Abstract
Reverse transcription-derived sequences account for at least half of the human genome. Although these retroelements are formidable motors of evolution, they can occasionally cause disease, and accordingly are inactivated during early embryogenesis through epigenetic mechanisms. In the mouse, at least for endogenous retroviruses, important mediators of this process are the tetrapod-specific KRAB-containing zinc finger proteins (KRAB-ZFPs) and their cofactor TRIM28. The present study demonstrates that KRAB/TRIM28-mediated regulation is responsible for controlling a very broad range of human-specific endogenous retroelements (EREs) in human embryonic stem (ES) cells and that it exerts, as a consequence, a marked effect on the transcriptional dynamics of these cells. It further reveals reciprocal dependence between TRIM28 recruitment at specific families of EREs and DNA methylation. It finally points to the importance of persistent TRIM28-mediated control of ERE transcriptional impact beyond their presumed inactivation by DNA methylation.
The human genome hosts some 100,000 human endogenous retroviruses (HERVs) or fragments thereof, more than 500,000 Long INterspersed Elements (LINEs, or L1), a million Alu repeats (a primate-specific subset of Short INterspersed Elements, or SINEs), and about 3600 copies of the hominoid-restricted SINE-VNTR-Alu (SVAs) (de Koning et al. 2011). These endogenous retroelements (EREs) can recombine, disrupt, or mobilize genes and alter their expression, and as such are essential contributors to evolution (Cordaux and Batzer 2009). Endogenous retroviruses (ERVs) and LINEs have the ability to replicate autonomously through the copy-and-paste mechanism typical of this class of genetic elements, whereas SINEs and SVAs depend for this on LINE-provided trans-acting functions such as reverse transcriptase (Finnegan 2012). To date, only an estimated 100 LINEs, less than a thousand Alu repeats, and between 20 and 50 SVAs are still mobilization-competent, yet these collectively account for thousands of somatic insertions and about one new germ line integrant every 20 human births, sometimes causing disease (Kaer and Speek 2013).
EREs also provide their host organism with a reservoir of cis-acting transcription modulators, as initially discovered with maize transposons (McClintock 1956). ERV-contained sequences regulate vertebrate development (Wang et al. 2007; Bourque et al. 2008; Kunarso et al. 2010; Mey et al. 2012; Schmidt et al. 2012) and contributed, for instance, to the evolutionary diversification of the placenta (Chuong et al. 2013). Moreover, ERVs can serve as tissue-specific promoters or enhancers for cellular genes, and control of some ERV-cellular gene pairs is coordinated notably in ES cells (Buzdin et al. 2006; Karimi et al. 2011; Macfarlan et al. 2011, 2012; Rebollo et al. 2011). However, most EREs are silenced by histone methylation, histone deacetylation, and DNA methylation in early embryos (Rowe and Trono 2011).
In the mouse, at least for a subset of EREs, important mediators of this process are the tetrapod-specific KRAB-ZFPs and their cofactor TRIM28 (also known as KAP1 or TIF1B). Trim28 knockout is embryonic lethal, and a maternal allele deletion leads to epigenetic instability during the mouse oocyte to embryo transition (Cammas et al. 2000; Messerschmidt et al. 2012). In murine embryonic stem cells, this protein is important for self-renewal (Hu et al. 2009; Seki et al. 2010). TRIM28 is also essential for the silencing of endogenous and some exogenous retroviruses in murine embryonic cells (Wolf and Goff 2007, 2009; Matsui et al. 2010; Rowe et al. 2010), where its KRAB-ZFP-mediated docking triggers the formation of heterochromatin, notably through the recruitment of SETDB1, the histone methyltransferase responsible for depositing the H3K9me3 repressive mark (Schultz et al. 2002; Ivanov et al. 2007; Frietze et al. 2010), and of DNA methyltransferases, the action of which extends to adjacent CpG islands (Quenneville et al. 2011, 2012; Zuo et al. 2012). A major consequence of the TRIM28-mediated repression of ERVs is the preservation of the transcription dynamics of murine ES cells, as repressive chromatin marks at murine ERVs are replaced upon Trim28 knockout by histone modifications typically found on active enhancers, which results in inducing the expression of nearby cellular genes, notably those harboring bivalent promoters (Rowe et al. 2013b).
The present study reveals that human-specific EREs are controlled by TRIM28 in human embryonic stem cells and that the range of TRIM28-controlled EREs, hence the effect of this regulatory system on the transcriptional landscape of these cells, is very broad. Furthermore, it brings to light reciprocal dependence between DNA methylation and TRIM28-induced ERE repression. Finally, itstrongly suggests that, irrespective of the DNA methylation status of EREs themselves, TRIM28-induced chromatin modifications at these loci are crucial to regulate their local transcriptional impact.
Results
TRIM28 controls a broad range of EREs in human ES cells
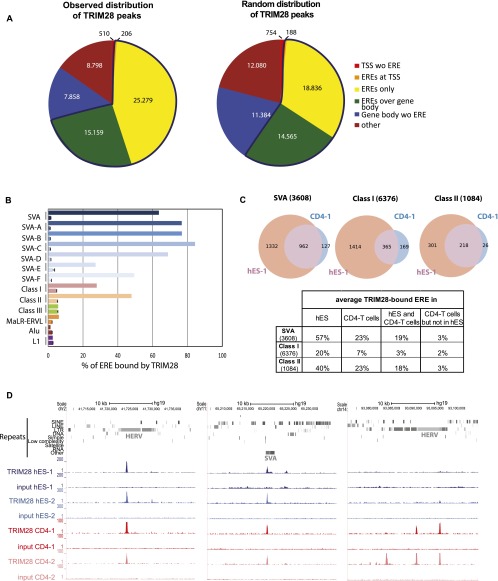
In order to investigate the functions of TRIM28-mediated regulation in human ESC, we first combined TRIM28-specific chromatin immunoprecipitation with deep sequencing (ChIP-seq) (Supplemental Table S1). More than 57,000 TRIM28 peaks were detected, close to three-quarters of which were on EREs, much more than expected by chance (Fig. 1A). TRIM28-bound EREs comprised, on average, about two-thirds of SVAs, one-fifth of Class I, and half of Class II HERVs, but much smaller fractions of Class III HERVs, Alu and L1 repeats (Fig. 1B,C; Supplemental Fig. S1A). Within SVAs, TRIM28 was significantly (Cochran-Armitage test for trend, P-value < 2.2 × 10−16) more associated with older family members (types A–D) than with their younger, human-restricted counterparts (types E and F), despite equivalent mapping efficiency for members of the two subgroups. On average, about half of Class II HERVs, one-third of SVAs, and one-sixth of Class I HERVs marked by TRIM28 in ES cells still bore the co-repressor in primary human CD4+ T-lymphocytes, where some EREs appeared to be newly recognized (Fig. 1C,D; Supplemental Fig. S1B).
Figure 1.
TRIM28 is associated with Class I and II HERVs and SVAs in hESC. (A) Observed and expected distribution of TRIM28 ChIP-seq peaks among indicated entities as annotated on the UCSC Genome Browser. Only HERVs containing both LTR and internal sequences were included in the analysis and counted as one full-length single element (see repeats analysis in Methods for details). Number of peaks for each category is indicated. The blue line delineates the fraction with ERE elements. (TSS) Transcription start site ±500 bp; (ERE) endogenous retroelement. (B) Relative TRIM28 occupancy of different ERE families. For each ERE (as defined in A), both the values observed (top bar) and expected by chance (bottom bar, as obtained from averaging a 10-times randomization of TRIM28 locations) are shown. For SVAs, data for both the whole family and its different subgroups are given. The abundant and heterogeneous MaLR-ERVL elements were monitored separately from the HERV class III. ChIP-seq data from an independent biological replicate are illustrated in Supplemental Figure S1A. (C) Comparison of TRIM28 binding to indicated EREs in human ES cells and CD4+ T-lymphocytes. In parentheses, numbers of EREs as defined in A are given. The ChIP was performed on T-cells pooled from three independent donors. The Venn diagrams represent data generated from one hES and one CD4+ T-cell ChIP-seq (hES-1 and CD4-1). Similar results were obtained from hESC and T-cell biological replicates (hES-2 and CD4-2) (Supplemental Fig. S1B). Diagrams obtained from replicates are illustrated in Supplemental Figure S1B. The table gives the average of the values obtained with all the replicates. (D) ChIP-seq maps of TRIM28 providing examples of EREs bound both in ES and CD4+ T-cells or in CD4+ T-cells only. Two biological replicates are illustrated in each case (hES-1, hES-2 and CD4-1, CD4-2) and displayed on the UCSC Genome Browser with the RepeatMasker function disabled to visualize EREs. Control maps obtained for the sequencing of the respective inputs are shown (set to the same vertical scale).
In ES cells, a large fraction of TRIM28-enriched EREs, notably almost all TRIM28-bearing SVAs and about half of the Class I and Class II HERVs, were also adorned with H3K9me3 (Supplemental Fig. S2A). ChIP-qPCR using consensus-specific primers on WT and TRIM28 knockdown cells (Supplemental Fig. S3) confirmed the TRIM28-dependent enrichment in SETDB1 and this repressive mark at HERVs and SVAs (Supplemental Fig. S2B). Accordingly, RNA-seq revealed the up-regulation of some ERE families upon TRIM28 depletion, notably from the Class I and Class II HERVs (Supplemental Fig. S2C). This was confirmed by quantitative RT-PCR with primers specific to members of these families, as exemplified by the class II HERVK14CI, the up-regulation of which was prevented by expression of a shRNA-resistant form of TRIM28 (Supplemental Figs. S2D, S4).
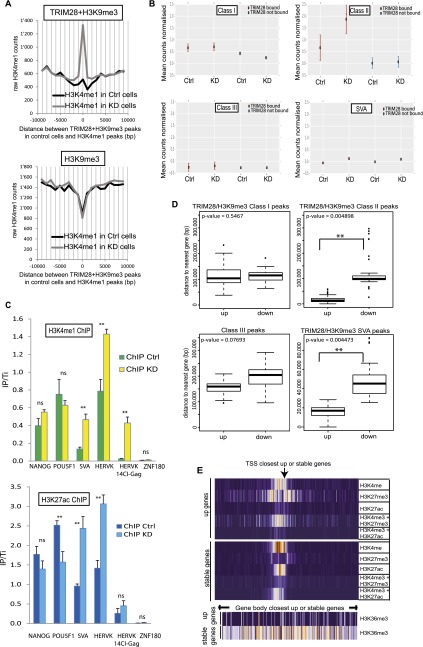
About 6% of the genomic SETDB1- and H3K9me3-rich loci gained H3K4me1 enhancer marks upon TRIM28 depletion, while this was not the case at TRIM28-devoid H3K9me3-enriched regions (Fig. 2A). This pattern was significantly observed for EREs predominantly from the Class II ERV and SVA groups which also gain H3K27ac, another active enhancer mark, suggesting that these EREs contained enhancers normally occluded by TRIM28 and revealed upon its depletion (Fig. 2B,C). Furthermore, 617 and 738 cellular genes were more than two times up- and down-regulated in TRIM28 KD cells, respectively (Supplemental Fig. S5A). Remarkably, genes up-regulated upon TRIM28 depletion were far closer to TRIM28-bound, H3K9me3-bearing Class II HERVs or SVAs than genes down-regulated in this setting (Fig. 2D). In contrast, no statistically significant relationship was noted between distance to similarly marked Class I or to Class III HERVs and a particular pattern of de-regulation. Interestingly, chromatin marks borne at baseline by SVA- or Class II HERV-close genes that were up-regulated following TRIM28 knockdown differed markedly from those found at genes similarly close to these retroelements but unaffected by the depletion (Fig. 2E; Supplemental Fig. S5B). These indeed harbored marks of active transcription, with H3K4me3 and H3K27ac at their promoter and H3K36me3 over their transcribed region, whereas the promoters of genes up-regulated following TRIM28 removal were depleted in H3K27ac, and bore the combination of active H3K4me3 and repressive H3K27me3 marks typical of bivalent promoters, and no H3K36me3 over their gene body.
Figure 2.
TRIM28 prevents enhancer activation marks at EREs and modulates the local transcriptional landscape in hESC. (A) ChIP-Cor analysis demonstrating that H3K9me3 at TRIM28-bearing loci (top panel) is replaced by H3K4me1 upon TRIM28 KD, while this does not occur at TRIM28-independent H3K9me3-positive sites (bottom panel). (B) H3K4me1 ChIP-seq analysis on sh-NS control (Ctrl) and TRIM28 KD cells, giving counts associated with TRIM28-bound or -devoid EREs. The total number of reads per 106 bases of ERE is normalized to the total number of reads in the ChIP-seq and to the number of reads found in the respective inputs. TRIM28-bound Class II and SVA elements significantly gain H3K4me1 after TRIM28 KD (Wilcoxon test gave P-values < 0.0001 for Class II and SVA but no significance for Class I and Class III HERVs). (C) H3K4me1 (top panel) and H3K27ac (bottom panel) ChIP-qPCR on control and TRIM28 KD cells using either generic primers detecting SVA and HERVK families or primers detecting HERVK14CI members. Primers targeting the 3′ end of ZNF180 and those targeting NANOG and POU5F1 promoters are used as negative and positive controls, respectively. Values were normalized to their respective total inputs (IP/Ti) and to the positive control GAPDH. Bars represent the mean and SD of technical replicates (n = 3). T-tests were used to compare controls and TRIM28-depleted samples. ([**] P-value ≤ 0.01). (D) Average distance between genes up- or down-regulated upon TRIM28 depletion and indicated genomic features. Up-regulated genes had a statistically higher chance of being close to a TRIM28/H3K9me3 bearing Class II HERVs or SVAs. Wilcoxon Mann-Whitney nonparametric test P-values are indicated. (E) Indicated histone marks ChIP-seq data from the ENCODE Project found within 10 kb (in 100-bp bins; orange and purple correspond to high and low relative occupancy, respectively) of the TSS (top) or over the transcribed region (bottom) of TRIM28-bound, H3K9me3-bearing, SVA-close genes, either up-regulated (up) or unaffected (stable) upon TRIM28 KD.
Interdependence between TRIM28 ERE recruitment and DNA methylation
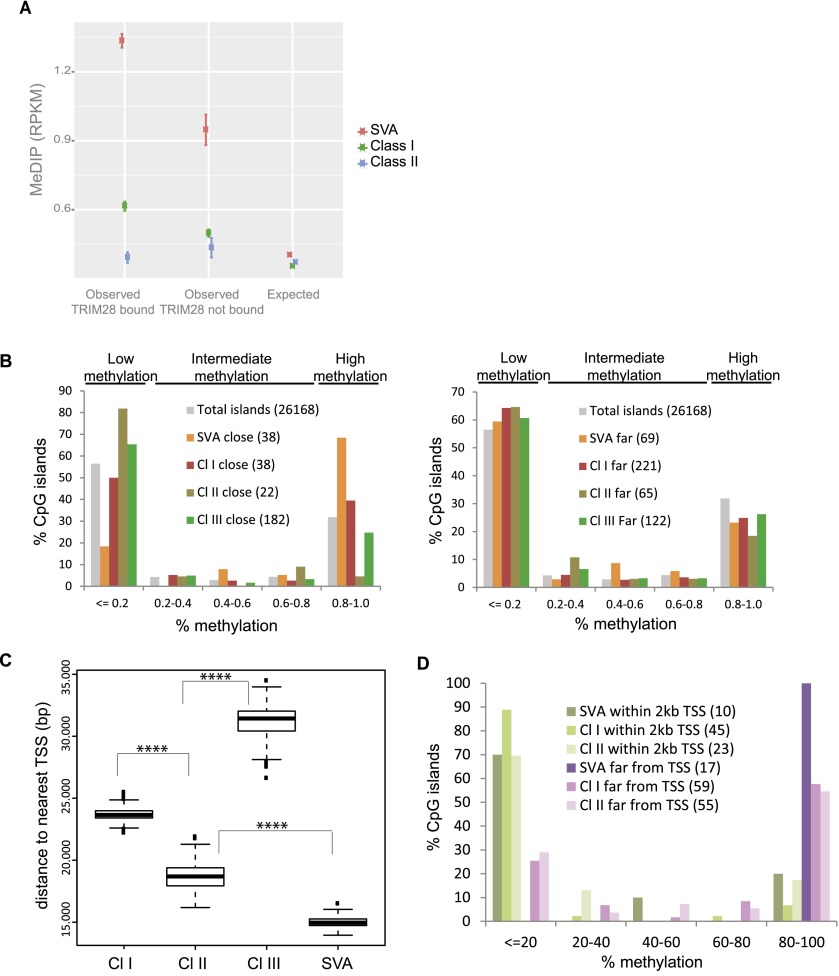
In murine ES cells, TRIM28-induced repression leads to DNA methylation, which is thought to result in permanent silencing of EREs (Quenneville et al. 2012; Rowe et al. 2013a). We thus examined the methylation status of EREs in human ES cells by correlating publicly available ENCODE (Encyclopedia of DNA Elements) Project DNA methylation data with our TRIM28 ChIP-seq results. TRIM28-bound SVA elements were markedly more CpG methylated than their TRIM28-free counterparts (Fig. 3A). Class I HERVs, although less GC-rich than SVAs, also displayed TRIM28-associated DNA methylation. This pattern was not detectable on Class II HERVs, whereas the TRIM28-devoid Class III elements did not exhibit any enrichment in CpG methylation compared to random sites (data not shown).
Figure 3.
TRIM28 recruitment to ERE leads to nearby CpG methylation. (A) DNA methylation on TRIM28-bound versus -unbound EREs, using publicly available MeDIP sequences. The observed mean RPKM for bound and unbound ERE is shown. For the expected values, we repositioned each ERE in each chromosome 1000 times at random. T-tests comparing bound and unbound elements gave P-values < 0.0001 for Class I and SVA EREs and P-value = 0.08 for Class II EREs. (B) Distribution of CpG islands according to methylation status and distance to indicated EREs, whether close (<2 kb for SVAs and 5 kb for HERVs to analyze similar number of CpG islands, left panel) or far (between 2 and 4 kb for SVAs, 15 and 30 kb for HERVs, right panel), using total CpG islands genome-wide as a control. In parentheses are numbers of CpG islands corresponding to each case. (C) Average distance between indicated EREs and closest TSS. P-values < 0.0001 (****) (Wilcoxon Mann-Whitney nonparametric test) are highly significant between all groups. (D) Distribution of CpG islands close to ERE, according to methylation status, whether ERE is close to (<2 kb) or far from (>20 kb) a TSS. In parentheses are numbers of CpG islands corresponding to each case.
To explore further the impact of ERE-mediated TRIM28 recruitment on local chromatin configuration, we examined the methylation status of CpG islands (Lister et al. 2009) located at various distances from TRIM28-bound EREs in hES cells (Fig. 3B). Reminiscent of a recent observation on murine retroelements (Quenneville et al. 2012), CpG islands located within a few kb of a TRIM28-bound SVA or Class I HERV had a significantly greater chance of being heavily methylated (>80% metCpG) than islands situated farther away from these elements or elsewhere in the genome. However, surprisingly, we noted the inverse for TRIM28-bound HERV Class II-close CpG islands, of which 80% were <20% methylated, while the methylation status of CpG islands located near TRIM28-poor Class III HERVs was identical to that of CpG islands genome-wide. Because the binding of transcription factors can prevent CpG methylation (Stadler et al. 2011), we measured the average distance between given ERE families and the nearest gene transcriptional start site (TSS). We found it to be significantly shorter for Class II HERVs and SVAs (median 19 kb and 15 kb, respectively) than for Class I (24 kb) or Class III (32 kb) HERVs (Fig. 3C). When we incorporated this parameter into our analysis, we found that CpG island hypermethylation was triggered efficiently by Class I HERVs, Class II HERVs, and mainly SVAs, but that it was blocked for all three types of retroelements if these were integrated within 2 kb of a TSS (Fig. 3D).
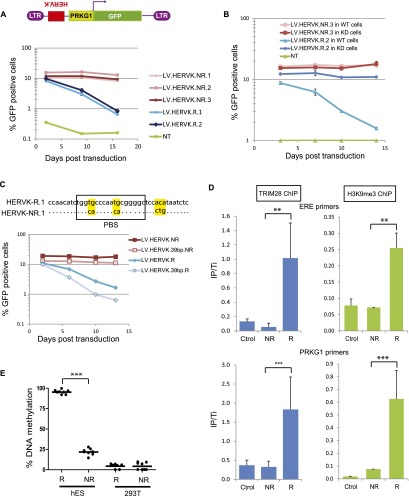
Confirming the TRIM28 dependence of their silencing in human ES cells, the TRIM28-binding fragment (as determined by ChIP-seq) of HERVKs up-regulated upon TRIM28 knockdown (HERVK-R [Repressed]), but not the corresponding sequence of HERVKs unaffected in this setting (HERVK-NR [Non-repressed]), could induce the repression of an adjacent PRKG1-GFP cassette when introduced in human ES cells by LV-mediated transduction, in a TRIM28-dependent fashion (Fig. 4A,B). For one TRIM28-repressed HERVK, we narrowed down the cis-acting repressive element to 39 nucleotides (nt), which overlapped with the primer binding site sequence (PBS) (Fig. 4C). This interestingly correlates with the silencing of murine leukemia virus (MLV) in murine embryonic cells, which stems from the ZFP809/TRIM28-mediated recognition of the primer binding site PBSPro (Wolf and Goff 2007, 2009; Wolf et al. 2008). Noteworthy, the hereby-identified HERVK repressor sequence was not active in murine ES cells, probably owing to the absence in this species of a KRAB-ZFP ortholog capable of recognizing its sequence (Supplemental Fig. S6). Silencing was also absent in human 293T cells, suggesting ES cell-restricted expression for this putative regulator (Supplemental Fig. S6). We could also demonstrate that HERV-derived repressor sequences triggered the rapid recruitment of TRIM28, which induced not only H3K9me3-mediated silencing of an adjacent promoter (Fig. 4D) but also its DNA methylation in human ES, but not in 293T, cells (Fig. 4E).
Figure 4.
TRIM28 recruitment on HERVK leads to repression and methylation of an adjacent promoter in hES. (A) Sequences corresponding to TRIM28-bound regions of two (R1 and R2) HERVKs only expressed when TRIM28 was depleted, or the corresponding 600-bp-long fragment of three (NR1–NR3) HERVKs expressed in WT cells were cloned in the antisense direction in depicted LV. The resulting LVs were used to transduce hESC in duplicates, and GFP expression was monitored over time by FACS. The average and SD of the duplicates are shown. (NT) Nontransduced. (B) Similar experiment in LV-shE transduced (WT) versus TRIM28 KD cells. Repression is TRIM28-dependent. The average and SD of the duplicates are shown. (C) Similar experiment with a 39-bp-long, PBS-encompassing HERVK fragment, sufficient to induce TRIM28-mediated repression. Alignment of PBS R1- and NR1-derived sequences is on top, with mismatches highlighted in yellow. (D) ChIP-qPCR (4 d post-transduction) showing that TRIM28 is recruited and H3K9me3 deposited on ERE (top panel) and PRKG1 (bottom panel) sequences of proviruses from the repressed LV-HERVK-R but not from the repression-resistant LV-HERVK-NR. Immunoprecipitates were normalized to their respective total input and enrichment on the positive control ZNF180. Results represent two independent experiments with technical replicates (n = 3) for TRIM28 ChIP and technical replicates (n = 3) for H3K9 ChIP. (**) P-value ≤ 0.01, (***) P-value ≤ 0.001. (Ctrol) LV devoid of ERE fragment. (E) Human ES or 293T cells were transduced with LV.HERVK.R or NR, and 2 wk later, percentage of de novo methylation at each tested CpG (n = 8) of the PRKG1 promoter was measured by bisulfite quantitative pyrosequencing. (***) P-value < 0.0001.
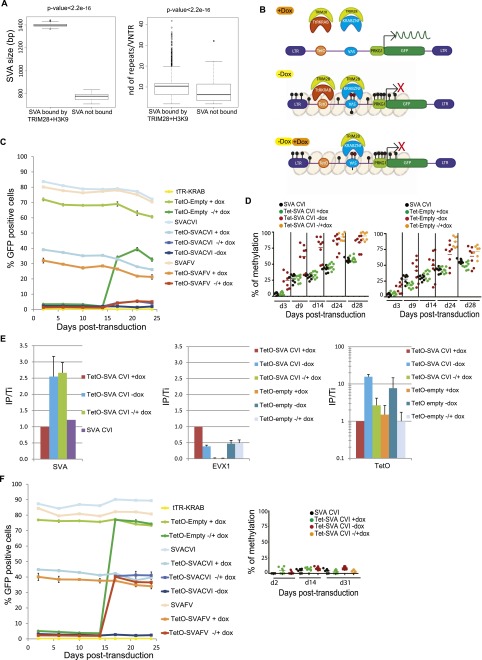
In spite of their marked TRIM28 enrichment in ES cells, SVAs were not induced in knockdown cells in our RNA-seq (and as confirmed by quantitative RT-PCR in Supplemental Fig. S7A), perhaps due to their high levels of DNA methylation (Fig. 3A; Supplemental Fig. S7B). The region within SVAs responsible for recruiting TRIM28 could not be determined precisely by ChIP-seq, but we observed that TRIM28-bound elements were longer and contained a significantly higher number of repeats (10 versus 7, 40 bp long, on average) in the central VNTRs (variable number of tandem repeats) than their unbound counterparts (Fig. 5A). Still, full-length SVA sequences cloned from several TRIM28-bound members of this family of EREs did not induce silencing of an adjacent PRKG1-GFP cassette in human ES cells (data not shown). We thus reasoned that TRIM28 recruitment to SVAs in human ES cells might be methylation-dependent, analogous to the ZFP57-mediated tethering of the co-repressor to imprinting control regions (ICRs) (Quenneville et al. 2011). To test this hypothesis, we capitalized on our previous demonstration that tethering to Escherichia coli Tet operator-derived TetO motifs of a fusion protein in which KRAB is linked to the DNA-binding domain of the Tet repressor (tTR-KRAB) results in the progressive DNA methylation of adjacent sequences in ES cells (Quenneville et al. 2012). We thus engineered a new set of lentivectors containing an array of TetO sites upstream or not of various SVA inserts placed antisense, 5′ of the PRKG1-GFP cassette (Fig. 5B). The 1245-bp-long SVA-CVI element is a canonical SVA including an Alu, VNTR, and HERVK-10 homolog fragments but lacking the 5′ hexamer repeats usually found in this class of EREs. SVA-FV is a smaller, 644-bp-long element, with only one CCCTCT 5′ hexamer followed by VNTRs. TRIM28-induced silencing and progressive methylation of the SVA elements and PRKG1 promoter in all TetO-containing lentivectors could be obtained in ES cells expressing the tTR-KRAB fusion repressor (Fig. 5C,D; Supplemental Fig. S8A). Noteworthy, the kinetic of PRKG1 methylation was faster when a SVA insert was present. In the absence of an SVA insert, both binding of TRIM28 and repression of the PRKG1-GFP unit could be reverted by the addition of doxycycline, which sequesters the tTR-KRAB protein away from its DNA target. In contrast, when SVA inserts were present, TRIM28 recruitment and PRKG1 silencing were irreversible (Fig. 5C,E). This supported a model whereby methylation of the SVA-derived sequences made them competent for TRIM28 recruitment. To ascertain this point, we stably knocked down the de novo DNA methyltransferases DNMT3A and 3B by LV-mediated shRNA interference (82% and 77% of the respective mRNAs were depleted in the double knockdown cells). This was expected to modify neither baseline levels of genome methylation, since the maintenance DNA methyltransferase DNMT1 was still present, nor tTR-KRAB-dependent TRIM28 recruitment on the proviruses. However, it resulted in fully dox-reversible transcriptional repression of the PRKG1-GFP cassette even from the SVA-containing vectors (Fig. 5F). Of note, we surprisingly observed that expression from the PRKG1 promoter did not correlate with the methylation status of the eight of its CpG nucleotides included in our analysis. Indeed, following 14 d without dox, that is, of tTR-KRAB-mediated repression, methylation of these CpG was progressively induced even without the SVA insert and did not significantly revert after addition of doxycycline, even though expression was reinstated (Fig. 5C,D). It indicates that transcriptional activity and CpG methylation are uncoupled for this promoter, at least within the region examined. More in line with our expectations, silencing was nearly completely dox-reversible in 293T whether or not a SVA sequence was placed upstream of the PRKG1 promoter (Supplemental Fig. S8B), consistent with a cellular environment in which TRIM28-induced repression was previously demonstrated not to result in DNA methylation (Quenneville et al. 2012).
Figure 5.
Interdependence between TRIM28-mediated SVA recruitment and DNA methylation. (A) TRIM28-bound SVAs are larger (left) and contain significantly more repeat units (right) than their TRIM28-devoid counterparts (10 versus 7 on average, P-value: Wilcoxon nonparametric test). (B) Working model to probe the DNA methylation dependence of TRIM28 recruitment to SVAs. In the presence of dox (top), tTR-KRAB is sequestered away from its TetO DNA target, and no repression occurs. In the presence of dox (middle), tTR-KRAB-induced heterochromatin formation (pink balls) leads to DNA methylation of adjacent sequences (black circles), including the SVA insert, which is then recognized by a putative SVA-specific KRAB-ZFP, and of the PRKG1 promoter, which is silenced. When dox is added (bottom), tTR-KRAB is removed, but the SVA-bound KRAB-ZFP maintains transcriptional repression. (C) tTR-KRAB-expressing hES cells were transduced in duplicate with lentivectors containing only TetO (TetO-Empty), only SVA (SVACVI or SVAFV), or both sequences upstream of a PRKG1-GFP cassette. Cells were maintained off (−Dox), on (+Dox), or 14 d off followed by 14 d on Dox (±Dox), and GFP expression was monitored over time by FACS. Results are representative of three independent experiments with similar results up to 31 d post-transduction. (D) Methylation of the PRKG1 promoter, contained in indicated vectors and conditions detailed in C, was monitored by pyrosequencing. Each dot represents a CpG (n = 8 except in ± conditions where n = 7). (E) TRIM28-specific ChIP-qPCR on chromatin harvested from indicated settings, with primers specific for the vector-contained SVA and TetO motifs or for the cellular EVX1 gene as a negative control. Bars represent mean and SD of two independent experiments (with n = 3 technical replicates). Immunoprecipitates were normalized to the total input (IP/Ti) and enrichment on the positive control ZNF180. (F) SVA-induced repression is de novo DNA methylation-dependent. (Left) Same as in C but in DNMT3A- and DNMT3B-depleted hES cells. The silencing of the PRKG1 promoter is reversible upon dox addition whether or not a SVA sequence is present. (Right) Pyrosequencing demonstrating the absence of de novo DNA methylation on the promoter in DNMT3A/DNMT3B KD cells.
Discussion
The present work demonstrates that TRIM28-mediated repression controls a wide spectrum of endogenous retroelements hosted in the human genome, and as such is key to the transcriptional dynamics of human embryonic stem cells. We first found that about three-quarters of TRIM28 binding sites on the genome of ES cells reside in EREs and coincide with SETDB1 recruitment, deposition of the H3K9me3 repressive mark and, in many cases, DNA methylation that extends to nearby CpG islands. Upon TRIM28 knockdown, repressive marks are replaced on a subset of EREs by chromatin modifications typically found on active enhancers, and adjacent genes are often activated, in particular if they harbor bivalent promoters. Sequences could be isolated from TRIM28-bound EREs, which could induce in cis the TRIM28-dependent repression and DNA methylation of an adjacent PRKG1 promoter in a lentiviral vector system. While an HERVK-derived, PBS-overlapping fragment achieved this effect without modification, with SVA-derived sequences, TRIM28 binding and repression required prior DNA methylation of the retroelement insert. This corroborates our observation that most TRIM28-bound SVAs are methylated in ES cells and suggests a role for tethering factors, the DNA binding of which is methylation-dependent, as for ZFP57 at ICRs. Establishment of naive human stem cells as recently described (Gafni et al. 2013; Leitch et al. 2013) could help in addressing further the role of methylation on TRIM28 recruitment on various EREs.
At least over the 8-CpG examined, we surprisingly observed that the percentage of CpG methylation of the PRKG1 promoter did not correlate with its expression potential. Indeed, methylation of these CpG di-nucleotides was established after 2 wk of tTR-KRAB-induced repression and did not revert once the trans-repressor was removed from its DNA target by addition of dox, irrespective of the presence or absence of an intervening SVA sequence. Yet, PRKG1-driven transcription resumed unless a TRIM28-tethering SVA-derived insert was located next to the promoter. While exploring further this phenomenon will require more extensive studies of the DNA methylation or hydroxymethylation status and kinetics of all CpG units contained in the PRKG1 and other promoters in this and other settings, these results already indicate that promoter methylation and transcriptional silencing should not be considered as always strictly coupled.
We do not think that our analysis identified all EREs ever bound by TRIM28 during human early embryogenesis. Human ES cells, under the conditions used in our study, are thought to correspond to so-called epi-stem cells, a relatively late predifferentiation stage. Their analysis thus gives only a snapshot of this developmental period, and it is likely that other elements are bound by TRIM28 earlier or later in the embryo. Supporting this hypothesis, many imprinting control regions (ICRs), which are bona fide TRIM28 targets, were no longer bound by TRIM28 in our human ES cells where ZFP57, the protein responsible for tethering TRIM28 to these elements, was expressed only at very low levels. It will be interesting to compare the ERE recruitment of TRIM28 in these and more naive ES cells. In addition, our data also suggest that the presence or absence of TRIM28 on given EREs is influenced by their age. We found that the youngest, human-restricted SVAs (group E and F), which are <3.5 M yr old (Wang et al. 2005), were less frequently associated with TRIM28 than their older counterparts. This could be because not enough time elapsed since they invaded the genome for KRAB-ZFPs or other TRIM28-tethering proteins recognizing their sequence to have been selected. Conversely, Class III HERVs encompass the oldest identifiable HERVs, which can be traced back to some 100 Mya, correlating their paucity around TSSs indicative of extensive purifying selection. Accordingly, their absence of recognition by TRIM28 more probably reflects the accumulation of mutations alleviating the need for transcriptional control. Finally, Class II HERVs, which include retroviruses endogenized after humans and chimpanzees diverged <7 Mya, are controlled by TRIM28.
That only a fraction of TRIM28-bound EREs became up-regulated when the co-repressor was depleted could reflect at least four nonmutually exclusive mechanisms. First, residual TRIM28 expression might suffice to prevent their de-repression. The CRISPR technology would allow us to knock out TRIM28 in hES cells, but the observed rapid lethality of the Trim28 deletion in murine ESC suggests that only a very efficient or an inducible methodology would provide enough cells for many of the analyses to be performed. Second, the transcript-generating potential of many EREs might be inactivated by mutations. Third, transcription factors recognizing their promoters may be missing from ES cells. Last, their DNA might already be irreversibly silenced by methylation, abrogating the effects of histone-restricted changes resulting from TRIM28 removal. However, the presence of TRIM28 at these retroelements implies that maintaining their repressive histone marks is functionally important irrespective of their DNA methylation status, even if not for their own control. This is confirmed by the observed up-regulation of genes located near TRIM28- and H3K9me3-bearing SVAs or Class II HERVs, when the co-repressor is depleted, in particular, if these genes harbor bivalent promoters and hence are primed for transcription.
Relatedly, our finding that many EREs targeted by TRIM28 in human ES cells still bear the co-repressor in adult lymphocytes argues against a simple model, whereby recognition of EREs during the early embryonic period leads to their complete inactivation, alleviating the need for persistent control. KRAB-independent binding of TRIM28 at specific genomic loci was reported to occur in human HEK293 cells, based on the study of a RBCC domain-deleted TRIM28 mutant (Iyengar et al. 2011). This mutant was not detected at EREs, suggesting that TRIM28 is tethered to these elements by KRAB-ZFPs in somatic cells as well. Nevertheless, our results suggest that a wide array of tethering factors, whether KRAB-ZFPs or not, are expressed in somatic tissues and serve to anchor the TRIM28-associated repressor machinery to ERE-based enhancers or promoters, thus likely contributing to the tissue specificity of these regulatory sequences.
Methods
Plasmids and LV production
LV-tTRKRAB, pLKO-shTRIM28, and pRRL.PRKG1.GFP-derived LV-TetO doxycyclin-inducible vectors were previously described (Quenneville et al. 2012). Genomic sequences encompassing SVA elements (chr2: 99604662-99605906 and chr4: 108998393-108999046) were synthesized by GenScript, cloned antisense by EcoRV and XhoI in pRRL.PRKG1.GFP, and transferred into EcoRV in LV-TetO derivatives. pLVHM-shDNMT3A and –shDNMT3B were obtained by inserting sh-oligonucleotides in the pLVTHM vector where the TetO sequence was previously removed, and the EEF1A1 promoter replaced by hPRKG1 and GFP by blasticidin or neomycin, respectively (see http://tronolab.epfl.ch/lentivectors for LV details). Lentivectors used as controls are identical except that there is no sh in them. HA-hTRIM28 partially codon-optimized sequence was synthetized by GenScript and cloned in a lentivector (pFUT-HA.hu.TRIM28optim). HERVK 5′ UTR was cloned using primers specific for the HERVK14CI consensus (http://www.girinst.org/repbase/; Jurka et al. 2005) from WT or KD hES cDNA. LV production protocols are detailed at http://tronolab.epfl.ch. LV backbones are available at Addgene (http://www.addgene.org/). Sequences of all primers are given in Supplemental Material.
Stem cell culture and transduction
H1 ESCs (WA01, WiCell) were maintained in mTesRI (StemCell Technologies) on hES-qualified Matrigel (BD Biosciences). For transduction, hES cells were detached using TryPLE Express (Invitrogen), split at 40,000 cells/cm2 and maintained for at least 12 h with ROCK inhibitor (Y-27632) to increase viability, transduced at MOI of 0.25–4 and selected with hygromycin (100 μg/mL for 4 d, then 50 μg/mL), blasticidin (10 μg/mL), or neomycin (200 μg/mL) when relevant. Pluripotent gene expression was monitored in routine by FACS analysis with the BD Human Pluripotent Stem Cells Transcription Factor Analysis kit.
Chromatin immunoprecipitation
Chromatin was prepared from 107 H1 cells or primary CD4+ T-cells isolated from two donors as previously described (Rowe et al. 2013b), and ChIP performed with rabbit anti-TRIM28 (SY 3267-68, 60 µL per IP, Tronolab) or Abcam anti-TRIM28 (for the hES-2 ChIP-seq, ab10483, 5 µL per IP), anti-H3K9me3 (pAb-056-050, 1 µg/µL, 10 µL per IP, Diagenode), anti-SETDB1 (a kind gift from F. Rauscher, 5 µL per IP), and anti-H3K4me1 (pAb-037-050, 5 µL per IP, Diagenode) antibodies. All qPCRs were performed in triplicate with SYBR Green mix (Applied Biosystems) on IPed material and Total input (Ti) using primers listed in the Supplemental Material. For sequencing, Ti and ChIP libraries were prepared using 10 ng of material with gel selection of 100- to 300-bp fragments and checked by Bioanalyzer (Agilent). The 80- or 100-base single-end or paired-end reads from the Illumina Genome Analyzer II were mapped to the human genome (hg19 assembly) using the short read aligner program Bowtie (Langmead et al. 2009), allowing up to three mismatches and a maximum of five repeats. Peaks for TRIM28 and H3K9me3 ChIP-seq were defined using the Model-based Analysis of ChIP-seq algorithm (Zhang et al. 2008) and normalized to Ti. Genomic region analyses were done using BEDTools (Quinlan and Hall 2010). Correlation analyses between ChIP peaks and genomic features were done with the ChIP-Cor analysis module (http://ccg.vital-it.ch/chipseq/chip_cor.php). Public data for 36-base single-end reads were downloaded from the ENCODE Project (GSM733657 [H3K4me3 H1 hES], GSM733748 [H3K27me3 H1 hES], GSM733718 [H3K27ac], and GSM733725 [H3K36me3 H1 hES]) and remapped to the human genome (hg19), allowing one mismatch. Enriched regions were defined using the ChIP-Part analysis module from the ChIP-seq analysis suite (http://ccg.vital-it.ch/chipseq/).
RNA sequencing and RT-qPCR
RNA was extracted (with the Qiagen RNeasy kit and On-Column DNase treatment) from controls or KD H1 cells 14 d after TRIM28 depletion for cDNA library preparation. The 76-base single-end reads from the Illumina Genome Analyzer II were mapped to the human transcriptome (hg19) using the Bowtie short read aligner, and counts were normalized to the transcript length and to the total number of reads. All RT-qPCR reactions were performed with independent biological duplicates using random hexamers, and each cDNA was tested in triplicate with SYBR Green mix (Applied Biosystems) and primers listed in the Supplemental Material. Negative controls without reverse transcription enzyme were processed in parallel.
Repeats analysis
Repeat annotations and coordinates were downloaded from the UCSC Genome Browser and consensus sequences from the Repbase human version 17.09 (http://www.girinst.org/repbase/ (Jurka et al. 2005). HERVs were further grouped in three classes based on previous classifications (Mager and Medstrand 2005). Since the binding of TRIM28 on exogenous and endogenous ERVs has been shown to bind on internal ERE sequences, and since we saw that this was mainly the case in our ChIP-seqs, we decided to remove solo LTRs from our analysis and to take into account only elements which contain LTRs with internal fragments. A table with the list of element coordinates in the respective classes is provided as Supplemental Table S2. Individual HERV repeats in the same orientation and flanked by two LTRs presumably represent one element and were then counted as one single element. Then, to attribute names to these elements and establish a list of TRIM28-bound elements group by group, only internal regions were used and only elements with fragmented pieces belonging to the same group were taken (Supplemental Table S3). Two lists of SVAs were generated: one with SVAs as annotated on the UCSC RepeatMasker tracks or one with fragmented elements removed whenever closer than 100 bp, in the same orientation, and from the same family, since we could not determine if these were truly different elements or not. The results were always similar in both cases, and we decided to work with the first full list. Because of very high redundancy and heterogeneity, MaLR-ERVLs were removed from Class III HERV in the whole analysis. The number of VNTRs per SVA was obtained with an in-house perl program using the coordinates and consensus sequence of the repeat found with the highest copy number in each SVA using Tandem Repeats Finder with the following parameters: 2, 7, 7, 80, 10, 35, 50 (http://tandem.bu.edu/trf/trf.html).
Data access
ChIP-seq and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE57989.
Acknowledgments
We thank J. Marquis for help with the ChIP protocol, Evarist Planet for the bioinformatics support, the staff of the CIG Technologies facility for library preparation and sequencing, and Vital-IT for computing. This work was financed through grants from the Swiss National Science Foundation and the European Research Council to D.T. (ERC 268721).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.172833.114.
References
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. . 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. 2006. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol 80: 10752–10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. 2000. Mice lacking the transcriptional corepressor TIF1B are defective in early postimplantation development. Development 127: 2955–2963 [DOI] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45: 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ 2012. Retrotransposons. Curr Biol 22: R432–R437 [DOI] [PubMed] [Google Scholar]
- Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. 2010. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS ONE 5: e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. . 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286 [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. . 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ 3rd, Farnham PJ. 2011. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol 31: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kaer K, Speek M. 2013. Retroelements in human disease. Gene 518: 231–241 [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. . 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. . 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. . 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. . 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DLM, Medstrand P. 2005. Retroviral repeat sequences. In Encyclopedia of life sciences. Wiley, New York. [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- McClintock B 1956. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 21: 197–216 [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502 [DOI] [PubMed] [Google Scholar]
- Mey A, Acloque H, Lerat E, Gounel S, Tribollet V, Blanc S, Curton D, Birot AM, Nieto MA, Samarut J. 2012. The endogenous retrovirus ENS-1 provides active binding sites for transcription factors in embryonic stem cells that specify extra embryonic tissue. Retrovirology 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. . 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Reports 2: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. . 2011. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet 7: e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. . 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013a. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. 2013b. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M. 2010. TIF1B regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci 107: 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. . 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. 2005. SVA elements: a hominid-specific retroposon family. J Mol Biol 354: 994–1007 [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci 104: 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Hug K, Goff SP. 2008. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci 105: 12521–12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. . 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. . 2012. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem 287: 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]