Abstract
In this paper, we present a novel framework for microscopic image analysis of nuclei, data management, and high performance computation to support translational research involving nuclear morphometry features, molecular data, and clinical outcomes. Our image analysis pipeline consists of nuclei segmentation and feature computation facilitated by high performance computing with coordinated execution in multi-core CPUs and Graphical Processor Units (GPUs). All data derived from image analysis are managed in a spatial relational database supporting highly efficient scientific queries. We applied our image analysis workflow to 159 glioblastomas (GBM) from The Cancer Genome Atlas dataset. With integrative studies, we found statistics of four specific nuclear features were significantly associated with patient survival. Additionally, we correlated nuclear features with molecular data and found interesting results that support pathologic domain knowledge. We found that Proneural subtype GBMs had the smallest mean of nuclear Eccentricity and the largest mean of nuclear Extent, and MinorAxisLength. We also found gene expressions of stem cell marker MYC and cell proliferation maker MKI67 were correlated with nuclear features. To complement and inform pathologists of relevant diagnostic features, we queried the most representative nuclear instances from each patient population based on genetic and transcriptional classes. Our results demonstrate that specific nuclear features carry prognostic significance and associations with transcriptional and genetic classes, highlighting the potential of high throughput pathology image analysis as a complementary approach to human-based review and translational research.
Keywords: Glioblastoma, large-scale image analysis, survival analysis, phenotype-genotype integration, translational research
I. Introduction
Multi-platform molecular analyses of high throughput sequencing, gene expression, epigenetic and genetic alterations have dramatically improved our understanding of the molecular underpinnings of brain tumors. As a result, significant advances have been made in the development of cancer therapies that target specific pathways in glioblastoma (GBM), a high-grade astrocytoma with a dismal prognosis and molecularly distinct subsets [1], [2], [3], [4]. By contrast, little effort has been made on extracting tissue phenotypic information from imaging data and integrating features with clinical and molecular characterizations.
Pathologists make diagnostic and prognostic decisions primarily by visual assessments of tissue samples in histologic sections. As a result, it is rational to assume that pathology imaging data contains phenotypic information that could be measured precisely and be linked to underlying molecular profiles and clinical outcome. Thus, high-throughput pathology image analysis could serve as a valuable vehicle to assist clinical diagnosis and tissue-based research. To strengthen our ability to extract such phenotypic information embedded in tissue, we have made significant advances in large-scale pathology image analysis within the In Silico Brain Tumor Research Center (ISBTRC) at Emory University. Using mostly the emerging data from The Cancer Genome Atlas (TCGA) Project [5], we have developed novel approaches and tools to uncover fundamental aspects of GBM tumor biology by interrogating whole-slide digitized pathology images, molecular data and clinical outcome [6], [7], [8], [9], [10], [11].
In this paper, we present our large-scale pathology image analysis pipeline and infrastructure specifically developed for high throughput nuclei analysis. The overall schema of this work is illustrated in Fig. 1. We also demonstrate a methodology for identifying imaging features that have prognostic values or are related to transcriptional subtypes, gene expression, genetic alterations, and epigenetics. To enable such a large-scale image and integration analysis, we propose a database solution to model, manage, and query image analysis results in a systematic and efficient manner. We also present our High Performance Computing (HPC) paradigm that leverages a grid-computing environment consisting of machines equipped with multi-core CPUs and Graphic Processor Units (GPUs) for efficient analyses of imaging data. With our experiments, we demonstrate that large-scale microscopy image analysis of pathologic features can uncover clinically meaningful molecular correlates.
Fig. 1.
Architecture of pathology image analysis and integration framework.
II. Data Acquisition
Multiple types of data for analysis were downloaded from the TCGA portal [12] and the Memorial Sloan-Kettering Cancer Center (MSKCC) [13], including digitized microscopic images of TCGA GBM pathology slides, clinical information, and molecular data.
In this study, we used 416 whole-slide microscopic images from 159 GBM patients within the TCGA dataset. These digitized slides were Hematoxylin and Eosin (H&E) stained permanent sections of tissues that were formalin-fixed and paraffin-embedded. All slides were scanned at 20× magnification with a high-resolution, high-throughput digitized scanner. Tissue details, even at the cellular level, are visually perceivable, making it possible for application of machine-based image analyses. The size of the complete image data set for study is approximately 175 Gigabytes with JPEG compression ratio of 5.11.
Numerous aspects of clinical data from TCGA dataset were captured in multiple data files, including drug treatment, radio- and chemo-therapy, examination results, experimental protocols, demographic data, surgery information, and tissue biopsy data. We parsed out all clinical data grouped by patients, and recorded the following information for survival analysis: survival and right censored status. Molecular data, including epigenetic DNA methylation, genetic alterations (somatic mutations, and chromosome alterations), and gene expression from multiple platforms were also obtained from TCGA and MSKCC portals. Based on a recent study of TCGA GBMs, four clinically relevant transcriptional subtypes have been defined based on unsupervised hierarchical clustering of gene expression, namely, Proneural (PN), Neural (NR), Classical (CL) and Mesenchymal (MS) [4]. Each subtype is defined by a characteristic gene expression profile, but also has characteristic genetic alterations, including mutations and chromosomal changes (amplification/deletion). For our study, transcriptional subtypes were obtained from the supplementary information of the work in [4]. Subtypes for samples absent from this set were determined with Prediction Analysis of Microarray (PAM) software version 2.21 using RMA normalized Affymetrix HT-HGU133 mRNA expression platform data. A sample expression average was computed for samples with multiple corresponding arrays. Unlogged expression was filtered to remove probes with a fold change less than 1.5 or an expression range less than 20. Analyzed somatic mutation data for gene IDH1, PTEN, TP53, and copy number data for gene CDK4, EGFR, MDM2, NF1, and PDGFRA were obtained from MSKCC. Moreover, analysis of the DNA sequencing data from TCGA GBM samples has led to identification of a CpG island methylator phenotype (G-CIMP) that is associated almost exclusively with PN-subtype cases and secondary GBMs with IDH1 mutantation [14]. Following the analysis procedure in [14], we obtained a set of G-CIMP GBMs. In addition, we downloaded TCGA mRNA expressions from RNA sequencing data via the MSKCC portal.
III. Nuclei Analysis with GBM Pathology Images
Of the large number of potential pathologic features in GBM, we focussed our analysis on individual tumor nuclei as they are the dominant feature, carry important clinical information, and are critical in the morphologic diagnosis of numerous diseases.
Segmentation of all nuclei within the digital slides is the first step in the nuclear analysis. We started segmentation with a recognition module for non-tissue or red blood cell regions. The percentage of area occupied either by blank spaces or red blood cells as indicated by color was computed to determine whether a given image region contains sufficient neoplastic tissue for analysis. With a priori knowledge on cell histology, nuclei are known as compact, round-to-oval, and regular-shaped objects with dark color on H&E stains. However, nuclei identification still presents serious challenges in that many other histological structures and artifacts in microscopy images can appear similar to nuclei. To remedy this problem, we need to reduce noise to an acceptable level and enhance nuclei contrast. Meanwhile, it is readily noticeable in microscopy images that nuclei, even for those in close proximity to each other, may have variable intensities or colors resulting from a large number of factors, ranging from variations in tissue section thickness to heterogeneous tissue responses to chemical stains. As a consequence, no single cutoff is available to identify nuclei regions from their surrounding areas.
One effective solution to this challenge is to normalize image background using the morphological reconstruction [15], [10], a shape-based mathematical morphology operation widely used in image processing. Morphological reconstruction is essentially a composition of a series of morphological modules tightly coupled with the concept of connectivity [16]. With this technique, true foreground objects, i.e. nuclei in our study, can be uncovered from image background severely corrupted by noise signals through local image “normalization”.
Two image morphological components, namely marker Φ and mask Ψ image, are involved in a morphological reconstruction operation, which can be expressed as follows:
| (1) |
where is a function recursively defined as:
| (2) |
In (1), n* is the smallest positive number such that , and ρ represents the structural element with which marker image Φ is recursively dilated. In addition, ‘⊕’ represents a fundamental morphological operation, known as dilation. With this morphological operation, the state of any given pixel in the output image is determined by applying the “max” rule to the corresponding pixel and its neighbors in the input image. To be more specific, dilation for either binary or grayscale image can be defined as follows:
| (3) |
where I(x, y) is the input image, and ρ(x, y) represents the neighborhood region of the structural element centered at pixel (x, y). Users can specify the size and shape of neighborhood region within the structural element to capture objects with specific shapes and scales in image I(x, y).
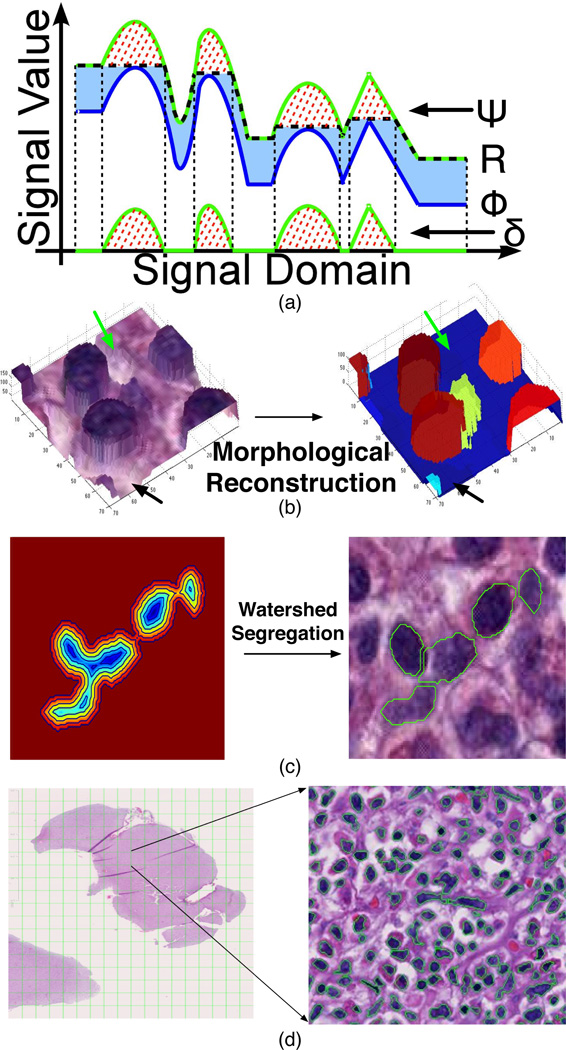
For better presentation, an illustration visualizing a typical reconstruction process on a one-dimensional signal is given in Fig. 2a, where the marker and mask signal are represented in blue and green curves, respectively. Over the iterations of the reconstruction operation in (2), the marker gradually arises across the light blue area and converges to the final reconstructed signal represented by the black dashed curve. It is notable that the reconstructed signal only differs from the mask at places where peaks reside. As a result, the merit of “normalizing” the background variation with morphological reconstruction plays a critical role in mitigating severe noise and artifacts that make nuclei identification a challenging problem. In Fig. 2b, we present a three-dimensional surface of a small image region before (left) and after (right) the application of morphological reconstruction operation. When subtracting the reconstructed image from the mask image Ψ, the difference image (Fig. 2b right) consists of a near zero-level background, and a group of enhanced foreground peaks, each representing an object of interest. Bumpy areas in background (green and black arrows) in Fig. 2b (left) are flattened in the difference image after the morphological reconstruction in Fig. 2b (right), therefore improving the contrast between the background and foreground objects.
Fig. 2.
Nuclei segmentation modules. (a) One dimensional illustration of the morphological reconstruction process with a marker signal (blue) recursively dilated and suppressed with the mask (green) until no change occurs between two successive iterations. (b) Three dimensional view of the effect of the morphological reconstruction. (c) Clumped nuclei are segregated with watershed segmentation. (d) A segmented image region is shown.
To conduct the morphological reconstruction operation, we converted the color image to a gray level image by complementing its first color channel. This is simply supported by the domain knowledge that nuclei tend to have lower intensities as compared with those of background regions due to the Haematoxylin stain and by the fact that the first color channel yields the best contrast between nuclear and non-nuclear regions after careful tests. As morphological reconstruction operation can readily normalize background regions degraded by artifacts arising from tissue preparation and the scanning process, it enables us to separate the foreground objects from the normalized background with a user-provided threshold.
As a pathology image is a 2-D representation of a 3-D tissue biopsy, it is common to see a large number of clumped nuclei in digital slides. It can be challenging to assign nuclear boundaries to a specific group of overlapped nuclei, as these are often ambiguous in microscopy images. An effective way to solve this problem is to think of a set of overlapped nuclei as a group of basins in the image domain, where the ridges between basins are the borders that isolate nuclei from each other. This is exactly the idea behind the watershed algorithm [17]. In our application, we computed the distance transformation of the binary mask image Γ(x, y) as follows:
| (4) |
where ∂Γ represents the boundary of a foreground object. Watershed algorithm is subsequently applied to the distance map D(x, y) where ridges between pairs of connected nuclear regions are detected as separating boundaries [18], [19]. In Fig. 2c, we present an example where the nuclear boundaries of a group of connected nuclei are determined with the watershed method. The distance transformation map superimposed with iso-contours is illustrated on the left. Ridges (in green) detected by watershed algorithm over distance transformation map are superimposed on the original color image in Fig. 2c (right).
After the aforementioned steps, some false positive nuclei, usually small in size and irregular in shape, could be retained in image domain. Therefore, we removed these false positive objects that were either too small or too irregular to be nuclei with size and shape filters. The final segmentation result on a typical image region is shown in Fig. 2d.
After segmentation, a diverse set of features was computed to characterize segmented nuclei. To fully describe distinct nuclear features from complementary perspectives, we computed 23 features from four categories for each segmented nucleus, namely nuclear morphometry, region texture, image intensity and gradient statistics, as shown in Table I. In the nuclear morphometry feature class, the degree of elongation, size, and regularity of nuclei are characterized, as nuclear morphology has proved to correlate with the expression of oligodendrocyte specific genes, and carry discriminating information between astrocytoma and oligodendroglioma nuclei [11]. Nuclear texture information is also captured with multiple texture descriptors, as nuclei of distinct categories exhibit variable degrees of uneven staining that result from heterogeneous clumping of chromatin. Features relevant to cytological intensity and degree of inhomogeneity as depicted by gradient are included in the feature suite as well.
TABLE I.
NUCLEAR FEATURES.
| Feature Group | Feature Names |
|---|---|
| Nuclear Morphometry | Area, Perimeter, Eccentricity, Circularity, MajorAxisLength, MinorAxisLength, Extent |
| Texture | Entropy, Energy, Skewness, Kurtosis |
| Intensity Statistics | AvgIntensity, StdIntensity, MaxIntensity, MinIntensity |
| Gradient Statistics | AvgGradientMagnitude, StdGradMag, Enropy-GradMag, EnergyGradMag, SkewnessGradMag, KurtosisGradMag, Sum of Canny Edge Pixels, Mean of Canny Edge Pixels |
IV. Large-scale Pathology Image Representation, Management, and Queries
With recent advance in technology, modern slide scanners can now efficiently produce high-resolution images of whole tissue slides. With systematic whole slide image analysis, a vast amount of morphological information can be extracted from various biological scales. However, the large scale of derived data and the complexity of spatial data correlation pose a major challenge to data management infrastructures appropriate for managing and querying analytical results and annotations of whole slide images.
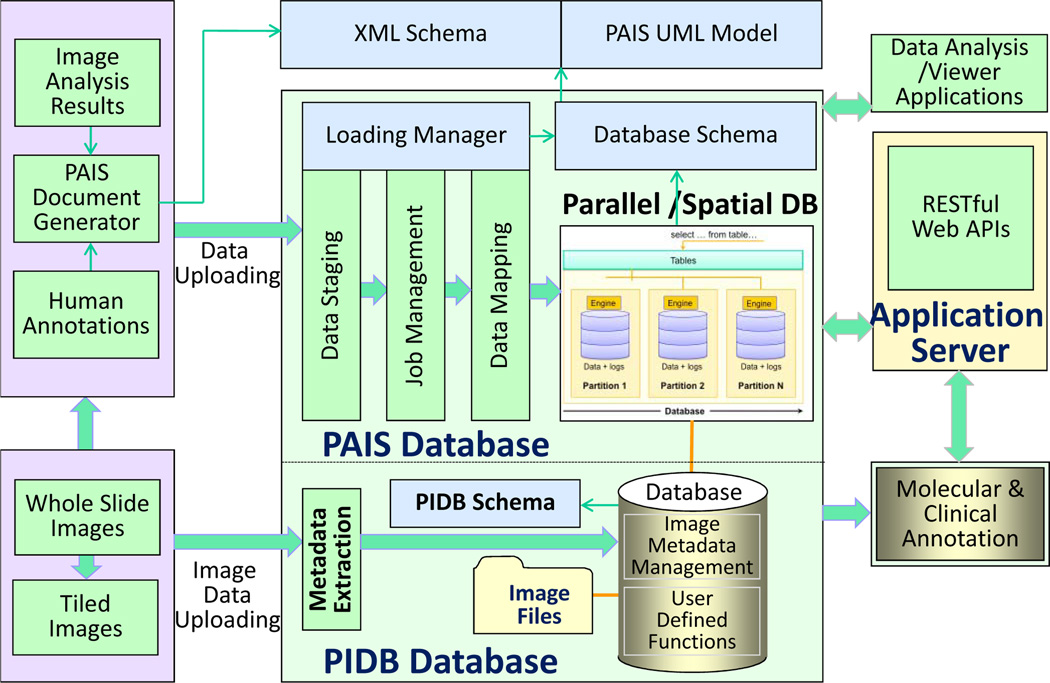
To remedy this problem, we have developed the Pathology Analytical and Imaging Standards (PAIS) model [20] to provide a flexible, efficient, and semantically enabled data model for pathology image analysis and characterization. The data model represents virtual slide related image reference, annotation, markup and feature information. This set of information includes 1) context relating to patient data, specimen preparation, special stains, among others; 2) image reference information that describes an image or a group of images used as the base for markups and annotations, including resolutions, regions; 3) human observations involving pathology characteristics; and 4) algorithm and human-described segmentations, features and classifications. Moreover, it supports the provenance of the markups and annotations through a description of the computation and an identification of input and output datasets. The logical model of PAIS consists of 62 classes and associations across them. As information from pathology images is spatially related, PAIS provides a Markup class to delineate a spatial region in images and represents a set of values derived from pixels, which can be in the form of geometric shapes, surfaces, and fields. Modeling such markup information provides the foundation for supporting powerful spatial queries. We have also developed an XML based schema for representing and exchanging PAIS data as PAIS XML documents. To reduce the storage size of PAIS documents, we compress PAIS documents as zip files by default.
Based on the data model, we have designed and implemented a relational database infrastructure with spatial database extension to manage image analysis results and human annotations (Fig. 3). The architecture includes components for result validation, data representation, data uploading and mapping, data management and queries, parallel database component, Web APIs for queries and application integration, and integration with pathology image data management component (PIDB database) and relevant clinical and molecular annotations. The PAIS data management component encapsulates the database, the data loading and query subcomponents. We have used IBM DB2 with Spatial Extender [21] in our implementation. The spatial extension provides support of spatial tables for managing geometric shapes, and the Structure Query Language (SQL) for writing spatial queries. Efficient spatial query support is through a spatial index engine, i.e., a grid based index in DB2 by partitioning space into many small grid cells [20], [21]. The database is designed to support queries on both metadata and spatial features for data retrieval, comparative data analysis, and algorithm validation. The query types include: 1) queries involving combinations of image and algorithm metadata to retrieve analysis results; 2) queries involving combinations of image and algorithm metadata to retrieve analysis results; 3) spatial queries, such as those used to assess relative prevalence of features or classified objects in slides or to assess spatial coincidence of combinations of features or objects.
Fig. 3.
Pathology analytical imaging data management architecture.
V. High Performance Computing for Large-scale Pathology Image Analysis
To accommodate the massive data and computational demands of image analysis algorithms, we employed high-performance techniques to accelerate our image analysis procedure with the large TCGA image dataset. The high-performance version of our application has been built targeting modern hybrid computing systems equipped with multi-core CPUs and graphics processing units (GPUs). The computation power of GPUs has been rapidly increased in the last few years. Contemporary GPUs provide very fast memories and massive multi-processing capabilities, which typically exceed those of CPUs. As a consequence, GPUs have been successfully used as general purpose processing devices by many applications [22]. The efficient use of machines equipped with CPUs and GPUs, however, is very challenging. A programmer needs to consider the distribution of work not only across cluster nodes in a distributed memory machine, but also among processors (CPU cores and GPUs) within each machine. Additionally, the acceleration attained by different application operations ported to GPUs may vary according to their suitability for parallel execution. This variability should also be taken into account during the scheduling of operations for execution with a CPU or GPU in order to achieve maximum performance.
The overall parallelization strategy we followed is organized into three steps: (i) efficient implementation of application individual operations for CPUs and GPUs; (ii) multinode parallelization of the application targeting distributed memory systems; and, (iii) coordination of the execution within each of the hybrid nodes, which includes performance aware assignment of operations to CPUs and GPUs available in a machine. To have an efficient implementation of the individual operations to CPUs and GPUs, we tried as much as possible to reuse codes from other research groups or publicly available libraries. For those operations with no preexisting efficient implementation, however, we have created our own implementation, which uses C++ for CPU-based operations or CUDA (Computing Unified Device Architecture) for GPU-based implementations. In our implementations, we have identified the common computation patterns used by the operations, which were further ported to GPUs and used as building blocks for implementing the required operations [23], [24], [25].
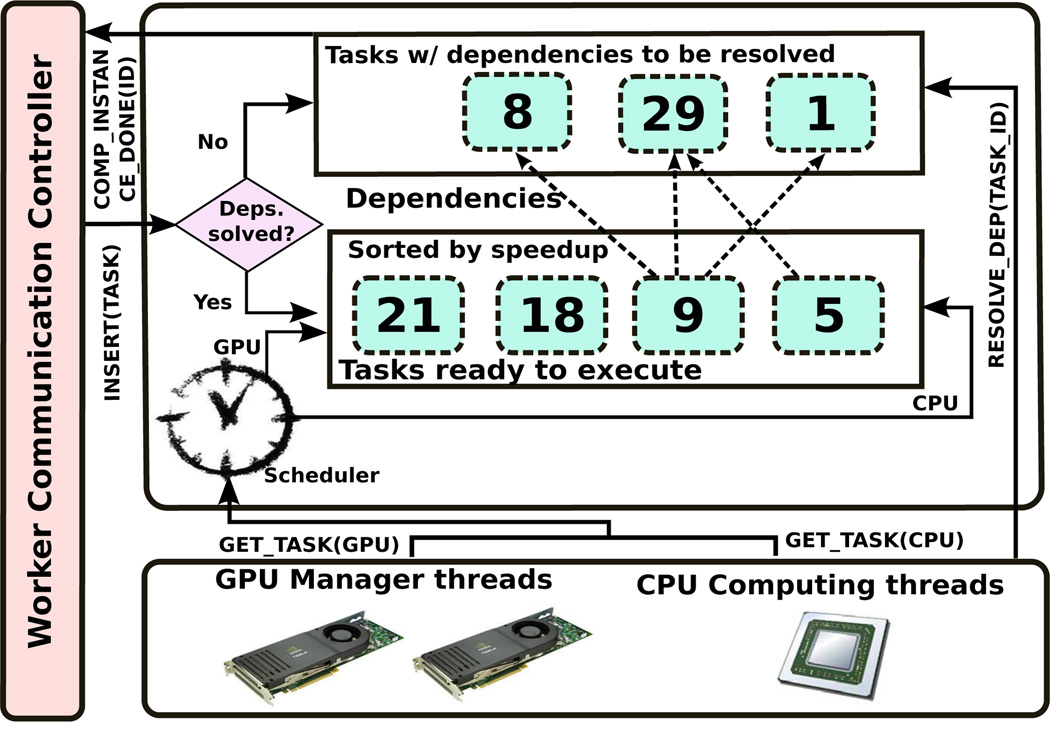
The multi-node parallelization approach employed combines the coarse-grain dataflow pattern with the bag-of-tasks pattern in order to facilitate the implementation of the analysis application from a set of operations on data. In further detail, the application is described as a pipeline with “Nuclei Segmentation” and “Feature Extraction” as the coarse-grain stages to be applied to each of the input image sub-regions. These coarse-grain computation stages are assigned to nodes of a distributed memory machine in a demand-driven basis as their dataflow dependencies are satisfied. Each of the coarse-grain stages could then be described as another dataflow of fine-grain operation, which can be executed using either CPU or GPU. This hierarchical dataflow representation lends itself to a separation of concerns and enables the use of different scheduling approaches at each level. In order to maximize the performance in hybrid machines, the scheduling decision on the assignment of fine-grain operations to CPUs and GPUs is made upon whether operations are expected to have accelerations on a GPU. In our scheduling, fine-grain operations are maintained into a sorted list by the expected GPU acceleration (speedup), and those operations with highest expected speedups are assigned to GPUs, while CPUs execute operations with lowest expected speedups. Fig. 4 presents an overview of the environment that coordinates the execution in each machine.
Fig. 4.
An overview of the parallel job execution environment.
VI. Clinical and Molecular Correlates of Nuclear Features Derived from Microscopy Image Analysis
Microscopic features of cancer, such as tumor cell morphology, are measurable and have biologic, diagnostic and therapeutic significance. In our study, the derived nuclear features of hundreds of millions of nuclei from 416 whole-slide pathology images processed by image analysis algorithms with HPC and parallel computation infrastructure support were used to correlate with clinical and molecular data. As there are millions of nuclei in slides of each patient, we calculated mean and standard deviation of each nuclear features from each patient. Therefore, each patient is represented by 46 nuclear feature statistics. Multiple integrative experiments were conducted to explore the potential prognostic values and molecular links embedded in nuclear features.
A. Nuclear Features with Prognostic Significance
We first interrogated clinical correlates of nuclear features by regularized least-squares regression using Least Absolute Shrinkage and Selection Operator (LASSO) [26] and Cox proportional hazards regression [27]. LASSO is a quadratic programming problem penalizing the number of regression coefficients:
| (5) |
where yi is the survival, xi is the nuclear feature vector, β is the coefficient vector, and λ is a nonnegative regularization parameter. As we increase λ, we force more predictor coefficients to zero. In our experiment, we set it to 0.58, excluding only highly correlated nuclear features while retaining fitting Mean Squared Error (MSE) low.
After exclusion of highly confounded nuclear feature statistics in LASSO screening process, we next used Cox proportional hazards regression method to identify those features with prognostic value. The Cox proportional hazard model can be described as:
| (6) |
where hazard H(t) is the multiplication of a baseline hazard H0(t) and an exponential factor determined by the sum of a set of nuclear features x weighted by coefficients b.
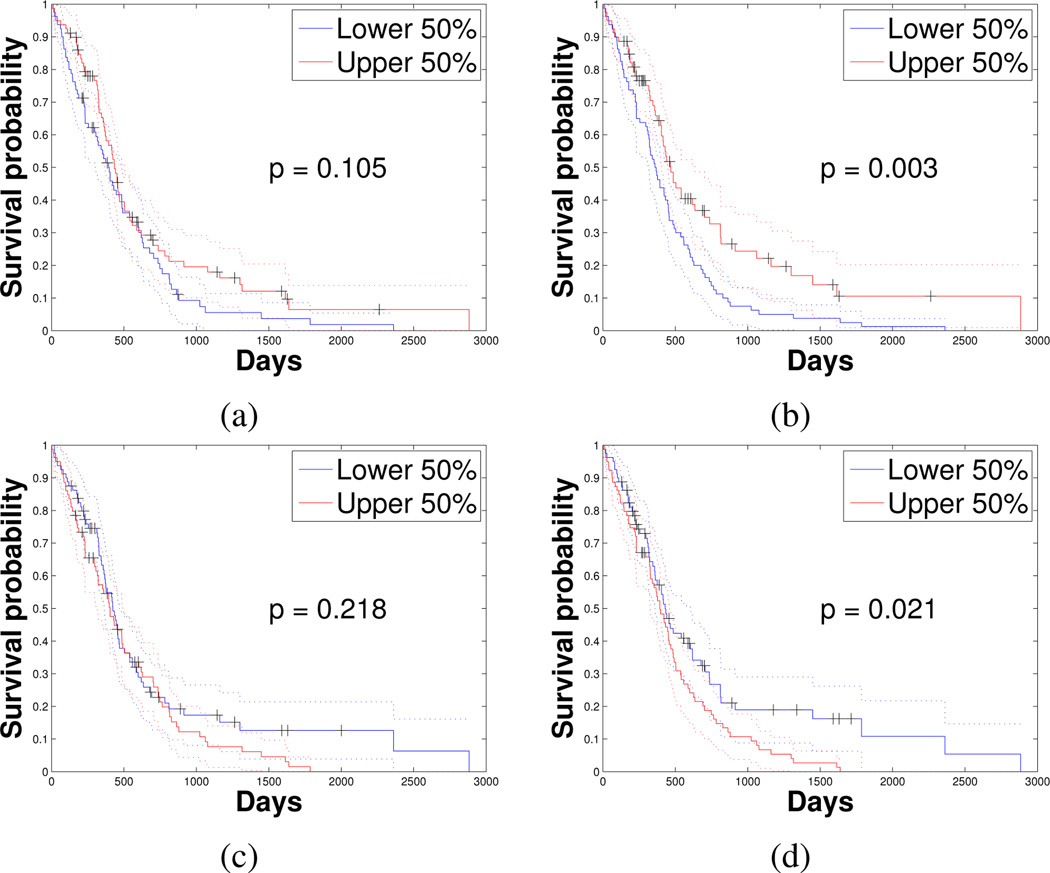
We found statistics of four specific nuclear features that were strongly correlated with patient survival, as summarized in Table II. Interestingly, the mean of nuclear Circularity was positively correlated with survival. This could be related to the fact that Circularity reflects an oligodendroglioma population (or a degree of oligodendroglioma differentiation) within the GBM, which is composed mostly of tumor cells with astrocytic differentiation. Nuclei of oligodendroglioma are rounder and smaller with relatively uniform nuclear textures, in contrast to astrocytoma nuclei. Clinically, oligodendroglioma morphology presence is associated with prolonged survival as compared to astrocytoma typically enriched with elongated and irregular nuclei. As a result, GBMs with a mixed morphology have a better prognosis than pure astrocytic neoplasms. While taking an average operation could reduce the true signal strength substantially, we were still able to demonstrate the significant role of nuclear Circularity as an important phenotypic feature on predicting survival, suggesting a strong correlation between morphometry features and survival. To further demonstrate the clinical correlates of nuclear features, we illustrate in Fig. 5 the survival difference between two equally numbered patient groups divided by the four nuclear feature statistics (the upper and lower 50%). The associated p-values with logrank test [28] were 1.05e-1 for mean of Circularity, 2.98e-3 for mean of MinIntensity, 2.18e-1 for standard deviation of MajorAxisLength, and 2.08e-2 for standard deviation of MeanIntensity. Although mean of Circularity and standard deviation of MajorAxisLength do not reach significance on survival analysis in the log-rank test, the Cox proportional hazards model agrees with all four Kaplan-Meier plots in Fig. 5, where the upper 50% of patients identified by nuclear features with negative Cox proportional hazards regression coefficients (i.e. means of Circularity and MinIntensity) have more favorable survival than the lower 50% (Fig. 5a and b). For standard deviations of MajorAxisLength and MeanIntensity (Fig. 5c and d), the upper 50% of patients have shorter survival than the lower 50% as these two features have positive Cox proportional hazards regression coefficients.
TABLE II.
STATISTICS OF NUCLEAR FEATURES WITH PROGNOSTIC SIGNIFICANCE IDENTIFIED FROM COX PROPORTIONAL HAZARD REGRESSION.
| Feature | Statistics | Regression Coeff. (b) | p value |
|---|---|---|---|
| Circularity | Mean | −41.43 | 1.34e-2 |
| MinIntensity | Mean | −24.69 | 2.32e-2 |
| MajorAxisLength | Standard Deviation | 3.29 | 3.80e-2 |
| MeanIntensity | Standard Deviation | 156.21 | 4.88e-2 |
Fig. 5.
Kaplan-Meier plot of upper and lower 50% of GBMs divided by (a) mean of nuclear Circularity; (b) mean of MinIntensity features; (c) standard deviation of MajorAxisLength; and (d) standard deviation of MeanIntensity.
B. Correlation of Nuclear Features with Molecular Data
We next investigated whether nuclear features were correlated with GBM molecular signatures. For patients divided by each type of molecular data, we carried out one-way analysis of variation (ANOVA) to test if group means of nuclear features were significantly distinct from each other [29]. For those nuclear feature means significantly different across groups, we further applied a multiple comparison procedure to identify which specific pairs of means were significantly different [30]. There are numerous ways to partition patients in terms of molecular signature affinity. We present molecular criteria for patient stratification and the associated findings as follows.
Four transcriptional subtypes of GBM have been recognized: Proneural (PN), Neural (NR), Classical (CL) and Mesenchymal (MS). Results: Eccentricity mean in PN is lower than that in MS and CL (p=3.81e-4). MinorAxisLength mean in PN is greater than that in MS and CL (p=8.87e-3). Extent mean in PN is greater than that in MS (p=3.20e-2). These results are most compatible with PN GBMs having rounder nuclei than those of the other transcriptional classes.
GBM CpG island methylator phenotype (G-CIMP). Results: Standard deviations of Area, Perimeter, MinorAxisLength, and means of Entropy and MeanGradMag of G-CIMP group are greater than those of non-G-CIMP group (p=9.78e-3, 7.36e-3, 2.71e-3, 1.63e-4, and 9.60e-3, respectively). Mean of Energy in G-CIMP GBM nuclei is less than that in non-G-CIMP group (p=2.28e-5). G-CIMP GBMs appear to have more nuclear variability in size than the non-G-CIMP tumors.
Chromosome 1p/19q co-deletion status. Results: Means of Circularity and Extent of 1p/19q deleted GBMs are greater than those of 1p/19q intact GBMs (p=1.16e-2 and 1.92e-2, respectively). Means of Eccentricity and MajorAxisLength of the 1p/19q deleted GBM group are less than those of the 1p/19q intact group (p=3.89e-2 and 1.44e-2, respectively). Thus, nuclei of 1p/19q deleted GBMs are rounder and smaller than GBMs with 1p/19q intact.
Somatic mutation status for signature genes, including gene IDH1, PTEN and TP53. Results: Standard deviations of MinorAxisLength and means of Entropy and MeanGradMag in IDH1 mutant GBMs are greater than those in the wild type (p=2.40e-2, 6.46e-3, and 4.44e-2, respectively). Mean of Energy in IDH1 mutant tumors is less than that in the wild type (p=2.75e-3). Means of Circularity and Extent in PTEN mutant GBMs are less than those in the wild type (p=9.68e-3 and 1.76e-2, respectively). In TP53 mutant tumors, the mean of Extent is less than that of wild type tumors (p=3.77e-2). Thus, PTEN and TP53 mutations are likely associated with nuclei that are less regular and round.
Signature gene copy number data, including gene CDK4, EGFR, MDM2, NF1, and PDGFRA. Results: Standard deviation of Area and MinorAxisLength and mean of Perimeter in CDK4 amplification group are greater than those in the wild type group (p=3.27e-2, 1.29e-2, and 4.78e-2, respectively). Means of Area, Perimeter, and MinorAxisLength in MDM2 amplified GBMs are greater than those in the wild type group (p=3.43e-2, 2.93e-2, and 1.42e-2, respectively). Means of Eccentricity and MeanCanny in EGFR amplified tumors are greater than those in EGFR wild type tumors (p=4.38e-2 and 4.90e-2). PDGFRA amplified tumors have lower mean of Eccentricity (p=2.31e-2) and higher mean of Extent (p=3.19e-2) than PDGFRA wild type tumors.
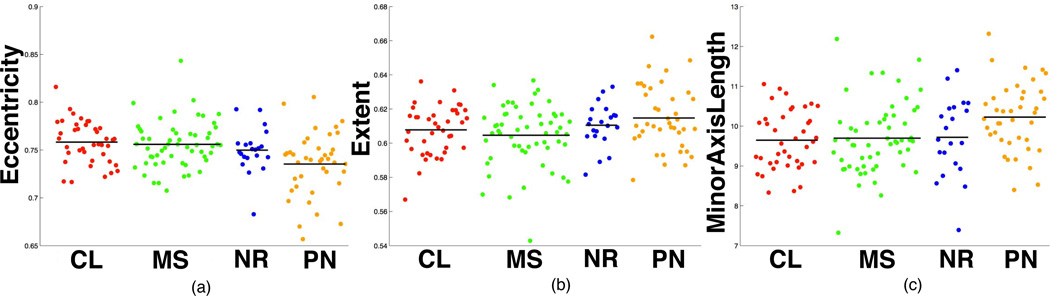
These quantitative results conform to neuropathologists subjective descriptions regarding the relationship between nuclear morphometry and molecular signatures. For example, GBMs in PN subtype are enriched in cases with PDGFRA amplification, which has been associated with a greater oligodendroglioma morphology (more nuclear regularity and roundness). In our experiments, we found that GBMs of PN subtype had the smallest mean of nuclear Eccentricity and the largest mean of nuclear Extent, and MinorAxisLength (Fig. 6), all supporting the typical nuclear features of oligodendroglial differentiation.
Fig. 6.
Scatter plots of nuclear feature measures for four transcriptional subtypes of GBMs. Nuclear features include mean of (a) Eccentricity; (b) Extent; and (c) MinorAxisLength.
C. Gene Expression Correlates of Nuclear Features
To explore the potential biological links between morphology and molecular data, we next correlated nuclear features with gene expression related to specific GBM properties: 1) stem cell markers, SOX2, MYC, and NANOG; 2) the glial differentiation marker GFAP; 3) the hypoxia markers carbonic anhydrase IX (CAIX) and VEGFA; and 4) the cell proliferation markers PCNA and MKI67. We correlated mRNA expression of these signature genes with nuclear feature statistics by the Spearmans rank correlation method [31]. A heat map of the correlation result is shown in Fig. 7, where we find that 1) MYC gene expression is negatively correlated with the mean of nuclear Circularity (ρ = −0.306); and 2) gene expression of MKI67 is positively correlated with the means of nuclear Perimeter and MajorAxisLength (ρ = 0.388 and 0.369) and negatively correlated with Circularity and Extent (ρ = −0.525 and −0.355, respectively). These results suggest that MYC expression is lower in cells with round nuclei and that cell proliferation is greater in GBMs with elongated and larger nuclei, typical of more anaplastic cells with pure astrocytic differentiation.
Fig. 7.
Heat map of Spearmans Rank correlation coefficients of nuclear feature statistics (y-axis) and biologically relevant gene expression (x-axis).
D. Identification of Representative Nuclei
To inform human-based pathologic review, we found representative nuclear instances from GBMs of specific molecular classes. In this manner, we could establish a nuclear morphometry reference library to assist neuropathologists to standardize nuclear instances associated with specific molecular signatures. In our experiments, we grouped patients based on transcriptional subtypes, 1p/19q co-deletion status, and genetic alterations. With each molecular criterion for patient stratification, we carried out one-way analysis of variation (ANOVA) to test if group means of nuclear features are significantly distinct. For each nuclear feature with a significant difference across patient groups, we computed its mean and standard deviation within each group and queried for the most typical nuclear instances with feature values of μ − 3σ, μ − 2σ, μ − σ, μ, μ + σ, μ + 2σ, and μ + 3σ along the feature distribution. For example, we present reference nuclei examples from the four transcriptional subtypes of TCGA GBMs in Fig. 8. This analysis demonstrates the potential to develop a large-scale quantitative image analysis workflow, high performance computing infrastructure, database solution for large-scale pathology imaging information and multidimensional data integration platform capable of informing human-based pathologic review.
Fig. 8.
Spectrum of representative nuclei from the nuclear reference library for each of four transcriptional subclasses of GBM. Nuclei are queried based on the location along the morphologic distribution of four nuclear features.
VII. Conclusion
In this paper, we present a new framework for computer-based microscopy image analysis and large-scale integrative research, enabled by a pathology analytical imaging database (PAIS) and high performance computing using multi-core CPUs and Graphical Processor Units (GPUs). We applied our image analysis workflow to whole slide pathology images of glioblastomas from The Cancer Genome Atlas dataset. With integrative studies, we found statistics of four specific nuclear features were significantly associated with patient survival. We correlated nuclear features with molecular data and found interesting results that support pathologic domain knowledge. Additionally, gene expressions of stem cell marker MYC and cell proliferation maker MKI67 were found correlated with nuclear features. To inform pathologists, we also queried the most representative nuclear instances from each patient population determined by molecular classes. These results suggest that the developed framework holds promise for building an integrative model supported by the high throughput nuclear morphology analysis pipeline that can complement human-based pathologic review and improve ongoing large-scale translational research.
Contributor Information
Jun Kong, Email: jun.kong@emory.edu.
Fusheng Wang, Email: fusheng.wang@emory.edu.
George Teodoro, Email: george.teodoro@emory.edu, teodoro@gatech.edu.
Lee Cooper, Email: lee.cooper@emory.edu.
Carlos S. Moreno, Email: cmoreno@emory.edu.
Tahsin Kurc, Email: tkurc@emory.edu.
Tony Pan, Email: tony.pan@emory.edu.
Joel Saltz, Email: jhsaltz@emory.edu.
Daniel Brat, Email: dbrat@emory.edu.
References
- 1.Brat D, Perry A. Practical surgical neuropathology: A diagnostic approach. Philadelphia: Churchill-Livingstone; 2010. [Google Scholar]
- 2.Kleihues P, Cavenee W. World Health Organization Classification of Tumours. 2nd ed. Lyon, France: IARC Press; 2000. Pathology and generics of tumours of the nervous system. [Google Scholar]
- 3.Kolles H, Niedermayer I, Feiden W. Grading of astrocytomas and oligodendrogliomas. Pathologe. 1998;19(4):259–268. doi: 10.1007/s002920050282. [DOI] [PubMed] [Google Scholar]
- 4.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TCGA workgroup. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong J, Cooper L, Wang FS, Gutman D, Gao JJ, Chisolm C, Sharma A, Pan T, Van Meir EG, Kurc T, Moreno C, Saltz J, Brat DJ. Integrative, Multi-modal Analysis of Glioblastoma Using TCGA Molecular Data, Pathology Images and Clinical Outcomes. IEEE Trans. on Biomed. Eng. 2011 Dec;58(12):3469–3474. doi: 10.1109/TBME.2011.2169256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper L, Kong J, et al. An Integrative Approach for In Silico Glioma Research. IEEE Trans. on Biomed. Eng. 2010;57(10):2617–2621. doi: 10.1109/TBME.2010.2060338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong J, Cooper L, Moreno C, Wang F, Kurc T, Saltz J, Brat D. In Silico Analysis of Nuclei in Glioblastoma using Large-scale Microscopy Images Improves Prediction of Treatment Response; Int. Conf. of the IEEE Eng. in Med. and Bio. Society; 2011. pp. 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper L, Kong J, Gutman D, Wang FS, Gao JJ, Appin C, Cholleti S, Pan T, Sharma A, Scarpace L, Mikkelsen T, Kurc T, Moreno C, Brat D, Saltz J. Integrated morphologic analysis for the identification and characterization of disease subtypes. Am Med Inform Assoc. 2012;19:317–323. doi: 10.1136/amiajnl-2011-000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong J, Cooper L, Kurc T, Brat D, Saltz J. Towards Building Computerized Image Analysis Framework for Nucleus Discrimination in Microscopy Images of Diffuse Glioma; Int. Conf. of the IEEE Eng. in Med. and Bio. Society; 2011. pp. 6605–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong J, Cooper L, Wang F, Gao J, Teodoro G, Scarpace L, Mikkelsen T, Moreno C, Saltz J, Brat D. A novel paradigm for determining molecular correlates of tumor cell morphology in human glioma whole slide images. Scientific Meeting and Education Day of the Society for Neuro-Oncology. 2013 [Google Scholar]
- 12.NCI. The Cancer Genome Atlas. 2001 http://cancergenome.nih.gov/
- 13.Memorial Sloan-Kettering Cancer Center. http://www.mskcc.org/mskcc/ [Google Scholar]
- 14.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent L. Morphological Grayscale Reconstruction in Image Analysis: Applications and Efficient Algorithms. IEEE Transactions on Image Processing. 1993;2(2):176–201. doi: 10.1109/83.217222. [DOI] [PubMed] [Google Scholar]
- 16.Robinson K, Whelan PF. Efficient Morphological Reconstruction: A Downhill Filter. Pattern Recognition Letters. 2004;25(15):1759–1767. [Google Scholar]
- 17.Fernand M. Topographic distance and watershed lines. Signal Processing. 1994;38:113–125. [Google Scholar]
- 18.Heinz B, Gil J, Kirkpatrick D, Werman M. Linear Time Euclidean Distance Transform Algorithms. IEEE Trans. on Pattern Analysis and Machine Intelligence. 1995;17(5):529–533. [Google Scholar]
- 19.Roerdink J, Meijster A. The watershed transform: definitions, algorithms, and parallelization strategies. Fundamenta Informaticae. 2000;41:187–228. [Google Scholar]
- 20.Wang F, Kong Jun, Cooper L, et al. A Data Model and Database for High-resolution Pathology Analytical Image Informatics. J. of Pathology Informatics. 2011;2(32) doi: 10.4103/2153-3539.83192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.IBM DB2 Spatial. Available from: http://www-01.ibm.com/software/data/spatial/ [Google Scholar]
- 22.NVIDIA. GPU Accelerated Applications. 2013 http://www.nvidia.com/object/gpu-accelerated-applications.html. [Google Scholar]
- 23.Teodoro G, Pan T, Kurc TM, Kong J, Cooper LA, Podhorszki N, Klasky S, Saltz J. High-throughput Analysis of Large Microscopy Image Datasets on CPU-GPU Cluster Platforms; IEEE Int’l Parallel and Distributed Processing Symposium; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teodoro G, Kurc T, Pan T, Cooper L, Kong J, Widener P, Saltz J. Accelerating Large Scale Image Analyses on Parallel, CPU-GPU Equipped Systems; IEEE Int’l Parallel and Distributed Processing Symposium; 2012. pp. 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teodoro G, Pan T, Kurc T, Kong J, Cooper L, Saltz J. Efficient Irregular Wavefront Propagation Algorithms on Hybrid CPU-GPU Ma- chines. Parallel Computing. 2013;39(4–5):189–211. doi: 10.1016/j.parco.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software. 2010;33(1) [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen E. Multivariate survival analysis using Cox’s regression model. Hepatology. 1987;7:1346–1358. doi: 10.1002/hep.1840070628. [DOI] [PubMed] [Google Scholar]
- 28.David H. Linear Rank Tests in Survival Analysis: Encyclopedia of Biostatistics. Wiley Interscience; 2005. [Google Scholar]
- 29.Moore DS, McCabe GP. Introduction to the Practice of Statistics. 4th ed. W H Freeman & Co.; 2003. [Google Scholar]
- 30.Milliken GA, Johnson DE. Analysis of Messy Data, Volume 1: Designed Experiments. Boca Raton, FL: Chapman & Hall/CRC Press; 1992. [Google Scholar]
- 31.Corder GW, Foreman DI. Nonparametric Statistics for Non-Statisticians: A Step-by-Step Approach. Hoboken, NJ: Wiley; 2009. [Google Scholar]