogawa H. Involvement of mast cells in IL-12/23 p40 production is essential for survival from polymicrobial infections. Blood. 2007;109:4846–4855. doi: 10.1182/blood-2006-09-045641. [DOI] [PubMed] [Google Scholar]
- 45.O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO. J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picard C, Fieschi C, Altare F, Al-Jumaah S, Al-Hajjar S, Feinberg J, Dupuis S, Soudais C, Al-Mohsen IZ, Genin E, Lammas D, Kumararatne DS, Leclerc T, Rafii A, Frayha H, Murugasu B, Wah LB, Sinniah R, Loubser M, Okamoto E, Al-Ghonaium A, Tufenkeji H, Abel L, Casanova JL. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am. J. Hum. Genet. 2002;70:336–348. doi: 10.1086/338625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirhonen J, Matikainen S, Julkunen I. Regulation of virus-induced IL-12 and IL-23 expression in human macrophages. J. Immunol. 2002;169:5673–5678. doi: 10.4049/jimmunol.169.10.5673. [DOI] [PubMed] [Google Scholar]
- 48.Rudner XL, Happel KI, Young EA, Shellito JE. Interleukin-23 (IL-23)-IL-17 cytokine axis in murine Pneumocystis carinii infection. Infect. Immun. 2007;75:3055–3061. doi: 10.1128/IAI.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 50.Schuetze N, Schoeneberger S, Mueller U, Freudenberg MA, Alber G, Straubinger RK. IL-12 family members: Differential kinetics of their TLR4-mediated induction by Salmonella enteritidis and the impact of IL-10 in bone marrow-derived macrophages. Intern. Immunol. 2005;17:649–659. doi: 10.1093/intimm/dxh247. [DOI] [PubMed] [Google Scholar]
- 51.Smits HH, Beelen AJvan, Hessle C, Westland R, Jong Ede, Soeteman E, Wold A, Wierenga EA, Kapsenberg ML. Commensal Gram-negative bacteria prime human dendritic cells for enhanced IL-23 and IL-27 expression and enhanced Th1 development. Eur. J. Immunol. 2004;34:1371–1380. doi: 10.1002/eji.200324815. [DOI] [PubMed] [Google Scholar]
- 52.Suhara W, Yoneyama M, Kitabayashi I, Fujita T. Direct involvement of CREB-binding protein/p300 in sequencespecific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 2002;277:22304–22313. doi: 10.1074/jbc.M200192200. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, Bhatia M. Epigenetic regulation of tumor necrosis factor alpha. Mol. Cell. Biol. 2007;27:5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, Powrie F. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 57.Utsugi M, Dobashi K, Ishizuka T, Kawata T, Hisada T, Shimizu Y, Ono A, Mori M. Rac1 negatively regulates lipopolysaccharide-induced IL-23 p19 expression in human macr-phages and dendritic cells and NF-kappaB p65 trans activation plays a novel role. J. Immunol. 2006;177:4550–4557. doi: 10.4049/jimmunol.177.7.4550. [DOI] [PubMed] [Google Scholar]
- 58.Vanden Eijnden S, Goriely S, Wit DDe, Goldman M, Willems F. Preferential production of the IL-12(p40)/IL-23 (p19) heterodimer by dendritic cells from human newborns. Eur. J. Immunol. 2006;36:21–26. doi: 10.1002/eji.200535467. [DOI] [PubMed] [Google Scholar]
- 59.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Wada Y, Lu R, Zhou D, Chu J, Przewloka T, Zhang S, Li L, Wu Y, Qin J, Balasubramanyam V, Barsoum J, Ono M. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood. 2007;109:1156–1164. doi: 10.1182/blood-2006-04-019398. [DOI] [PubMed] [Google Scholar]
- 61.Wahl SM, Katona IM, Stadler BM, Wilder RL, Helsel WE, Wahl LM. Isolation of human mononuclear cell subsets by counterflow centrifugal elutriation (CCE). II. Functional properties of B-lymphocyte-, T-lymphocyte-, and monocyte-enriched fractions. Cell. Imunol. 1984;85:384–395. doi: 10.1016/0008-8749(84)90252-1. [DOI] [PubMed] [Google Scholar]
- 62.Waibler Z, Kalinke U, Will J, Juan MH, Pfeilschifter JM, Radeke HH. TLR-ligand stimulated interleukin-23 subunit expression and assembly is regulated differentially in murine plasmacytoid and myeloid dendritic cells. Mol. Immunol. 2007;44:1483–1489. doi: 10.1016/j.molimm.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 64.Yang CS, Song CH, Lee JS, Jung SB, Oh JH, Park J, Kim HJ, Park JK, Paik TH, Jo EK. Intracellular network of phosphatidylinositol 3-kinase, mammalian target of the rapamycin/70 kDa ribosomal S6 kinase 1, and mitogen-activated protein kinases pathways for regulating mycobacteria-induced IL-23 expression in human macrophages. Cell. Microbiol. 2006;8:1158–1171. doi: 10.1111/j.1462-5822.2006.00699.x. [DOI] [PubMed] [Google Scholar]
- 65.Zelante T, Luca ADe, Bonifazi P, Montagnoli C, Bozza S, Moretti S, Belladonna ML, Vacca C, Conte C, Mosci P, Bistoni F, Puccetti P, Kastelein RA, Kopf M, Romani L. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur. J. Immunol. 2007;37:2695–2706. doi: 10.1002/eji.200737409. [DOI] [PubMed] [Google Scholar]
- 66.Zhou L, Nazarian AA, Xu J, Tantin D, Corcoran LM, Smale ST. An inducible enhancer required for IL-12b promoter activity in an insulated chromatin environment. Mol. Cell. Biol. 2007;27:2698–2712. doi: 10.1128/MCB.00788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]