SUMMARY
The hippocampus, as part of the cerebral cortex, is essential for memory formation and spatial navigation. Although it has been extensively studied, especially as a model system for neurophysiology, the cellular processes involved in constructing and organizing the hippocampus remain largely unclear. Here, we show that clonally related excitatory neurons in the developing hippocampus are progressively organized into discrete horizontal, but not vertical, clusters in the stratum pyramidale, as revealed by both cell type-specific retroviral labeling and mosaic analysis with double markers (MADM). Moreover, distinct from those in the neocortex, sister excitatory neurons in Cornu Ammonis 1 region of the hippocampus rarely develop electrical or chemical synapses with each other. Instead, they preferentially receive common synaptic input from nearby fast-spiking (FS), but not non-FS, interneurons and exhibit synchronous synaptic activity. These results suggest that shared inhibitory input may specify horizontally clustered sister excitatory neurons as functional units in the hippocampus.
INTRODUCTION
The hippocampus together with the neocortex comprises most of the cerebral cortex. Arising from the dorsal telencephalon or the pallium, the hippocampus and the neocortex become anatomically distinct parts of the cortex. The neocortex consists of six layers of neurons, with excitatory neurons occupying layers II to VI. In contrast, the hippocampus contains mostly a single layer with densely packed pyramidal neurons – the stratum pyramidale – that is divided into two major regions, Cornu Ammonis 1 (CA1) and CA3, and a small transitional region, CA2. The CA regions are capped by the dentate gyrus (DG) (Nauta and Feirtag, 1986). As the most inferior part of the hippocampal formation, the subiculum connects CA1 with the entorhinal and other cortices.
Besides their structural differences, the circuit organization of the hippocampus and the neocortex are also distinct. The thalamus relays incoming sensory input into the neocortex and mainly targets layer IV neurons, which project up to the superficial layer II/III neurons. Layer II/III neurons project down to the deep layer V and VI neurons, which project primarily out of the neocortex, e.g. to the thalamus, brainstem and spinal cord (Douglas and Martin, 2004). On the other hand, the entorhinal cortex (EC), located in the parahippocampal gyrus, provides the major input to the hippocampus, either to the DG and the CA3 regions or to the CA1 and the subiculum. The flow of information within the hippocampus is mostly unidirectional, starting in the DG, then moving to the CA3, the CA1, the subiculum, and finally out of the hippocampus to the EC (van Strien et al., 2009). Given that the hippocampus and the neocortex are derived from neural progenitors expressing similar transcription factors including Pax6 and Emx1/2 (Hebert and Fishell, 2008), how they adopt fundamentally different structural and functional organization, especially at the cellular level, remains an intriguing question.
Previous histological, genetic and lineage tracing studies have provided a comprehensive understanding of the construction of the neocortex. Proliferation of neuroepithelial cells in the neuroectoderm produces radial glial cells (RGCs), a transient but pivotal cell population in neocortical development (Alvarez-Buylla et al., 2001). With the cell bodies located in the ventricular zone (VZ) lining the ventricle, RGCs display a bipolar morphology with one short apical process that reaches the luminal surface of the VZ (i.e. the ventricular endfoot) and another long basal process that extends to the pial surface (i.e. the radial glial fiber). In addition to their well-characterized role in supporting radial migration of newborn neurons (Hatten, 1990; Rakic, 1971), RGCs are mitotically active and responsible for producing nearly all neocortical excitatory neurons either directly or indirectly through transient amplifying progenitors, such as intermediate progenitors (IPs, also called basal progenitors) (Anthony et al., 2004; Englund et al., 2005; Haubensak et al., 2004; Malatesta et al., 2000; Miyata et al., 2004; Noctor et al., 2001; Noctor et al., 2004; Stancik et al., 2010; Tamamaki et al., 2001). Newborn neurons then migrate radially to constitute the future neocortex. Successive waves of newly generated neurons migrate past the existing early-born neurons and occupy more superficial positions, creating neocortical layers in an “inside-out” fashion (Angevine and Sidman, 1961).
Moreover, clonal analyses in the developing neocortex have led to the “radial unit hypothesis” (Rakic, 1988). Interestingly, we recently found that radially aligned sister excitatory neurons preferentially form electrical synapses with each other, which facilitates the development of specific chemical synapses between sister neurons and the emergence of a functional columnar organization in the neocortex (Li et al., 2012; Yu et al., 2009; Yu et al., 2012). These studies demonstrate that clonal analyses of neuronal production and organization can provide fundamental insights into the structural and functional development of brain structures. To date, while the specifying signals and patterning events of hippocampal development have been extensively explored (Lee et al., 2000; Mangale et al., 2008; Nielsen et al., 2007; Tole et al., 1997; Xie et al., 2010; Zhao et al., 1999), a systematic and definitive clonal analysis of the structural and functional development of the hippocampus is still missing. Previous lineage analyses of hippocampal development have been limited to coarse embryonic studies using mouse chimeras or mosaic transgene expression (Martin et al., 2002; Soriano et al., 1995), and a retroviral study in rats showing that clonally related neurons can be found in more than one cytoarchitectonic region (Grove et al., 1992).
In this study, we genetically labeled individual dividing progenitor cells in the VZ of the developing hippocampal primordium using two different cell type-specific approaches: retrovirus-mediated gene transfer and mosaic analysis with double markers (MADM), and investigated lineage-dependent structural and functional organization of the hippocampus.
RESULTS
Hippocampal pyramidal neuron clonal clusters labeled by retrovirus infection
Similar to their counterparts in the neocortex, excitatory neurons in the hippocampus are produced locally by progenitors in the VZ of the developing hippocampal primordium (Angevine, 1965; Bayer, 1980; Nowakowski and Rakic, 1981), whereas inhibitory interneurons originate distantly in the ventral telencephalon (Pleasure et al., 2000; Tricoire et al., 2011). To selectively label excitatory neuron progenitors, we took advantage of the exquisite fidelity of the subgroup A avian sarcoma and leukosis virus (ASLV) – receptor interaction and combined it with mouse genetics (Brown et al., 2011; Seidler et al., 2008). Low-titer avian RCAS (Replication-Competent ALSV long terminal repeat with a Splice acceptor) retrovirus expressing enhanced green fluorescence protein (EGFP) was injected into the lateral ventricle of Emx1-Cre;R26LSL-TVAiLacZ/+ mouse embryos, in which ALSV receptor TVA was specifically expressed in progenitors of the dorsal telencephalon with the Cre-loxP strategy (Gorski et al., 2002; Seidler et al., 2008). Injections were made on embryonic day (E) 11, 12 and 14, and brains were recovered at postnatal day (P) 7–30 for serial sectioning, immunohistochemistry and three-dimensional (3D) reconstruction to recover all the labeled cells in the hippocampus.
Discrete clusters of labeled pyramidal neurons with characteristic morphology were frequently observed (Figure 1A), suggesting that spatially isolated neuronal clusters originate from individual dividing progenitors and are thereby clones. To ensure reliable identification of “true” individual clones, we serially diluted the injected retrovirus to label on average just about one excitatory neuronal cluster per hippocampus (Figures 1A–1C; Movie S1). Similar hippocampal excitatory neuron clonal clusters were also readily labeled using Moloney murine leukemia retrovirus (Figures 1D and 1E). To further confirm the clonal relationship between neurons within individual clusters, we injected a mixture of low-titer retroviruses expressing either EGFP or red fluorescence protein mCherry, and examined the probability of observing spatially isolated clusters containing cells expressing the same fluorescent protein versus those containing cells expressing different fluorescent proteins in the hippocampus. We found that nearly all spatially isolated clusters (256 out of 260) were comprised of neurons expressing either EGFP or mCherry (Figures 1D and 1E; Movie S2), as further confirmed quantitatively by the Nearest Neighbor Distance (NND) analysis (Figure 1F). This clear segregation of neurons expressing EGFP or mCherry into individual clusters strongly argues that neurons in individual clusters are clonally related siblings.
Figure 1. Clonal pyramidal neuron clusters in the hippocampus labeled by retrovirus.
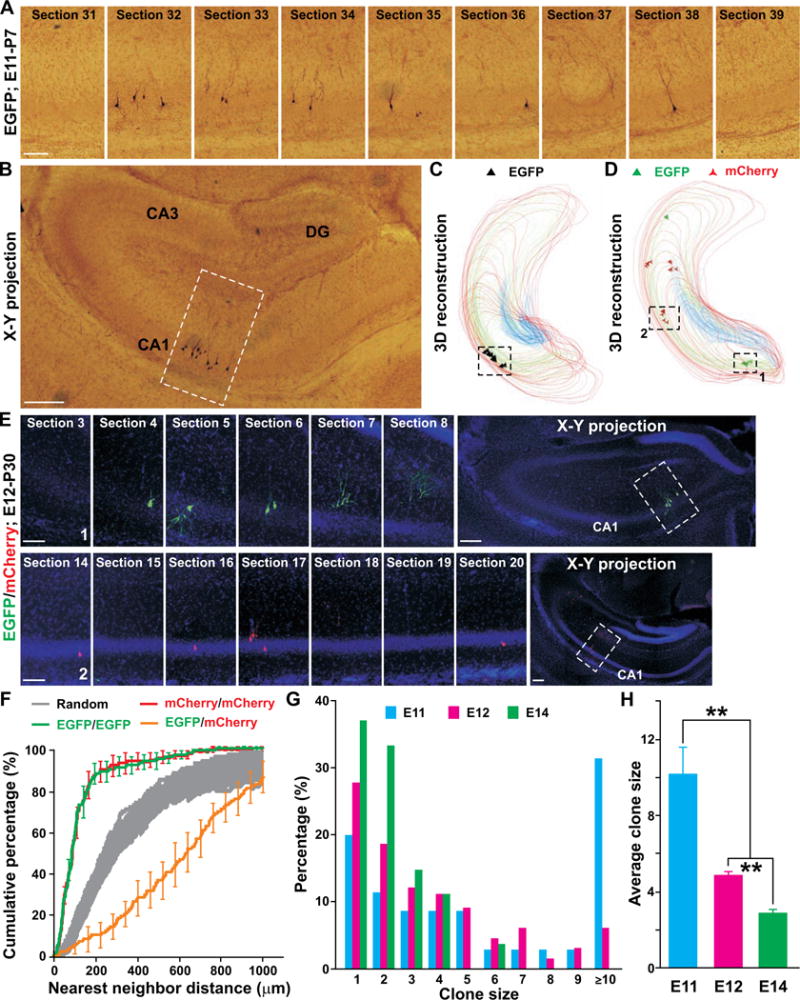
(A) Confocal images of consecutive sections harboring a single pyramidal neuron clonal cluster in the hippocampus of a P30 Emx1-Cre;R26LSL-TVAiLacZ/+ mouse infected with RCAS-EGFP virus at E11. Scale bar: 100 μm. (B) Projection image of the clone. Broken lines indicate the location of the clone. Scale bar: 200 μm. (C) 3D reconstruction image of the clone. Lines indicate the contours of the hippocampal subregions (red, CA1; green, CA3; blue, DG). Triangles represent the cell bodies of labeled pyramidal neurons and are approximately five times the actual cell body size. Similar symbols and displays are used in subsequent figures. (D) 3D reconstruction image of a P30 hippocampus infected with a mixture of retroviruses expressing EGFP (green) or mCherry (red) at E12. Broken lines indicate a green fluorescent neuron cluster (1) and a red fluorescent neuron cluster (2). (E) Confocal images of consecutive sections harboring clone 1 (top) and 2 (bottom) in D. Projection images are shown to the right. Broken lines indicate the locations of the clones. Scale bars: 100 μm and 200 μm. (F) NND analysis of retroviral-labeled neurons. Colored lines represent the cumulative frequency of NNDs of the same color-labeled (EGFP/EGFP, green or mCherry/mCherry, red) neurons, the different color-labeled (EGFP/mCherry, orange) neurons and the random datasets simulated 100 times (gray). (G) Histogram of clonal cluster size labeled at different embryonic stages (E11, n=35; E12, n=198; E14, n=27). (H) Average size of clones labeled at different embryonic stages (**, P<0.01). Data are represented as mean±s.e.m.
We next analyzed the size of individual clones labeled at different embryonic stages (Figures 1G and 1H). A small fraction (<20%) of clones labeled at E11 were composed of a single neuron while the rest had two or more neurons. The single neuron clones represent retroviral integration into a postmitotic neuron arising from asymmetric neurogenic division. On the other hand, the large clones likely represent retroviral integration into a self-renewing progenitor that undergoes symmetric amplification division(s) and/or multiple rounds of asymmetric neurogenic division. The progressive increase in the percentage of single neuron clones and the concomitant decrease in the percentage of large clones suggest that, as development proceeds, the frequency of asymmetric division increases and the frequency of symmetric division decreases in the developing hippocampal primordium.
Hippocampal pyramidal neuron clonal clusters labeled by MADM
To corroborate our findings with retroviral labeling, we exploited the MADM system (Bonaguidi et al., 2011; Hippenmeyer et al., 2010; Zong et al., 2005). To achieve selective labeling of excitatory neuron progenitors and their progeny (i.e. clones), we used the Emx1-CreERT2 mouse line (Kessaris et al., 2006). A single dose of tamoxifen (TM) was administered to timed pregnant Emx1-CreERT2;MADM dams at E10, E11, E12 or E13 via intraperitoneal injection. Brains were recovered and analyzed at P10-30.
In the absence of TM treatment, we found no labeled cells (n=5 brains). With TM treatment, we observed on average one discrete cluster of excitatory pyramidal neurons in each hippocampus. As predicted, individual clusters contained either a combination of green and red fluorescent neurons (Figures 2A, 2B, and S1A; Movie S3) or yellow fluorescent neurons (Figures 2C, 2D, and S1B; Movie S4). We did not observe any clusters with a mixture of green/red neurons with yellow neurons (0 out of 26). These results suggest that individual clusters are derived from a single progenitor and thereby represent clones. The size and progressive changes of the clonal clusters labeled by MADM were consistent with those labeled by retrovirus. These two independent approaches clearly demonstrate that sister excitatory neurons originating from individual progenitors form spatially isolated clusters in the stratum pyramidale of the hippocampus. Notably, glial cells were also observed in some clones (Figures 2A, 2C and S1, arrowheads), suggesting that progenitors in the VZ of the hippocampal primordium are capable of generating glia in addition to neurons.
Figure 2. Clonal pyramidal neuron clusters in the hippocampus labeled by MADM.
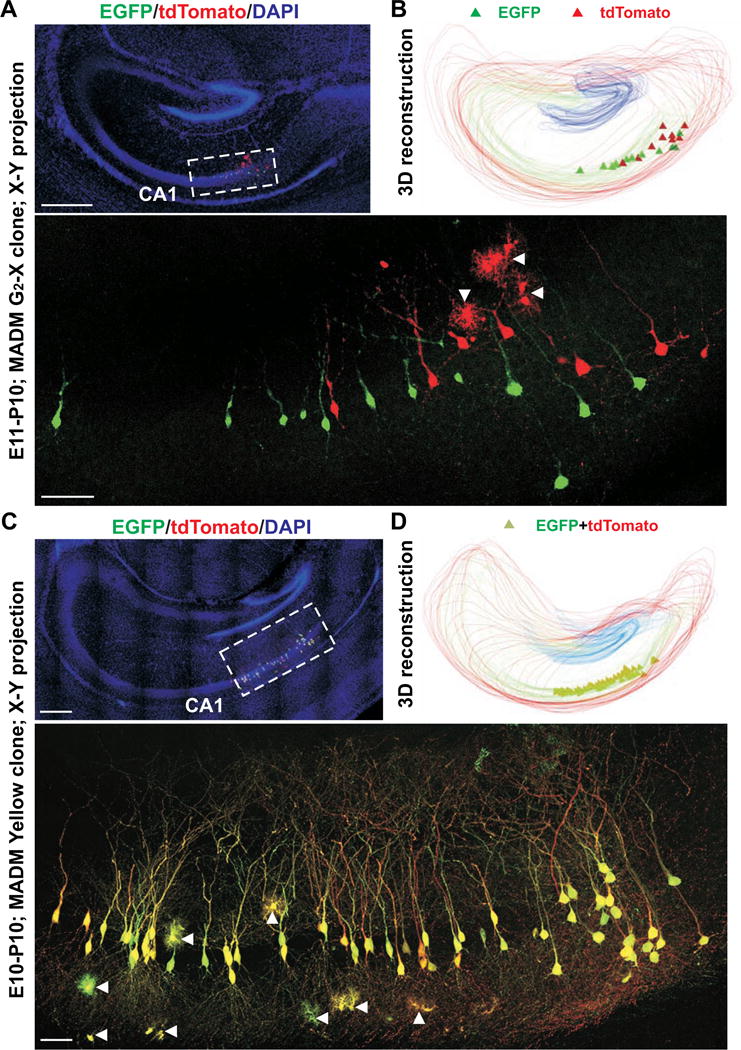
(A) Confocal projection images of a single G2-X EGFP (green)/tdTomato (red) clone labeled at E11. High magnification image (broken lines) is shown at the bottom. Arrowheads indicate glial cells. Scale bars: 50 μm and 400 μm. (B) 3D reconstruction image of the clone. (C) Confocal projection images of a single yellow clone labeled at E10. High magnification image (broken lines) is shown at the bottom. Scale bars: 50 μm and 400 μm. (D) 3D reconstruction image of the clone. See also Figure S1.
Progressive development of hippocampal pyramidal neuron clones
To gain more insight into hippocampal clone formation and organization, we examined the progressive development of embryonic clones. We carried out in utero intraventricular injections of serially diluted retrovirus expressing EGFP at E12 to label dividing progenitors in the VZ of the hippocampal primordium at clonal density. At 24 hours after injection (i.e. E12-E13), labeled cells were predominantly single, and either resembled RGCs with cell bodies in the VZ or were multipolar short-process cells with cell bodies located above the VZ. We also observed clusters that comprised two cells: one RGC-like (arrow) and another multipolar short-process cell (filled arrowheads) contacting the radial glial process (open arrowheads) (Figure 3A, left). To confirm the identity of RGC-like cells, we performed immunohistochemistry and found that RGC-like cells were positive for Pax6 (Figure 3B, arrows), Nestin (Figure S2A), and brain-lipid-binding protein (BLBP) (Figure S2B) – the specific markers expressed by radial glial progenitors (RGPs). Moreover, we found that RGC-like cells were mitotically active, as they possessed condensed DNA (Figure S2C) and incorporated bromodeoxyuridine (BrdU) (Figure S2D). Additionally, we observed cells expressing Tbr2, an IP cell marker, in clonal clusters (Figure 3C, open arrows). Indeed, our live imaging analysis of individual clones revealed RGP divisions at the VZ surface (Figure 3D, top and Movie S5) and IP divisions away from the VZ surface (Figure 3D, bottom and Movie S6).
Figure 3. Development of hippocampal pyramidal neuron clones.
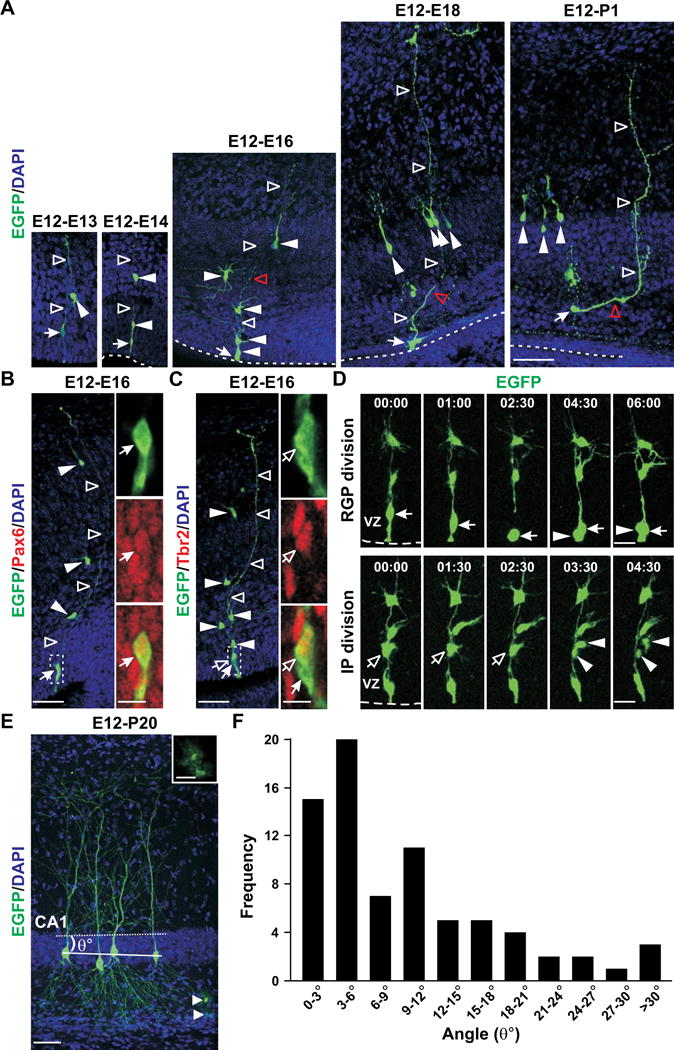
(A) Images of spatially isolated hippocampal clones 1 day (E12-E13), 2 days (E12-E14), 4 days (E12-E16), 6 days (E12- E18) and 8 days (E12-P1) after EGFP-expressing (green) retrovirus injection at E12 stained with DAPI (blue). Arrows indicate the RGC and filled arrowheads indicate short-process daughter cells of individual clones. Open arrowheads indicate the long radial process. Broken lines indicate the VZ surface. Scale bars: 50 μm. (B) Images of an E16 EGFP-expressing hippocampal clone (green) labeled at E12 and stained with the antibody against Pax6 (red), an RGP marker, and DAPI (blue). Arrows indicate the RGC that is positive for Pax6 and possesses a short ventricular endfoot and a long radial process (open arrowheads). Filled arrowheads indicate the short-process daughter cells arrayed along the radial glial fiber. High-magnification images (broken lines) are shown to the right. Scale bars: 50 μm and 10 μm. (C) Images of an E16 EGFP-expressing hippocampal clone (green) labeled at E12 and stained with the antibody against Tbr2 (red), an IP marker, and DAPI (blue). Open arrows indicate the IP that is positive for Tbr2 and arrows indicate the RGP. High-magnification images (broken lines) are shown to the right. Scale bars: 50 μm and 10 μm. (D) Time lapse imaging analysis of RGP (top, arrows) and IP (bottom, open arrows) divisions at and away from the VZ surface, respectively. Time (hours:minutes) is shown at the top. Arrowheads indicate the progeny. Broken lines indicate the VZ surface. Scale bars: 20 μm. (E) Image of a P20 clone labeled by retrovirus expressing EGFP (green) at E12 stained with DAPI (blue). The solid line indicates the mid-plane of the 3D clone and the broken line indicates the plane of the stratum pyramidale contour. The angle between the two planes (θ°) reflects the spatial distribution of the clone. Arrowhead indicates glial cells and a higher magnification image is shown in inset. Scale bars: 50 μm and 25 μm. (F) Quantification of the spatial distribution of clones with more than three neurons in the stratum pyramidale of the mature hippocampus. See also Figure S2 and S3.
At 48 hours after injection (i.e. E12-E14), we observed radially arrayed clones comprising an RGP (arrow) with 1–3 daughter cells (2.7±0.2 cells per cluster, n=23; filled arrowheads) arrayed along the radial glial process (open arrowheads) (Figure 3A). At 96 hours after injection (i.e. E12-E16), typical clones consisted of an RGP and on average 4–5 daughter cells (4.7±0.3 cells per cluster, n=37) that remained largely organized vertically. Some daughter cells at the top of the clusters acquired the typical morphology of a developing hippocampal pyramidal neuron, likely representing the earliest born neurons in the clone. Six days after injection (i.e. E12-E18), nearly all daughter cells acquired the typical pyramidal neuron morphology. Moreover, the daughter cells (filled arrowheads) no longer arrayed vertically along the radial glial process (open arrowheads); instead, they became horizontally organized (i.e. parallel to the VZ surface, broken lines) in the future stratum pyramidale. Interestingly, coinciding with this spatial organization change, the radial glial processes of RGPs displayed a bending near the future stratum pyramidale (Figure 3A, red open arrowheads). As time proceeded (E12-P1), the bend in the radial glial process and the horizontal orientation of daughter cell distribution became more prominent. In comparison, neocortical clones examined at the same developmental stages remained largely vertical and there was little persistent bending in the radial glial processes (Figure S3A). Together, these results suggest that hippocampal clones exhibit distinct developmental behavior from neocortical clones in transforming from an initially vertical organization to a horizontal organization relative to the VZ surface. Consistent with this, live imaging experiments showed that below the bend of the radial glial process, the progeny of hippocampal RGPs were in close contact with the mother progenitor and migrated radially along the radial glial process. However, around the bend, the progeny progressively departed the mother progenitor, acquired the pyramidal cell morphology and became horizontally distributed in the future stratum pyramidale (Figure S3B and Movie S7). Related to this, while neurons in the neocortex exhibited a distinct birth date–dependent inside-out radial organization, neurons in the hippocampus did not (Figure S2E). In addition, consistent with that RGPs undergo consecutive rounds of asymmetric division, sister neurons within individual clones were not all born at the same time (Figure S2F).
To further confirm that sister pyramidal neurons are oriented horizontally but not vertically in the stratum pyramidale, we systematically characterized the spatial organization of clonal clusters in the relatively mature hippocampus at P10-30 (Figures 3E, 3F, and S3C). The vast majority of clones (72 out of 75) were indeed organized parallel to the stratum pyramidale. Notably, the average size of clones labeled at E12 was similar when examined at E16, E18, P1 or P10-30 (E16: 4.7±0.3, n=37; E18: 4.8±0.5, n=17; P1: 4.9±0.5, n=7; P10-30: 4.8±0.3, n=143), suggesting that hippocampal pyramidal neuron neurogenesis is largely complete by E16. Moreover, these data indicate that the clones identified in the mature hippocampus are complete and the overall dispersion of clonally related pyramidal neurons is limited.
No preferential electrical or chemical synapse formation between sister pyramidal neurons in CA1
The clustering of sister pyramidal neurons in the hippocampus raises the intriguing possibility that specific synapses are formed between them, as observed in the neocortex (Yu et al., 2009; Yu et al., 2012). To address this, we performed quadruple whole-cell patch clamp recordings to simultaneously record from two EGFP-expressing sister pyramidal neurons and two nearby non-EGFP-expressing pyramidal neurons in the stratum pyramidale of CA1 region (Figures 4A–4D). Clones were labeled at E12 and recordings were performed at E17-P33.
Figure 4. No preferential synaptic connectivity between sister pyramidal neurons in CA1.
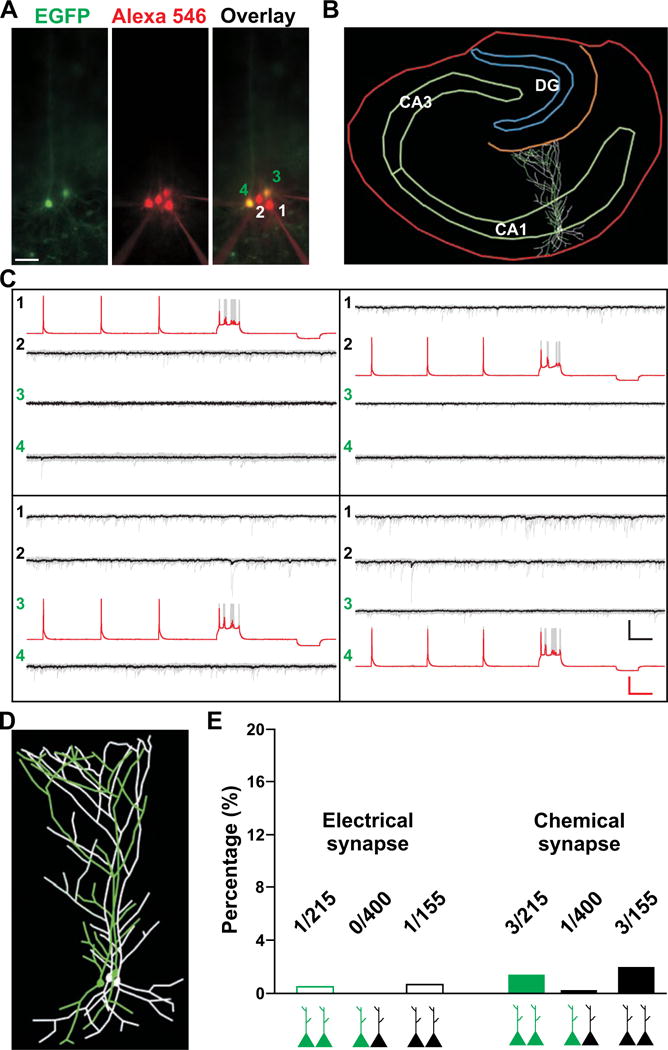
(A) Images of a quadruple recording of two EGFP-expressing sister pyramidal neurons (3 and 4, green) and two nearby non-EGFP-expressing pyramidal neurons (1 and 2) in CA1 filled with Alexa 546-conjugated biotin (red). Scale bar: 50 μm. (B) Morphological reconstruction of the slice and recorded neurons shown in A. (C) Sample traces of action potentials and hyperpolarization (red traces) triggered in the presynaptic neurons and responses (black traces) recorded in the postsynaptic neurons. The bold traces represent the average and the grey traces represent individual recordings. Scale bars: 50 mV, 20 pA and 100 msec. (D) High magnification image of morphological reconstruction of neurons recorded in A. (E) Quantification of the rate of synaptic connectivity. See also Figure S4.
We recorded a total of 215 pairs of sister pyramidal neurons and found that only one was connected by electrical synapses and three pairs were connected by chemical synapses (Figures 4E and S4A). Similarly, only 1 out of 400 non-sister (i.e. one EGFP-expressing and one non-EGFP-expressing) pyramidal neuron pairs was connected by chemical synapses and none were connected by electrical synapses. In addition, only 1 out of 155 pairs and 3 out of 155 pairs of similarly situated non-EGFP-expressing pyramidal neuron pairs were connected by electrical and chemical synapses, respectively. Similar results were obtained with sister pyramidal neurons labeled by MADM (Figures S5A and S5B). Together, these results suggest that sister pyramidal neurons in CA1 do not preferentially form electrical or chemical synapses with each other. The overall local synaptic connectivity rate between hippocampal pyramidal neurons is very low, as shown in previous studies (Knowles and Schwartzkroin, 1981; MacVicar and Dudek, 1981; Schmitz et al., 2001).
Synchronous synaptic activity between sister pyramidal neurons in CA1
Interestingly, while sister pyramidal neuron pairs in CA1 do not preferentially develop synapses with each other, they exhibited synchronous activity. Prominent spontaneous currents were frequently detected in EGFP-expressing sister pyramidal neurons and their nearby non-EGFP-expressing pyramidal neurons (Figures S4B and S4C). However, unexpectedly, more synchronous currents were observed between sister pyramidal neuron pairs (Figure 5A, red asterisks). To quantitatively assess this, we computed the cross-correlogram of spontaneous currents and found that there was a significant increase in the activity around 0 millisecond (msec) between sister (Figure 5B, red arrow and inset), but not non-sister (Figure 5B), neuron pairs. These results suggest that sister pyramidal neurons preferentially exhibit synchronous (within 1 ms) spontaneous activity.
Figure 5. Preferential synchronization of spontaneous activity between sister pyramidal cells in CA1.
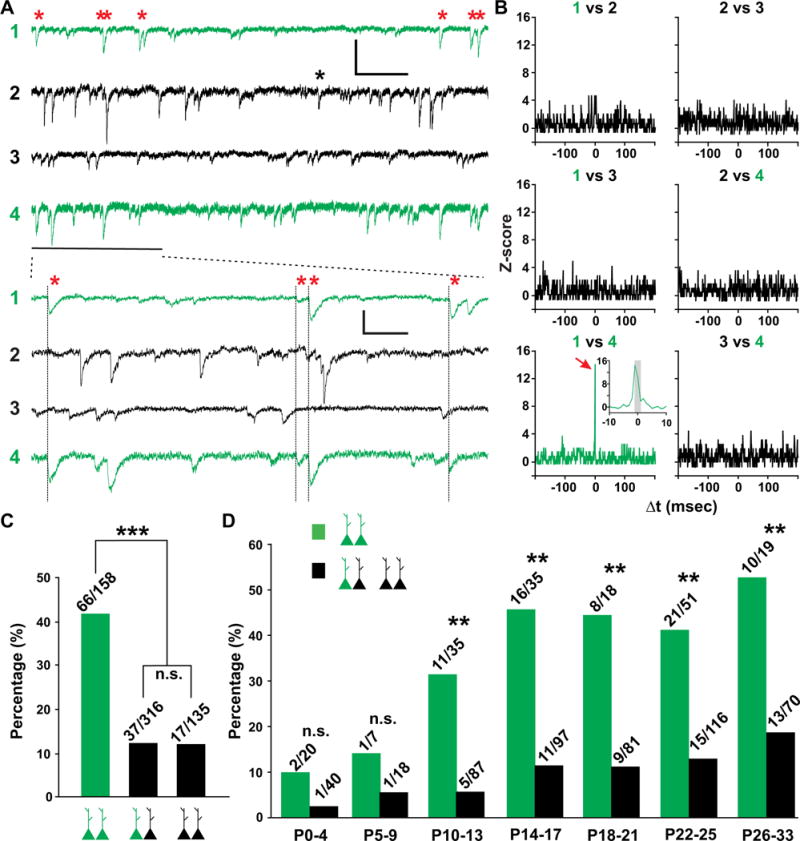
(A) Sample traces of spontaneous activity of two EGFP-expressing sister pyramidal neurons (1 and 4, green) and two nearby non-EGFP-expressing pyramidal neurons (2 and 3, black). High temporal resolution displays of a segment of recordings (thick line) are shown at the bottom. Red asterisks and thin lines indicate synchronized events between sister pyramidal neurons. Black asterisk indicates synchronized events between non-EGFP-expressing pyramidal neurons. Scale bars: 20 pA, 400 msec and 100 msec. (B) Normalized cross-correlogram (Z-score) for neuron pairs in A. Bin size is 1 msec. Note that the frequency of events is significantly increased around 0 msec (red arrow and inset) for the sister pyramidal neuron pair (1 vs. 4, green), but not for any other neuron pairs. The grey region in inset corresponds to −1 msec ≤ Δt ≤ 1 msec. Similar symbols and displays are used in subsequent figures. (C) Percentage of pyramidal neuron pairs from P10-33 mice exhibiting synchronized spontaneous activity (***, P<0.0001; n.s., not significant). (D) Percentage of pyramidal neuron pairs exhibiting synchronized spontaneous activity at different developmental stages (**, P<0.01; n.s., not significant). See also Figure S5.
Of 158 EGFP-expressing sister pyramidal neuron pairs from P10-33 mice, more than 40% (66 out of 158) showed robust synchronous spontaneous activity (Figures 5C and S4D). In contrast, only ~12% (37 out of 316) of non-sister pyramidal neuron pairs and only ~12% (16 out of 135) of similarly situated non-EGFP-expressing pyramidal neuron pairs showed similar synchronicity. Intriguingly, this synchronicity in spontaneous activity appeared to be developmentally regulated (Figure 5D). While the rate of finding a synchronous pair of sister pyramidal neurons was low prior to P10 (2 out of 20 at P0-4 and 1 out 7 from P5-9), it increased dramatically around the second postnatal week (11 out of 35 at P10-13, and 16 out of 35 at P14-17) (Figure 5D, green bars), suggesting that the second postnatal week is a critical period for the development of synchronous activity between sister pyramidal neurons. This time window coincides with the critical time window of chemical synapse formation in the hippocampus (Fiala et al., 1998). A small increase in non-sister pyramidal neuron synchronous activity was also observed (Figure 5D, black bars). Notably, synchronous activity was also preferentially found between sister pyramidal neuron pairs labeled by MADM (Figures S5C–S5E).
Common inhibitory synaptic inputs between sister excitatory neurons in CA1
To test whether the observed synchronous activity is indeed synaptic activity, we performed pharmacological experiments using tetrodotoxin (TTX, 1 μM), a Na+ channel blocker that prevents action potential generation (Figure 6A). We found that TTX treatment eliminated a vast majority of spontaneous activity; moreover, sister excitatory neurons no longer exhibited synchronous activity (Figures 6B and 6C). These results suggest that synchronous activity between sister pyramidal neurons are synaptic events triggered by spontaneous action potentials.
Figure 6. Synchronized activity between sister pyramidal neurons is mediated by synaptic GABA-A receptors.
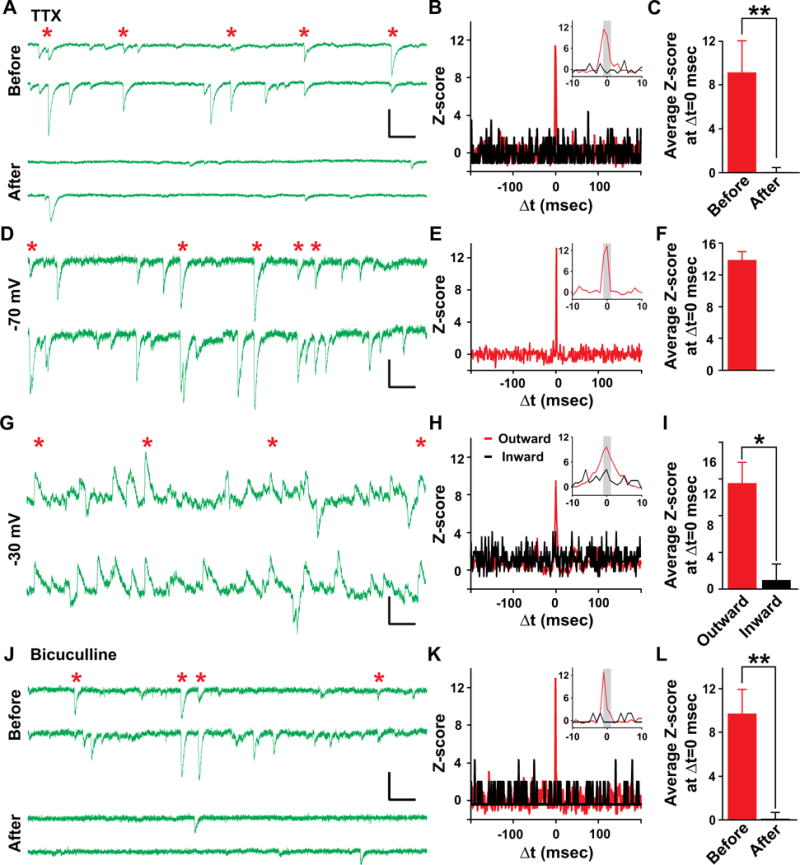
(A–C) Synchronized activity is blocked by TTX treatment. (A) Sample traces of spontaneous activity of a sister pyramidal neuron pair before and after TTX treatment. Red asterisks indicate synchronized events. (B) Normalized cross-correlogram before (red) and after (black) TTX treatment. (C) Average Z-score at Δt=0 msec before (red) and after (black) TTX treatment (n=7; **, P<0.01). (D–I) Synchronized activity is reversed around −36 mV. (D–F) Sample traces of spontaneous activity (D), normalized cross-correlogram (E) and average Z-score at Δt=0 msec (F) of sister pyramidal neuron pairs recorded at −70 mV (n=3). (G–I) Sample traces of spontaneous activity (G), normalized cross-correlogram (H) and average Z-score at Δt=0 msec (I) from the same pairs of sister pyramidal neurons recorded at −30 mV. Note that outward (red), but not inward (black), events are synchronized (n=3; *, P<0.05). (J–L) Synchronized activity is blocked by bicuculline treatment. Sample traces of spontaneous activity (J), normalized cross-correlogram (K) and average Z-score at Δt=0 msec (L) of sister pyramidal neuron pairs before (red) and after (black) bicuculline treatment (n=5; **, P<0.01). Scale bars: 20 pA and 100 msec. Data are represented as mean±s.e.m. See also Figure S6.
Pyramidal neurons in CA1 receive both glutamatergic excitatory and γ-aminobutyric acid (GABA)-ergic inhibitory synaptic inputs. To uncover the nature of synchronous activity, we took advantage of the distinct reversal potentials of glutamate receptor-mediated excitatory postsynaptic currents (EPSCs) and GABA-A receptor-mediated inhibitory postsynaptic currents (IPSCs). We identified sister pyramidal neuron pairs that exhibited robust synchronous activity when neurons were clamped at −70 mV (Figures 6D–6F). As predicted, when the same neurons were clamped at −30 mV, both outward and inward currents were observed (Figure 6G). Interestingly, only outward, but not inward, currents exhibited clear synchronization (Figures 6H and 6I), suggesting that synchronized synaptic activity is mediated by GABA-A, but not glutamate, receptors.
To further confirm this, we examined the effects of bicuculline (10 μM), a specific GABA-A receptor inhibitor, and D-APV (50 μM) and NBQX (5 μM), the inhibitors of N-methyl-D-aspartate (NMDA)- and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors, respectively. As expected, bicuculline treatment completely eliminated the synchronous activity between sister pyramidal neurons (Figures 6J–6L). In contrast, D-APV and NBQX had no significant effect (Figure S6). Together, these results demonstrate that sister pyramidal neurons exhibit synchronous GABA-A receptor-mediated inhibitory synaptic activity.
FS interneurons selectively provide common synaptic input to sister excitatory neurons in CA1
A diverse group of inhibitory interneurons provides synaptic inhibition to excitatory neurons in the hippocampus (Freund and Buzsaki, 1996; Klausberger and Somogyi, 2008; McBain and Fisahn, 2001). Synchronous synaptic inhibitory activity may arise from common inhibitory synaptic input received by sister pyramidal neurons. To test this, we explored the circuitry mechanism underlying the synchronicity. Quadruple whole-cell recordings were performed onto two EGFP-expressing sister pyramidal neurons, one adjacent non-EGFP-expressing pyramidal neuron, and one nearby inhibitory interneuron at P14-30 (Figure 7A). The identity of recorded neurons as excitatory pyramidal neurons or inhibitory interneurons was confirmed by their firing properties (Figure 7B) and morphological reconstructions (Figures 7E and 7F). If an inhibitory interneuron co-innervates two or more pyramidal neurons, synchronous input should be detected. Indeed, action potentials in a fast-spiking (FS) inhibitory interneuron (cell 1) simultaneously elicited faithful postsynaptic currents (blue arrows) in two EGFP-expressing sister pyramidal neurons (green cells 3 and 4), but not in the adjacent non-EGFP-expressing control pyramidal neuron (cell 2) (Figures 7C and 7D).
Figure 7. FS interneurons selectively provide common synaptic input to sister pyramidal neurons in CA1.
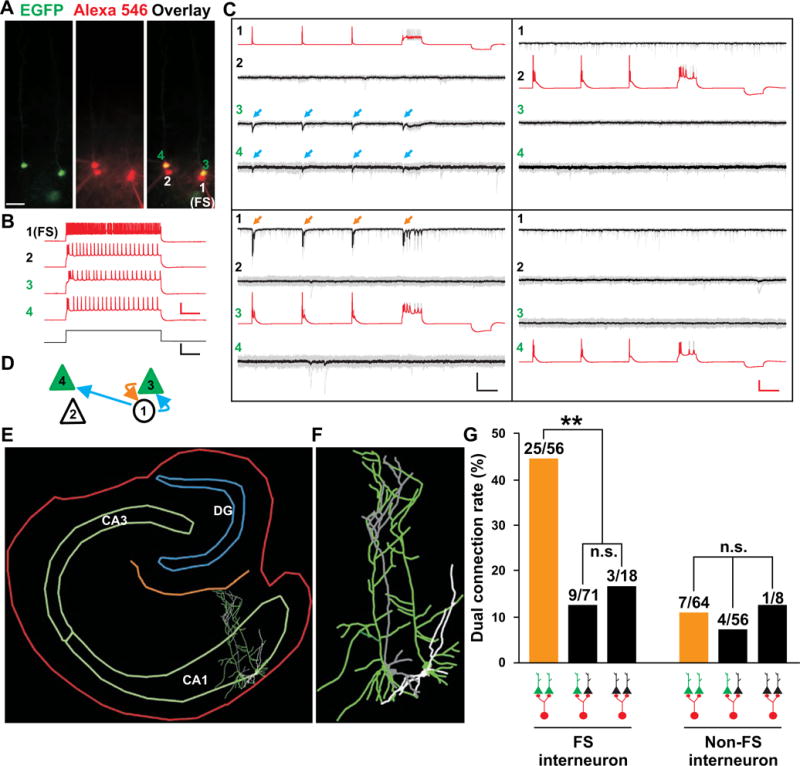
(A) Images of a quadruple recording of two EGFP-expressing sister pyramidal neurons (3 and 4, green), one adjacent non-EGFP-expressing pyramidal neuron (2) and one nearby FS interneuron (1) filled with Alexa 546-conjugated biotin (red). Scale bar: 50 μm. (B) Firing patterns of the four neurons in A. Sample traces of neurons in response to current injection (bottom). Scale bars: 50 mV, 500 pA and 200 msec. (C) Sample traces of the connectivity between the four neurons. Blue arrows indicate postsynaptic responses in sister neurons 3 and 4 triggered by action potentials in neuron 1 and orange arrows indicate reciprocal postsynaptic responses in neuron 1 triggered by action potentials in neuron 3. Scale bars: 50 mV, 20 pA and 200 msec. (D) Schematic of the connectivity among the four neurons. (E, F) Morphological reconstruction images of the four neurons and the slice bearing them. (G) Rate of dual connectivity between FS or non-FS interneurons and sister or non-sister pyramidal neurons (**, P<0.01; n.s., not significant). See also Figure S7.
Interestingly, we also observed a strong interneuron subtype specificity in providing common synaptic input selectively to sister pyramidal neurons. Around 45% (25 out of 56) of FS interneurons simultaneously innervated nearby sister pyramidal neuron pairs, whereas only about 11% (7 out of 64) of non-FS interneurons co-innervated nearby EGFP-expressing sister excitatory neurons (Figures 7G, yellow bars, and S7). Moreover, these shared inhibitory inputs from FS interneurons were specific to sister pyramidal neuron pairs, as only 12.7% (9 out of 71) of FS interneurons co-innervated nearby non-sister pyramidal neuron pairs (Figures 7G, black bars, and S7). In addition, only about 16.7% (3 out of 18) of FS interneurons co-innervated nearby similarly situated non-EGFP-expressing pyramidal neuron pairs. The rate of non-FS interneurons co-innervating nearby non-sister pyramidal neuron pairs or non-EGFP-expressing pyramidal neuron pairs was 7.1% and 12.5%, respectively. The overall connectivity between interneurons and pyramidal neurons was ~30.5% (105 out of 344 pairs), slightly higher than previously reported (Knowles and Schwartzkroin, 1981; Lacaille et al., 1987). Together, these results suggest that sister excitatory neurons in the hippocampus preferentially receive common inhibitory synaptic input selectively from nearby FS, but not non-FS, interneurons, which would contribute to the synchronous activity of sister pyramidal neurons.
DISCUSSION
Here, we used two powerful and specific genetic labeling methods, including avian retroviral labeling in conjunction with mouse genetics and MADM, and performed a systematic and quantitative clonal analysis of excitatory pyramidal neuron production and organization in the developing hippocampus. We found that progenitors in the VZ of the hippocampal primordium are RGPs with characteristic morphological, molecular and mitotic features. Moreover, the composition, organization and development of hippocampal clones at early embryonic stages are highly reminiscent of those of neocortical clones (Noctor et al., 2001). However, as time proceeded, we began to observe interesting differences in clonal organization between the hippocampus and the neocortex that have not been appreciated previously. While the daughter cells of neocortical clones remain predominantly arrayed radially/vertically along the radial glial process/fiber, those of hippocampal clones become progressively horizontally distributed along the future stratum pyramidale. This transition from a radial to horizontal organization of daughter cells is accompanied by a dramatic morphological change in hippocampal but not neocortical RGPs. The radial glial fiber of hippocampal RGPs exhibits a progressive bending near the future stratum pyramidale. Interestingly, this bend in the radial glial process has been noted previously in the fetal monkey hippocampus (Nowakowski and Rakic, 1979). The basis of this bending remains to be determined and is likely linked to the overall expansion and folding of the hippocampal formation. Nonetheless, our data suggest that this special morphological change likely facilitates the sequential deposition of the neuronal progeny of individual RGPs along the stratum pyramidale and the formation of a predominantly one-layered hippocampus. A new mode of neuronal migration in the hippocampus has recently been reported (Kitazawa et al., 2014) and may also contribute to the horizontal organization of hippocampal pyramidal neurons.
As a consequence of this radial to horizontal shift in clonal organization, sister excitatory neurons in the mature hippocampus are organized into discrete horizontal clusters, but not radial arrays as suggested previously (Martin et al., 2002). Our clonal labeling of a single cluster in the entire hippocampus allows unequivocal characterization of the distribution of sister pyramidal neurons. Clones exhibit considerable uniformity in distribution. The spread of a clone is roughly proportional to the number of neurons it contains. Similar observations have been made in a previous retroviral study (Grove et al., 1992). However, another study using retroviruses with distinguishable tags has suggested a wide spread of hippocampal clones (Reid et al., 1995). Our collective observations of a single cluster with no scattered neurons in individual hippocampi, no mixture of neurons originating from differentially labeled progenitors, and similarly sized clonal clusters observed at E16-18 and P10-30 do not support a wide dispersion.
Importantly, hippocampal clones are distinct from neocortical clones not only in structural organization, but also in functional organization. Previously we found that vertically aligned sister excitatory neurons in the neocortex preferentially develop electrical and chemical synapses with each other (Yu et al., 2009; Yu et al., 2012), which contributes to the functional columnar organization of the neocortex (Li et al., 2012). In contrast, despite their relative proximity, the connectivity of sister pyramidal neurons in the CA1 region of the hippocampus is extremely sparse. Recent studies suggest that the intra-neocortical connections effectively amplify thalamic input and thereby reinforce the sensory representation (Li et al., 2013a; Li et al., 2013b; Lien and Scanziani, 2013). On the other hand, a limited local connectivity permits dynamic cooperation of densely packed hippocampal pyramidal neurons for a large number of representations and population coding (Klausberger and Somogyi, 2008; Leutgeb et al., 2005).
In fact, GABAergic interneurons have been postulated to contribute to the dynamic selection and control of cell assemblies in the hippocampus (Klausberger and Somogyi, 2008). Remarkably, we found that even though sister pyramidal neurons in CA1 are not directly synaptically connected, they preferentially receive common synaptic input from nearby FS interneurons. Related to this, they exhibit preferential synchronous spontaneous and evoked synaptic activity mediated by GABA-A receptors. While additional mechanisms may contribute to synchronous activity between sister pyramidal neurons, the existence of this precise local microcircuit is likely critical for sister pyramidal neuron development and function. Moreover, FS interneurons are powerful at controlling the timing and probability of firing in pyramidal neurons (Hasenstaub et al., 2005). Previous studies have demonstrated that in the hippocampus, common inhibitory synaptic inputs can effectively entrain pyramidal neuron activity and synchronize their firing via the interaction of GABA-A receptor-mediated hyperpolarizing synaptic events with intrinsic membrane conductance in pyramidal neurons (Cobb et al., 1995; Hilscher et al., 2013). Unlike the neocortex, in which correlated inhibitory synaptic activity between nearby pyramidal neurons is common due to extensive connections from interneurons to adjacent pyramidal neurons (Pfeffer et al., 2013; Sippy and Yuste, 2013), synchronized inhibitory synaptic events between non-sibling hippocampal pyramidal neurons are far less common, likely due to fewer innervations from nearby interneurons (Knowles and Schwartzkroin, 1981; Lacaille et al., 1987; Lacaille and Schwartzkroin, 1988). We thus predict that sister pyramidal neurons in the hippocampus are preferentially synchronized in their activity, which will be fundamental for the operation of hippocampal neuronal networks. For example, preferential synchronization of sister excitatory neurons may allow them to function as potential units to encode similar place fields or memory information. Notably, anatomically organized clusters of hippocampal neurons with similar place fields have been found in previous studies (Dombeck et al., 2010; Eichenbaum et al., 1989; Hampson et al., 1999). It will be interesting to test whether these clusters of functionally similar neurons are indeed lineage-related by exploiting recent advances in in vivo functional imaging of hippocampal neurons at cellular resolution in behaving animals (Dombeck et al., 2010; Ziv et al., 2013).
We also observed recurrent excitation from pyramidal neurons to FS interneurons, suggesting a potential feedback loop in synchronizing sister pyramidal neurons in the hippocampus. The precise identity of FS interneurons that provide common input selectively to sister pyramidal neurons remains to be determined. Potential candidates include parvalbumin-positive basket and axo-axonic cells located in the stratum oriens and the stratum pyramidale (Kawaguchi et al., 1987; Sik et al., 1995) that are capable of robustly driving synchronous activity in pyramidal neurons (Cobb et al., 1995). Moreover, FS interneurons exhibit specific firing patterns and contribute to different aspects of network oscillations in vivo (Klausberger et al., 2003; McBain and Fisahn, 2001). Therefore, they may control sister pyramidal neurons in a temporally distinct and brain state-dependent manner (Gentet et al., 2010; Somogyi and Klausberger, 2005).
The mechanisms underlying the development of this lineage-dependent fine-scale inhibitory connectivity remain to be investigated. FS interneurons in the hippocampus are produced in the medial ganglionic eminence (MGE) (Pleasure et al., 2000; Tricoire et al., 2011). They arrive at the hippocampus at the embryonic stage and occupy predominantly the stratum pyramidale and the stratum oriens (Tricoire et al., 2011), where RGPs and newborn pyramidal neurons reside. It is possible that interactions between local FS interneurons and nearby RGPs lead to the emergence of preferential connectivity between FS interneurons and the neuronal progeny of RGPs, which closely associate with the mother RGPs during their production and initial migration. While we did not observe any obvious transient or lasting synaptic connectivity between sister pyramidal neurons, the formation of this precise microcircuit is likely regulated by activity-dependent processes.
In summary, we found that excitatory pyramidal neuron clones in the hippocampus are organized into discrete horizontal clusters. They preferentially share common inhibitory synaptic input from nearby FS interneurons and exhibit synchronous activity. In comparison, excitatory neuron clones in the neocortex are organized into discrete radial clusters. They preferentially develop synapses with each other and share similar physiological properties (Li et al., 2012; Yu et al., 2009; Yu et al., 2012). Together, these findings demonstrate that the lineage relationship of excitatory neurons fundamentally influences the structural and functional organization of two different regions of the cerebral cortex with distinct manifestations.
EXPERIMENTAL PROCEDURES
In utero infection and MADM labeling
In utero intraventricular injection was performed as previously described (Yu et al., 2009). For MADM labeling, Emx1-CreERT2;MADM-11GT/GT mice were crossed with MADM-11TG/TG mice. Pregnant dams were injected intraperitoneally with a single dose of tamoxifen (25–50μg/g body weight, Sigma) dissolved in corn oil (Sigma) at E10, E11, E12 and E13.
Serial sectioning, immunohistochemistry, imaging and three-dimensional reconstruction
Serial coronal or transverse sections (70 μm) of the brain were prepared using a vibratome (Leica Microsystems) and processed for immunohistochemistry. Z-stack images were taken using a confocal laser scanning microscope (FV1000, Olympus) and were further analyzed using FluoView (Olympus) and Photoshop (Adobe). For 3D reconstruction, each section was analyzed in sequential order from rostral to caudal using Neurolucida and StereoInvestigator (MicroBrightField, Inc.). Data are presented as mean±s.e.m., and statistical differences were determined using non-parametric Mann–Whitney–Wilcoxon and Kruskal–Wallis tests.
Electrophysiology and analysis
Transverse slices (250–350 μm) from E17-P33 mice were prepared using a vibratome (Leica Microsystems) in choline solution bubbled with 95% O2/5% CO2. Slices were kept in artificial cerebro-spinal fluid (ACSF) at 32°C for 1 hour and then at room temperature before recording. For recordings, slices were transferred to a recording chamber perfused with warm bubbled ACSF at 32°C. Multiple electrodes of whole-cell recordings were made using an Axon Multiclamp 700B amplifier and pCLAMP10.2 (Molecular Devices). Normalized cross-correlograms of spontaneous events were analyzed as previously described (Yu et al., 2012). Peaks in the cross-correlogram were considered significant if individual bins exceeded any adjacent bins within 200 msec by three standard deviations (i.e. the difference of Z scores was >3). The statistical differences between groups were determined by Chi-squared test.
Supplementary Material
HIGHLIGHTS.
Clonally related pyramidal neurons form horizontal clusters in the hippocampus.
Sister pyramidal neurons do not preferentially develop synapses with each other.
Sister pyramidal neurons exhibit synchronous synaptic activity.
Sister pyramidal neurons receive common inputs from nearby FS interneurons.
Acknowledgments
We thank Drs. Nicoletta Tekki-Kessaris and Dieter Saur for providing the Emx1-CreERT2 and R26LSLTVA-iLacZ mouse lines, Dr. Shaoyu Ge for sharing retroviral vectors, and Dr. Eric Holland for initially supporting our avian retroviral engineering. We are grateful to Drs. Yasunori Hayashi, Alexandra L. Joyner, Mary E. Hatten and Mu-ming Poo, and Ildiko Lily Erdy for critical reading of the manuscript. This work was supported by grants from NIH (R01DA024681, R01MH101382 and P01NS048120) and the Simons Foundation (GC221194).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.-T.X. and S.-H.S. conceived the project. Z.H. and K.H. performed quantitative analysis on 3D datasets and helped with synchronization analysis. P.G., W.Shi, and O.K. prepared MADM samples. S.H. performed embryonic electrophysiological recordings. Z.L. prepared retroviruses. W.Shao provided technical advice on live imaging experiments. K.N.B. prepared RCAS-infected samples. H.-T.X. performed all the other experiments and analysis. H.-T.X. and S.-H.S. wrote the manuscript with comments from all of other authors.
References
- Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Angevine JB., Jr Time of neuron origin in the hippocampal region. An autoradiographic study in the mouse. Exp Neurol Suppl. 1965;7(Suppl 2):1–70. [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with 3H-thymidine autoradiography. J Comp Neurol. 1980;190:87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb SR, Buhl EH, Halasy K, Paulsen O, Somogyi P. Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature. 1995;378:75–78. doi: 10.1038/378075a0. [DOI] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA. Neuronal circuits of the neocortex. Annu Rev Neurosci. 2004;27:419–451. doi: 10.1146/annurev.neuro.27.070203.144152. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Wiener SI, Shapiro ML, Cohen NJ. The organization of spatial coding in the hippocampus: a study of neural ensemble activity. J Neurosci. 1989;9:2764–2775. doi: 10.1523/JNEUROSCI.09-08-02764.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Gentet LJ, Avermann M, Matyas F, Staiger JF, Petersen CC. Membrane potential dynamics of GABAergic neurons in the barrel cortex of behaving mice. Neuron. 2010;65:422–435. doi: 10.1016/j.neuron.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove EA, Kirkwood TB, Price J. Neuronal precursor cells in the rat hippocampal formation contribute to more than one cytoarchitectonic area. Neuron. 1992;8:217–229. doi: 10.1016/0896-6273(92)90289-p. [DOI] [PubMed] [Google Scholar]
- Hampson RE, Simeral JD, Deadwyler SA. Distribution of spatial and nonspatial information in dorsal hippocampus. Nature. 1999;402:610–614. doi: 10.1038/45154. [DOI] [PubMed] [Google Scholar]
- Hasenstaub A, Shu Y, Haider B, Kraushaar U, Duque A, McCormick DA. Inhibitory postsynaptic potentials carry synchronized frequency information in active cortical networks. Neuron. 2005;47:423–435. doi: 10.1016/j.neuron.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Riding the glial monorail: a common mechanism for glial-guided neuronal migration in different regions of the developing mammalian brain. Trends Neurosci. 1990;13:179–184. doi: 10.1016/0166-2236(90)90044-b. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher MM, Leao KE, Leao RN. Synchronization through nonreciprocal connections in a hybrid hippocampus microcircuit. Front Neural Circuits. 2013;7:120. doi: 10.3389/fncir.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, Luo L. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Katsumaru H, Kosaka T, Heizmann CW, Hama K. Fast spiking cells in rat hippocampus (CA1 region) contain the calcium-binding protein parvalbumin. Brain Res. 1987;416:369–374. doi: 10.1016/0006-8993(87)90921-8. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa A, Kubo K, Hayashi K, Matsunaga Y, Ishii K, Nakajima K. Hippocampal Pyramidal Neurons Switch from a Multipolar Migration Mode to a Novel “Climbing” Migration Mode during Development. J Neurosci. 2014;34:1115–1126. doi: 10.1523/JNEUROSCI.2254-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Magill PJ, Marton LF, Roberts JD, Cobden PM, Buzsaki G, Somogyi P. Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature. 2003;421:844–848. doi: 10.1038/nature01374. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles WD, Schwartzkroin PA. Local circuit synaptic interactions in hippocampal brain slices. J Neurosci. 1981;1:318–322. doi: 10.1523/JNEUROSCI.01-03-00318.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaille JC, Mueller AL, Kunkel DD, Schwartzkroin PA. Local circuit interactions between oriens/alveus interneurons and CA1 pyramidal cells in hippocampal slices: electrophysiology and morphology. J Neurosci. 1987;7:1979–1993. doi: 10.1523/JNEUROSCI.07-07-01979.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaille JC, Schwartzkroin PA. Stratum lacunosum-moleculare interneurons of hippocampal CA1 region. II. Intrasomatic and intradendritic recordings of local circuit synaptic interactions. J Neurosci. 1988;8:1411–1424. doi: 10.1523/JNEUROSCI.08-04-01411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Moser MB, Moser EI. Place cells, spatial maps and the population code for memory. Curr Opin Neurobiol. 2005;15:738–746. doi: 10.1016/j.conb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Li LY, Li YT, Zhou M, Tao HW, Zhang LI. Intracortical multiplication of thalamocortical signals in mouse auditory cortex. Nat Neurosci. 2013a;16:1179–1181. doi: 10.1038/nn.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486:118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YT, Ibrahim LA, Liu BH, Zhang LI, Tao HW. Linear transformation of thalamocortical input by intracortical excitation. Nat Neurosci. 2013b;16:1324–1330. doi: 10.1038/nn.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien AD, Scanziani M. Tuned thalamic excitation is amplified by visual cortical circuits. Nat Neurosci. 2013;16:1315–1323. doi: 10.1038/nn.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar BA, Dudek FE. Electrotonic coupling between pyramidal cells: a direct demonstration in rat hippocampal slices. Science. 1981;213:782–785. doi: 10.1126/science.6266013. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LA, Tan SS, Goldowitz D. Clonal architecture of the mouse hippocampus. J Neurosci. 2002;22:3520–3530. doi: 10.1523/JNEUROSCI.22-09-03520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Fisahn A. Interneurons unbound. Nat Rev Neurosci. 2001;2:11–23. doi: 10.1038/35049047. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Nauta WJH, Feirtag M. Fundamental neuroanatomy. New York: Freeman; 1986. [Google Scholar]
- Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development. 2007;134:1133–1140. doi: 10.1242/dev.000265. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nowakowski RS, Rakic P. The mode of migration of neurons to the hippocampus: a Golgi and electron microscopic analysis in foetal rhesus monkey. J Neurocytol. 1979;8:697–718. doi: 10.1007/BF01206671. [DOI] [PubMed] [Google Scholar]
- Nowakowski RS, Rakic P. The site of origin and route and rate of migration of neurons to the hippocampal region of the rhesus monkey. J Comp Neurol. 1981;196:129–154. doi: 10.1002/cne.901960110. [DOI] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Rakic P. Guidance of neurons migrating to the fetal monkey neocortex. Brain Res. 1971;33:471–476. doi: 10.1016/0006-8993(71)90119-3. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Reid CB, Liang I, Walsh C. Systematic widespread clonal organization in cerebral cortex. Neuron. 1995;15:299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Schmitz D, Schuchmann S, Fisahn A, Draguhn A, Buhl EH, Petrasch-Parwez E, Dermietzel R, Heinemann U, Traub RD. Axo-axonal coupling. a novel mechanism for ultrafast neuronal communication. Neuron. 2001;31:831–840. doi: 10.1016/s0896-6273(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci U S A. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sik A, Penttonen M, Ylinen A, Buzsaki G. Hippocampal CA1 interneurons: an in vivo intracellular labeling study. J Neurosci. 1995;15:6651–6665. doi: 10.1523/JNEUROSCI.15-10-06651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy T, Yuste R. Decorrelating action of inhibition in neocortical networks. J Neurosci. 2013;33:9813–9830. doi: 10.1523/JNEUROSCI.4579-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano E, Dumesnil N, Auladell C, Cohen-Tannoudji M, Sotelo C. Molecular heterogeneity of progenitors and radial migration in the developing cerebral cortex revealed by transgene expression. Proc Natl Acad Sci U S A. 1995;92:11676–11680. doi: 10.1073/pnas.92.25.11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- Tole S, Christian C, Grove EA. Early specification and autonomous development of cortical fields in the mouse hippocampus. Development. 1997;124:4959–4970. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- Tricoire L, Pelkey KA, Erkkila BE, Jeffries BW, Yuan X, McBain CJ. A blueprint for the spatiotemporal origins of mouse hippocampal interneuron diversity. J Neurosci. 2011;31:10948–10970. doi: 10.1523/JNEUROSCI.0323-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- Xie Z, Ma X, Ji W, Zhou G, Lu Y, Xiang Z, Wang YX, Zhang L, Hu Y, Ding YQ, et al. Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc Natl Acad Sci U S A. 2010;107:6510–6515. doi: 10.1073/pnas.0912315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, He S, Chen S, Fu Y, Brown KN, Yao XH, Ma J, Gao KP, Sosinsky GE, Huang K, et al. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 1999;284:1155–1158. doi: 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


