Abstract
High-resolution microscopic magnetic resonance imaging (μMRI) and diffusion tensor imaging (DTI) were performed to characterize brain structural abnormalities in a mouse model of mucopolysaccharidosis type VII (MPS VII). μMRI demonstrated a decrease in the volume of anterior commissure and corpus callosum and a slight increase in the volume of the hippocampus in MPS VII vs. wild-type mice. DTI indices were analyzed in gray and white matter. In vivo and ex vivo DTI demonstrated significantly reduced fractional anisotropy in the anterior commissure, corpus callosum, external capsule and hippocampus in MPS VII vs. control brains. Significantly increased mean diffusivity was also found in the anterior commissure and corpus callosum from ex-vivo DTI. Significantly reduced linear anisotropy was observed from the hippocampus from in-vivo DTI, whereas significantly decreased planar anisotropy and spherical anisotropy were observed in the external capsule from only ex-vivo DTI. There were corresponding morphological differences in the brains of MPS VII mice by hematoxylin and eosin staining. Luxol fast blue staining demonstrated less intense staining of the corpus callosum and external capsule; myelin abnormalities in the corpus callosum were also demonstrated quantitatively in toluidine blue-stained sections and confirmed by electron microscopy. These results demonstrate the potential for μMRI and DTI for quantitative assessment of brain pathology in murine models of brain diseases.
Keywords: Diffusion tensor imaging, Live animal imaging, Lysosomal storage diseases, Microscopic MRI, Mucopolysaccharidosis, Myelination, Neuropathology
Introduction
Lysosomal storage diseases (LSDs) constitute a group of inherited disorders characterized by an accumulation of undigested or partially digested macromolecules that result in cellular dysfunction and clinical abnormalities (1-3). Progressive lysosomal storage and distension of cells are hallmarks of the pathological changes in LSDs. Major subgroups of LSDs are the mucopolysaccharidoses, which involve degradation of glycosaminoglycans (4). Central nervous system pathology in both human and animal models of mucopolysaccharidoses include swelling of the neurons and glial cells, neurodegeneration, abnormalities in neural stem cells, and changes in gene expression in multiple cellular processes (5-7).
Magnetic resonance imaging (MRI) has been used to demonstrate brain abnormalities in various types of metabolic disorders including mucopolysaccharidoses (MPS) in humans (8-10), but the small number of patients and difficulty in obtaining age matched controls, make quantitative analysis difficult (11). There are no reports on murine models of MPS using MRI methods to evaluate and monitor alterations in brain structure. However, high-field-strength magnets can be used to assess parameters such as distribution of stem cells that are labeled with paramagnetic agents in the mouse brain (12, 13). Diffusion tensor imaging (DTI) is a non-invasive imaging technique that allows investigation of the microstructural changes in gray and white matter regions of the brain (14, 15). Fractional anisotropy (FA) and mean diffusivity (MD), which are based on the rotational invariant eigenvalues, are the most commonly used DTI parameters for differentiating normal from various pathological conditions (16). FA is a measure indicating the overall directionality of water diffusion that is greater in organized white matter tracts and lower in CSF and disorganized fibers. MD is a DTI-derived parameter, rotationally invariant, which quantifies water diffusion within tissue (14). The rotationally variant indices give a more accurate measure of diffusion anisotropy than do the rotationally invariant measures, which tend to underestimate the true anisotropy. Rotationally invariant indices contract the tensor to one scalar value and do not provide the directional variation of the diffusion anisotropy. For example, a cigar-shaped and a pancake-shaped ellipsoid can have equal FA although their shapes are very different (17). A model of diffusion anisotropy that is based on a set of 3 basic diffusion tensor metrics and 3 major shapes of the diffusion tensor ellipsoid expressed by FA are linear anisotropy (CL), planar anisotropy (CP), and spherical anisotropy (CS), such that CL + CP + CS = 1 (17). Thus, these 3 metrics parameterize a barycentric space in which 3 shape extremes (linear, planar, and spherical) are at the corner of a triangle. These 3 metrics of tensor basics are then normalized to obtain the shape of the tensor, which provide information about the linearity and anisotropy of the tissue (18). Along with FA and MD, these geometrical DTI indices can also be helpful in characterization of shape of the diffusion tensor along with orientation and integrity of the brain tissues. (17, 19, 20).
Animal disease models are critical for developing non-invasive imaging methods to assess CNS pathology in live patient brains (11). In the current study, we evaluated the efficacy of MRI in characterizing a widely studied mouse model of MPS, i.e. MPS type VII, caused by a genetic mutation and deficiency of β-glucuronidase. β-glucuronidase is lysosomal acid hydrolase involved in the stepwise degradation of glucuronic acid-containing glycosaminoglycans (dermatan sulfate, heparan sulfate and chondroitin sulfate) (1, 21). The affected MPS VII mouse closely reflects the human disease and the pathology has been well characterized (22-24). Although the MPS diseases are not classified as leukodystrophies, it was recently reported that a number of myelin-associated genes are significantly downregulated in the MPS VII mouse brain (6). This aberrant gene expression may affect structural integrity of the white matter structures in the MPS VII mouse brain, but myelin is notoriously difficult to quantify in histological sections. DTI has been used for detecting changes in myelination in the developing brain (25-28), and for assessing tissue abnormalities in demyelinating diseases (29, 30) and other neurological disorders (31).
Hence, we evaluated the microstructural abnormalities in MPS VII mouse brain by ex vivo high-resolution microscopic MRI (μMRI) to determine the morphological changes in MPS VII mice and then in vivo and ex vivo DTI were performed to characterize the microstructural integrity of the brain tissue by using the diffusion properties of the brain water in this model.
Materials and Methods
Experimental Animals
Wild-type (WT) and MPS VII-affected C3H/HeOuJ mice were bred in the animal facility at Abramson Research Center, Children's Hospital of Philadelphia. The study was approved by the Institutional Animal Care and Use Committees of the University of Pennsylvania or Children's Hospital of Philadelphia. Age/sex-matched pairs of MPS VII (GusB-/-)-affected mice and WT littermates on a C3H/HeOuJ background (23) were used at 5 to 6 months of age in all assays; this is an age when there are severe pathologic abnormalities in the CNS (5, 6, 23, 24, 32). The following numbers of mice were used: μMRI, 5 WT, 5 MPS VII; in vivo and ex vivo DTI, 5 WT, 6 MPS VII; and quantitative histology of the corpus callosum, 4 WT, 4 MPS VII. The genotypes of the mice were determined at birth using a polymerase chain reaction assay for the mutation (33).
Ex Vivo μMRI
Animals were anesthetized with an intraperitoneal injection of ketamine. The mice were then transcardially perfused with phosphate saline buffer, followed by perfusion with a solution of 4% paraformaldehyde (PFA). After perfusion fixation, the brains were removed and stored at 4°C in 4% PFA solution for 7 days. After fixation, the brains were immersed in 4% PFA solution containing 2% gadopentetate dimeglumine (Gd-DTPA) for 4 days to reduce the T1 and T2 relaxation times of the tissue enabling high-resolution imaging in short time (34). The brain sample was then placed in a plastic tube with proton-free solution (Fomblin, Ausimont, Thorofare, NJ). A custom-built solenoid coil (20-mm inner-diameter) was used to acquire the data using a Varian 9.4T, 8.9 cm vertical bore magnet equipped with a 45-mm inner-diameter 100 gauss/cm gradient tube and interfaced to a Varian Direct Drive console (Agilent Technologies, Palo Alto, CA) operating the Varian software version vnmrj 2.3.C. A 3-dimensional gradient echo pulse sequence was used to acquire the μMRI data using the following parameters: time of repetition (TR) = 50 ms, echo time (TE) = 5 ms, number of acquisitions = 2, field of view (FOV) = 20 mm × 10 mm × 10 mm, and acquisition image matrix = 512 × 256 × 256, resulting in 39 μm isotropic resolution in an acquisition time of 1 hour and 50 minutes. After completion of ex vivo imaging, brain samples were transferred in a fresh solution of 4% PFA and stored at 4°C.
In Vivo DTI
For the in vivo MRI studies, animals were anesthetized with 3% isoflurane in oxygen. To minimize motion induced artifacts during the imaging studies, the head of the mouse was secured using ear pins and a nose cone using an in-house developed restraining device. Subdural needle electrodes were placed in the forelimbs, a respiration pillow was placed on the dorsal side of the body, a thermistor was inserted into the rectum and the animal was placed on a cradle. The cradle with the animal in position was then inserted into a 20-mm internal-diameter transmit-receive quadrature birdcage coil (M2M, Cleveland, OH) and the coil was placed in the center of the magnet. Core body temperature, heart rate and respiratory rate of the animal were monitored using an MR compatible vital signs monitoring unit (SA Instruments, Stony Brook, NY). In vivo DTI studies were performed on a 4.7T, 50-cm horizontal bore magnet (Magnex Scientific Inc., Abingdon, Oxon, UK) equipped with a 12-cm, 25 G/cm gradient set interfaced to a Varian Direct Drive console (Agilent Technologies) operating the Varian software version vnmrj 2.3.C. After initial scout and T2-weighted images in the axial plane, DTI images were acquired using a multi-slice diffusion-weighted spin echo sequence. The diffusion weighting was applied along 6 directions optimally selected for anisotropy measurement using a b-value of the 996.713 s/mm2. A total of 15 imaging slices in the axial plane were acquired to cover most of the brain parenchyma. Imaging parameters included: TR = 2000 ms; TE = 33 ms; FOV = 20 mm × 20 mm, number of averages = 2, interleaved slice orientation with 0.8-mm slice thickness and matrix size = 128 × 128, resulting in 156 × 156 × 800 μm resolution, with a total acquisition time of approximately 2 hours. During the scan, anesthesia was maintained between 1%-1.5% isoflurane and the animal body temperature was regulated at 37(±1)°C by blowing warm air into the magnet bore via a hose connected to a thermostatically controlled warm air device (SA Instruments). Following the in vivo DTI scan, the mice were killed by means of transcardiac perfusion and were fixed using 4% PFA and the brains were extracted and stored at 4°C for ex vivo DTI studies.
Ex Vivo DTI
Ex vivo DTI images were acquired on a Varian 9.4T, 8.9-cm vertical bore magnet and a specially designed loop gap resonator probe with 20-mm inner-diameter using a 3-dimensional multi-echo pulsed-gradient spin echo sequence (35, 36). The sequence parameters were as follows: TR =800 ms; TE = 29.50 ms; FOV = 17 mm × 8.5 mm × 10 mm; acquisition matrix size = 136 × 68 × 80; resulting in 125 × 125 × 125 μm isotropic resolution, number of acquisitions = 6 and b-value = 902 s/mm2. The diffusion-weighted images were acquired with diffusion weighting in 6 non-collinear directions in a total acquisition time of 13 hours and 19 minutes per brain sample. At the end of ex vivo scan the brains were stored at 4°C in 4% PFA for histology.
Image Processing and Data Quantification
Image reconstruction from acquired varian FID files was performed using in-house custom software routines in the IDL programming environment (ITT Visual Information Solutions, Boulder, CO). For the μMRI, high-resolution images from each individual brain were aligned to a template using in-house developed IDL based routines. Volumetric analysis of the μMR images was performed using ITK-SNAP (University of Pennsylvania, Philadelphia, www.itksnap.org) (37). Regions of interest (ROIs) were manually drawn in the anterior commissure, corpus callosum and hippocampus and the measured volumes were normalized to the overall brain volume in both MPS VII and WT mice.
The in vivo and ex vivo DTI data were processed to images and saved in DTI studio format using IDL routines. A Gaussian filter (width 0.5) was used to smooth the data and remove some noise from the images. DTIstudio software (Johns Hopkins School of Medicine, available at: www.mristudio.org) (38) was used to compute different DTI indices including FA, MD, CL, CP and CS maps from both in vivo and ex vivo DTI data.
To assess the changes in diffusivity further, a sub-analysis of the axial and radial diffusivity (AD and RD) values from the corpus callosum was also performed from the both in vivo as well as ex vivo DTI data (Supplementary Table). Despite the restraining device used, motion induced artifacts precluded the use of in vivo DTI data from all 15 slices; the central 3 to 4 slices were always free of any motion artifacts and were used to select the ROIs for data analysis (39).
The FA-weighted color maps were used to draw ROIs from different brain areas. We drew 2 ROIs bilaterally (left and right hemisphere) for the external and internal capsule and the cerebral cortex, caudate putamen, and hippocampus for the DTI images. For the corpus callosum, a single ROI was drawn on the mid axial slice at the level of the dorsal 3rd ventricle. Separate ROIs were drawn on the in vivo and ex vivo DTI data sets. Representative ROIs are shown in Figure 1A. Additionally, a single ROI was also placed on the anterior commissure at the level of lateral ventricles in both in vivo and ex vivo DTI images (Fig. 1B). The selection of ROIs was based on the known pathological abnormalities reported in various regions of the brain in MPS VII (40) and the availability of motion-free DTI data.
Figure 1.
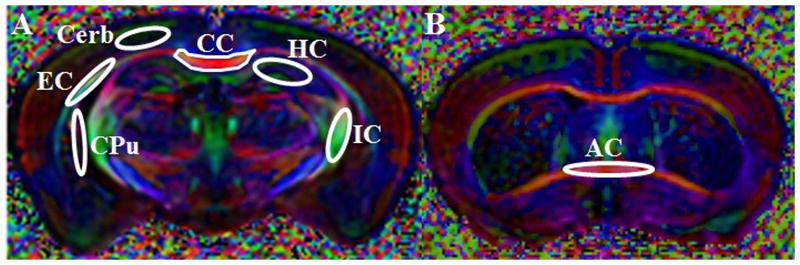
Region-of-interests (ROIs) in gray and white matter used for ex vivo and in vivo diffusion tensor imaging (DTI) data analysis. The ROIs are overlaid on fractional anisotropy-weighted color maps in which red is right-left, green is anterior-posterior, and blue denotes the superior-inferior direction. AC, anterior commissure; CC, corpus callosum; Cereb, cerebral cortex; Cpu, caudate putamen; HC, hippocampus; EC, external capsule; IC, internal capsule.
Separate cohorts of animals were used for volumetric measurement from different regions of the brain using μMRI. To confirm volumetric differences seen on μMRI, the volumes of the anterior commissure, corpus callosum and hippocampus regions were also measured from in vivo and ex vivo DTI data and normalized to the total brain volume. The b0 images from the DTI data set were used for volume measurements since they are similar to T2-weighted images and are co-registered to DTI parametric maps, thereby ensuring an accurate transposition of the ROIs from FA maps to these images. The ROIs were initially drawn on FA maps as described above and the segmented ROI was saved as an image, which was then overlaid on the b0 image to measure the volume of the particular region of the brain.
Histology
MPS VII (n = 3) and WT (n = 3) brain samples were randomly chosen from the cohort that underwent the μMRI experiment for hematoxylin and eosin staining. A separate cohort of brain samples [MPS VII (n = 2) and WT (n = 2)] was randomly selected for Luxol fast blue (LFB) staining from the samples used for DTI experiments. The brain samples were cut into 2 mm thick axial blocks with 5-6 blocks per brain. All tissue blocks were embedded in paraffin and sectioned in a series of 5 μm thick axial sections. Hematoxylin and eosin and LFB staining was performed according to standard methods (40). Stained slides imaged using a Leica dissecting microscope and digital camera (Leica Microsystems Inc., Buffalo Grove, IL). The digital images used identical intensity scales for all images.
To evaluate the white matter changes quantitatively, midline sagittal sections through the corpus callosum were prepared. Anesthetized mice (4 WT, 4 MPS VII) were transcardially perfused and fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). The tissues were quickly removed, then post-fixed in perfusate solution for at least 4 hours at 4°C; the samples were then osmicated and dehydrated and treated twice with propylene oxide for 5 minutes, incubated sequentially with Embed 812: propylene oxide mixtures (1:1 and 2:1), and incubated in pure Embed 812 mixture (Electron Microscopy Sciences, Hatfield, PA) overnight. Tissues were then embedded in fresh Embed 812 mixture at 60°C for 48 hours. For light microscopy, cross-sections were cut at a thickness of 1 μm on a Reichert Ultracut ultramicrotome, and stained with alkaline toluidine blue. Slides containing between 6 and 9 sections from each mouse were scanned using an Aperio ScanScope OS under oil at 83x. Callosal thickness and Feret diameter (length) were determined using Aperio Image Scope software. Callosal thickness was determined at a point just rostral to where the fornix abuts the corpus callosum (4, 41). The measurements were averaged and compared between genotypes by Student t-test.
To enumerate numbers of axons, images were acquired using a Leica DM 6000 microscope (Leica Microsystems, Inc.), a 100x oil objective, and a Leica DFC 360 monochrome camera. Representative ROIs from the corpus callosum of each animal were obtained from the dorsal aspect of the genu, the middle region rostral of the fornix (isthmus), and the dorsal aspect of the splenium. The image was first “flattened” using a 20-pixel feature width, which decreased the intensity of background pixels to even out the lighting; then the “Higauss” filter was used for sharpening; and finally, a Roberts edge filter was used to detect the fine edges in the image. These images were analyzed using Image Pro-Plus 6.1 after spatial filtration to count axons. A count of the objects in the Roberts images was conducted by automatic detection of bright objects subject to the following empirically derived parameters for detecting axons: area 1–20 μm2; roundness 0–1.5; and Feret diameter between 0.6–4.0 μm. The area and number of objects in each image and the mean diameters of the objects were determined. The objects that were counted were also overlaid on the original image to evaluate the fidelity of the capture process (4). Significance was calculated using Student t-test.
A representative electron micrograph was obtained on a transverse, midline thin section form the corpus callosum of a MPS VII brain. The brain tissue was trimmed into 2-mm blocks and fixed in 2% glutaraldehyde in 0.1M cacodylate buffer, pH 7.4, then post-fixed in 1% OsO4. Thin sections were stained with toluidine blue and uranyl acetate-lead citrate Images were acquired using a JEOL-1010 transmission electron microscope (JEOL USA, Inc. Peabody, MA) by the Penn Perelman School of Medicine Electron Microscopy Resource Laboratory.
Statistical Analysis
Volumetric comparisons of anterior commissure, corpus callosum and hippocampus as well as the whole brain were performed between MPS VII and WT mice from μMRI data along with in vivo and ex vivo DTI data. In vivo and ex vivo DTI data were also used to compare DTI indices from different gray and white matter regions between MPS VII and WT mice. An independent Student t-test was performed to assess significant differences in volume and DTI indices. All statistical computations were performed using statistical package for social sciences (SPSS, version 16.0 SPSS, Inc., Chicago, IL).
Results
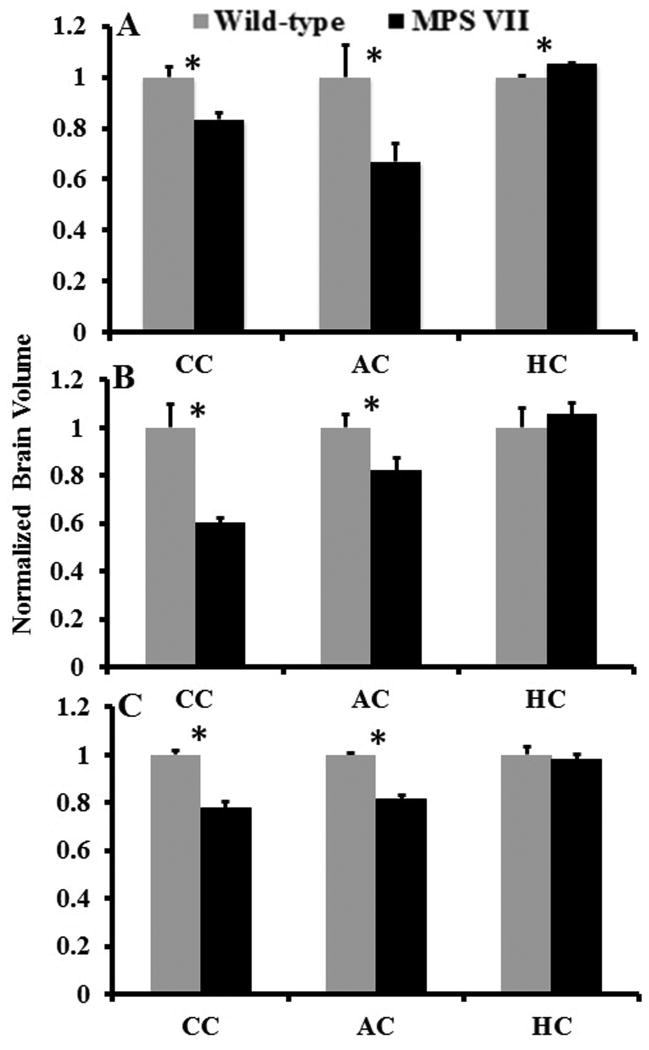
No significant differences in overall forebrain volume were noted between MPS VII and WT brains (p = 0.165). Hence, the volumes of the segmented regions were normalized to the total brain volume of the individual animal. μMRI revealed significantly lower anterior commissure (p = 0.042) and corpus callosum (p = 0.012) volumes in the MPS VII vs., the WT brains (Fig. 2A). In contrast, the hippocampal volume was significantly higher in MPS VII than normal controls (p = 0.002). Both the in vivo and ex vivo DTI data also demonstrated significantly lower anterior commissure (p = 0.048 and p = 0.041) and corpus callosum (p = 0.024 and p = 0.027) volume normalized to the whole brain volume in MPS VII compared to WT mice (Fig. 2B, C). However, in vivo and ex vivo DTI did not detect differences in hippocampal volumes (p = 0.742 and p = 0.614) (Fig. 2B, C).
Figure 2.
Bar graph showing normalized volumes from the segmented regions of the brain from mucopolysaccharidosis type VII (MPS VII) (black, n = 5 in microscopic magnetic resonance imaging [μMRI] and n = 6 for in vivo and ex vivo diffusion tensor imaging [DTI]) and wild-type mice (gray, n = 5) brains. Error bars represent SEM. (A) Regional volumes normalized to the overall brain volume using μMRI. (B, C) Normalized volume changes from the segmented regions from in vivo (B) and ex vivo (C) DTI data, respectively. (*) Asterisk represents significant differences with a p value of <0.05. AC, Anterior commissure; CC, corpus callosum; HC, hippocampus.
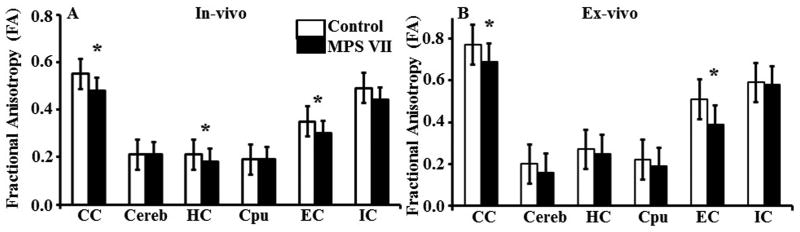
The brains of live animals were analyzed for several DTI parameters, i.e. FA, MD, CL, CP and CS. FA describes the directionality or coherence of diffusion and provides information about the magnitude of diffusion anisotropy; FA thus indirectly evaluates the integrity of white matter tracts and cellular microstructure (42). In vivo DTI data demonstrated significantly lower FA from the anterior commissure (p = 0.050), corpus callosum (p = 0.024), external capsule (p = 0.025), and hippocampus (p = 0.035) in the MPS VII vs. WT brains (Fig. 3A). MD, the trace of the diffusion matrix, is an average measure of molecular water diffusion and is affected by cellular size and integrity of the tissue (43). The gray and white matter regions did not show significant differences in MD values between the 2 groups in vivo (Table 1). Geometrical DTI indices provide additional information with respect to tissue microstructural integrity (18). Of these, CL describes diffusion mainly in the direction corresponding to the largest eigenvalues; we observed significantly lower CL in the hippocampi of MPS VII vs. WT mice (p = 0.043). CP is an index of diffusion restricted to a plane spanned by the 2 eigenvectors corresponding to the 2 largest eigenvalues. No significant differences in CP values were observed from any of the gray or white matter areas between MPS VII and WT mice. CS is the measurement of isotropic diffusion. Similar to CP, no significant differences in CS values were observed between MPS VII and WT mice.
Figure 3.
(A, B) Bar graph showing fractional anisotropy values from gray and white matter regions of the brain from in vivo (A) and ex vivo (B) diffusion tensor imaging (DTI) data, from mucopolysaccharidosis type VII (MPS VII) (black bars, n = 6) and wild type littermates (white bars, n = 5). Error bars represent SEM. (*) Asterisk represents significant differences with a p value of <0.05. CC, corpus callosum; Cereb, cerebral cortex; Cpu, caudate putamen; HC, hippocampus; EC, external capsule; IC, internal capsule.
Table 1. In Vivo Diffusion Tensor Imaging Parameter Values from White and Gray Matter Regions of the Brain of Wild-Type and Mucopolysaccharidosis VII Model Mice.
| Region | White matter | |||||
|---|---|---|---|---|---|---|
| FA | MD | CL | CP | CS | ||
| AC | Wild-type | 0.50 ± 0.05 | 0.62 ± 0.11 | 0.42 ± 0.06 | 0.20 ± 0.06 | 0.31 ± 0.07 |
| MPS VII | 0.40 ± 0.08 | 0.64 ± 0.07 | 0.38 ± 0.10 | 0.16 ± 0.04 | 0.46 ± 0.07 | |
| p value | 0.050* | 0.748 | 0.280 | 0.224 | 0.008* | |
| CC | Wild-type | 0.55 ± 0.05 | 0.59 ± 0.09 | 0.52 ± 0.05 | 0.16 ± 0.05 | 0.32 ± 0.06 |
| MPS VII | 0.48 ± 0.05 | 0.59 ± 0.09 | 0.48 ± 0.04 | 0.14 ± 0.03 | 0.39 ± 0.06 | |
| p value | 0.024* | 0.921 | 0.124 | 0.487 | 0.089 | |
| EC | Wild-type | 0.35 ± 0.05 | 0.60 ± 0.10 | 0.25 ± 0.07 | 0.27 ± 0.04 | 0.48 ± 0.06 |
| MPS VII | 0.30 ± 0.04 | 0.59 ± 0.11 | 0.20 ± 0.05 | 0.28 ± 0.06 | 0.52 ± 0.06 | |
| p value | 0.025* | 0.830 | 0.085 | 0.624 | 0.140 | |
| IC | Wild-type | 0.49 ± 0.09 | 0.63 ± 0.10 | 0.48 ± 0.06 | 0.17 ± 0.05 | 0.37 ± 0.08 |
| MPS VII | 0.44 ± 0.09 | 0.62 ± 0.10 | 0.41 ± 0.09 | 0.17 ± 0.04 | 0.42 ± 0.09 | |
| p value | 0.177 | 0.758 | 0.176 | 0.935 | 0.166 | |
| Gray matter | ||||||
| Cereb | Wild-type | 0.21 ± 0.04 | 0.61 ± 0.08 | 0.17 ± 0.06 | 0.17 ± 0.03 | 0.66 ± 0.06 |
| MPS VII | 0.21 ± 0.03 | 0.54 ± 0.09 | 0.21 ± 0.04 | 0.14 ± 0.05 | 0.66 ± 0.05 | |
| p value | 0.788 | 0.102 | 0.171 | 0.079 | 0.971 | |
| HC | Wild-type | 0.21 ± 0.03 | 0.64 ± 0.11 | 0.19 ± 0.05 | 0.14 ± 0.04 | 0.68 ± 0.05 |
| MPS VII | 0.18 ± 0.02 | 0.67 ± 0.16 | 0.15 ± 0.03 | 0.14 ± 0.02 | 0.66 ± 0.18 | |
| p value | 0.035* | 0.540 | 0.043* | 0.843 | 0.741 | |
| Cpu | Wild-type | 0.19 ± 0.05 | 0.73 ± 0.22 | 0.16 ± 0.03 | 0.15 ± 0.05 | 0.68 ± 0.06 |
| MPS VII | 0.19 ± 0.05 | 0.69 ± 0.14 | 0.16 ± 0.06 | 0.16 ± 0.02 | 0.68 ± 0.06 | |
| p value | 0.954 | 0.637 | 0.989 | 0.901 | 0.992 | |
FA = Fractional anisotropy, MD = Mean diffusivity (x10-3mm2/s), CL = Linear anisotropy, CP = Planner anisotropy, CS = Spherical anisotropy. AC =Anterior-commissure, Cereb = Cerebral cortex, CC = Corpus callosum, Cpu = Caudate putamen, HC = Hippocampus, EC = External capsule, IC =Internal capsule.
Asterisk represents significant differences between mucopolysaccharidosis (MPS) VII and wild type littermates.
The ex vivo DTI data demonstrated similar results as in vivo DTI data (Table 2). Specifically, significantly lower FA from the anterior commissure (p = 0.024), corpus callosum (p = 0.047), and external capsule (p = 0.016) was noted in the MPS VII mice (Fig. 3B). MD values were significantly higher in the anterior commissure (p = 0.048) and corpus callosum (p = 0.031) of MPS VII mice. There were no significant differences in the CL values. CS values were significantly higher in the anterior commissure of MPS VII brains (p = 0.005), and significantly lower in the external capsule compared to WT mouse brains (p = 0.048). In contrast to the in vivo measurements, the CP values were significantly lower in the external capsule of the MPS VII brains (p = 0.045).
Table 2. Ex Vivo Diffusion Tensor Imaging Parameter Values from White and Gray Matter Regions of the Brain of Wild-Type and Mucopolysaccharidosis VII Model Mice.
| Regions | White matter | |||||
|---|---|---|---|---|---|---|
| FA | MD | CL | CP | CS | ||
| AC | Wild-type | 0.75 ± 0.08 | 0.21 ± 0.05 | 0.70 ± 0.09 | 0.15 ± 0.06 | 0.13 ± 0.03 |
| MPS VII | 0.65 ± 0.01 | 0.28 ± 0.04 | 0.64 ± 0.03 | 0.15 ± 0.04 | 0.21 ± 0.03 | |
| p-value | 0.024* | 0.048* | 0.236 | 0.903 | 0.005* | |
| CC | Wild-type | 0.77 ± 0.06 | 0.23 ± 0.03 | 0.70 ± 0.11 | 0.32 ± 0.25 | 0.14 ± 0.06 |
| MPS VII | 0.69 ± 0.05 | 0.31 ± 0.06 | 0.63 ± 0.08 | 0.20 ± 0.09 | 0.18 ± 0.06 | |
| p-value | 0.047* | 0.031* | 0.245 | 0.348 | 0.275 | |
| EC | Wild-type | 0.51 ± 0.07 | 0.32 ± 0.01 | 0.30 ± 0.07 | 0.40 ± 0.06 | 0.30 ± 0.08 |
| MPS VII | 0.39 ± 0.05 | 0.36 ± 0.11 | 0.27 ± 0.08 | 0.32 ± 0.06 | 0.14 ± 0.08 | |
| p-value | 0.016* | 0.624 | 0.601 | 0.045* | 0.048* | |
| IC | Wild-type | 0.59 ± 0.06 | 0.30 ± 0.08 | 0.59 ± 0.12 | 0.17 ± 0.05 | 0.24 ± 0.07 |
| MPS VII | 0.58 ± 0.07 | 0.35 ± 0.14 | 0.56 ± 0.09 | 0.15 ± 0.02 | 0.30 ± 0.08 | |
| p-value | 0.980 | 0.451 | 0.714 | 0.358 | 0.296 | |
| Gray matter | ||||||
| Cereb | Wild-type | 0.20 ± 0.05 | 0.41 ± 0.14 | 0.14 ± 0.02 | 0.16 ± 0.05 | 0.70 ± 0.05 |
| MPS VII | 0.16 ± 0.01 | 0.41 ± 0.15 | 0.15 ± 0.02 | 0.12 ± 0.01 | 0.73 ± 0.01 | |
| p-value | 0.094 | 0.993 | 0.681 | 0.129 | 0.156 | |
| HC | Wild-type | 0.27 ± 0.03 | 0.39 ± 0.14 | 0.23 ± 0.04 | 0.17 ± 0.04 | 0.60 ± 0.06 |
| MPS VII | 0.25 ± 0.04 | 0.43 ± 0.16 | 0.24 ± 0.04 | 0.17 ± 0.04 | 0.63 ± 0.06 | |
| p-value | 0.494 | 0.692 | 0.422 | 0.837 | 0.443 | |
| Cpu | Wild-type | 0.22 ± 0.05 | 0.50 ± 0.26 | 0.18 ± 0.06 | 0.15 ± 0.01 | 0.67 ± 0.06 |
| MPS VII | 0.19 ± 0.07 | 0.51 ± 0.31 | 0.20 ± 0.11 | 0.13 ± 0.02 | 0.67 ± 0.13 | |
| p-value | 0.542 | 0.712 | 0.752 | 0.140 | 0.988 | |
FA = Fractional anisotropy, MD = Mean diffusivity (x10-3mm2/s), CL = Linear anisotropy, CP = Planner anisotropy, CS = Spherical anisotropy. AC = Anterior-commissure, Cereb = Cerebral cortex, CC = Corpus callosum, Cpu = Caudate putamen, HC = Hippocampus, EC = External capsule, IC = Internal capsule.
Asterisk indicates significant differences between mucopolysaccharidosis (MPS) VII and wild type littermates.
A sub-analysis of the axial (AD) and radial (RD) diffusivities was performed from the CC ROI of both the in vivo and ex vivo DTI data to characterize further the differences in the FA value observed in MPS VII mice. In vivo DTI demonstrated significantly greater RD values (p = 0.038) with no significant differences in AD values in MPS VII affected mice compared to WT animals (p = 0.566) (Supplementary Table 1). The ex vivo data also showed similar results with increased RD value (p = 0.032) and no significant difference in AD value (p = 0.321) from CC region of MPS VII mice (Supplementary Table 1) indicating myelin loss.
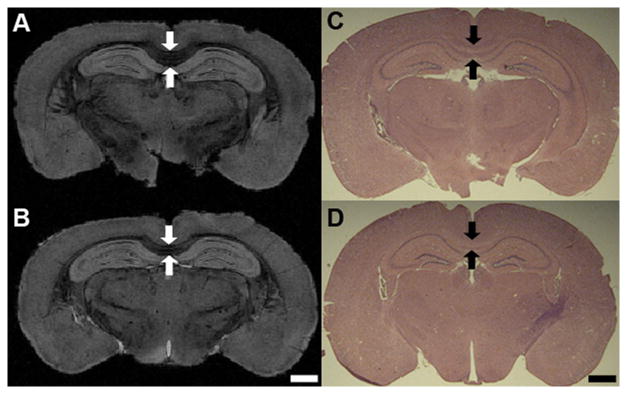
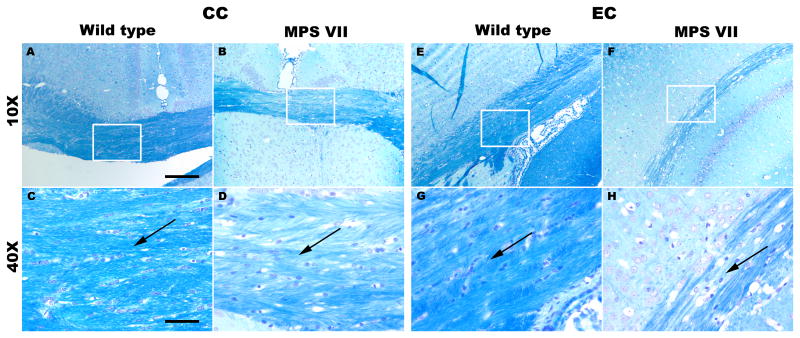
Histological staining showed reduced corpus callosum thickness in MPS VII brains (Fig. 4). By comparison, μMRI revealed the corpus callosum thickness as 476.2 ± 6.3 μm in WT mice (Fig. 4A, C) and only 332.7 ± 15.9 μm in MPS VII brains (Fig. 4B, D, p = 0.011). LFB staining of randomly selected samples demonstrated results that were consistent with the MRI findings from the corpus callosum and external capsule, i.e. the LFB-stained sections from these regions demonstrated a decrease in myelin staining and less densely packed cells with dispersed white matter fiber bundles in MPS VII mice (Fig. 5B, D, F, H) in comparison to brain sections from WT mice (Fig. 5A, C, E, G).
Figure 4.
(A-D) Microscopic magnetic resonance (μMR) images and hematoxylin and eosin staining from the corpus callosum and hippocampus region of wild-type (A, C) and mucopolysaccharidosis type VII (MPS VII) (B, D) mouse brains illustrating lower thickness of the corpus callosum (arrows) in the MPS VII brain (B, D). Scale bars for both μMR images and histology micrographs are 1 mm.
Figure 5.
(A-H) Luxol fast blue (LFB)-stained brain sections from the CC (A-D) and EC (E-H) regions of the brain showing myelin staining in wild type (A, C, E, G) and mucopolysaccharidosis type VII (MPS VII) (B, D, F, H) mice. The rectangular boxes on sections at magnification of 10x (scale bar = 200 μm) are zoomed at 40x (scale bar = 50 μm) (C, D, G, H). Black arrows indicate splitting of the fibers and degree of compactness of the white matter fiber bundles in wild-type and MPS VII mice. CC, corpus callosum; EC, external capsule.
Myelin of the MPS VII and WT brains was evaluated quantitatively in sagittal midline sections through the corpus callosum, using semi-thin plastic sections stained with toluidine blue (Table 3; Fig. 6). The corpus callosum of the MPS VII mouse was significantly thinner and shorter than the WT mice, leading to a significantly decreased cross sectional area (Table 3). Automated counting of 100x oil images detected a significant decrease in the number of myelinated axons in MPS VII mice (Table 3; Fig. 6A, B), but the mean size and distribution of sizes of the myelinated axons were not significantly different (Table 3). The decrease in myelinated axons was consistent with electron photomicrographs, which showed numerous non-myelinated axons in the MPS VII brain (Fig. 6C).
Table 3. Corpus Callosum Dimensions and Myelinated Axon Numbers and Sizes in Midline Sagittal View.
| Callosal dimensions | Number of myelinated axons per ROI | Mean myelinated axon diameter | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Minimum Thickness (mm) | Length (Feret) (mm) | Area (mm2) | Genu | Mid-region | Splenium | Genu (μm) | Mid-region (μm) | Splenium (μm) |
| Wild type (n=4) | 0.18 ± 0.04 | 3.74 ± 0.18 | 912.6 ± 36.3 | 3578 ± 365.8 | 3881 ± 471.0 | 3860 ± 709.4 | 0.81 ± 0.25 | 0.85 ± 0.55 | 0.81 ± 0.32 |
| MPS VII (n=4) | 0.12 ± 0.02 | 3.30 ± 0.15 | 542.3 ± 27.0 | 2551 ± 499.5 | 2276 ± 252.7 | 2434 ± 335.5 | 0.80 ± 0.36 | 0.84 ± 0.21 | 0.73 ± 0.16 |
| p value | <0.03 | <0.01 | <0.001 | <0.03 | <0.01 | <0.02 | >0.9 | >0.6 | >0.1 |
MPS, mucopolysaccharidosis; ROI, region of interest.
Figure 6.
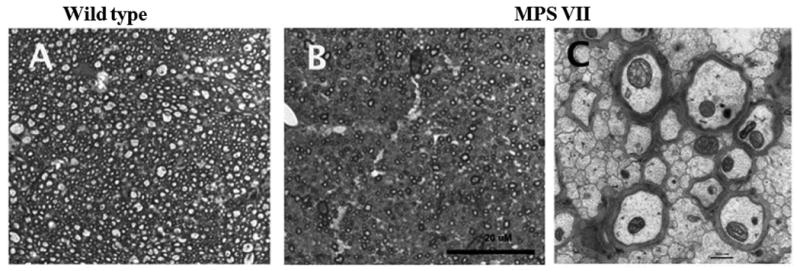
Differences in white matter structure in midline sagittal sections through the corpus callosum. (A, B) Semi-thin plastic sections (1 μm) from wild type (A) and mucopolysaccharidosis type VII (MPS VII) (B) mice were stained with toluidine blue. There was a reduction in numbers of myelinated axons in the MPS VII section compared to the wild type brain. (C) Electron microscopy image (25,000x) from the MPS VII corpus callosum illustrating the paucity of myelinated axons and the dense packing of non-myelinated fibers. Scale bars: B = 20 μm; C = 50 nm.
Discussion
Our group has recently reported that a number of genes expressed in oligodendrocytes are significantly downregulated in the MPS VII mouse brain, suggesting they may have defects in myelination and/or abnormal development of brain white matter structures (6). The MPS disorders have lesions throughout the brain but they are seen primarily in gray matter cells. Some human case reports have found changes in brain white matter areas (3, 8, 10), but it is not known if they are secondary to other severe changes in the brain such as hydrocephalus, neurodegeneration, or neuroinflammation. Because histopathologic stains are unreliable for quantifying white matter areas, we evaluated whether DTI methods could be used as a surrogate to detect differences in white matter structures in the MPS VII mouse brain compared to normal phenotype, age-matched siblings.
The finding that the overall brain volumes (measured by μMRI) of the MPS VII and normal brains were the same is of interest because the shape of the skull is abnormal in MPS diseases. This is due to the abnormalities in enchondral bone formation, including bones of the skull (22, 23). In the mouse this results in a brain that is shorter and wider than that in normal littermates and which requires modified stereotaxic coordinates to target homologous structures in the normal vs. diseased brain in the adult (44). Because of the differences in brain shape, as well as variation between individual MPS VII animals, there have been no previous quantitative studies on the total brain volume or on stereological counts of cells in specific brain nuclei in age-matched MPS VII and normal mice. Interestingly, after resection from the skull, the MPS VII brains relaxed into a shape similar to that of the normal brains (not shown). This suggests that the cytoarchitectonic relationships within the brain are likely similar even with a misshapen gross structure, which is consistent with histological analyses of, for example, experimental treatments (32, 44, 45).
We observed that the volume of the anterior commissure and the corpus callosum were smaller in the MPS VII brains, and these structures also exhibited significantly higher MD values along with significantly lower FA values in MPS VII vs. WT mice. The imaging observations were confirmed histologically by quantitative measures of the cross-sectional size of the corpus callosum. Reduced volume and FA, along with abnormal myelination of the white matter fibers in the CC and other white matter areas, is a novel finding in this model. Indeed, brain diseases in the MPS group (i.e. Hurler, Hunter, Sanfilippo, Morquio, Sly) are not classified as leukodystrophies (4). The significant reduction in myelinated fibers in the CC indicates there likely are abnormalities in the functional connectivity across the brain hemispheres. Our findings also indicate that in vivo DTI can be used to quantitatively assess the severity of disease including monitoring therapeutic responses in future studies. Significantly lower FA and higher radial diffusivity with no significant change in axial diffusivity in the CC of MPS VII mice is indicative of myelin loss (25), which was confirmed by LFB stain, semi-thin sections and electron microscopy (Figs. 5, 6).
Whereas significant differences in FA were observed, they were not reflected in CL, CP and CS measures. This can be explained by the fact that FA is a summary measure of the microstructural changes (integrity) of the fibers and not the direction of diffusivity, which is reflected by CL, CP or CS. As evident from histological studies, the density of white matter fibers seems to be lower in MPS mice, which can lead to lower FA values, but the direction of fibers appeared to be unchanged; this is consistent with the CL, CP and CS values being similar to the normal brain. Figure 6C also shows that the integrity of the cell wall is altered, which may be reflected in FA but not in the directional measures of CL, CP or CS.
The differences in volume and DTI indices of MPS VII mice correspond to the reduced number of myelinated axons in the diseased brain and are consistent with the changes observed in oligodendrocyte-associated genes (6). In the MPS VII mouse brain, astrogliosis, neurodegeneration, neuroinflammation and ventricular dilatation have been reported by histology, quantitative immuno-histochemistry, and transcriptomics (6, 24, 32), all of which may contribute to CNS dysfunction. To the best of our knowledge, the current study is the only quantitative imaging study to use MRI and DTI to assess the abnormalities in both the gray and white matter regions in any of the MPS mouse models. In an earlier MRI study of human MPS II (Hunter disease), decreased brain volume that correlated with cognitive impairment was reported (46). The intellectual disability likely arises from complex mechanisms initiated by the progressive lysosomal accumulation of glycosaminoglycans in neurons and glia (3, 6, 47). Human studies are, however, complicated by the degree of variability in the genetic mutation and the disease status and age of the patients; these parameters can all be controlled in a genetically defined mouse model, as reported in this study.
When white matter is involved in a neurodegenerative disease, gray matter structures may also be affected. Thus, it is difficult to determine whether the changes in white matter structures are primary, secondary to gray matter loss, or occur concurrently with and are underpinned by the same pathophysiologic mechanisms as the gray matter changes (47). DTI measures of anisotropy of the white matter areas are mainly determined by axonal directions although the concentration of the myelin, fiber size and cell types also play a role in the measurement of FA (48). The significantly reduced FA in the corpus callosum in MPS VII mice appeared to be due to a decrease in the number of myelinated axons (Table 3), which was also evident in the semi-thin sections and electron microscopy (Fig. 6). Changes in neuronal and glial cells are tightly coupled in both gray and white matter through several interaction and communication pathways. For example, myelinating glia can regulate axon diameter and even the survival of axons (49) and axons that become demyelinated can degenerate, leading to neuronal death (50). Thus, changes in white matter may contribute secondarily to changes in gray matter regions, such as the hippocampus, as observed in the present study. A decrease in FA and an increase in MD from the anterior commissure, corpus callosum and external capsule suggests increased diffusivity due to an increase in free water and loss of the integrity of the myelin sheaths and axons in these regions (51).
The linear and planar shape anisotropy, measured by DTI, emphasizes different white matter features. The CL value reflects the uniformity of white matter tract direction within an imaging voxel and correlates with coherent neural fiber structures. The significantly reduced CL from the external capsule of the MPS VII mice may indicate a decrease in axonal length and integrity of white matter fibers. Myelin staining by LFB also exhibited relatively lower density of white matter fibers in the MPS VII vs. WT mice (Fig. 5). Quantification of myelinated axons in the corpus callosum cross-sections showed that there were many fewer myelinated axons and that the loss occurred throughout the structure (Fig. 6B). The clusters of un-myelinated fibers between myelinated fibers demonstrated by electron microscopy in the MPS VII corpus callosum (Fig. 6C) correlate well with our DTI findings. Although the biophysical basis for the difference in the anisotropy of water diffusion in white matter of MPS VII mice is unclear, the reduced FA along with increased MD in the white matter of MPS VII mice might be due to aberrant expression of oligodendrocyte genes (5), abnormality in myelin, altering the micro-structural integrity of the white matter tracts.
Increased MD in the anterior commissure and corpus callosum was only observed from ex vivo DTI, while no significant changes in MD were observed from any region of the brain from in vivo DTI data. Because MD can be affected by tissue water content that can vary with fixation, this could account for the differences between in vivo and ex vivo results. In contrast, FA is unaffected by tissue fixation (52), and the differences in the FA values of MPS VII and control mice were consistent between the in vivo and ex vivo DTI. These findings indicate the potential added value of DTI parameters in the characterization of the brain disease models as opposed to a simple diffusion-weighted imaging scheme, which only provides information on MD.
The present findings also provide a potential means for assessing progression of disease and the effects of therapies in the brains of live mice. These studies have a high clinical and translational relevance because DTI is routinely used in the clinic. If validated in an animal model, such studies could be performed in humans for early diagnosis and possibly prognosis of various MPS disorders.
Care should be observed when comparing our results with other in vivo and ex vivo DTI studies in mouse models because our study has some limitations. A commonly used ROI analysis approach was used in this study to compare the DTI and volume differences between MPS VII and WT mice. While great care was used in drawing the ROIs, there is a potential bias in the selection of these regions because the gray matter regions selected in this study may have had some contribution from white matter and vice versa. Despite the use of a head-restraining device, in vivo DTI images suffered from motion-induced effects, which limited the analysis to a subset of slices rather than the entire brain. While cardiac gating in the in vivo rat brain DTI studies has been reported to increase the reliability of DTI data (53), we were unable to achieve a consistent cardiac and respiratory rate in the anesthetized mice. Furthermore, the MPS VII mice are metabolically fragile and do not tolerate deep anesthesia. Unlike clinical DTI studies, rodent brain DTI studies are typically performed with spin echo based DTI sequences because the susceptibility artifacts at higher fields severely limit the use of echo-planar based DTI sequences. The use of spin echo based DTI sequences results in increased acquisition time and thus most rodent brain DTI studies are performed using a 6-direction DTI sequence. The measured FA values from the CC region of WT mice were in agreement with earlier studies that also used a 6-direction DTI sequence (28, 54). Therefore, the values using these sequences are likely to differ from values obtained in studies using higher order diffusion directions or fast-spin echo DTI sequences. Thus, care should be taken to account for the differences in acquisition parameters when comparing the FA values between studies.
In conclusion, we have demonstrated the potential of the DTI in delineating microstructural brain abnormalities in a neurogenetic disease model, the MPS VII mice. The results demonstrate that μMRI and DTI hold promise as non-invasive, quantitative parameters to assess the microstructural integrity of brain tissue and may be used as surrogate imaging biomarkers for studying lysosomal storage diseases.
Supplementary Material
Acknowledgments
We thank S. Scherer and J. Grinspan for helpful discussions about myelin pathology and assays, and T. Clarke and K. Sanchez for excellent technical assistance. The studies used the Small Animal Imaging Facility of the University of Pennsylvania.
This study was supported in part by the NIH grants R21-HD058237 to HP; R01-NS38690 and R01-NS56243 to JHW; and the Institute for Translational Medicine and Therapeutics (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics to HP and JHW (UL1RR024134).
References
- 1.Neufeld EF, Muenzer J. The mucopolysaccharidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic & Molecular Basis of Inherited Disease. New York: McGraw-Hill; 2001. pp. 3421–52. [Google Scholar]
- 2.Neufeld EF. Lysosomal storage diseases. Annu Rev Biochem. 1991;60:257–80. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 3.Vedolin L, Schwartz IV, Komlos M, et al. Brain MRI in mucopolysaccharidosis: effect of aging and correlation with biochemical findings. Neurology. 2007;69:917–24. doi: 10.1212/01.wnl.0000269782.80107.fe. [DOI] [PubMed] [Google Scholar]
- 4.Steelman AJ, Thompson JP, Li J. Demyelination and remyelination in anatomically distinct regions of the corpus callosum following cuprizone intoxication. Neurosci Res. 2012;72:32–42. doi: 10.1016/j.neures.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogler C, Birkenmeier EH, Sly WS, et al. A murine model of mucopolysaccharidosis VII. Gross and microscopic findings in beta-glucuronidase-deficient mice. Am J Pathol. 1990;136:207–17. [PMC free article] [PubMed] [Google Scholar]
- 6.Parente MK, Rozen R, Cearley CN, et al. Dysregulation of gene expression in a lysosomal storage disease varies between brain regions implicating unexpected mechanisms of neuropathology. PLoS One. 2012;7:e32419. doi: 10.1371/journal.pone.0032419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohmi K, Greenberg DS, Rajavel KS, et al. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc Natl Acad Sci U S A. 2003;100:1902–7. doi: 10.1073/pnas.252784899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seto T, Kono K, Morimoto K, et al. Brain magnetic resonance imaging in 23 patients with mucopolysaccharidoses and the effect of bone marrow transplantation. Ann Neurol. 2001;50:79–92. doi: 10.1002/ana.1098. [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Dineen TE, Brack M, et al. The mucopolysaccharidoses: characterization by cranial MR imaging. AJNR Am J Neuroradiol. 1993;14:1285–92. [PMC free article] [PubMed] [Google Scholar]
- 10.Murata R, Nakajima S, Tanaka A, et al. MR imaging of the brain in patients with mucopolysaccharidosis. AJNR Am J Neuroradiol. 1989;10:1165–70. [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe JH, A P, Poptani H, Vite CH. Molecular imaging of gene therapy for neurogenetic diseases. San Diego, California: Academic Press; 2006. [Google Scholar]
- 12.Watson DJ, Walton RM, Magnitsky SG, et al. Structure-specific patterns of neural stem cell engraftment after transplantation in the adult mouse brain. Hum Gene Ther. 2006;17:693–704. doi: 10.1089/hum.2006.17.693. [DOI] [PubMed] [Google Scholar]
- 13.Magnitsky S, Walton RM, Wolfe JH, et al. Magnetic resonance imaging detects differences in migration between primary and immortalized neural stem cells. Acad Radiol. 2008;15:1269–81. doi: 10.1016/j.acra.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bihan D, Mangin JF, Poupon C, et al. Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging. 2001;13:534–46. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 15.Kumar M, Rathore RK, Srivastava A, et al. Correlation of diffusion tensor imaging metrics with neurocognitive function in Chiari I malformation. World Neurosurg. 2011;76:189–94. doi: 10.1016/j.wneu.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- 17.Vilanova AZS, Kindlmann G, Laidlaw D. An Introduction to Visualization of Diffusion Tensor Imaging and Its Applications. In: Hagen JWaH., editor. Visualization and Processing of Tensor Fields. Vol. 37. Berlin: Springer Berlin Heidelberg; 2006. pp. 121–53. [Google Scholar]
- 18.Peled S, Gudbjartsson H, Westin CF, et al. Magnetic resonance imaging shows orientation and asymmetry of white matter fiber tracts. Brain Res. 1998;780:27–33. doi: 10.1016/s0006-8993(97)00635-5. [DOI] [PubMed] [Google Scholar]
- 19.Kumar M, Gupta RK, Nath K, et al. Can we differentiate true white matter fibers from pseudofibers inside a brain abscess cavity using geometrical diffusion tensor imaging metrics? NMR Biomed. 2008;21:581–8. doi: 10.1002/nbm.1228. [DOI] [PubMed] [Google Scholar]
- 20.Alexander AL, Hasan K, Kindlmann G, et al. A geometric analysis of diffusion tensor measurements of the human brain. Magn Reson Med. 2000;44:283–91. doi: 10.1002/1522-2594(200008)44:2<283::aid-mrm16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Sly WS, Quinton BA, McAlister WH, et al. Beta glucuronidase deficiency: report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J Pediatr. 1973;82:249–57. doi: 10.1016/s0022-3476(73)80162-3. [DOI] [PubMed] [Google Scholar]
- 22.Birkenmeier EH, Barker JE, Vogler CA, et al. Increased life span and correction of metabolic defects in murine mucopolysaccharidosis type VII after syngeneic bone marrow transplantation. Blood. 1991;78:3081–92. [PubMed] [Google Scholar]
- 23.Casal ML, Wolfe JH. Variant clinical course of mucopolysaccharidosis type VII in two groups of mice carrying the same mutation. Lab Invest. 1998;78:1575–81. [PubMed] [Google Scholar]
- 24.Levy B, Galvin N, Vogler C, et al. Neuropathology of murine mucopolysaccharidosis type VII. Acta Neuropathol. 1996;92:562–8. doi: 10.1007/s004010050562. [DOI] [PubMed] [Google Scholar]
- 25.Song SK, Sun SW, Ramsbottom MJ, et al. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17:1429–36. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- 26.Neil JJ, Shiran SI, McKinstry RC, et al. Normal brain in human newborns: apparent diffusion coefficient and diffusion anisotropy measured by using diffusion tensor MR imaging. Radiology. 1998;209:57–66. doi: 10.1148/radiology.209.1.9769812. [DOI] [PubMed] [Google Scholar]
- 27.Nair G, Tanahashi Y, Low HP, et al. Myelination and long diffusion times alter diffusion-tensor-imaging contrast in myelin-deficient shiverer mice. Neuroimage. 2005;28:165–74. doi: 10.1016/j.neuroimage.2005.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aggarwal M, Mori S, Shimogori T, et al. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn Reson Med. 2010;64:249–61. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo AC, Petrella JR, Kurtzberg J, et al. Evaluation of white matter anisotropy in Krabbe disease with diffusion tensor MR imaging: initial experience. Radiology. 2001;218:809–15. doi: 10.1148/radiology.218.3.r01mr14809. [DOI] [PubMed] [Google Scholar]
- 30.Larsson EM, Englund E, Sjobeck M, et al. MRI with diffusion tensor imaging post-mortem at 3.0 T in a patient with frontotemporal dementia. Dement Geriatr Cogn Disord. 2004;17:316–9. doi: 10.1159/000077162. [DOI] [PubMed] [Google Scholar]
- 31.Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J Mol Neurosci. 2008;34:51–61. doi: 10.1007/s12031-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 32.Heuer GG, Passini MA, Jiang K, et al. Selective neurodegeneration in murine mucopolysaccharidosis VII is progressive and reversible. Ann Neurol. 2002;52:762–70. doi: 10.1002/ana.10373. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe JH, S M. In: Murine mucopolysaccharidosis type VII: A model system for somatic gene therapy of the central nervous system. Lowenstein PR, E L, editors. New York: Wiley; 1996. pp. 263–74. [Google Scholar]
- 34.Kim S, Pickup S, Hsu O, et al. Enhanced delineation of white matter structures of the fixed mouse brain using Gd-DTPA in microscopic MRI. NMR Biomed. 2009;22:303–9. doi: 10.1002/nbm.1324. [DOI] [PubMed] [Google Scholar]
- 35.Mori S, van Zijl PC. A motion correction scheme by twin-echo navigation for diffusion-weighted magnetic resonance imaging with multiple RF echo acquisition. Magn Reson Med. 1998;40:511–6. doi: 10.1002/mrm.1910400403. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Pickup S, Fairless AH, et al. Association between sociability and diffusion tensor imaging in BALB/cJ mice. NMR Biomed. 2012;25:104–12. doi: 10.1002/nbm.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, van Zijl PC, Kim J, et al. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81:106–16. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Wada R, Tifft CJ, Proia RL. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc Natl Acad Sci U S A. 2000;97:10954–9. doi: 10.1073/pnas.97.20.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Martins I, Wemmie JA, et al. Functional correction of CNS phenotypes in a lysosomal storage disease model using adeno-associated virus type 4 vectors. J Neurosci. 2005;25:9321–7. doi: 10.1523/JNEUROSCI.2936-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barazany D, Basser PJ, Assaf Y. In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain. 2009;132:1210–20. doi: 10.1093/brain/awp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huisman TA. Diffusion-weighted imaging: basic concepts and application in cerebral stroke and head trauma. Eur Radiol. 2003;13:2283–97. doi: 10.1007/s00330-003-1843-6. [DOI] [PubMed] [Google Scholar]
- 43.Hasan KM, Narayana PA. Retrospective measurement of the diffusion tensor eigenvalues from diffusion anisotropy and mean diffusivity in DTI. Magn Reson Med. 2006;56:130–7. doi: 10.1002/mrm.20935. [DOI] [PubMed] [Google Scholar]
- 44.Cearley CN, Wolfe JH. A single injection of an adeno-associated virus vector into nuclei with divergent connections results in widespread vector distribution in the brain and global correction of a neurogenetic disease. J Neurosci. 2007;27:9928–40. doi: 10.1523/JNEUROSCI.2185-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passini MA, Lee EB, Heuer GG, et al. Distribution of a lysosomal enzyme in the adult brain by axonal transport and by cells of the rostral migratory stream. J Neurosci. 2002;22:6437–46. doi: 10.1523/JNEUROSCI.22-15-06437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan Z, Styner M, Muenzer J, et al. Correlation of automated volumetric analysis of brain MR imaging with cognitive impairment in a natural history study of mucopolysaccharidosis II. AJNR Am J Neuroradiol. 2010;31:1319–23. doi: 10.3174/ajnr.A2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walterfang M, Fahey M, Abel L, et al. Size and shape of the corpus callosum in adult Niemann-Pick type C reflects state and trait illness variables. AJNR Am J Neuroradiol. 2011;32:1340–6. doi: 10.3174/ajnr.A2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark CA, Le Bihan D. Water diffusion compartmentation and anisotropy at high b values in the human brain. Magn Reson Med. 2000;44:852–9. doi: 10.1002/1522-2594(200012)44:6<852::aid-mrm5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 49.Nave KA. Myelination and support of axonal integrity by glia. Nature. 2010;468:244–52. doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- 50.Dutta R, Trapp BD. Pathogenesis of axonal and neuronal damage in multiple sclerosis. Neurology. 2007;68:S22–31. doi: 10.1212/01.wnl.0000275229.13012.32. discussion S43-54. [DOI] [PubMed] [Google Scholar]
- 51.Lebel C, Beaulieu C. Longitudinal development of human brain wiring continues from childhood into adulthood. J Neurosci. 2011;31:10937–47. doi: 10.1523/JNEUROSCI.5302-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D'Arceuil H, de Crespigny A. The effects of brain tissue decomposition on diffusion tensor imaging and tractography. Neuroimage. 2007;36:64–8. doi: 10.1016/j.neuroimage.2007.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Pickup S, Poptani H. Effects of cardiac pulsation in diffusion tensor imaging of the rat brain. J Neurosci Methods. 2010;194:116–21. doi: 10.1016/j.jneumeth.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y, Johnson GA. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage. 2011;56:1235–43. doi: 10.1016/j.neuroimage.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.