Abstract
Despite extensive studies for nearly three decades, lateral distribution of molecules in cholesterol/phospholipid bilayers remains elusive. Here we present a statistical mechanical model of cholesterol/phospholipid mixtures that is able to rationalize almost every critical mole fraction (Xcr) value previously reported for sterol superlattice formation as well as the observed biphasic changes in membrane properties at Xcr. This model is able to explain how cholesterol superlattices and cholesterol-phospholipid condensed complexes are inter-related. It gives a more detailed characterization of the LGI region (a broader region than the liquid disordered – liquid ordered mixed phase region), which is considered to be a sludge-like mixture of fluid phase and aggregates of rigid clusters. A rigid cluster is formed by a cholesterol molecule and phospholipid molecules that are condensed to the cholesterol. Rigid clusters of similar size tend to form aggregates, in which cholesterol molecules are regularly distributed into superlattices. According to this model, the extent and type of sterol superlattices, thus the lateral distribution of the entire membrane, should vary with cholesterol mole fraction in a delicate, predictable and non-monotonic manner, which should have profound functional implications.
Introduction
The effect of sterol content on cholesterol/phospholipid mixtures has been studied extensively, especially in the range 20-50 mol%, which is the cholesterol content typically found in mammalian cell plasma membranes. During the last two decades, a number of experiments have shown that spectroscopic properties of membrane probes and catalytic activities of membrane-associated enzymes as well as some other membrane-related events exhibit biphasic changes with sterol content at specific mole fractions (Xcr,obs) in fluid sterol/phospholipid mixtures (and reviewed in 1-4). Here “sterols” refers to cholesterol, ergosterol, dehydroergosterol (DHE) and other sterols with similar structures and “phospholipids” refers to diacyl phospholipids. The Xcr,obs values were found to be at or very close to the critical sterol mole fractions (Xcr) theoretically predicted for the formation of hexagonal or centered rectangular superlattices in the plane of the membrane 5-7. In the range of 19-53 mol% cholesterol, there are six theoretical Xcr values, i.e., 20.0, 22.2, 25.0, 33.3, 40.0, and 50.0 mol%. Although sometimes another critical mole fraction has been observed at 28.6 mol% 8 the excellent correlation between Xcr,obs and Xcr has been used as evidence for sterol superlattice formation in fluid sterol/phospholipid mixtures 5-6 and reviewed in 3-4. It has been proposed that sterol molecules tend to be maximally separated into regularly distributed superlattices in order to minimize the exposure of the hydrophobic region of the sterol molecules to water (the umbrella effect 9-10 and to reduce the deformation to the matrix lipid lattice caused by the rigid and bulky steroid ring 5-7.
The sterol superlattice model 5 proposes that not the entire membrane surface is covered by superlattices. At any given sterol mole fraction, regularly distributed superlattices always coexist with irregularly distributed areas; however, the extent of sterol superlattice reaches a local maximum at Xcr. This concept was first realized in the study of superlattices in 1-palmitoyl-2-(10-pyrenyl)decanoyl-sn-glycerol-3-phosphatidylcholine (Pyr-PC)/L-α-dimyristoylphosphatidylcholine (DMPC) mixtures. It was found that the excimer (E)-to-monomer (M) intensity ratio of pyrene fluorescence drops abruptly at critical Pyr-PC mole fractions predicted for maximal superlattice formation due to maximal separation of pyrene-labeled acyl chains, but does not go to zero due to the coexistence of regular and irregular regions 11. The coexistence of regular and irregular regions was subsequently revealed by Monte Carlo simulations 12. Furthermore, based on the data of nystatin partitioning into membranes, the area covered by sterol superlattices (Areg) was calculated to be ~71-89% at Xcr; and, Areg dropped abruptly when the sterol mole fractions were slightly (e.g., ~ 1 mol%) deviated from Xcr 13-14.
Sterol/phospholipid mixtures have also been described in terms of the formation of condensed complexes between sterol (C) and phospholipid (P) (reviewed in 15). This model considers the reaction: nqC + npP ↔ CnqPnp, where n is the cooperativity parameter, and p and q are relatively prime numbers. The relative stoichiometry q/(p + q) (e.g., 33.3 mol% sterol) can be determined from the position of the sharp cusp in the phase diagram 15-17. The critical sterol mole fractions theoretically predicted for maximal superlattice formation (Xcr) coincide with the relative stoichiometries q/(p + q) due to C-P complex formation. The condensed complex model has other features similar to those proposed in the sterol superlattice model. Both models contend that stability is greater and molecular order is higher at critical sterol mole fractions 17-20. The amount of condensed complex is a maximum at the relative stoichiometry (e.g., 33.3 mol% sterol), at which membranes have special properties 15-17. Because of these similarities, it has been speculated that condensed complexes and superlattices may share the same physical origin and may just occur at different times 21.
While direct visualization of sterol superlattices and condensed complexes are not feasible at present, much of our understanding of these structures can be obtained from computer simulations and modeling calculations. Monte Carlo simulations have been used to generate sterol superlattices at several Xcr values and revealed the underlying driving forces without the assumption of complex formation 9,22-23. Molecular dynamics (MD) simulations are able to simulate two-component systems 24 with fixed lateral distribution. After giving a relatively long simulation times (> 200 ns), lateral distribution of cholesterol in the superlattice bilayers was found to be more stable than that in the random bilayer 24. However, it is not certain whether the time used for MD simulation was long enough to capture the timescale of the lateral diffusion of the molecules and to give an account of the realistic lateral distribution of the molecules in the cholesterol/phospholipid mixtures. In a multi-component membrane with inhomogeneous lateral distribution the relaxation times for different components to sample conformations and orientations relative to each other are orders of magnitude longer than the nanosecond time scale sampled by MD 25.
In this work we develop a statistical mechanical model of cholesterol/phospholipid bilayers that is able to rationalize almost every Xcr value and the observed biphasic changes in membrane properties (such as Areg) at Xcr. This model indicates that cholesterol superlattices and cholesterol-phospholipid condensed complexes are interrelated. The occurrence of condensed complexes and sterol superlattices as delineated in this model provides new insight into the molecular details of the LGI region (liquid – gel region) in the phase diagram of cholesterol/phospholipid mixtures.
Model
On the condensing effect of cholesterol
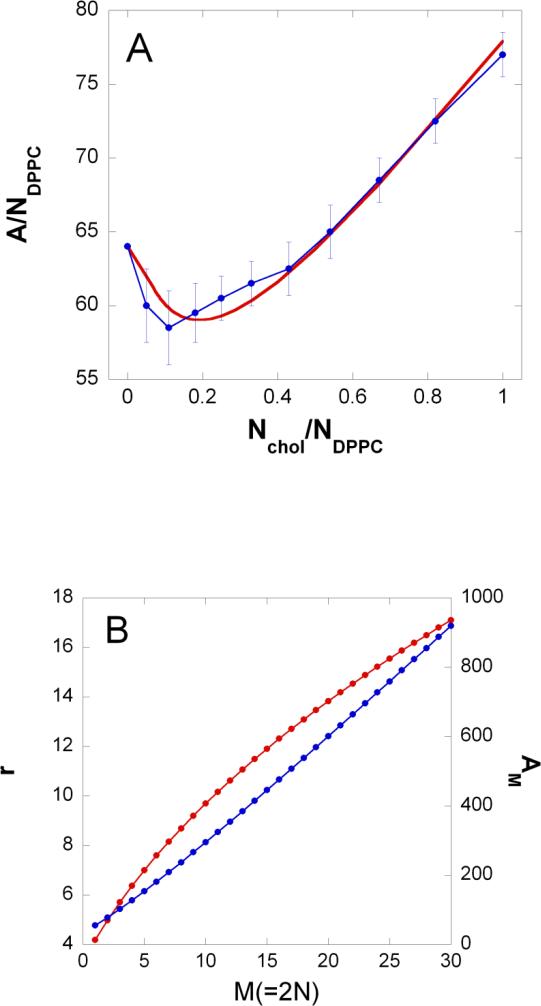
Molecular dynamical simulations 26 pointed out the condensing effect of cholesterol on phospholipid molecules in two-component cholesterol/phospholipid bilayers, i.e., the cross sectional area of the phospholipid molecules decreases with increasing proportion of the cholesterol molecules. The simulations determined the total membrane area divided by the number of DPPC (L-α-dipalmitoylphosphatidylcholine) molecules ( A/ NDPPC ), versus Nchol / NDPPC = Xc /(1 – Xc ), where Xc is the cholesterol mole fraction and Nchol is the number of cholesterol (see blue dots in Figure 1a). Based on this finding we assumed the following exponential relationship for the cross-sectional area of a phospholipid molecule, Ap(r) situated at a distance r from the center of the cholesterol molecule:
| (1) |
where rc is the radius of the cholesterol molecule, and is the cross sectional area of the phospholipid molecule in gel and fluid phase, respectively, and τ is a factor characterizing the strength of the condensation (see values of these parameters and refs. in Table 1). According to this expression, the cross sectional area of the phospholipid molecule increases from to as the distance from the center of the cholesterol molecule increases from rc to ∞. The number of phospholipid molecules surrounding the cholesterol within a radius r can be calculated from the following integral:
| (2) |
Figure 1. Condensing effect of cholesterol.
a) The total membrane surface area, A divided by the number of DPPC molecules, NDPPC versus the number of cholesterol molecules, Nchol divided by the number of DPPC molecules. Blue dots: from molecular dynamics calculations 44; Red line: calculated from Eq.2 (see text) by using parameter values τ=5Å and Ac = 34 Å2. The membrane surface area, A is given in Å2.
b) Red and blue curve is the radius, r and cross sectional area, AM(= r2π), respectively of a rigid cluster as a function of the number of hydrocarbon chains condensed to a cholesterol, M(=2N). These curves were calculated from Eq.2 (see text) by using parameter values τ = 5Å and Ac = 34 Å2. r and AM is given in Å and Å2, respectively.
Table 1.
List of parameters of the cholesterol/phospholipid model
| Parameter – symbol | Parameter - name | Parameter - value | Comments and references |
|---|---|---|---|
| z | Coordination number in triangular lattice | 4 | |
| Cross sectional area of DMPC in gel phase | 40 Å2 | 27 | |
| Cross sectional area of DMPC in fluid phase | 64 Å2 | 27 | |
| Ac | Cross sectional area of cholesterol | 34 Å2 | fitted (Fig.1) |
| rc = [Ac / π]1/2 | Radius of cholesterol's cross section | 3.29 Å | |
| τ | Condensation factor | 5 Å | fitted (Fig.1) |
| Δ HDMPC | DMPC transition enthalpy | 6000cal/mol | 27 |
| Δ SDMPC | DMPC transition entropy | 20.4cal/(mol·K) | 27 |
| euu | u-u interaction per unit length | −160.5cal/(mol· Å) | fitted (Fig.3) |
| eus | u-s interaction per unit length | −207cal/(mol· Å) | fitted (Fig.3) |
| s-s interaction per unit length at M >> 2 | −303.53cal/(mol· Å) | fitted (Fig.3) | |
| γ | −10952.48 Å/mol | fitted (Fig.3) | |
| T | absolute temperature | 310 K | 13 |
Note that Eq.2 assumes centro-symmetrical distribution of phospholipid molecules around the cholesterol molecule, an assumption that is not so good at small N. Let us imagine one layer of a bilayer that is built from similar units each containing one cholesterol and N(r) DPPC molecules. For this system Nchol / NDPPC = 1/ N(r) and A/ NDPPC = r2π/ N(r). Thus by using our simple model of condensation we can calculate the r2π/ N(r) versus 1/ N(r) curve (red curve in Figure 1a). This red curve is similar to the blue curve (obtained from MD simulations) at the following fitted parameter values: τ = 5Å and cholesterol cross sectional area . It is important to note that the fitted cross sectional area of the cholesterol molecule, Ac is close to the value obtained from cholesterol crystal data . By using the fitted τ and Ac values and Eq.2 one can calculate the N(r) function. Figure 1b (red curve) shows the inverse of 2N(r) function.
Modeling cholesterol/phospholipid mixture – qualitative description
We intend to model cholesterol/phospholipid mixtures close to the critical mole fractions. Our fluorescence/enzyme activity experimental data show that at each critical mole fraction a densely and a loosely packed phase coexist in the bilayer 13-14. In general an inhomogeneous system that is in thermal equilibrium with its surrounding is in a configuration that minimizes its free energy. This configuration is an optimal balance between low energy/low entropy and high entropy/high energy phases. As an analog case one may mention the gel-fluid mixed phase region of DMPC/DSPC bilayers 27.
In our model the high entropy/high energy phase contains cholesterol and phospholipid molecules where the phospholipid molecules are in fluid state and both cholesterol and phospholipid molecules are able to diffuse laterally (similar to liquid disordered phase 28). The low energy/low entropy phase, is represented by relatively rigid clusters (similar to condensed complexes 15). At critical mole fraction each rigid cluster is formed by phospholipid molecules that are condensed to a central cholesterol molecule. Within a cluster of strongly interacting components, the lateral diffusion of the molecules is negligible. However the rigid clusters are able to diffuse laterally in the loose phase and tend to aggregate with clusters of similar size. Within the aggregate of rigid clusters, the cholesterol molecules are regularly distributed, i.e., a ‘superlattice’ of cholesterol is formed.
The definition of the rigid cluster is a simplification of the real situation in the following two aspects: 1) At the 1st critical mole fraction the rigid cluster contains M=1 acyl chain, at the 2nd critical mole fraction the rigid cluster contains M=2 acyl chains, etc. The relationship between M and the respective critical mole fraction is:
| (3) |
Thus at Xcr = 0.2, M is equal to 8, so it should be the 8th critical mole fraction.
In reality a rigid cluster at the M-th critical mole fraction, may also contain more than or less than M/2 phospholipid molecules; however, M/2 is the most probable number. 2) If M is an odd number, M/2 refers to a non-integer number of phospholipid molecules. For example, M=9 refers to a cluster containing 4.5 phospholipid molecules or 9 acyl chains. This is impossible since in this study always two hydrocarbon chains belong to one phospholipid molecule. In reality, at the 9th critical mole fraction, two size rigid clusters, containing 4 and 5 phospholipid molecules are present at about the same concentration. However, as we will see, our model is able to handle only one size rigid clusters. Thus every time when M is an odd number the two size rigid clusters are substituted by one size rigid clusters each containing the average of M-1 and M+1 phospholipid molecules. The validity of this substitution is discussed in Supporting Information (part 1).
Modeling cholesterol/phospholipid mixture – statistical mechanical description
Defining the lattice
Let us use the following notations: n is the number of cholesterol molecules, m is the number of hydrocarbon chains, Ntot = n + m/2 is the total number of molecules, M is the number of hydrocarbon chains within a rigid cluster, Xc = n/Ntot is the cholesterol mole fraction, Ns is the number of rigid clusters (= the number of cholesterol molecules in rigid clusters), is the mole fraction of cholesterol molecules situated in rigid clusters.
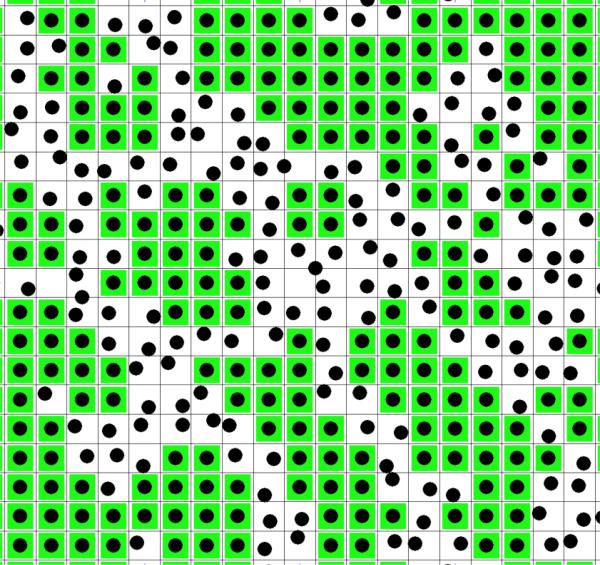
We develop a lattice model of the cholesterol/phospholipid two-component bilayer. A layer of the bilayer is represented by a lattice where each lattice unit is a square (see Figure 2). The surface area belonging to a lattice unit, AM is equal to the cross section of a rigid cluster formed by a cholesterol and M hydrocarbon chains condensed to the cholesterol. Figure 1b (blue curve) shows the calculated cross sectional area of the rigid cluster, AM as a function of the number of hydrocarbon chains condensed to the cholesterol, M.
Figure 2. Lattice model of phospholipid/cholesterol membrane.
A layer of the membrane is represented by units of squares (black lines). The surface area of a unit is equal with the surface area of a rigid cluster, AM (see Figure 1b). A unit represents either a rigid cluster (green unit with black dot at the center) or part of the fluid phase (white unit with randomly distributed black dot). Black dot: cholesterol. Green square: phospholipid molecules condensed to the central cholesterol.
There are two types of lattice units: Ns of them represent the rigid clusters (in Figure 2 these are green units with black dot at their center), while the remaining Nu lattice units represent the fluid phase of the system (in Figure 2 these are white units with randomly distributed black dots). Note that in the lattice model rigid clusters are represented by squares, rather than circles, in order to get tight packing within the aggregates. Thus Nu is equal with the surface area of the fluid phase divided by the surface area of a lattice unit (AM):
| (4) |
Free energy of the lattice
The free energy of the lattice has several energy and entropy terms. The internal energies of the s and u state lattice units Es and Eu are:
| (5) |
| (6) |
where εc and are the intramolecular energy of a cholesterol and the intramolecular energy of a phospholipid molecule in the fluid phase, respectively. is the average intramolecular energy of a phospholipid molecule in a rigid cluster. The interaction energy between the lattice units, Ei is calculated by assuming periodic boundary conditions (2nd equality in Eq.7; see Supporting Information part 2):
| (7) |
where Nuu , Nss and Nus is the number of nearest neighbor lattice units existing in u-u, s-s and u-s state, respectively. εuu, εss and εus is the interaction energy between nearest neighbor lattice units existing in u-u, s-s and u-s state, respectively, w = εus – (εuu + εss)/2 is the cooperativity energy and z=4 is the coordination number of the lattice, and . In the third equality the number of u-s nearest neighbor lattice units, Nus is approximated by zNuNs/(Nu + Ns) [Bragg-Williams approximation 29].
The lattice entropy contains four terms: 1) and 2) the internal entropy of the u and s state lattice units Su and Ss, respectively, 3) the mixing entropy of the molecules within the fluid phase and 4) the mixing entropy of the u and s state lattice units .
| (8) |
| (9) |
where σc and are the intramolecular entropy of a cholesterol and the intramolecular entropy of a phospholipid molecule in the fluid phase, respectively. is the average intramolecular entropy of a phospholipid molecule in a rigid cluster. The mixing entropy of the n – Ns cholesterol molecules and the phospholipid molecules in the fluid phase is:
| (10) |
where k = 1.987cal /(mol · K ) is the Boltzmann constant, and in the second equality Stirling's approximation was utilized. Note that both cholesterol and phospholipid molecules belong to each u state lattice unit and thus each u state lattice unit has mixing entropy. is the sum of the mixing entropies of the u state lattice units. On the other hand, the mixing entropy of the molecules belonging to an s state lattice unit is zero because the cholesterol is assumed to be located exactly at the center of the rigid cluster and thus the number of microstates within a rigid cluster is 1.
The s and u state lattice units can be arranged ong the lattice on different ways and thus the mixing entropy of the s and u state lattice units is:
| (11) |
where in the second equality Stirling's approximation was again utilized. By means of Eqs.5-11 the free energy, F of the lattice is:
| (12) |
where T is the absolute temperature.
On the energy and entropy parameters of the model
Our model will be applied to DMPC/cholesterol mixtures. In spite of the fact that the values of τ and Ac have been determined above for DPPC/cholesterol mixtures we will use the same parameter values for DMPC/cholesterol mixtures. Because of the close similarity of DMPC and DPPC molecules it is expected that an MD simulation of DMPC/cholesterol mixture would produce a similar curve to the one presented in Figure 1a and the value of the respective τ parameter would be similar too. DMPC's adjusted cross sectional area is 7% smaller than DPPC's adjusted cross sectional area 30 and thus one can expect a slight downward shift of the blue curve in Figure 1a. MD simulated electron density profiles 31-32 hint that cholesterol-DMPC interaction is similar to cholesterol-DPPC interaction because the DMPC and DPPC hydrocarbon chains are equal and longer than the length of the cholesterol, respectively. Thus we expect that the shape of the blue curve in Figure 1a will not change#. Since a lattice unit represents several molecules, the parameters associated with it depend on the actual number of the molecules and their physical and geometrical properties.
Let us consider first the intramolecular energy difference: . In an s state lattice unit the condensing effect of the cholesterol brings the nearby phospholipid molecules close to the gel state. Thus if the rigid cluster contains only one DMPC molecule, i.e., M=2 the energy difference is where ΔHDMPC is the gel-to-fluid transition enthalpy of DMPC bilayers 27. In the case of M>2 the average state of the phospholipid molecules in the rigid cluster is between the fluid and gel state. This average state can be characterized by the decrease of the cross sectional area of the phospholipid molecules as a consequence of the condensing effect of the cholesterol. Thus we assume that
| (13) |
One can assume a similar equation for the intramolecular entropy difference:
| (14) |
where ΔSDMPC is the gel-to-fluid transition entropy of DMPC bilayers.
Similarly, the interaction energy between nearest neighbor lattice units depends on the geometrical and physical properties of the interacting units. Since the size of a unit (either u or s state) is defined by the size of a rigid cluster, the interaction free energies should be proportional to the circumference or the radius of a rigid cluster, r(M). Thus
| (15) |
| (16) |
| (17) |
where proportionality factors euu and eus (the u-u and u-s interaction energies per unit length) are constant. However, in the case of s-s interaction the interaction energy per unit length changes with the peripheral density of the phospholipid molecules in the rigid clusters. In Eq. 17 the peripheral density is proportional to 1/ Ap(r) and γ is the proportionality constant. With decreasing M this density increases and the interaction per unit length increases, too. Note that the proportionality factors euu and eus are considered to be constant because the fluid phase is loosely packed and the short range van der Waals interactions between u-u and u-s units are slightly affected by the cholesterol content of the fluid phase. On the other hand the peripheral density of the rigid clusters depends on the cholesterol content of the rigid phase and has significant effect on the interaction of the closely packed s-units.
Table 1 lists the constants utilized by this model.
Results
The free energy function in Eq.12 can be used when the membrane is close to the critical cholesterol mole fractions, where the fundamental assumption of our model is most applicable, i.e, the system contains one type of rigid cluster each containing M/2 phospholipid molecules condensed to one cholesterol molecule. At any given cholesterol mole fraction Xc, which is close to a critical mole fraction , one can find the average number of the s state lattice units, . For large Ntot the position of the minimum of the free energy function well approximates the average (maximum term method 33). By using this average and Eq.4 one can calculate the average number of u state lattice units . Finally, at given Xc and M, the proportion of the densely packed area (i.e., the area covered by the s state lattice units) of the cholesterol/phospholipid bilayer is:
| (18) |
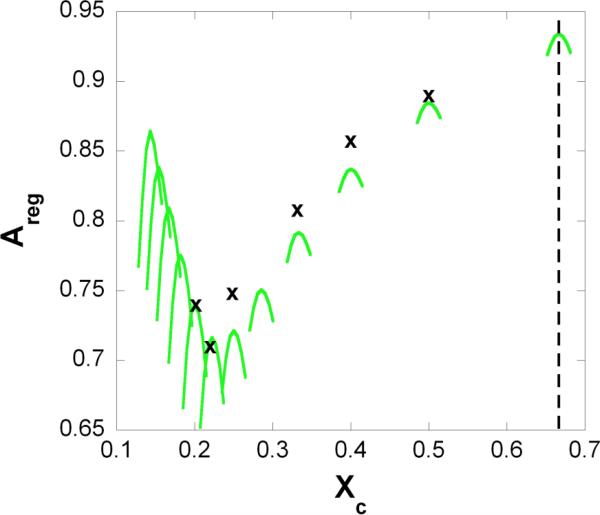
The proportion of the densely packed area has been measured at different cholesterol mole fractions in cholesterol/DMPC and cholesterol/1-palmitoyl-2-oleoyl-sn-3-phosphocholine (POPC) mixtures 13-14. Figure 3 shows the measured Areg values at six critical mole fractions (x) and calculated Areg vs. Xc curves at several critical mole fractions (green lines). We emphasize the following points: (1) the calculated Areg (green curves) yields a biphasic change with Xc , showing a local maximum at Xcr , similar to the Areg curves determined from nystatin measurements 13-14 and (2) the local maximum points of the calculated curves (green curves) match with the local maxima (x) of the Areg curves determined from nystatin fluorescence 13-14.
Figure 3. Proportion of regularly packed membrane area.
Regular area fraction, Areg is plotted against the cholesterol mole fraction, Xc. x: measured local maxima of the regular area fraction 13; green lines: the curve of regular area fractions calculated by Eq.18. Dashed line: cholesterol precipitates from the bilayer above this mole fraction (as noted in the text, this actual limit is characteristic for cholesterol/PC bilayers). The values of the model parameters are listed in Table 1.
The critical mole fraction, is listed in Table 2 at different M values that were used in our calculations. We also listed the cooperativity energy of the aggregation of the rigid clusters w (defined at Eq.7) for each M value. These cooperativity parameters were calculated from our model by using Eqs.15-17 and the model parameters in Table 1 and Figure 1b.
Table 2.
Cluster characteristics
| M | w (cal/mol) a | reference b | |
|---|---|---|---|
| 1 | 0.666 | 268.2 | |
| 2 | 0.5 | 280.1 | 13 |
| 3 | 0.4 | 289.6 | 13 |
| 4 | 0.333 | 297.8 | 13 |
| 5 | 0.286 | 305.3 | 8 |
| 6 | 0.25 | 312.4 | 13 |
| 7 | 0.222 | 319.2 | 13 |
| 8 | 0.2 | 325.9 | 13 |
| 9 | 0.182 | 332.4 | 8 detected dip at Xcr = 0.175 |
| 10 | 0.166 | 338.9 | 2 |
| 11 | 0.154 | 345.4 | 19 |
| 12 | 0.143 | 351.8 | 19 |
Cooperativity energy defined by w = εus – (εuu + εss)/2.
The listed references refer to ergosterol/DMPC and cholesterol/DMPC mixtures. A complete list of critical concentration measured on different cholesterol/phospholipid mixtures can be found at Ref.2.
So far we calculated the properties of the coexisting densely and a loosely packed phase at 310K, where the majority of the fluorescence/enzyme activity measurements of the biphasic changes were performed 13. However, by using the same model one can calculate the system's properties at different temperatures as well. The average enthalpy per phosholipid molecule can be calculated from the following equation:
| (19) |
where the 3rd term in the square bracket is the average number of gel state phospholipid molecules. By means of Eq.19 one can calculate the temperature, T95%ΔH where 95% of the phospholipids are in fluid state, i.e. the temperature at which .
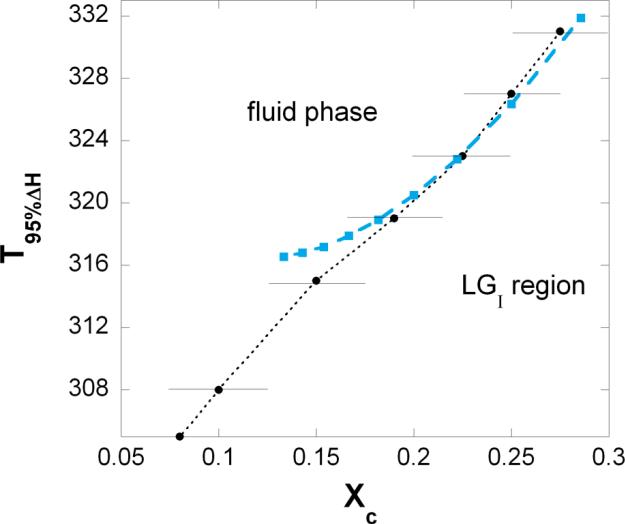
Above T95%ΔH the coexisting densely and loosely packed phase (called LGI region) turns into fluid phase. In Figure 4 these temperatures are shown at critical cholesterol mole fractions, (blue squares). Representative data points from FRAP and ESR experiments are shown with error bars 34. Detected phase boundary is marked by dotted line.
Figure 4. Calculated and measured phase boundary.
Close to the critical cholesterol mole fractions one can calculate the temperature, T95%ΔH where 95% of the phospholipids are in fluid state, i.e. the temperature where (see Eq.19). Above T95%ΔH the LGI phase turns into fluid phase. This calculated temperature is plotted against the critical mole fraction, (blue squares). Representative experimental data points are shown with error bars (black dots from 34). Detected phase boundary is marked by dotted line.
Discussion
Liquid disordered – liquid ordered mixed phase or LGI region
As a result of a subtle difference in the interpretation of the experimental data two rather different phase diagrams of cholesterol/phospholipid mixtures have been proposed. The first and more frequently cited phase diagram of cholesterol/phospholipid mixtures (see Fig.6a in 35) was proposed by Ipsen and his coworkers 28. Based on DSC, NMR, EPR and micromechanical studies, a phase diagram, typical for eutectic mixtures, has been constructed. Near the transition temperature of the phospholipid component the following phases were identified: liquid-crystalline phase, referred to as liquid disordered (ld phase), at low cholesterol mole fractions ( Xc ≤ 0.08); at high cholesterol mole fractions ( Xc ≥ 0.25) the phase was referred to as liquid ordered (lo phase); while these two phases coexist at intermediate cholesterol mole fractions. The nomenclature of liquid ordered phase was coined to describe lipids with ordered and extended acyl chains, but which still exhibit the fast lateral diffusion and high rotational mobility that is characteristic of a liquid.
The second and less frequently cited phase diagram (see Fig.6b in 35) is based on DSC 36, NMR 37 and FRAP 38-39 data. According to this phase diagram, near the transition temperature of the phospholipid component, the system is in fluid phase up to about Xc = 0.08 and then as the cholesterol mole fraction increases (from 0.08 to 0.6) the fluid phase gradually converts to lo phase. Thus in contrast to the first type of phase diagram there is no phase boundary at Xc = 0.25. Also, micromechanical studies 40 showed that at 0.5 ≥ Xc ≥ 0.125 the mixture behaved as a liquid with no surface shear rigidity but with greatly reduced membrane-area compressibility and no sharp change in the micromechanical properties was found at Xc = 0.25. In the literature this gradually changing phase has no generally accepted name. It was called lo phase 35, LGI region, i.e. liquid-gel type phase 37, and Loα + Loβ mixed phase 36. Based on our fluorescence/enzyme activity data we prefer to use the second type of the phase diagram and call the above discussed phase LGI region. Although, a specific behavior detected close to Xc = 0.25 should be mentioned here: the local maxima of the Areg curves have minimal value at (see Figure 3).
Critical mole fractions in LGI region
When developing the above mentioned two types of phase diagrams the fluorescence/enzyme activity data from cholesterol/phospholipid (or cholesterol analogue/phospholipid) mixtures were not examined using small sterol mole fraction increments. It was in 1994 when the first fluorescence data on dehydroergosterol/DMPC mixtures with small sterol mole fraction increments (~0.3 mol%) were published 5. Since then, fluorescence properties, such as intensity, anisotropy, quenching rate constant, and lifetime, measured at many different sterol mole fractions in a variety of sterol/phospholipid mixtures showed maxima (or minima) at certain critical mole fractions, Xcr 's (reviewed in 1). This phenomenon was also observed when using infrared spectroscopy and non-fluorescence based enzyme assays (reviewed in 2).
In the light of the model described in this paper one can give a more detailed characterization of the LGI region that is able to rationalize the biphasic changes in fluorescence and enzymatic activity at Xcr as well. Thus the LGI region is considered to be a mixture of fluid phase and rigid clusters. A rigid cluster is formed by a cholesterol molecule and phospholipid molecules that are condensed to the cholesterol. The composition of a rigid cluster, agrees with a measured critical mole fraction, Xcr , i.e. where M is the number of acyl chains condensed to the cholesterol molecule. The shape of an isolated rigid cluster is assumed to be cylindrical, while the radius of the cylinder depends on the number of condensed acyl chains, M (see Eq.2 and Figure 1b). Rigid clusters of similar size tend to aggregate. Within each aggregate of closely packed rigid clusters the cholesterol molecules are regularly distributed (see Figure 2). This is similar to superlattice theories of critical mole fractions. However, in contrast to superlattice theories, in our model the acyl chains surrounding each cholesterol molecule do not sit on a separate lattice point, but rather a whole rigid cluster belongs to a lattice point. As a consequence of these differences our model is able to predict more critical concentrations than Virtanen's superlattice theory 7 in which the guest molecules are assumed to follow hexagonal order. For example Xcr = 0.333 is equal to in our model, while Virtanen's superlattice theory is unable to predict this critical concentration. However, it should be mentioned that, in the extended hexagonal sterol superlattice theory 5 where a cholesterol molecule is allowed to occupy two hexagonal lattice points when the cholesterol cross sectional area is larger than the cross sectional area of one phopholipid acyl chain, Xcr = 0.333 can be predicted. However, non of the superlattice theories are able to predict the critical mole fraction observed at 28.6 mol% 8, while this is the 5th critical mole fraction, according to our model.
Characterization of the LGI region
Since the cooperativity energy of aggregation of rigid clusters, w is smaller than the thermal energy unit, kT (see Table 2) there are numerous aggregates with a broad size distribution 41. The aggregates are not percolated, i.e., there is no stable aggregate with size comparable with the bilayer surface area.
The above characterization of the LGI region is consistent with all the available experimental results (DSC, NMR, FRAP, micromechanics, fluorescence spectroscopy). Namely, the presence of condensed phospholipid molecules increases the acyl chain order, the rigid clusters decreases the membrane lateral compressibility, and finally because of the microscopic size of the aggregates of the rigid clusters the membrane remains mechanically fluid-like. The fluid-like membrane with numerous aggregates of the rigid clusters reminds us to the spongy lumps of drift ice called sludge and one may call the LGI region as a ‘sludge phase’ of phospholipid/cholesterol mixtures.
Comparing the model with experimental results
The model predicts critical mole fractions at any positive integer of M by Eq.3. The first critical concentration, and consequently a maximum of Areg is expected to be measured at (see Figure 3). However this should be a half-maximum because the cholesterol starts to precipitate from the phosphatidylcholine lipid matrix above this critical mole fraction. It is important to note that the solubility limit depends on the lipid, e.g. for PEs, the solubility limit is 0.50 42. In this case the maximum at is not measurable at all, while only the half-maximum is measurable at . Thus the actual solubility limit marks the upper limit of the applicability of our model. All critical mole fractions listed in Table 2 were found experimentally, although and were detected only rarely 2.
With increasing M the predicted critical mole fractions become closer to each other and their reliable detection becomes increasingly difficult. Also, the condensing effect of the cholesterol should be weaker on phospholipid molecules that are farther away. Once the condensing effect is comparable with the thermal energy unit we reach the upper bound of the size of a rigid cluster, and the respective cholesterol mole fraction marks the lower limit of the applicability of our model. The limit mole fraction of the applicability increases with increasing temperature. At T=318K the limit mole fraction is 0.1666 where the calculated phase boundary starts deviating from the detected phase boundary (see Figure 4). This limit mole fraction should be lower at T=308K where critical mole fraction was detected even at 0.085,43. The model was unable to predict the critical mole fractions occasionally detected at 0.265 and 0.468,43. However, the dips/peaks at 0.265 and 0.46 were observed only in rare cases which may reflect their instability.
The model works close to each critical mole fraction where the existence of one type of rigid cluster is assumed. We plan to eliminate this limitation of the model in the future.
In spite of the fact that our model is a massive simplification of the real system it is able to explain and reproduce most of its detected features. It is also encouraging that the values of the model parameters (listed in Table 1) are within a physically meaningful range. Finally, it is important to mention that the model provides the same results (Figures 3,4) at different coordination numbers, z as long as the values of the following products remain the same: zess , zeus and zeuu .
Comparison with the theory of condensed complexes
The model of condensed complexes of cholesterol and phospholipids developed by Radakrishnan and McConnell 16 is the closest to the model presented in this work. Both models consider the same three components: (1) phospholipid, (2) cholesterol (these components form the fluid phase) and (3) condensed complex (that we call rigid cluster). Rigid clusters represent a subset of condensed complexes, where q=1. The model of condensed complexes applies the regular solution theory, while our model uses the rather similar lattice theory. The fundamental difference is, that based on experimental and molecular dynamics simulations, in our model the excluded area interactions between the s and u units are explicitly taken into account, while in the condensed complexes model the components are considered as points and their interactions are represented by three fitted model parameters: α12 , α13 , and α23. These model parameters depend on the composition of the condensed complex and should be fitted again when the composition of the condensed complex is changing. On the other hand the parameters in our model are the same for any composition of the rigid clusters.
Conclusions
The statistical mechanical model described above indicates that phospholipid-cholesterol condensed complexes and sterol superlattices are inter-related. The condensed complexes (rigid clusters) are the “precursors” of sterol superlattices (aggregates of rigid clusters), which occur at a later time. A rigid cluster is formed by a cholesterol molecule and phospholipid molecules that are condensed to the cholesterol. Rigid clusters of similar size tend to form closely packed aggregates, in which cholesterol molecules are regularly distributed into superlattices. Our model rationalizes almost every critical sterol mole fraction for superlattice formation and shows that a biphasic change in membrane properties such as Areg at the critical mole fractions is plausible from the statistical thermodynamics point of view. Using this model, the LGI region of the phase diagram can be considered as a mixture of fluid phase and superlattices (aggregates of rigid clusters). The extent and type of superlattices should vary with cholesterol mole fraction in a predictable, non-monotonic manner. Consequently, membrane properties (e.g., phase diagram and membrane packing) and functions (e.g., solute permeability and surface enzyme activities) should change with cholesterol content in the same manner.
Supplementary Material
Acknowledgments
We are very thankful for Professor Olle Edholm for the MD simulations on DMPC/cholesterol mixtures. I.P.S. also acknowledges Professors Stuart Sealfon, Pál Jedlovszky and Sri Chinmoy's help. This work was supported by contract NIH/NIAID HHSN272201000054C. P.L.G.C. acknowledges the support from NSF (DMR1105277).
Footnotes
Just recently, Professor Olle Edholm ran MD simulations at 303K for DMPC/cholesterol mixtures and obtained a similar curve, with a slight downward shift, to the one presented in Figure 1a (unpublished result).
References
- 1.Chong PLG, Sugar IP. Chemistry and Physics of Lipids. 2002;116:153. doi: 10.1016/s0009-3084(02)00025-7. [DOI] [PubMed] [Google Scholar]
- 2.Chong PLG, Olsher M. Soft Materials. 2004;2:85. [Google Scholar]
- 3.Chong PLG, Zhu WW, Venegas B. Biochimica Et Biophysica Acta-Biomembranes. 2009;1788:2. doi: 10.1016/j.bbamem.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Somerharju P, Virtanen JA, Cheng KH, Hermansson M. Biochimica Et Biophysica Acta-Biomembranes. 2009;1788:12. doi: 10.1016/j.bbamem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Chong PLG. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10069. doi: 10.1073/pnas.91.21.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virtanen JA, Ruonala M, Vauhkonen M, Somerharju P. Biochemistry. 1995;34:11568. doi: 10.1021/bi00036a033. [DOI] [PubMed] [Google Scholar]
- 7.Virtanen JA, Somerharju P, Kinnunen PK. J. Journal of Molecular Electronics. 1988;4:233. [Google Scholar]
- 8.Liu F, Chong PLG. Biochemistry. 1999;38:3867. doi: 10.1021/bi982693q. [DOI] [PubMed] [Google Scholar]
- 9.Huang JY, Feigenson GW. Biophysical Journal. 1999;76:2142. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang JY. Johnson ML, Holt JM, Ackers GK, editors. Methods in Enzymology: Biothermodynamics,Vol 455, Part A. 2009;455:329. doi: 10.1016/S0076-6879(08)04219-5. [DOI] [PubMed] [Google Scholar]
- 11.Tang D, Chong PLG. Biophysical Journal. 1992;63:903. doi: 10.1016/S0006-3495(92)81672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugar IP, Tang DX, Chong PLG. Journal of Physical Chemistry. 1994;98:7201. [Google Scholar]
- 13.Wang MM, Sugar IP, Chong PLG. Biochemistry. 1998;37:11797. doi: 10.1021/bi980290k. [DOI] [PubMed] [Google Scholar]
- 14.Wang MM, Olsher M, Sugar IP, Chong PLG. Biochemistry. 2004;43:2159. doi: 10.1021/bi035982+. [DOI] [PubMed] [Google Scholar]
- 15.McConnell HM, Radhakrishnan A. Biochimica Et Biophysica Acta-Biomembranes. 2003;1610:159. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan A, McConnell HM. Biophysical Journal. 1999;77:1507. doi: 10.1016/S0006-3495(99)76998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radhakrishnan A, McConnell HM. Journal of the American Chemical Society. 1999;121:486. [Google Scholar]
- 18.Venegas B, Wolfson MR, Cooke PH, Chong PLG. Biophysical Journal. 2008;95:4737. doi: 10.1529/biophysj.108.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Sugar IP, Chong PLG. Biophysical Journal. 1997;72:2243. doi: 10.1016/S0006-3495(97)78868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan A, McConnell H. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12662. doi: 10.1073/pnas.0506043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venegas B, Sugar IP, Chong PLG. Journal of Physical Chemistry B. 2007;111:5180. doi: 10.1021/jp070222k. [DOI] [PubMed] [Google Scholar]
- 22.Parker A, Miles K, Cheng KH, Huang J. Biophysical Journal. 2004;86:1532. doi: 10.1016/S0006-3495(04)74221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helrich CS, Schmucker JA, Woodbury DJ. Biophysical Journal. 2006;91:1116. doi: 10.1529/biophysj.105.076281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Q, Cheng KH, Vaughn MW. Journal of Physical Chemistry B. 2007;111:11021. doi: 10.1021/jp070487z. [DOI] [PubMed] [Google Scholar]
- 25.Vaz WLC, Melo ECC, Thompson TE. Biophysical Journal. 1989;56:869. doi: 10.1016/S0006-3495(89)82733-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkowitz ML. Biochimica Et Biophysica Acta-Biomembranes. 2009;1788:86. doi: 10.1016/j.bbamem.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Sugar IP, Thompson TE, Biltonen RL. Biophysical Journal. 1999;76:2099. doi: 10.1016/S0006-3495(99)77366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ipsen JH, Karlstrom G, Mouritsen OG, Wennerstrom H, Zuckermann MJ. Biochimica Et Biophysica Acta. 1987;905:162. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 29.Huang K. Statistical Mechanics. Wiley, John & Sons; New York: 1990. [Google Scholar]
- 30.Nagle JF, Tristram-Nagle S. Biochimica Et Biophysica Acta-Reviews on Biomembranes. 2000;1469:159. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jedlovszky P, Mezei M. Journal of Physical Chemistry B. 2003;107:5311. [Google Scholar]
- 32.Smondyrev AM, Berkowitz ML. Biophysical Journal. 1999;77:2075. doi: 10.1016/S0006-3495(99)77049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill TL. Dover; New York: 1987. [Google Scholar]
- 34.Almeida PFF, Vaz WLC, Thompson TE. Biochemistry. 1992;31:6739. doi: 10.1021/bi00144a013. [DOI] [PubMed] [Google Scholar]
- 35.Veatch SL, Keller SL. Biochimica Et Biophysica Acta-Molecular Cell Research. 2005;1746:172. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 36.McMullen TPW, McElhaney RN. Biochimica Et Biophysica Acta-Biomembranes. 1995;1234:90. doi: 10.1016/0005-2736(94)00266-r. [DOI] [PubMed] [Google Scholar]
- 37.Huang TH, Lee CWB, Dasgupta SK, Blume A, Griffin RG. Biochemistry. 1993;32:13277. doi: 10.1021/bi00211a041. [DOI] [PubMed] [Google Scholar]
- 38.Rubenstein JR, Smith BA, McConnell HM. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:15. doi: 10.1073/pnas.76.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alecio MR, Golan DE, Veatch WR, Rando RR. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79:5171. doi: 10.1073/pnas.79.17.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans E, Needham D. Faraday Discussions. 1986;81:267. doi: 10.1039/dc9868100267. [DOI] [PubMed] [Google Scholar]
- 41.Sugar IP. Journal of Physical Chemistry B. 2008;112:11631. doi: 10.1021/jp800945j. [DOI] [PubMed] [Google Scholar]
- 42.Huang JY, Buboltz JT, Feigenson GW. Biochimica Et Biophysica Acta-Biomembranes. 1999;1417:89. doi: 10.1016/s0005-2736(98)00260-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu F, Chong PLG. Biophysical Journal. 1997;72:MP266. doi: 10.1016/S0006-3495(97)78868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edholm O, Nagle JF. Biophysical Journal. 2005;89:1827. doi: 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.