Abstract
In laser photostimulation, small clusters of neurons in brain slices are induced to fire action potentials by focal glutamate uncaging, and synaptic connectivity between photoexcited presynaptic neurons and individual postsynaptic neurons is assessed by intracellular recording of synaptic events. With a scanner, this process can be repeated sequentially across a patterned array of stimulus locations, generating maps of neurons’ local sources of synaptic inputs. Laser scanning photostimulation (LSPS) based on patterned glutamate uncaging offers an efficient, quantitative, optical-electrophysiological way to map synaptic circuits in brain slices.
INTRODUCTION
Less than 20 years ago Callaway and Katz (Callaway and Katz 1993) demonstrated that synaptic circuits in brain slices could be mapped by recording from a single postsynaptic neuron at one location and ‘photoexciting’ – through UV photolysis of caged glutamate – a small number of presynaptic neurons at another. Since then, photostimulation by UV uncaging of glutamate has been used to map synaptic circuits across much of the central nervous system, including spinal cord (Kato et al. 2007), brainstem (Kim and Kandler 2003), superior colliculus (Pettit et al. 1999), thalamus (Lam and Sherman 2005; Deleuze and Huguenard 2006), hippocampus (Brivanlou et al. 2004), thalamocortical pathways (Bureau et al. 2006; Llano et al. 2009), and more. A number of neocortical areas have been examined with LSPS, particularly visual (Dantzker and Callaway 2000; Sawatari and Callaway 2000) and barrel cortex (Schubert et al. 2001; Shepherd et al. 2003; Bureau et al. 2006; Brill and Huguenard 2009; Xu and Callaway 2009), and recently also auditory (Barbour and Callaway 2008) and motor cortex (Weiler et al. 2008; Yu et al. 2008). LSPS has also proven useful for characterizing cortical ‘circuitopathies’ in neurological disease models (Jin et al. 2006; Bureau et al. 2008; Wood et al. 2009). The efficacy of glutamate based photostimulation for circuit mapping (in contrast to electrical stimulation) derives from the ability to stimulate neurons with high precision and speed and without stimulating axons of passage. The interested reader is referred to the primary literature for further specific information about circuit mapping in different brain regions and species, and for detailed descriptions of recent technical innovations, including 2-photon based LSPS (Nikolenko et al. 2007; Matsuzaki et al. 2008), high-speed scanning (Boucsein et al. 2005; Shoham et al. 2005), and LSPS combined with imaging of activity-dependent intrinsic signals (Llano et al. 2009). This chapter presents a general approach and set of guidelines for quantitative circuit mapping using ‘standard’ LSPS methods based on ‘1-photon’ glutamate uncaging involving a UV laser, a pair of scanning mirror galvanometers, a patch clamp set-up, and open-source data acquisition software.
RELATED INFORMATION
For further details of the LSPS system described here, see (Shepherd et al. 2003; Shepherd et al. 2005; Shepherd and Svoboda 2005; Shepherd 2006; Weiler et al. 2008). For an early general review, see (Katz and Dalva 1994).
IMAGING SETUP
LSPS Hardware
Patterned glutamate uncaging entails the control of a focused beam of light in space and time. A standard approach is to use voltage-controlled mirror galvanometers (Model 6210; Cambridge Technology, Inc.) to scan the beam of an ultraviolet (UV) laser (3500 series; DPSS Lasers, Inc.). To assemble the LSPS setup, place a laser and an upright microscope (BX series; Olympus) on a vibration isolation table. Place in the beam path (in order): Pockels cell (Conoptics), mechanical shutter (VMM-D1; Uniblitz, Vincent Associates), neutral density filter wheel (Edmund), partial reflector (e.g. glass slide) deflecting a small fraction of the beam to a UV-sensitive photodiode (Edmund), scan mirrors, scan lens (focal length 100 mm, UV-grade, UV-coated, planoconvex lens, Edmund Optics), and dichroic mirror (380DRLP; Chroma Technology). The dichroic, mounted in a video adapter port (U-DPTS; Olympus) on top of the microscope, reflects the UV beam into the optical axis of the microscope, and transmits visible light for capturing bright-field or epifluorescence images by a video camera mounted on the adapter. A camera with a large CCD array such as the Kodak 2020 chip (e.g. Retiga 2000 series; Q-Imaging) provides a suitable field of view for mapping circuits in cortical slices. Replace the microscope’s tube lens with a UV transmissive lens (focal length 180 mm, UV-grade, UV-coated, custom lens; CVI Melles Griot). Attach a low-magnification objective lens (4x, 0.16 NA, UPlanApo; Olympus). In this hardware configuration, the optics and microscope are fixed to the vibration isolation table, and the manipulators for the recording electrodes are attached to the stage, which is moved by an X-Y stage translator (translator, Danaher; controller, Sutter). Alternatively, the microscope can be mounted on a X-Y translator, with the beam coupled via an optical fiber; in this case the laser and other optical components ‘upstream’ to the scan mirrors can be moved off the vibration table. Install equipment for patch clamp recording (amplifier, micromanipulators), and computer interface boards (National Instruments) for controlling devices and sampling signals. Build a small-volume recirculating perfusion system as described elsewhere (Shepherd and Svoboda 2005).
Caution: Class 4 UV lasers should only be used by qualified operators.
LSPS Software
Download and install Ephus, an open-source suite of software tools for laser-scanning and electrophysiology experiments. Ephus is freely available at openwiki.janelia.org, along with detailed information on software installation, calibration, and experimental procedures related to patterned photostimulation.
MATERIALS
Reagents
Artificial cerebrospinal solution (ACSF) <R>
Carbogen (95% O2 – 5% CO2 mixture)
Cutting solution <R>
High-divalent ACSF <R>
Caged glutamate stock solution <R>
R-CPP (Tocris) or other NMDA antagonist
Equipment
Slice weights – short (2–4 mm) curved segments of gold wire (0.813 mm diameter; Alfa Aesar), slightly flattened with a ball peen hammer
Temperature-controlled water bath
Vibratome
METHODS
Prepare brain slices
-
1
Cut brain slices on vibratome (0.3 mm thick) in cutting solution.
-
2
Incubate slices in pre-warmed (35 °C) oxygenated ACSF, for 15–30 minutes.
-
3
Store slices at 22 °C until used.
Familiarity with brain slice procedures is assumed. Slice quality is important. The protocol described here is for mapping inputs to pyramidal neurons in mouse cortex, but should be easy to adapt for other vertebrate and invertebrate preparations.
Map a neuron’s synaptic inputs
-
4
Add 7.5 mL of high-divalent ACSF to the recirculating bath system, and add CPP (0.01 mM) or other NMDA antagonist, and MNI-glu (0.2 mM). Recordings are performed at room temperature (22 °C).
-
5
Transfer a slice to the microscope’s recording chamber, orient it approximately as desired for mapping, and anchor it with slice weights.
-
6
Using a high-power objective, select a neuron and establish a whole-cell recording. To record excitatory synaptic currents, put the amplifier in voltage clamp mode, and hold near the reversal potential for inhibitory conductances, typically near −70 mV.
To record inhibitory inputs, hold at the reversal potential for glutamatergic conductances, typically near 0 mV. To record synaptic potentials, use current clamp mode.
-
7
Carefully switch to a low-power objective.
It is important to keep objectives clean, as salt accretions can block UV.
-
8
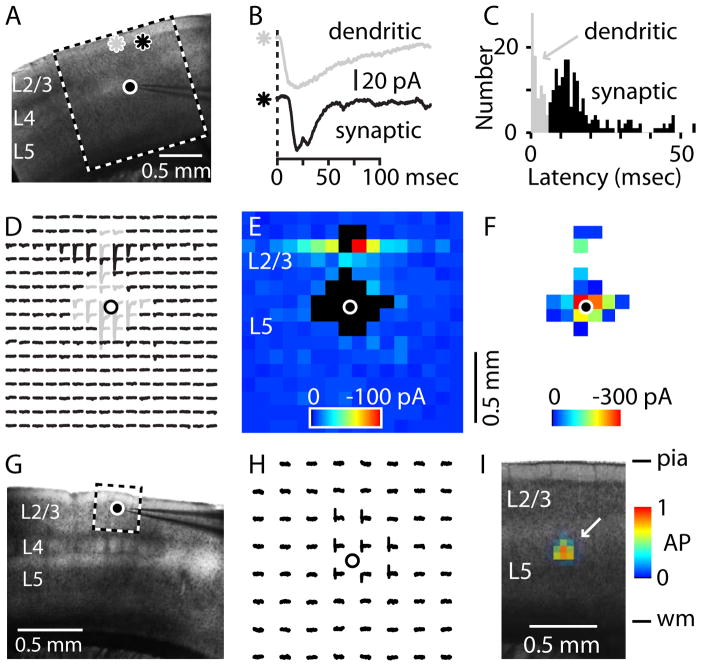
Using the stage controller, carefully position the slice within the camera’s field of view as desired for mapping (Fig. 1A).
-
9
Using software tools, select a 16-by-16 stimulus grid with 0.1 mm spacing between sites. Overlay it on the slice image, and align as desired with slice landmarks for mapping (Fig. 1A).
-
10
Deliver a 1.0-msec photostimulus to a site near the neuron’s own apical dendrites. This should generate a rapid-onset, large-amplitude response (Fig. 1A–C).
-
11
Deliver stimuli to other sites, away from the neuron’s dendrites. At some locations this should evoke synaptic inputs, arriving with longer onset latencies than dendritic responses (Fig. 1A–C).
-
12
Adjust the power to the desired mapping level (e.g. 20 mW in the specimen plane).
-
13
Set the interstimulus interval to 1.0 seconds and acquire a map (Fig. 1D, E, F).
-
14
Change the map sequence, and acquire a second map.
-
15
Change the sequence again, and acquire a third map. End of synaptic input map acquisition.
Figure 1. Synaptic input maps and excitation profiles.
(A) Arrangement for recording a synaptic input map from a L5 neuron in a mouse motor cortex brain slice. L, cortical layers. Circle, recorded neuron. Asterisks, photostimulus locations. Box, perimeter of 16-by-16 photostimulus grid, 100 μm spacing between sites.
(B) Dendritic (top, gray) and synaptic (bottom, black) responses to photostimuli at the locations indicated by the asterisks in panel A. Note shorter onset latency of dendritic than synaptic response.
(C) Histogram of event latencies. Gray, dendritic responses; black, synaptic inputs.
(D) Trace-map of responses to patterned uncaging. Gray, dendritic responses; black, synaptic or null responses.
(E) Pixel-map of synaptic input amplitude (black traces in D). Black pixels, dendritic-response sites.
(F) Pixel-map of dendritic responses (gray traces in D).
(G) Arrangement for recording an excitation profile from a L2/3 neuron in somatosensory barrel cortex; note L4 barrels. Circle, recorded neuron. Box, perimeter of 8-by-8 stimulus grid, 50 μm spacing.
(H) Excitation profile: trace-map showing that action potentials were evoked by stimuli close to the soma (circle).
(I) Average excitation profile for motor cortex pyramidal neurons (arrow), overlain on slice image (to scale), illustrating spatial scale and amplitude of photoexcitation of neurons at a stimulus location.
Map a neuron’s excitation profile
-
16
Using a high-power objective, select a neuron and establish a loose-seal (cell-attached) recording with the amplifier in voltage follower mode.
-
17
Carefully switch to a low-power objective.
-
18
Using the stage controller, carefully position the slice within the camera’s field of view as desired for mapping (Fig. 1G).
-
19
Using software tools, select an 8-by-8 stimulus grid with 0.05 mm spacing. Overlay it on the slice image, and center it on the soma of the recorded neuron (Fig. 1G).
-
20
Set the interstimulus interval to 1.0 seconds and acquire a map. This should show several peri-somatic sites of photoexcitability (Fig. 1H). End of excitation profile acquisition.
Analyze the traces
-
21
To analyze synaptic input map traces, first exclude traces with dendritic responses. For mouse cortical circuits this can be done by temporal windowing, because dendritic and synaptic response latencies are bimodally distributed (Fig. 1C).
-
22. Calculate the mean post-stimulus response over a short time interval (e.g. 50 msec). Pixels in synaptic input maps are color-coded based on a look-up table, or colormap, rendering dendritic responses as black pixels (Fig. 1E). Dendritic response maps (Fig. 1F) reflect the neuron’s dendritic topography.
Note that any parameter of interest can be mapped (e.g. peak, mean, latency, number of events).
23. <optional> Average the three maps to obtain a single map of the neuron’s inputs. Averaging is primarily used as a noise-reduction image analysis technique.
24. For excitation profile traces, use spike-detection routines to identify the number and timing of action potentials in the excitation profile traces.
25. Analyze excitation profile data to estimate the intensity and resolution of photostimulation (Shepherd et al. 2003; Shepherd and Svoboda 2005; Weiler et al. 2008).
26. <optional> Visualize the intensity and resolution of photostimulation by averaging a group of excitation profiles for a given cell type (e.g. pyramidal neurons) and superimposing this on a slice image (Fig. 1I).
TROUBLESHOOTING
Problem: Responses absent or weak
[Step 10–11]
Solutions: Ensure that there is adequate laser intensity in the specimen plane. At least 20 mW should be available for mapping. If not, either the laser output or UV transmission of the optical system should be increased. Ensure that slices are healthy.
Problem: Synaptic responses seem too strong
[Step 11]
Solutions: Ensure that synaptic driving is not causing reverberant excitatory activity in response to photostimulation, by performing excitation profiles and synaptic input maps for a range of stimulation intensities. The use of high divalent concentrations and NMDA blockade is usually sufficient to prevent over-excitation, but it may be necessary to work at low stimulus intensities if disynaptic events are evident.
Problem: Responses appear to be mis-mapped
[Step 10–11]
Solutions: Shifting of the locations of dendritic responses relative to the location of the soma indicates that the imaging system needs re-calibration. Use software tools to calibrate the correct voltage offsets and amplitudes for controlling the mirror angles.
Problem: No action potentials in excitation profiles
[Step 20]
Solutions: Ensure that the lack of action potentials is not a false negative due to a poor loose-seal recording. If no action potentials are evoked using the standard stimulus intensities, then increase the power and stimulate directly over the soma. If no action potentials are detected even at the highest powers, reject the neuron as a potentially false negative. If an action potential is recorded at higher but not standard powers, keep it as a true negative.
Problem: Excitation profiles indicate different efficacies of photostimulation for different experimental conditions.
[Step 20]
Solution: This is the reason for doing excitation profiles; if the efficacy of photostimulation differs for different conditions, such differences should be taken into consideration in the analysis. Otherwise, a change in apparent synaptic strength could be incorrectly ascribed to differences in synaptic connectivity, rather than photoexcitability. Excitation profiles should be acquired for any new set of conditions.
DISCUSSION
The expected outcome of LSPS mapping of synaptic inputs is a rapidly acquired, highly reproducible set of ‘images’ showing the local sources of synaptic input to individual neurons in brain slices. The expected outcome of excitation profile mapping is a data set that can be analyzed to obtain quantitative estimates for the resolution and intensity of photostimulation. LSPS microscopy based on patterned uncaging offers a unique tool for investigating the ‘electroanatomy’ of synaptic circuits. Advantages of LSPS include (1) high spatial resolution for stimulating neurons near their cell bodies, because axons of passage are not excited; (2) rapid sampling across multiple sites in a stimulus pattern; (3) does not require genetic manipulations. Limitations include (1) lack of cell-type selectivity, in that any neurons at the stimulus location are potentially photostimulated; (2) selective stimulation of individually targeted presynaptic neurons not routinely feasible; (3) limited use for higher-frequency trains of stimuli at the same location, due to glutamate receptor desensitization and caged compound depletion. The general mapping approach described can also be used for circuit mapping with 2-photon glutamate uncaging (Nikolenko et al. 2007; Matsuzaki et al. 2008). Photostimulation based on channelrhodopsin-2 (ChR2) (Nagel et al. 2003) offers another way to selectively photostimulate (transfected) neurons (Petreanu et al. 2007; Wang et al. 2007). Channelrhodopsin-2 assisted circuit mapping (CRACM) (Petreanu et al. 2007) is closely related to the methods described here. A key difference is that CRACM by design excites ChR2-expressing axons, whereas LSPS based on patterned glutamate uncaging avoids axons of passage.
RECIPES
Recipe: Cutting solution
| Stock: | |
|---|---|
| Reagent | Quantity (for 2 L) |
| sodium bicarbonate | 84.01 g |
| sodium phosphate monohydrate monobasic, NaH2PO4 1H2O | 137.99 g |
| potassium chloride, KCl, 3M stock | 1.667 mL |
| calcium chloride, CaCl2, 1 M stock | 1 mL |
| magnesium chloride, MgCl2 6H2O, 1 M stock | 14 mL |
| glucose | 9.01 g |
| Storage: 4 °C. Shelf life: up to 1 week | |
| User aliquot: | |
|---|---|
| Reagent | Quantity (for 0.5 L) |
| cutting solution stock | 0.5 L |
| choline choride | 7.68 g |
| ascorbic acid | 1.15 g |
| pyruvic acid | 0.17 g |
| Storage: 4 °C. Shelf life: up to 1 week | |
Recipe: ACSF
| Stock: | |
|---|---|
| Reagent | Quantity (for 2 L) |
| sodium chloride, NaCl | 14.844 g |
| sodium bicarbonate, NaHCO3 | 4.2 g |
| Na phosphate monohydrate monobasic, NaH2PO4 1H2O | 0.345 g |
| potassium chloride, KCl | 0.373 g |
| glucose | 9.01 g |
| Storage: 4 °C. Shelf life: up to 1 week | |
| User aliquots with standard divalent cation concentrations (2 mM Ca2+, 1 mM Mg2+): | |
|---|---|
| Reagent | Quantity (for 0.5 L) |
| ACSF solution stock | 0.5 mL |
| calcium chloride, CaCl2, 1 M stock solution | 1.0 mL |
| magnesium chloride, MgCl2, 1 M stock solution | 0.5 mL |
| Storage: 4 °C. Shelf life: up to 1 week | |
Recipe: High-divalent ACSF
| User aliquots with high divalent cation concentrations (4 mM Ca2+, 4 mM Mg2+): | |
|---|---|
| Reagent | Quantity (for 0.5 L) |
| ACSF solution stock | 0.5 mL |
| calcium chloride, CaCl2, 1 M stock solution | 1.0 mL |
| magnesium chloride, MgCl2, 1 M stock solution | 0.5 mL |
| Storage: 4 °C. Shelf life: up to 1 week | |
Recipe: Caged glutamate stock solution
Add H20 to the vial of MNI-caged glutamate (Tocris) to a concentration of 10 mM. Important: Prepare at room temperature, as the compound will precipitates if vials are placed on ice. Important: The formula weight is batch-specific. Aliquot into single-use aliquots.
Storage: −20 °C. Shelf life: months to years.
Acknowledgments
Funded by the International Rett Syndrome Foundation, National Institutes of Health (grants NS061963 and EB001464), Simons Foundation, and Whitehall Foundation.
References
- Barbour DL, Callaway EM. Excitatory local connections of superficial neurons in rat auditory cortex. J Neurosci. 2008;28(44):11174–11185. doi: 10.1523/JNEUROSCI.2093-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucsein C, Nawrot M, Rotter S, Aertsen A, Heck D. Controlling synaptic input patterns in vitro by dynamic photo stimulation. J Neurophysiol. 2005;94(4):2948–2958. doi: 10.1152/jn.00245.2005. [DOI] [PubMed] [Google Scholar]
- Brill J, Huguenard JR. Robust short-latency perisomatic inhibition onto neocortical pyramidal cells detected by laser-scanning photostimulation. J Neurosci. 2009;29(23):7413–7423. doi: 10.1523/JNEUROSCI.6098-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou IH, Dantzker JL, Stevens CF, Callaway EM. Topographic specificity of functional connections from hippocampal CA3 to CA1. Proc Natl Acad Sci U S A. 2004;101(8):2560–2565. doi: 10.1073/pnas.0308577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau I, Shepherd GM, Svoboda K. Circuit and plasticity defects in the developing somatosensory cortex of FMR1 knock-out mice. J Neurosci. 2008;28(20):5178–5188. doi: 10.1523/JNEUROSCI.1076-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau I, von Saint Paul F, Svoboda K. Interdigitated Paralemniscal and Lemniscal Pathways in the Mouse Barrel Cortex. PLoS Biol. 2006;4(12):e382. doi: 10.1371/journal.pbio.0040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci U S A. 1993;90(16):7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzker JL, Callaway EM. Laminar sources of synaptic input to cortical inhibitory interneurons and pyramidal neurons. Nat Neurosci. 2000;3(7):701–707. doi: 10.1038/76656. [DOI] [PubMed] [Google Scholar]
- Deleuze C, Huguenard JR. Distinct electrical and chemical connectivity maps in the thalamic reticular nucleus: potential roles in synchronization and sensation. J Neurosci. 2006;26(33):8633–8645. doi: 10.1523/JNEUROSCI.2333-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Prince DA, Huguenard JR. Enhanced excitatory synaptic connectivity in layer v pyramidal neurons of chronically injured epileptogenic neocortex in rats. J Neurosci. 2006;26(18):4891–4900. doi: 10.1523/JNEUROSCI.4361-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G, Kawasaki Y, Ji RR, Strassman AM. Differential wiring of local excitatory and inhibitory synaptic inputs to islet cells in rat spinal lamina II demonstrated by laser scanning photostimulation. J Physiol. 2007;580(Pt.3):815–833. doi: 10.1113/jphysiol.2007.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Dalva MB. Scanning laser photostimulation: a new approach for analyzing brain circuits. J Neurosci Methods. 1994;54(2):205–218. doi: 10.1016/0165-0270(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Kim G, Kandler K. Elimination and strengthening of glycinergic/GABAergic connections during tonotopic map formation. Nat Neurosci. 2003;6(3):282–290. doi: 10.1038/nn1015. [DOI] [PubMed] [Google Scholar]
- Lam YW, Sherman SM. Mapping by laser photostimulation of connections between the thalamic reticular and ventral posterior lateral nuclei in the rat. J Neurophysiol. 2005;94(4):2472–2483. doi: 10.1152/jn.00206.2005. [DOI] [PubMed] [Google Scholar]
- Llano DA, Theyel BB, Mallik AK, Sherman SM, Issa NP. Rapid and sensitive mapping of long-range connections in vitro using flavoprotein autofluorescence imaging combined with laser photostimulation. J Neurophysiol. 2009;101(6):3325–3340. doi: 10.1152/jn.91291.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Kasai H. Three-dimensional mapping of unitary synaptic connections by two-photon macro photolysis of caged glutamate. J Neurophysiol. 2008;99(3):1535–1544. doi: 10.1152/jn.01127.2007. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100(24):13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat Methods. 2007;4(11):943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10(5):663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Pettit DL, Helms MC, Lee P, Augustine GJ, Hall WC. Local excitatory circuits in the intermediate gray layer of the superior colliculus. J Neurophysiol. 1999;81(3):1424–1427. doi: 10.1152/jn.1999.81.3.1424. [DOI] [PubMed] [Google Scholar]
- Sawatari A, Callaway EM. Diversity and cell type specificity of local excitatory connections to neurons in layer 3B of monkey primary visual cortex. Neuron. 2000;25(2):459–471. doi: 10.1016/s0896-6273(00)80908-3. [DOI] [PubMed] [Google Scholar]
- Schubert D, Staiger JF, Cho N, Kotter R, Zilles K, Luhmann HJ. Layer-specific intracolumnar and transcolumnar functional connectivity of layer V pyramidal cells in rat barrel cortex. J Neurosci. 2001;21(10):3580–3592. doi: 10.1523/JNEUROSCI.21-10-03580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GMG. Imaging cortical circuits with laser scanning photostimulation. In: Buzsáki G, editor. Visualizing Large-Scale Patterns of Activity in the Brain: Optical and Electrical Signals. Society for Neuroscience; Washington, D.C: 2006. [Google Scholar]
- Shepherd GMG, Pologruto TA, Svoboda K. Circuit analysis of experience-dependent plasticity in the developing rat barrel cortex. Neuron. 2003;38(2):277–289. doi: 10.1016/s0896-6273(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Shepherd GMG, Stepanyants A, Bureau I, Chklovskii DB, Svoboda K. Geometric and functional organization of cortical circuits. Nature Neuroscience. 2005;8(6):782–790. doi: 10.1038/nn1447. [DOI] [PubMed] [Google Scholar]
- Shepherd GMG, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. J Neurosci. 2005;25:5670. doi: 10.1523/JNEUROSCI.1173-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham S, O’Connor DH, Sarkisov DV, Wang SS. Rapid neurotransmitter uncaging in spatially defined patterns. Nat Methods. 2005;2(11):837–843. doi: 10.1038/nmeth793. [DOI] [PubMed] [Google Scholar]
- Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, Kasai H, Hall WC, Feng G, Augustine GJ. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104(19):8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler N, Wood L, Yu J, Solla SA, Shepherd GMG. Top-down laminar organization of the excitatory network in motor cortex. Nat Neurosci. 2008;11(3):360–366. doi: 10.1038/nn2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L, Gray NW, Zhou Z, Greenberg ME, Shepherd GM. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J Neurosci. 2009;29(40):12440–12448. doi: 10.1523/JNEUROSCI.3321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29(1):70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Anderson CT, Kiritani T, Sheets PL, Wokosin DL, Wood L, Shepherd GM. Local-circuit phenotypes of layer 5 neurons in motor-frontal cortex of YFP-H mice. Front Neural Circuits. 2008;2:6. doi: 10.3389/neuro.04.006.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]