Abstract
Chronic myeloproliferative neoplasms (MPNs) are a group of related conditions characterized by the overproduction of cells from one or more myeloid lineages. More than 95% of cases of polycythemia vera, and roughly half of essential thrombocythemia and primary myelofibrosis acquire a unique somatic 1849G>T JAK2 mutation (encoding V617F) that is believed to be a critical driver of excess proliferation1–4. We report here that JAK2V617F-associated disease is strongly associated with a specific constitutional JAK2 haplotype, designated 46/1, in all three disease entities compared to healthy controls (polycythemia vera, n = 192, P = 2.9 × 10−16; essential thrombocythemia, n = 78, P = 8.2 × 10−9 and myelofibrosis, n = 41, P = 8.0 × 10−5). Furthermore, JAK2V617F specifically arises on the 46/1 allele in most cases. The 46/1 JAK2 haplotype thus predisposes to the development of JAK2V617F-associated MPNs (OR = 3.7; 95% CI = 3.1–4.3) and provides a model whereby a constitutional genetic factor is associated with an increased risk of acquiring a specific somatic mutation.
The finding of JAK2V617F was a major step forward in understanding the pathogenesis of MPNs, but it remains unclear how this single abnormality gives rise to distinct clinical entities. Clinical phenotype is clearly associated with JAK2V617F dosage: in many PV and MF cases JAK2V617F is reduced to homozygosity as a consequence of acquired isodisomy (generally referred to as acquired uniparental disomy; aUPD) at chromosome 9p, but this is rare in ET2,5,6. Several lines of evidence indicate that other, largely uncharacterized, acquired abnormalities also have a role in specifying disease phenotype either in combination with or independently of JAK2V617F (refs. 7,8). Furthermore, both epidemiological data and family studies indicate that inherited factors may predispose to MPNs9,10 and it has also been suggested that inherited SNPs within JAK2 are associated with specific MPN subtypes11.
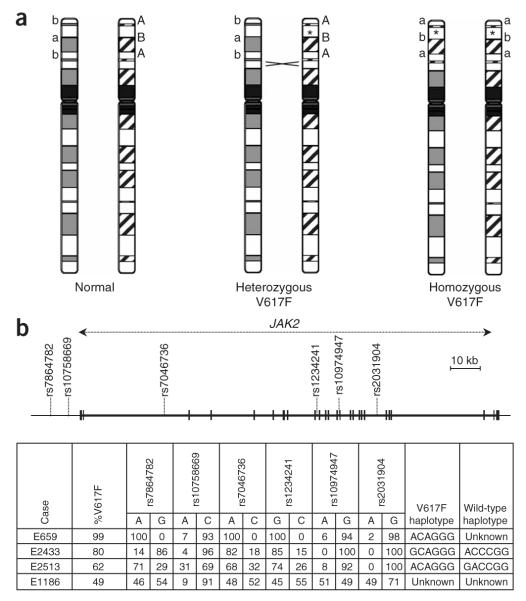
To determine the role of inherited factors, we initially analyzed six JAK2-spanning SNPs (rs7864782, rs10758669, rs7046736, rs12342421, rs10974947, rs2031904) in MPN cases with a homozygous JAK2V617F clone (%V617F allele >50%; n = 142) using pyrosequencing, which provides a quantitative readout of allele ratios. The mitotic recombination that gives rise to aUPD typically involves most of chromosome 9p2,6 and thus SNPs within this region are also reduced to homozygosity; consequently, the haplotype on which JAK2V617F arose could be read directly from allele ratios that were significantly greater than the expected value of 0.5. In most cases with 60–90% V617F, the residual haplotype (that is, the haplotype on which V617F had not arisen) could also be read by the finding of heterozygous allele ratios in the range 0.1–0.4. In cases with ≥90% V617F, however, information about alleles on the non-V617F chromosome was lost (Fig. 1 and Methods). Notably, of the 142 alleles that harbored V617F, 109 (77%) had an identical haplotype (subsequently designated 46/1) within the JAK2 gene, whereas this haplotype was seen for only 9 of the 74 (12%) residual wild-type alleles that could be read (P = 1.4 × 10−20, Fisher’s exact test, two-tailed). These results indicated that homozygosity for JAK2V617F was not random, but rather occurred preferentially when this mutation was present on a specific JAK2 haplotype.
Figure 1.
Allele distortions due to aUPD enable direct reading of JAK2 haplotypes. (a) The JAK2V617F mutation (indicated by an asterisk) and flanking SNPs are reduced to homozygosity in a proportion of cells following mitotic recombination. (b) SNPs and JAK2V617F were quantified by pyrosequencing. In many cases that harbored a homozygous JAK2V617F clone, it was possible to directly read the haplotype on which the mutation arose by the finding that one allele at each SNP predominated (allelic ratio ≥0.6, for example, cases E659, E2433 and E2513). In cases with a homozygous clone and %V617F <90%, it was usually possible to read the residual haplotype (that is, the haplotype of the chromosome that had not acquired JAK2V617F) by the finding of allelic ratios between 0.1–0.4 (for example, cases E2433 and E2513). Where the homozygous clone was small or nonexistent (most cases with %V617F <60%), neither the JAK2V617F nor wild-type haplotype could be read (for example, case E1186).
To explore this observation in more detail, we first determined the haplotype structure of JAK2 using 14 SNPs genotyped by the Wellcome Trust Case Control Consortium (WTCCC) in 1,500 healthy blood donors from the UK12. PHASE analysis13 inferred 92 haplotypes, of which nine accounted for 94% of JAK2 alleles (Fig. 2). Two haplotypes (numbers 46 and 1; referred to henceforth together as 46/1) were identical except for rs7864782 and had a combined frequency of 0.24. A tagged SNP (rs12340895) that was in complete linkage disequilibrium (LD) with 46/1 was then used to screen for this haplotype in further cases. In an initial analysis of 177 heterozygous JAK2V617F-positive MPNs, 46/1 occurred more frequently (135/354 alleles) than in 188 locally sourced healthy controls (92/376 alleles; P = 0.0001) as well as the WTCCC cohort (P = 3.3 10−8). The 46/1 haplotype was more frequent in all JAK2V617F-positive disease entities regardless of origin (UK or United States); however, there was no difference in the frequency of 46/1 between controls and cases with idiopathic erythrocytosis (Table 1). To determine whether JAK2V617F was in cis or trans to the 46/1 allele in cases that were heterozygous for both the mutation and the haplotype, we carried out allele-specific PCRs for JAK2V617F to amplify products that included a second 46/1 tag SNP (rs12343867) in intron 14. Sequencing of the products in 66 informative cases showed that 49 (74%) JAK2V617F alleles arose on a 46/1 allele, whereas only 17 (26%) residual wild-type alleles were 46/1 (P = 2.1 × 10−8).
Figure 2.

SNPs, haplotypes and LD around JAK2. (a) The nine most common JAK2 haplotypes in the UK population. The 14 SNPs in bold were analyzed by the WTCCC in 1,500 blood donors from which the frequencies were determined; asterisks indicate SNPs that tag 46/1, which are highlighted in gray. rs78644782, rs10124001 and rs10758669 are immediately upstream of JAK2; all other SNPs are within JAK2 introns. (b) LD in the JAK2 region (HapMap data release 23a/phase II March 2008).
Table 1.
Summary of genotyping results
| Category | Number of cases |
Number of 46/1 alleles |
Number of non-46/1 alleles |
Frequency 46/1 |
P value (versus UK local controls) |
OR (95% CI) |
P value (versus WTCCC controls) |
OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| JAK2V617F-positive PV | 192a | 197 | 171 | 0.54 | 2.88E–16 | 3.6 (2.6–4.9) | 7.56E–30 | 3.6 (2.9–4.6) |
| JAK2V617F-positive ET | 78 | 79 | 77 | 0.51 | 8.24E–09 | 3.2 (2.1–4.7) | 4.27E–12 | 3.2 (2.4–4.5) |
| JAK2V617F-positive MF | 41a | 37 | 40 | 0.48 | 8.00E–05 | 2.9 (1.7–4.7) | 6.12E–06 | 2.9 (1.9–4.6) |
| JAK2V617F-positive unclassified MPN | 124a | 135 | 106 | 0.56 | 3.33E–15 | 3.9 (2.8–5.6) | 3.40E–24 | 4.0 (3.1–5.3) |
| Idiopathic erythrocytosis | 76 | 41 | 111 | 0.27 | 5.80E–01 | 1.1 (0.7–1.7) | 4.37E–01 | 1.2 (0.8–1.7) |
| JAK2V617F-negative MPN (UK) | 47 | 36 | 58 | 0.38 | 9.00E–03 | 1.9 (1.2–3.1) | 2.00E–03 | 2.0 (1.3–3.0) |
| JAK2V617F-positive ET (GR) | 143 | 124 | 162 | 0.43 | 4.51E–07b | 2.4 (1.7–3.3)b | 9.18E–12 | 2.4 (1.9–3.1) |
| JAK2V617F-negative ET (GR) | 136 | 89 | 183 | 0.33 | 9.00E–02b | 1.5 (1.1–2.1)b | 2.00E–03 | 1.5 (1.2–2.0) |
| UK controls | 188 | 92 | 284 | 0.24 | – | – | 8.48E–01 | 1.0 (0.8–1.3) |
| WTCCC controls | 1,500 | 720 | 2,280 | 0.24 | 8.48E–01 | 1.0 (0.8–1.3) | – | – |
| GR controls | 108 | 55 | 161 | 0.25 | 8.43E–01 | 1.1 (0.7–1.6) | 6.22E–01 | 1.1 (0.8–1.5) |
PV, polycythemia vera; ET, essential thrombocythaemia; MF, myelofibrosis; MPN, myeloproliferative neoplasm (all samples from the first six categories were from the UK and United States). GR, Greek samples; UK, UK samples; WTCCC, Wellcome Trust Case Control Consortium analysis of 1,500 UK blood donors.
These groups included some cases with ≥90% V617F and thus the residual wild-type allele could not be assigned as 46/1 or not 46/1.
Values versus healthy Greek controls. All P values were calculated using Fisher’s exact test, two-tailed.
We have previously described a polycythemia vera pedigree in which JAK2V617F was not inherited but arose independently in two affected individuals14. Family members were analyzed for rs12340895: one affected individual (UPN 534) was heterozygous for 46/1 but the second (UPN 533) was negative for this haplotype (Fig. 3). Allele-specific PCR for UPN 534 showed that JAK2V617F had arisen on the 46/1 allele, confirming the association between this haplotype and the mutation. However, this pedigree illustrates that 46/1 is not solely responsible, at least in this family, for predisposition to polycythemia vera. Indeed, linkage of disease to 9p has not been described in any family with MPN.
Figure 3.

Familial polycythemia vera pedigree. The two affected individuals (UPNs 534 and 533) are shown as black circles. The genotype for rs12340895 is shown (G = 46/1 allele; C = non-46/1 allele), as is the %V617F in affected cases. Allele-specific PCR for UPN 534 showed that JAK2V617F arose on the 46/1 allele. All other individuals had normal blood counts and were negative for JAK2V617F, PRV1 overexpression and endogenous erythroid colony growth14.
We suggest two hypotheses to account for the association of MPNs with 46/1: (i) JAK2V617F may arise randomly on all haplotypes but 46/1 is in LD with an unknown constitutional functional variant that interacts with JAK2V617F in a manner that makes the development of clinically manifest disease more likely compared to JAK2V617F on a non-46/1 haplotype, or (ii) there is a specific mutational mechanism by which JAK2V617F preferentially arises on a 46/1 haplotype. These hypotheses are not necessarily mutually exclusive. Inspection of the HapMap data (Fig. 2) indicates that the entire JAK2 gene is contained within a 280-kb LD block that includes two other genes (INSL4 and INSL6) that are not expressed in hemopoietic cells, as verified by RT-PCR analysis. It is highly likely therefore that any functional variant within 46/1 directly affects JAK2. Notably, rs10758669, a SNP that also tags 46/1, was identified as significant in a recent genome-wide association study of Crohn’s disease15, thus supporting the hypothesis of a functional JAK2 variant on that allele. This SNP was also reported to be significantly associated with polycythemia vera but not with essential thrombocythemia or myelofibrosis11, a result that is presumably explained by the relatively high prevalence of JAK2V617F in polycythemia vera compared to the other two subtypes.
JAK2 is required for signaling by diverse myeloid cytokine receptors (for example, IL-3, G-CSF, GM-CSF, EPO) as well as other receptors in lymphoid and nonhemopoietic cells16. To investigate the possibility that JAK2 on the 46/1 haplotype is different functionally from other JAK2 alleles, we tested whether 46/1 influences myeloid colony formation in hematologically normal individuals (n = 56). In a prospective analysis, we counted the numbers of granulocyte-macrophage colony-forming units (CFU-GM) and erythroid burst-forming units (BFU-E) in peripheral blood and compared the results to JAK2 haplotype. Individuals that carried at least one 46/1 allele grew significantly fewer CFU-GM, consistent with the hypothesis that JAK2 on 46/1 is indeed functionally different from other JAK2 alleles. There was no effect, however, on BFU-E growth (Fig. 4).
Figure 4.
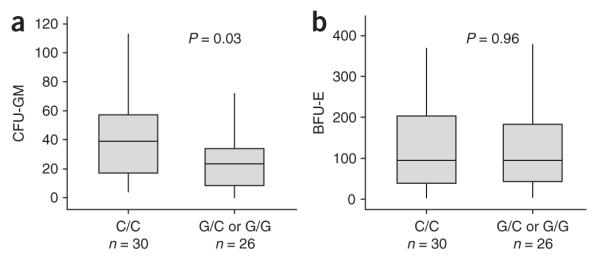
Association between JAK2 haplotype and numbers of hemopoietic colonies. CFU-GM and BFU-E colony growth per 4 × 105 peripheral blood mononuclear cells from 56 healthy controls that were 46/1 nullizygous (C/C at rs12340895, n = 30) or had at least one 46/1 allele (G/C, n = 21; or G/G, n = 5). Box plots illustrate the 95% range (vertical lines), median (horizontal lines) and interquartile range (boxes). Colony numbers were compared by the Mann-Whitney U test.
One possible reason for the observed association might be that JAK2 on 46/1 is expressed more or less than JAK2 on other haplotypes. To explore this possibility, we used pyrosequencing to quantify the allele ratios of two JAK2 exonic SNPs (rs10429491 and rs2230724) in matched cDNA and genomic DNA from control JAK2V617F-negative cases that were heterozygous for at least one of the exonic SNPs as well as heterozygous for 46/1 (n = 46). We found no differences in allele ratios in cDNA and genomic DNA with either SNP, indicating that 46/1 is not associated with either increased or decreased JAK2 expression, at least in peripheral blood leukocytes (Supplementary Fig. 1 online).
Finally, we explored the possibility that a functional variant might also be relevant to the pathogenesis of JAK2V617F-negative MPNs. We genotyped rs12340895 in JAK2V617F-negative essential thrombocythemia and myelofibrosis cases (n = 47) from the UK and found that 36 or 94 alleles were 46/1, significantly higher than the frequency in locally sourced controls and the WTCCC cohort (P = 0.009 and P = 0.002, respectively). However, we failed to confirm this association in an additional series of JAK2V617F-negative essential thrombocythemia cases (n = 136) and controls (n = 108) from Greece, although the P value of 0.09 and slightly elevated odds ratio suggests that the relevance of 46/1 to JAK2V617F-negative cases warrants further investigation (Table 1).
Our data thus demonstrate that both homozygous and heterozygous JAK2V617F-associated disease is preferentially associated with 46/1, and that this haplotype seems to be in LD with an as-yetuncharacterized functional variant. However, this does not exclude the possibility that JAK2 on 46/1 allele might be also be hypermutable. Whatever the mechanism, our data indicate that 46/1 is a strong predisposition factor for development of JAK2V617F-associated MPNs (OR = 3.7; 95% CI = 3.1–4.3; relative risk = 2.6; 95% CI = 2.3–2.9; n = 435 cases versus WTCCC controls). The counts of 46/1 alleles in cases and WTCCC controls and the population frequency from the WTCCC data (Table 1), suggest that 46/1 accounts for 28% of the population attributable risk17. A recent Swedish study demonstrated a relative risk of 5.7 in first-degree relatives of individuals with polycythemia vera10, corresponding to an attributable risk of 53%. Assuming no difference between the UK and Swedish populations, 46/1 thus accounts for slightly over 50% of the increased risk in first-degree relatives. For essential thrombocythemia and myelofibrosis, the contribution of 46/1 is less clear, as it is unknown what proportion of the risk in first-degree relatives10 is attributable to cases that are JAK2V617F positive and those that are JAK2V617F negative.
In addition to the specific association we describe here in MPNs, our findings may have wider relevance. Genome-wide association studies are identifying increasing numbers of loci that predispose to diverse malignancies18–20; our findings suggest that these loci should be considered as candidates for the acquisition of somatic mutations.
METHODS
Subjects
We analyzed a total of 775 subjects with MPN, of whom 183 had JAK2V617F-negative disease and 592 were JAK2V617F positive (PV, n = 203; ET, n = 224; MF, n = 41; unclassified MPN, n = 124). Subjects were recruited from clinics in the UK, United States and Greece. We also analyzed a previously described family with MPN from Germany14. For controls we analyzed healthy individuals from the UK (n = 188) and Greece (n = 108), and we also used data generated by the WTCCC from the UK blood donor cohort (n = 1,500)12. The study was approved by the relevant internal review boards and ethics committees and informed consent was provided according to the Declaration of Helsinki.
Genotyping
Total peripheral blood leukocyte DNA was analyzed by pyrosequencing for SNPs and JAK2V617F as described21. Primer sequences are provided in Supplementary Table 1 online. Because the allelic ratios (the ratio of allele A to allele B at any SNP) for any heterozygous SNPs were distorted away from the expected value of 0.5 in cases with a sizeable homozygous JAK2V617F clone, we adopted the following scoring criteria for all SNPs: (i) if one allele had an allelic ratio ≥0.9, the sample was scored as homozygous for that SNP; (ii) where allelic ratios were 0.11–0.89, samples were scored as heterozygous. These cutoffs were at least 3 s.d. more than background (that is, values read for allele B in healthy controls who were A/A homozygotes). Similarly, for homozygous JAK2V617F cases with V617F >50%, SNPs with allelic ratios ≥0.6 or ≤0.4 were considered to be derived from the V617F-mutated or residual wild-type alleles, respectively. For JAK2V617F homozygous cases in which the V617F was ≥90%, no information could be obtained about the residual wild-type JAK2 allele and thus these cases were considered to contribute only one allele to the analysis.
Allele-specific PCR
Allele-specific PCR was performed using forward primers that were specific for JAK2V617F (VF-ASF) or the corresponding wild-type sequence (WT-ASF) in combination with a common reverse primer (ASR), producing a 565-bp product that included the 46/1 tag SNP rs12343867. Amplification conditions were optimized on DNA from the HEL cell line (100% V617F) and normal healthy controls. Products were sequenced to determine whether JAK2V617F was on the 46/1 allele or not.
Colony analysis
Mononuclear cells (MNCs) from peripheral blood of hematopoietically normal controls were isolated by centrifugation over lymphoprep (Axis-Shield) and cultured in methylcellulose medium (H4434; Stem Cell Technologies) at a density of 4 × 105 cells per 30-mm plate (in a final volume of 1 ml), in triplicate, following the manufacturer’s instructions. Colonies comprising a minimum of 100 cells were counted on day 14, and characterized on the basis of morphology as either CFU-GM, CFU-GEMM, CFU-E or BFUE, as described by StemCell Technologies. JAK2 haplotype status was determined after colonies were counted using DNA extracted from the MNC or granulocyte cell fractions.
Expression analysis
RNA was extracted from peripheral blood leukocytes of JAK2V617F-negative MPD cases that were known to be heterozygous for rs10429491 and/or rs2230724 (JAK2 exonic SNPs) as well as heterozygous for 46/1, as determined by rs12340895 genotype. RNA was reverse transcribed with random hexamer primers and the ratio of the two alleles for each SNP in genomic DNA and cDNA was determined by specific Pyrosequencing assays (Supplementary Table 1).
Statistical analysis
The proportion of 46/1 alleles for each case subgroup was compared to controls using Fisher’s exact test (two-tailed). Colony numbers were compared to genotype using the Mann-Whitney U test. For the expression analysis, the mean and variance of SNP allelic rations were compared by t and F tests, respectively. Odds ratios (OR) were calculated as (number of 46/1 alleles in cases/number of 46/1 alleles in controls)/(number of non-46/1 alleles in cases/number of non-46/1 alleles in controls). Relative risk (RR) was calculated as (number of 46/1 alleles in cases/number of 46/1 alleles in cases plus controls)/(number of non-46/1 alleles in cases/number of non-46/1 alleles in cases plus controls). Population attributable risks were calculated as (f(RR − 1)/1 + f(RR − 1))100, where f is allele frequency17.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Leukaemia Research (UK) Specialist Programme Grant 0280 and makes use (in part) of data generated by the WTCCC. A full list of the investigators who contributed to the generation of the WTCCC data are available from www.wtccc.org.uk, funding for which was provided by the Wellcome Trust under award 076113. We are grateful to P. Strike (Salisbury Research and Development Support Unit) for statistical advice. A.R. was supported by the ’Deutsche José Carreras Leukämie-Stiftung e.V. - DJCLS R06/02, Germany.
Footnotes
AUTHOR CONTRIBUTIONS The study was designed by A.V.J., A. Chase., F.G. and N.C.P.C. A.V.J. performed the laboratory analysis. R.T.S., D.O., K.Z., Y.L.W., H.L.P., H.C. and A.R. provided clinical samples and associated information. A.V.J., A. Chase, A. Collins and N.C.P.C. analyzed the data. N.C.P.C. wrote the first draft of the manuscript and all authors contributed to and approved the final version.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 2.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 3.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Kralovics R, Guan Y, Prchal JT. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera. Exp. Hematol. 2002;30:229–236. doi: 10.1016/s0301-472x(01)00789-5. [DOI] [PubMed] [Google Scholar]
- 6.Jones AV, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 7.Pikman Y, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kralovics R. Genetic complexity of myeloproliferative neoplasms. Leukemia. 2008;22:1841–1848. doi: 10.1038/leu.2008.233. [DOI] [PubMed] [Google Scholar]
- 9.Rumi E, et al. Disease anticipation in familial myeloproliferative neoplasms. Blood. 2008;112:2587–2588. doi: 10.1182/blood-2008-05-160739. [DOI] [PubMed] [Google Scholar]
- 10.Landgren O, et al. Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24, 577 first-degree relatives of 11, 039 patients with myeloproliferative neoplasms in Sweden. Blood. 2008;112:2199–2204. doi: 10.1182/blood-2008-03-143602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardanani A, Fridley BL, Lasho TL, Gilliland DG, Tefferi A. Host genetic variation contributes to phenotypic diversity in myeloproliferative disorders. Blood. 2008;111:2785–2789. doi: 10.1182/blood-2007-06-095703. [DOI] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cario H, Goerttler PS, Steimle C, Levine RL, Pahl HL. The JAK2V617F mutation is acquired secondary to the predisposing alteration in familial polycythaemia vera. Br. J. Haematol. 2005;130:800–801. doi: 10.1111/j.1365-2141.2005.05683.x. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandberg EM, Wallace TA, Godeny MD, VonDerLinden D, Sayeski PP. Jak2 tyrosine kinase: a true jak of all trades? Cell Biochem. Biophys. 2004;41:207–232. doi: 10.1385/cbb:41:2:207. [DOI] [PubMed] [Google Scholar]
- 17.Khoury MJ, Beaty TH, Cohen BH. Fundamentals of Genetic Epidemiology. Oxford University Press; New York: 1993. [Google Scholar]
- 18.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 20.Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 21.Jones AV, et al. Minimal molecular response in polycythemia vera patients treated with imatinib or interferon alpha. Blood. 2006;107:3339–3341. doi: 10.1182/blood-2005-09-3917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.