Summary
Bacterial pathogens bearing capsular polysaccharides identical to mammalian glycans benefit from an additional level of protection from host immune response.
The increasing prevalence of antibiotic resistant bacteria portends an impending post-antibiotic age, characterized by diminishing efficacy of common antibiotics and routine application of multifaceted, complementary therapeutic approaches to treat bacterial infections, particularly multidrug-resistant organisms. The first line of defense for most bacterial pathogens consists of a physical and immunological barrier known as the capsule, commonly composed of a viscous layer of carbohydrates that are covalently bound to the cell wall in Gram-positive bacteria or often to lipids of the outer membrane in many Gram-negative bacteria. Bacterial capsular polysaccharides are a diverse class of high molecular weight polysaccharides contributing to virulence of many human pathogens in the gut, respiratory tree, urinary tract, and other host tissues, by hiding cell-surface components that might otherwise elicit host immune response. This review highlights capsular polysaccharides that are structurally identical or similar to polysaccharides found in mammalian tissues, including polysialic acid and glycosaminoglycan capsules hyaluronan, heparosan, and chondroitin. Such non-immunogenic coatings render pathogens insensitive to certain immune responses, effectively increasing residence time in host tissues and enabling pathologically relevant population densities to be reached. Biosynthetic pathways and capsular involvement in immune system evasion are described providing a basis for potential therapies aimed at supplementing or replacing antibiotic treatment.
Keywords: capsular polysaccharides, glycosaminoglycans, polysialic acid, bacterial pathogens, immune system evasion, combating antibiotic resistance
Introduction
Bacterial capsular polysaccharides (CPSs) are major virulence factors that confer protective effects to their bearers against a wide range of environmental pressures, most notably against the immune system during infection of their animal hosts. Although capsules are often associated with descriptions of pathogenic bacteria due to the large proportion of encapsulated invasive pathogens, non-pathogenic and commensal bacteria also benefit from the ability to envelope themselves with a capsule (Hafez et al., 2009; Dasgupta & Kasper, 2010). In Gram-negative bacteria, capsular polysaccharides are often attached to the outer membrane at their reducing end through covalently-linked lipids that are inserted into the lipid bilayer of the membrane. This provides a surface layer of water-saturated, high molecular weight polysaccharides that limit desiccation in the face of harsh environmental conditions, block infection by most bacteriophages, and thwart phagocytosis and other host immune responses by physically restricting access to cell surface antigens. These polysaccharide cloaks are likely rational targets for wide-spectrum therapeutic compounds aimed at replacing or supplementing antibiotic treatment of microbial infections, as removal of the capsule exposes bacteria to routine immune clearance pathways mediated frequently by activation of the complement system.
Historical perspective
The molecular compositions of CPSs vary extensively between organisms and even between strains within a single species, but, despite this diversity, some species from distinct orders have been shown to biosynthesize identical CPS structures (DeAngelis, 1999; DeAngelis & White, 2002). The existence of highly homologous biosynthetic machinery for production of identical polysaccharides between microbes suggests that capsular gene clusters have been acquired through horizontal gene transfer; conversely, non-homologous glycosyltransferases biosynthesize identical polysaccharides in disparate organisms (Vann et al., 1981; Finne et al., 1983; Korhonen et al., 1985; Jann & Jann, 1998), an occurrence that has likely developed through functional convergent evolution (DeAngelis, 2002a, b) facilitated by inter-kingdom coevolution of prokaryotic pathogens with their eukaryotic hosts (Gagneux & Varki, 1999; Chen & Varki, 2010). Capsule structure diversity was originally investigated as part of a broad effort to classify bacterial strains based upon their interaction with human serum; that is, bacteria were serotyped through differentiation of their cell surface antigens (Lancefield, 1933). Serotyping is critical for understanding pathogenicity from medical, diagnostic, and immunological perspectives and has remained the dominant method for classifying strains of capsular bacteria. While studies in the early twentieth century demonstrated that polysaccharides present in the capsules of both Gram-negative and Gram-positive bacteria were antigenically distinct from cellular protein fractions (Heidelberger & Avery, 1923), research throughout the following decades further differentiated and classified these polysaccharidic antigens and ultimately implicated the eponymous CPSs in elevated serum resistance and inhibition of granulocytic phagocytosis (Peterson et al., 1978; Horwitz & Silverstein, 1980).
Complementary serological and clinical studies during the latter half of the twentieth century identified a subset of streptococcal, staphylococcal, meningococcal, and Escherichia coli CPSs associated with increased virulence and widespread incidence in severe bacterial infections, provoking investigation of the relationship between CPS structure and immunogenicity (Robbins et al., 1974; Kaijser et al., 1977). Elucidation of the chemical structures of various K-antigens (an alternative name for the CPS of E. coli) and apparent demarcation based upon their physical properties prompted development of a K-antigen classification system (Orskov et al., 1977) that was revised over time to incorporate genetic and biomolecular evidence (Jann & Jann, 1997), ultimately being supplanted by a more robust grouping scheme based on genetic, biochemical, and molecular criteria (Whitfield & Roberts, 1999). Serotyping systems for other species were developed in a similar manner, but the relative ease of Gram-negative CPS structural characterization and the genetic tractability of E. coli enabled more rapid development of the E. coli antigen classification scheme. Owing to their antigenicity in mammals, most CPS structures elicit T lymphocyte-independent immune responses that induce IgM antibody production but fail to stimulate T cell-dependent IgM-IgG switching, an important attribute to ensure long-lasting immunity (Weintraub, 2003; Avci & Kasper, 2010). However, purified CPSs from some of the most commonly isolated strains were determined to be non-immunogenic due to structural identity with human glycans (Edwards et al., 1982; Johnson, 1991; Hérias et al., 1997). Capsule-deficient mutants of these strains generally exhibit decreased virulence, persistence, and serum sensitivity (Pluschke et al., 1983; Hérias et al., 1997). As discussed in greater detail later, antibody generation proved difficult against purified mammal-like bacterial CPSs composed of hyaluronan (HA), heparosan, or certain congeners of unsulfated chondroitin or polysialic acid (PSA).
It should be noted here that there are reports of antibodies generated against these CPSs under unique circumstances (Frosch et al., 1985; Jennings et al., 1985; Kabat et al., 1986; Kröncke et al., 1990; Finke et al., 1991; Troy 1992; Born et al., 1996). However, careful consideration should be given to possible antigenic determinants for antibodies generated in such experiments and whether access to the epitopes would result in protective responses in vivo. If serum-accessible portions of these CPSs are identical to mammalian glycans, it seems unlikely that antibodies could be elicited against these “self” epitopes. In some cases, antibodies were raised in autoimmune animal hosts, where self-protection capacity was diminished due to immune dysregulation (Bitter-Suermann et al., 1986). Immune response by healthy animal hosts requires other possible explanations to clarify this paradox:
The CPS possesses an exposed antigenic determinant not found in the corresponding mammalian glycan, such as a deacetylated amino sugar or terminal unsaturated bond generated by a lyase (a class of enzymes acting to cleave acidic polysaccharides through an eliminase mechanism, in contrast to hydrolyzing glycosidases) (Linhardt et al., 1986) or some other non-self chemical decoration.
CPS purification exposes a non-mammalian antigenic determinant, like an anchoring moiety composed of a phospholipid or a monosaccharide or oligosaccharide linker constituent not biosynthesized in mammals.
The antigenic determinant spans a self and non-self domain on the purified CPS, thereby cross-reacting with the self-domain.
The antibody is not specific for the CPS, but is cross-reacting due to structural similarity to the true antigen.
In one example, a human IgM class antibody reactive against PSA was shown to be non-specific due to the antibody's reactivity with polynucleotides, a scenario where cross-reactivity was enabled possibly due to similar surface charge distributions in these two negatively charged biopolymers (Kabat et al., 1986). An IgG antibody against PSA was also isolated in autoimmune NZB mice (Frosch et al., 1985). Nevertheless, evidence supports the conclusion that non-immunogenic bacterial CPSs sharing structural identity with host glycans confer an additional protective advantage over immunogenic, non-host CPSs.
Due to their poor immunogenicity, mammal-like CPSs were originally considered non-typeable by traditional means. However, “typing” of some non-immunogenic CPSs was later achieved by screening strains against phages only capable of attachment and subsequent infection when a specific CPS is displayed on the bacterial surface, such as E. coli K1 and K5-specific phages (Roberts et al., 1986; Scholl et al., 2001). With increasing genetic characterization of CPS-producing strains, polymerase chain reaction and sequencing-based methods such as multi-locus sequence typing (MLST) can now be used for fast and accurate molecular typing (Townsend et al., 2001; O'Hanlon et al., 2004; Durso et al., 2005; Kong et al., 2005; Zhu et al., 2012). Despite the utility of antibody-based serotyping, confounding variables at the immunological level make molecular diagnosis an attractive alternative.
Bacterial glycans
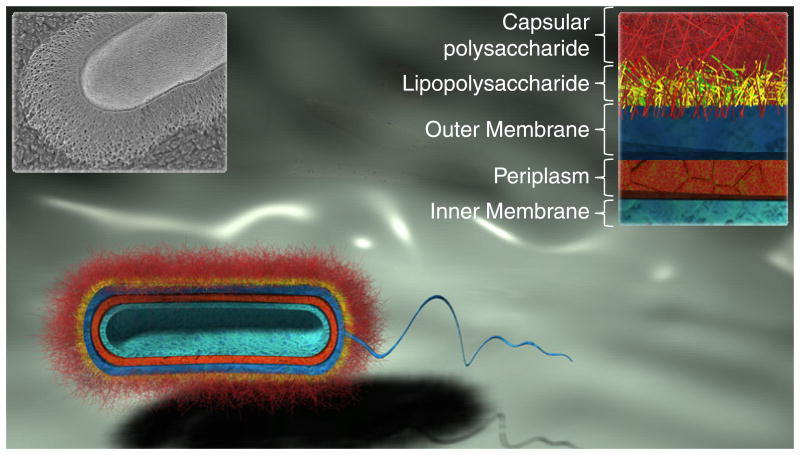
Bacteria produce an array of carbohydrates that are not limited to CPSs, and an understanding of these bacterial glycans is critical for appreciating the role of the CPS in the pathogenesis of infection. As depicted in Fig. 1, Gram-negative bacteria possess an external layer of long CPS chains that are covalently anchored by phospholipids to the outer leaflet of the outer membrane, an asymmetric lipid bilayer with an external layer composed primarily of lipid A (also known as endotoxin) and an internal phospholipid layer. Anchored to the outer membrane by lipid A, the lipopolysaccharide (LPS) serves as a hydrophilic barrier to natural hydrophobic antibiotics and is composed of three regions: (1) lipid A, a highly conserved region that possesses two phosphorylated, β-1,6 linked N-acetyl-d-glucosamine (GlcNAc) saccharide residues bearing variable numbers and lengths of fatty acyl chains; (2) the core oligosaccharide, which can be subdivided into the inner and outer cores. The inner core is covalently bound to lipid A and possesses a species-dependent or strain-dependent nonlinear oligosaccharide composed of 3-deoxy-d-mannooctulosonic acids (KDO), heptoses, and some non-glycan components such as pyrophosphoethanolamine (PPEtn). The outer core is linked to the terminal heptose of the inner core and possesses a more variable nonlinear structure of primarily hexose residues; (3) the last region of the LPS is known in E. coli as the O-antigen due to its distinct antigenicity from the K-antigen. The O-antigen is a repetitive glycan that varies in composition and length between species and strains, but it is typically masked from the environment by the K-antigen. Finally, an oligopeptide-cross-linked lattice of alternating β-1,4 linked GlcNAc and N-acetylmuramic acid (MurNAc) residues known as peptidoglycan is constrained within the periplasmic space between the outer and inner (cytoplasmic) membranes. In many species, peptidoglycan sugar moieties are further modified after installation (Vollmer, 2008). Water-soluble β-glucan polymers known as membrane-derived oligosaccharides (MDOs) are found near the inner membrane and are decorated with negatively charged ethanolamine, phosphoglycerol, and succinyl groups. These highly charged MDOs protect the inner cell membrane from low osmotic conditions (Esko et al., 2009). Thus, glycans are critical components of the cell wall that contribute to structural integrity and interaction with the environment (Comstock & Kasper, 2006) (Fig. 2). As Gram-positive bacteria do not possess an outer cell membrane, the peptidoglycan layer is thicker compared to Gram-negative bacteria. Lipoteichoic acids anchor the inner layers of peptidoglycan to a glycolipid extending from cytoplasmic membrane, while wall-associated teichoic acids tether additional outer layers together through covalently linkages to MurNAc residues. CPSs biosynthesized by Gram-positive bacteria can be anchored to the inner membrane, to oligopeptide cross-linkers within peptidoglycan, or to GlcNAc residues in the peptidoglycan lattice (Hanson & Neely, 2012).
Fig. 1.
Schematic cross-sectional representation of layers constituting the bacterial cell wall of a typical Gram-negative bacterium. The thick external CPS layer conceals the bacterium to prevent desiccation, bacteriophage infection, complement-mediated killing, and opsonophagocytosis. The black and white inset (top left) shows a quick-freeze, deep-etch scanning electron micrograph of the Gram-negative organism Bacteroides thetaiotaomicron (Martens et al., 2009); this SEM image was originally published in The Journal of Biological Chemistry. Martens EC, Roth R, Heuser JE & Gordon JI. Cover image. J Biol Chem. 2009; 284(27):cover. © the American Society for Biochemistry and Molecular Biology.
Fig. 2.
Glycan-centric schematic of typical Gram-negative cell wall components. Membrane proteins and other cell-wall constituents are neglected for simplicity. (a) Cell wall cross-section. (b) Lipopolysaccharide. (c) Capsular polysaccharide. Abbreviations are as follows: Lyso-PG = lyso-phosphatidylglycerol, GlcNAC = N-acetylglucosamine, MurNAc = N-acetylmuramic acid, GalNAc = N-acetylgalactosamine, KDO = 3-deoxy-D-mannooctulosonic acid, PPEtn = Pyrophosphoethanolamine, GlcA = glucuronic acid.
In addition to LPS and CPS, many species of Gram-negative and Gram-positive bacteria also biosynthesize and secrete an assortment of high-molecular weight glycopolymers known as exopolysaccharides, which have long been considered determinants of biofilm physicochemical properties (Costerton et al., 1987). Surprisingly, over the last decade exopolysaccharides from certain microbes have also been shown to inhibit biofilm formation by other microbial species (Valle et al., 2006; Kim et al., 2009; Nithya et al., 2010; Bendaoud et al., 2011; Jiang et al., 2011). As an example of the diversity of bacterial glycans, the glycocalyx of a strain of E. coli can simultaneously possess six exopolysaccharides. In addition to biosynthesis of covalently bound O-antigen and K-antigen, a linear heteropolysaccharide known as enterobacterial common antigen (ECA) is produced by all members of the Enterobacteriaceae family and is also frequently bound to the outer membrane. ECA consists of a conserved [→3)-α-Fuc4NAc-(1→4)-β-ManNAcA-(1→4)-α-GlcNAc-(1→]n repeating trisaccharide unit that can be bound by the reducing end of GlcNAc to the LPS core, anchored to the outer membrane by a phosphate bridge with diacylglycerophosphate, or secreted in a water-soluble, cyclized, and partially 6-O-acetylated (on GlcNAc) form with polymerization degree typically between 3 and 6 trisaccharide repeats (Erbel et al., 2003; Fregolino et al., 2012). Other exopolysaccharides, such as colanic acid (known as M-antigen), are also secreted into the environment by many E. coli strains. Although only some colanic acid (CA) remains loosely associated around the cell, especially when constrained within a biofilm during suboptimal growth, an E. coli strain has been isolated in which CA is ligated to the outer core of the LPS in place of the O-antigen (Meredith et al., 2007). Another exopolysaccharide of E. coli, commonly secreted by many other bacteria as well, is the adhesin poly-β-1,6-GlcNAc, a homopolymer that encourages adherence and biofilm formation (Wang et al., 2004, 2005). Finally, E. coli and other bacteria also biosynthesize and secrete an exopolysaccharide known as bacterial cellulose, or poly-β-1,4-glucan, as a component of the bacterial extracellular matrix (Zogaj et al., 2001). It can be inferred that dynamic regulation of this suite of bacterial extracellular glycans allows sampling of many glycocalyx states to adapt to a wide range of environments (Meredith et al., 2007).
While Gram-positive CPS structures are diverse and can be difficult to characterize, Gram-negative CPS structures are comparatively simple and have thus been more amenable to categorization (Whitfield, 2006). In spite of the depth and breadth of chemical and serological analysis of CPS structures, however, there remains a dearth of evidence regarding linkage of Gram-negative CPS to the cell. A very recent and fascinating report has resolved this longstanding enigma for a class of model E. coli and Neisseria meningitidis strains, demonstrating that capsules assembled through a common ATP-binding cassette (ABC) transporter pathway are biosynthesized on a nearly conserved lyso-phosphatidylglycerol anchoring moiety through an oligo-KDO linker, presumably guiding the translocation of such CPSs to the cell surface in a CPS- and organism-independent manner (Willis et al., 2013). CPSs in this category are known as Group 2 K-antigens in E. coli, but all characterized N. meningitidis strains possess a homologous transport system described below. Despite the variety of bacterial glycans, CPSs comprised of non-immunogenic polysaccharides are of primary medical interest due to their conspicuous ability to evade the immune system. Those sharing identity with human polysaccharides have been cataloged in a number of pathogenic species, but the most well characterized CPS structures are found in E. coli, N. meningitidis, Pasteurella multocida, and Streptococcus pyogenes. It is interesting to compare P. multocida, Avibacterium paragallinarum, and E. coli since strains of all species have been found to produce chondroitin and heparosan, while certain strains of P. multocida also possess HA capsules. Although P. multocida and A. paragallinarum are predominantly animal pathogens rather than human pathogens, the ability of these microorganisms to produce identical non-immunogenic capsules to E. coli through similar yet distinct genetic and enzymatic processes warrants inclusion of the species in this review. Moreover, the diseases caused by these organisms in livestock pose economic threats and cause concern regarding the contribution of antibiotic-laden livestock feed to the spread of antibiotic resistance. Identical PSA capsules are also produced between different species, including meningitis-causing strains of E. coli and N. meningitidis, while HA is found in the capsules of strep throat and necrotizing fasciitis-causing S. pyogenes and P. multocida, the etiological agent of fowl cholera and many other mammalian and bird diseases. Bacteria containing these CPSs are compiled in Table 1 and are included in the discussion where relevant.
Table 1. Pathogenic bacteria possessing non-immunogenic CPSs that are identical to human and animal glycans.
| CPS | GA G | Organism* | Serotype/Capsule Type | Disease(s) (Organism) | Reference(s) |
|---|---|---|---|---|---|
| Polysialic Acid | No | Escherichia coli | K1 | Meningitis, Urinary Tract Infection, diarrhea, septicemia (human) | (Silver et al., 1988) |
| Polysialic Acid | No | Neisseria meningitidis | B | Meningitis (human) | (Finne et al., 1983) |
| Polysialic Acid | No | Moraxella nonliquefaciens | Endopthlamitis, sepsis, meningitis, endocarditis (human) | (Bøvre et al., 1983; Devi et al., 1991; Rafiq et al., 2011) | |
| Polysialic Acid | No | Mannheimia (formerly Pasteurella) haemolytica | A2 | Bovine respiratory disease (bovine) | (Adlam et al., 1987; Rice et al., 2007) |
|
| |||||
| Chondroitin† | Yes | Escherichia coli | K4 | Urinary Tract Infection, diarrhea (human); diarrhea (bovine) | (Rodriguez et al., 1988; Orskov et al., 1985; Moxley & Francis, 1986) |
| Chondroitin | Yes | Pasteurella multocida | type F | Fowl cholera (avian) | (Rimler & Rhoades, 1987) |
| Chondroitin | Yes | Avibacterium paragallinarum | genotype I | Coryza (avian) | (Wu et al., 2010; Zhao et al., 2010) |
|
| |||||
| Heparosan | Yes | Escherichia coli | K5 | Urinary Tract Infection (human) | (Minshew et al., 1978; Zingler et al., 1990) |
| Heparosan | Yes | Pasteurella multocida | type D | Pneumonia (porcine) | (Ewers et al., 2006) |
| Heparosan | Yes | Avibacterium paragallinarum | genotype II | Coryza (avian) | (Wu et al., 2010) |
|
| |||||
| Hyaluronan | Yes | Streptococcus pyogenes | Scarlet fever, pharyngitis (human) | (Wessels et al., 1991; Ralph & Carapetis 2013) | |
| Hyaluronan | Yes | Streptococcus equi subsp. zooepidemicus | Septicemia, meningitis, endocarditis and arthritis (bovine, porcine, ovine, and canine) | (Wibawan et al., 1999; Wei et al., 2012) | |
| Hyaluronan | Yes | Streptococcus dysgalactiae subsp. equisimilis | Streptococcal Toxic Shock Syndrome (human) | (Calvinho et al., 1998; Hashikawa et al., 2004) | |
| Hyaluronan | Yes | Streptococcus uberis | Mastitis (bovine) | (Almeida & Oliver, 1993; Almeida et al., 2013) | |
| Hyaluronan | Yes | Streptococcus equi subsp. equi | Upper respiratory tract infection (equine) | (Anzai et al., 1999) | |
| Hyaluronan | Yes | Pasteurella multocida | type A | Respiratory disease (bovine, feline) | (Borrathybay et al., 2003b; Ewers et al., 2006) |
| Hyaluronan | Yes | Avibacterium paragallinarum | Coryza (avian) | (Sawata & Kume, 1983; Byarugaba et al., 2007) | |
In recent years, there has been much confusion regarding delineation of Streptococcus species and subspecies due to imprecise, muddled, and archaic classification systems (Jensen & Kilian, 2012). Similar scenarios occurred with Avibacterium and Mannheimia. Organism names are provided as originally reported unless a clear indication of misclassification was detected.
Fructosylated
Molecular mimicry and coevolution
In evaluating the ability of host-like CPSs to evade the immune system, it is important to understand which host tissues contain similar molecules and how pathogens coevolved with humans to enable such mimicry. The first capsule type addressed in this review is known as polysialic acid or PSA, consisting of N-acetylneuraminic acid (Neu5Ac) monomers joined with various glycosidic linkages. PSA capsules produced by strains of K1 E. coli, N. meningitides serotype B, Moraxella nonliquefaciens, and Mannheimia haemolytica (previously Pasteurella haemolytica) A2 are characterized by α-2,8 glycosidic linkages (Adlam et al., 1987; Devi et al., 1991), while PSA produced by other microorganisms possess α-2,9 glycosidic linkages (N. meningitidis serotype C strains) or alternating α-2,8 and α-2,9 glycosidic linkages (K92 E. coli strains) (Glode et al., 1977; Lifely et al., 1986). In mammals PSA is an α-2,8-linked polysaccharide on neural cell adhesion molecule (NCAM), which is found on the surface of neurons, glial cells, and natural killer cells, a type of lymphocyte functioning as an integral part of the innate or non-specific immune response (Rutishauser, 2008; Chang et al., 2009). Increased surface polysialylation leads to charge repulsion and is associated with decreased NCAM adhesion in animals (Rutishauser et al., 1985) and resistance to phagocytosis by PSA capsular bacteria (King et al., 2007). Structural identity of mammalian PSA with E. coli K1 and N. meningitidis type B CPSs—particularly embryonic NCAMs, which contain more than 50 α-2,8 Neu5Ac repeating units, compared to a much lower degree of polysialylation in adult NCAMs (Jann & Jann, 1998)—contributes to the neuroinvasiveness of these incredibly virulent neuropathogens (Robbins et al., 1974; Sarff et al., 1975). The PSA capsule is thought to enable traversal of the blood-brain barrier (Kim et al., 2003), thus leading to high rates of morbidity and mortality in neonatal meningitis and serious neurological conditions in survivors of the disease (Kaper et al., 2004).
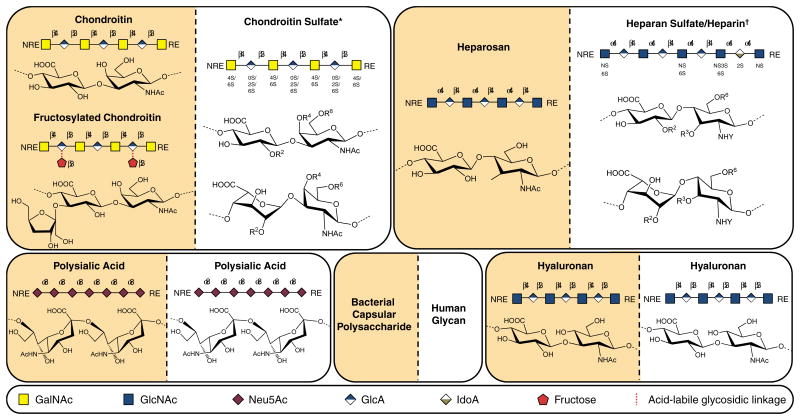
The other three host-like CPSs discussed in this review are considered glycosaminoglycans (GAGs), or negatively charged, linear polysaccharides identical to the backbones of GAGs found in animals and composed of a repeating core disaccharide unit, comprised of an uronic acid residue linked to an amino sugar (Höök et al., 1984). Although the monomeric sugar precursors constituting these core disaccharide units are conserved in GAGs, the disaccharide units in the animal GAGs heparan sulfate and chondroitin sulfate are not strictly repeating because they are variably sulfated and acetylated within a single chain. The dominant disaccharide unit in the polysaccharide and the distribution of specific disaccharide types, glycosidic linkage configurations, molecular weight, degree of sulfation and acetylation, and in some cases degree of epimerization define the class of GAG and contribute to heterogeneity within each class. GAGs exhibit their numerous biological activities by interacting with proteins including growth factors, chemokines, and adhesion molecules (Capila & Linhardt, 2002; Linhardt & Toida, 2004). The interactions between GAGs and pathogens can also represent the first line of contact between pathogen and host cell and are crucial to a pathogen's invasive potential (Kamhi et al., 2013). Symbolic representations of bacterial CPS structures and related animal glycan structures are illustrated in Fig. 3.
Fig. 3.
Symbolic representations and chemical structures of glycans described in this review. Non-immunogenic bacterial CPSs and structurally related animal glycans exhibited side-by-side to demonstrate similarity between backbones. In the case of chondroitin sulfate (CS) and heparan sulfate/heparin, bacterial CPS structures are identical to precursors of the mature human glycans depicted here. Of note, a related GAG known as dermatan sulfate also shares the unsulfated chondroitin backbone as a biosynthetic precursor, but, unlike CS, some glucuronic acid residues in the chain are epimerized to iduronic acid. CS type B possesses iduronic acid residues, so it is sometimes classified as dermatan sulfate. Conversely, HA and PSA structures are identical in microbial CPS and mature human GAGs. *R2,4,6 = H or SO3-; †R2,3,6 = H or SO3-, Y = SO3- or Ac (Ac = COCH3). Detailed disaccharide structures have been reported elsewhere (Sugahara & Mikami, 2007; Chang et al., 2012b).
In particular, the CPS produced by K4 E. coli and P. multocida type F strains are related to CS, a class of sulfated GAG characterized by a [→4) β-D-glucuronic acid (GlcA) (1→3) N-acetyl-β-d-galactosamine (GalNAc) (1→]n disaccharide repeat, where position and extent of sulfation are tissue and organism-dependent (Rodriguez et al., 1988; Volpi, 2007; Volpi et al., 2008). While K4 CPS GlcA residues are substituted with bisecting β-fructofuranose units between C2 of fructose and C3 of GlcA, the fructose is acid-labile, and K4 CPS is thought to exist as an unsubstituted backbone in certain low pH environments (Jann & Jann, 1990). Conversely, P. multocida type F CPS is identical to the unsulfated chondroitin precursor of animal CS (DeAngelis et al., 2002). Considering the limited patterns of O-sulfo group substitution within the disaccharide-repeating unit of animal CS (Sugahara & Mikami, 2007), it is apparent that the order of the sulfonation reactions is important in CS biosynthesis and that O-sulfo groups in certain positions can preclude the activity of downstream sulfotransferases (Schiraldi et al., 2010). CS occurs in animals as an O-linked glycan chain, covalently bound to serine residues of proteins, through a specific tetrasaccharide linkage, resulting in glycoconjugates known as proteoglycans (Esko, 2009). This class of GAG is found primarily in the extracellular matrix where one or more CS chain is attached to an array of core proteins affording proteoglycans with various structural and regulatory roles. Proteoglycans mediate a myriad of physiological interactions such as cellular recognition, communication, migration, adhesion, and proliferation (Thelin et al., 2013).
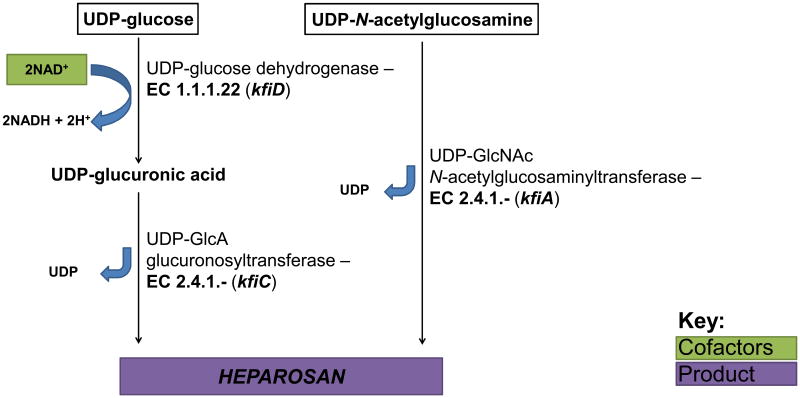
The CPS produced by K5 E. coli strains such as Bi 8773-41 and the probiotic strain Nissle 1917 (Lodinová-Žaadniková et al., 1992) and also by P. multocida type D strains is composed of heparosan (Vann et al., 1981; DeAngelis & White, 2002). Heparosan, comprised of [→4) β-D-GlcA (1→4) α-d-GlcNAc (1→]n repeating disaccharide units, is identical to the mammalian precursor for heparin and heparan sulfate (Ly et al., 2011) (Fig. 3). Heparan sulfate is typically found on the cell membrane and in the extracellular matrix as a component of proteoglycans (Gallagher, 1989). Heparin, a highly sulfated variant of heparan sulfate, is biosynthesized as an intracellular proteoglycan, serglycin (Li et al., 2012). The GAG heparin is a widely used anticoagulant pharmaceutical (Capila & Linhardt, 2002). Although the natural function of heparin is not well understood, its release from the granules of mast cells is localized to damaged tissue and contributes to wound healing and defense against opportunistic infection (Zehnder & Galli, 1999). The non-template-driven biosynthesis of heparan sulfate and heparin affords a diverse range of disaccharide units from a modest number of biosynthetic enzymes (Fig. 3).
The GAG known as hyaluronan or HA is structurally identical in animals and in capsules of S. pyogenes groups A and C, P. multocida type A, and some other species of bacteria (DeAngelis, 1999). HA GAG consists of an unmodified [→4) β-D-GlcA (1→3) β-D-GlcNAc (1→]n disaccharide repeating unit in both animals and bacteria. In animals, this high molecular weight polysaccharide is the predominant GAG in the extracellular matrix and is found in high quantities in skin, connective tissues, cartilage, synovial fluid, and the vitreous humor of the eye (Höök et al., 1984; Dougherty & van de Rijn, 1992).
The role of these host-like, or “self”, capsules enveloping the surfaces of invasive pathogens seems quite clear: molecular mimicry enables such pathogens to evade an immune system that has learned to ignore self molecules based on cell surface interactions. As glycans are ubiquitous on cell surfaces (Gallagher, 1989), they are likely the first molecules contacted by pathogens that utilize cellular adhesion during infection, so pathogens bearing these capsules have an evolutionary advantage. The question that remains is how humans and other animals have evolved to combat these camouflaged pathogens and how pathogens continually evolve to successfully colonize their animal hosts. In a series of papers, Ajit Varki argues that genetic evidence suggests pathogens—which evolve orders of magnitude faster than their hosts due to horizontal gene transfer, high mutation rates, fast growth, and short life spans—develop self-CPSs through convergent evolution, where the glycosyltransferase genes responsible for biosynthesizing the glycans in pathogen and host are not homologous in most cases (Gagneux & Varki, 1999). Considering that glycosyltransferases are highly conserved within the host and yet biosynthesize highly diverse glycan structures and distributions in different cell types and tissues, coupled with the combinatorial style of glycan interactions and the ability to maintain functional specificity when a participating glycan is modified, Varki also argues that sexual reproduction-enabled mutations in host glycosyltransferases and subsequent change in glycan profile allow these multicellular organisms to adapt to pathogenic pressure. Futhermore, Varki speculates that the coevolution of pathogens and their hosts has not only tailored the diversity of glycan structures and expression patterns, but that pathogenic pressures stemming from host-like capsules contribute significantly to speciation of multicellular organisms (Varki, 2006).
Immune response and clearance of encapsulated pathogens
Both evasion of complement-mediated killing and failure of being ingested by phagocytic cells enhance the virulence of the CPS. The polysaccharide capsules are effective physical barriers that protect the bacteria from being killed. The fact that bacteria capsules are commonly hydrophilic and negatively charged diminishes their removal through phagocytosis. The hydrophilic nature causes high-level hydration and reduces the surface tension at the phagocyte and bacterium interface (Kuberan & Linhardt, 2000). Additionally, the negatively charged polysaccharides on the bacterial surface repel the negatively charged surface of phagocytes, thus increasing the unfavorable interaction when phagocytosis or complement-mediated lysis occurs (Moxon & Kroll, 1990; Kuberan & Linhardt, 2000). According to van Oss and Gillman, the phagocytic cells such as polymorphonuclear leukocytes (PMNs), monocytes, and macrophages repel the encapsulated bacteria due to the net Lewis AB repulsion between the hydrophilic outer layers (Klainer & Geis, 1975; van Oss et al., 1975), which reduce the surface tension between the phagocytic cell and the bacterium (Moxon & Kroll, 1990). For example, the cell surface of Staphylococcus aureus became less hydrophilic after removing the capsule and its phagocytic uptake was enhanced (van Oss et al., 1975). A similar phenomenon was observed with the encapsulated strain of Salmonella typhimurium, which resists phagocytosis, but when unencapsulated it is readily phagocytized (Cunningham et al., 1975). Non-effective contact can often lead to the failure of phagocytic engulfment. More intuitive is simple charge-charge repulsion between the negative charge of the CPS and the glycocalyx of the phagocytic cell. The more highly charged the CPS the more likely a bacterium is to avoid opsonophagocytosis (Moxon & Kroll, 1990). Poor phagocytosis of a ‘smooth surface’ may directly result from the physical surface properties instead of biological interaction of capsules with phagocytic signaling and complement-mediated molecules. Direct experimental testing of this hypothesis remains challenging.
The interaction between the CPS of the bacterial surface and the host's complement system is also a key contributor to bacterial virulence. In the early stage of the immune response, the control and defense mechanism of the host are contingent on the classic and alternative complement pathways. The classic pathway is usually initiated by antigen-antibody binding. The C1 complement complex, which is a multi-molecular protease consisting of three subunits C1q, C1r and C1s, triggers the classical pathway of complement, first binding to the aggregated antibody molecule, then sequentially cleaving and activating the complement protein C4 and proenzyeme C2 to form a C3 convertase, C2bC4b (Jann & Jann, 1997). This process is regulated by C4-binding protein C4bp (Roberts, 1996) and is usually retarded during the encapsulated bacterial invasion. The C3 convertase then converts C3 to the activated C3b, which will be deposited on the bacterial cell surface. This process is controlled by factors B and H of the alternative pathway (Jann & Jann, 1997). The alternative pathway can be activated in the absence of antibody binding to the bacterial surface and therefore is very important in immunity to encapsulated or unencapsulated bacteria. In other words, the alternative pathway provides a way for the immune system to kill bacteria in the blood in the absence of specific antibodies. The alternative pathway utilizes the serum protein C3b, which is then activated by serum factor B, D and properdin (Moxon & Kroll, 1990), to form convertase C3bBb that amplifies the complement cascade for more C3 conversions and C3b deposition (Roberts, 1996). The activation of C3b results in a ligand targeting specific receptors on PMNs or macrophages. The binding of a C3b opsonized microbe to the complement receptor on PMNs or macrophages initiates phagocytosis and ultimately killing of the encapsulated bacteria. In addition, following C3b deposition, the sequential activations of C5 to C9 forms a membrane attack complex that directly leads to the lysis and death of some Gram-negative bacteria (Moxon & Kroll, 1990; Roberts, 1996).
This bacterial defense mechanism and the subsequent response of complement-mediated bacterial killing by the host can be blocked at numerous sites by CPSs avoiding serum-mediated killing and enhancing virulence. Some capsules protect the bacteria from being attacked by steric mechanisms. Bacteria such as pneumococci promote C3b deposit on the bacterial cell surface underneath the capsule, shielding it from recognition by the phagocytic cell (Winkelstein, 1981). Some bacterial capsules interrupt the binding of C3b to the bacterial surface by affecting regulatory proteins, such as factor B and H (Loos, 1985; Cross, 1990). Capsules that exert such a defense mechanism usually contain N-acetylneuraminic acid (Neu5Ac) since it contains a factor H binding site. The stimulation of H-C3b, correspondingly decreases the amplification convertase C3bBb, leading to failure of the complement cascade (Moxon & Kroll, 1990). Some capsules cannot bind to factor B, thus causing more H-C3b formation (Winkelstein, 1981). Strains such as E. coli K1, E. coli K92, N. meningitides types B and C and Group B Streptococcus polysaccharides have capsules that inhibit alternative complement activation by these mecahnisms (Stevens et al., 1978; Wessels et al., 1989).
The mimicry of the CPS structure to substances within the host serves as a virulence factor preventing bacteria phagocytosis. A CPS can mimic a similar structure found within the host representing “self”, and therefore both avoid recognition as foreign and circumvent triggering the host immune response (Kuberan & Linhardt, 2000). The CPS K1 has the same poly-α-2,8-Neu5Ac (PSA) structure as carbohydrate portion of NCAM, required for organogenesis and neural cell growth (Finne, 1982; Kuberan & Linhardt, 2000). Similarly, the CPS K5 strain of E. coli shares the same structure as mammalian heparosan (Navia et al., 1983). An X-ray diffraction study showed that the K4 capsule was poorly immunogenic due to its similar helix structure to CS. The removal of fructosyl linkage under low pH environment transforms K4 CPS into non-immunogenic chondroitin (Jann & Jann, 1997).
Capsular polysaccharide transport, genetics, biosynthesis, and role in immune system evasion
The chemical properties and immunogenicity of CPSs are dictated by variations in number, order, and diversity of monosaccharide constituents, anomeric centers (α- or β-), glycosidic linkage positions, absolute configuration (L or D), ring forms (pyranose or furanose), degree of chemical modification (O-acetylation, for example), and overall conformation (Mazmanian & Kasper, 2006). There is a wide range of capsule types among bacterial orders and even within a single species. For instance, strains belonging to one of the most well-studied CPS-producing species, E. coli, are known to biosynthesize approximately 80 CPS structures. The number of known capsule types increases dramatically when considering other genera, but capsules in other organisms are less well characterized due to limited biochemical studies and relative genetic recalcitrance. Nevertheless, studies in the model capsular species E. coli suggest that the capsule assembly pathways are comparatively limited in scope, where a diverse assortment of CPSs are assembled and translocated to the cell surface using identical strategies. Biochemical and genetic evidence in Gram-negative bacteria paints a picture of a veritable orchestra of catalytic enzymes, structural proteins, and transport proteins interacting in a transmembrane complex that spatially and temporally organizes biosynthesis and transport. The modularity of the cooperating sub-complexes allows distinct CPS biosynthetic enzymes, complexed at the inner membrane, to utilize identical transport systems for translocation of disparate CPS. Whitfield and coworkers recently showed an ABC-transporter dependent pathway common to some E. coli and N. meningitidis strains results in the biosynthesis of unique CPSs on a common anchor structure (Willis et al., 2013). This apparently ensures successful CPS transport and outer membrane attachment. Similarly, another commonly conserved transport system, known as the Wzy-dependent pathway, shares the ability to assemble CPSs with relaxed specificity for CPS structure. Although a wide range of bacteria utilize the ATP-dependent and Wzy-dependent pathways for CPS assembly, the majority of experimental evidence has been acquired in E. coli. Homologous genes between species have been identified by sequence similarity in many cases rather than by functional characterization. Hence this section of the review will focus on E. coli as a model system and draw comparisons between related bacteria where relevant.
Transport pathways
In E. coli, CPS structures have been classified into four groups. Group 1 and 4 CPS structures (as well as colanic acid) are found in enteropathogenic (EPEC), enterotoxigenic (ETEC), and enterohemorrhagic E. coli (EHEC) strains and are assembled through what is known as the Wzy-dependent pathway. This pathway is distinct from the so-called ABC-transporter dependent pathway that is responsible for assembly and transport of Group 2 and 3 CPSs and that is described in detail later. While uronic acid sugars are common to Group 1 CPS repeat units, Group 4 CPS repeats are characterized by the presence of acetamido sugars. Despite this apparent structural distinction between Group 1 and 4 CPSs, both are polymerized and transported to the cell surface in a similar manner. In the Wzy-dependent system, serotype-specific repeating units are assembled from cytosolic sugar precursors and linked to undecaprenyl diphosphate by glycosyltransferases, unique to the specific type of CPS being synthesized, which are embedded in the cytosplasmic membrane. Individual undecaprenyl diphosphate-linked repeating units are then transferred across the inner membrane to the periplasm by a flippase, Wzx, which also passes the repeat unit to an integral membrane protein known as Wzy. Wzy processively catalyzes addition of these individual Group 1 and 4 CPS repeat units to the reducing end of the growing polysaccharide chain, which elongates in the periplasm without being released by Wzy until chain termination (Yi et al., 2006). Wza, Wzb, and Wzc are responsible for control of chain length and export from the periplasmic face of the inner membrane to the cell surface. In Group 4 strains, longer polysaccharide chains can be incorporated into the LPS structure and effectively anchored by lipid A, although these K-antigens are classified as KLPS to distinguish their unique attachment mechanism (Whitfield, 2006). In Group 1 strains, shorter polysaccharide chains can also form KLPS, but longer chains are known to assemble capsules without covalent attachment to LPS. Although the outer-membrane protein Wzi had been implicated in attachment of Group 1 CPSs (specifically the K30 antigen) to the outer membrane (Rahn et al., 2003), the exact mechanism was unknown until recently. A paradigm shift in understanding CPS attachment resulted from a study that concluded K30 CPS remained associated with the outer surface of the cell due to interactions with an outer-membrane lectin, Wzi, that captures secreted CPS and serves as a nucleation site for further CPS recruitment (Bushell et al., 2013). Wzy-dependent capsules are also biosynthesized in Klebsiella pneumoniae, and much of the molecular insight for early steps in this pathway came from studies of Salmonella enterica O-antigen assembly. As CPSs in this class do not share identity with animal glycans, they elicit an immune response and are thus out of the scope of this review. The reader is directed to two excellent reviews compiling recent research in this area (Whitfield, 2006; Reid & Cuthbertson, 2012).
Group 2 and 3 E. coli CPSs are produced in strains commonly associated with extraintestinal infections (ExPEC), while all known E. coli CPS structures sharing identity with animal glycans belong to Group 2. It is also interesting to note that Group 2 and 3 E. coli CPSs share certain similar structure and assembly characteristics with strains of N. meningitidis, P. multocida, Haemophilus influenzae, and Campylobacter jejuni (Whitfield, 2006). In contrast to Group 1 and 4 CPSs, the repeat units of Group 2 and 3 CPSs exhibit extensive variation in structure. Similar to the Wzy-dependent transport system, the ABC-transporter dependent system expressed by Group 2 and 3 E. coli strains has relaxed specificity for CPS structure, successfully transporting very distinct structures across the cell wall. A striking difference compared to Wzy-dependent assembly is that Group 2 and 3 CPSs, assembled by ABC-transporter dependent pathways, are completely polymerized in the cytoplasm and then transported across the cell wall to the outside of the cell. CPSs of this class are elongated by processive, CPS-specific glycosyltransferases that are co-localized to the cytoplasmic surface of the inner membrane with other proteins belonging to the coordinated biosynthetic-transport complex. Details regarding polymerization initiation are not fully resolved, but CPSs from N. meningitidis group B, E. coli K1, and E. coli K5 strains (all Group 2 type capsules) were recently shown (Willis et al., 2013) to be linked to a well-conserved lyso-phosphatidylglycerol (lyso-PG) terminus by a poly-β-KDO linker. It should be noted that slight variation in fatty acyl chain length and number of KDO repeats was measured within single cultures and between organisms. For instance, the single fatty acyl chain of lyso-PG in most cultures varied between saturated C16 (palmitoyl-PG) or monounsaturated C18 (oleoyl-PG), but one culture produced diacyl-PG with either two C16 chains (dipalmitoyl-PG), two C18 chains (dioleoyl-PG), or one C16 adjacent to a C18 chain (palmitoyl-oleoyl-PG). Furthermore, the number of KDO monomers exhibited inter-strain and intra-strain variation between 5 and 9 KDO repeats. This discovery suggests that the common glycolipid carrier is the anchor by which the ABC-transporter guides Group 2 CPSs from the cytoplasm to the outer membrane. However, the mechanism by which the glycolipid carrier is assembled and attached to the nascent polysaccharide remains undetermined. Comparatively little is known about assembly and transport of Group 3 CPS, but high sequence homology with Group 2 transport machinery suggests that the two groups share a common transport mechanism. Studies on Group 3 strains, none of which are known to produce animal-like glycans, are reviewed elsewhere (Barrett et al., 2002).
Genetics
Genes involved in CPS biosynthesis and transport are typically organized within a so-called capsular gene cluster. Gene products participating in transport are generally more well conserved, while capsule biosynthetic enzymes are specific to capsule type, again suggesting an organization in which CPS transport proteins interface with CPS biosynthetic enzymes in a modular, interchangeable fashion. In fact, episomal expression of CPS biosynthetic enzymes from one Group 2 strain has been shown to lead to functional capsule “transplantation” in another acapsular Group 2 strain, where CPS is secreted by the common transport complex as expected (Zhang et al., 2012). General characteristics of capsular gene clusters include distinct GC-content compared to the rest of the chromosomal DNA, lending additional evidence that these genes were acquired through horizontal transfer. Furthermore, CPS gene clusters are often encoded within regions of the genome known as genomic (also pathogenicity-associated) islands (Sun et al., 2005; Wiles et al., 2008), or segments of the genome prone to horizontal gene transfer that often encode virulence factors (Ostblom et al., 2011). Since the pioneering experiment in which Silver and coworkers cloned and heterologously expressed the E. coli K1 capsule—the first study to clone an entire CPS gene cluster—Group 2 K-antigen assembly systems have become the prototype for genetic and biochemical characterization of ABC-transporter dependent pathways (Silver et al., 1981).
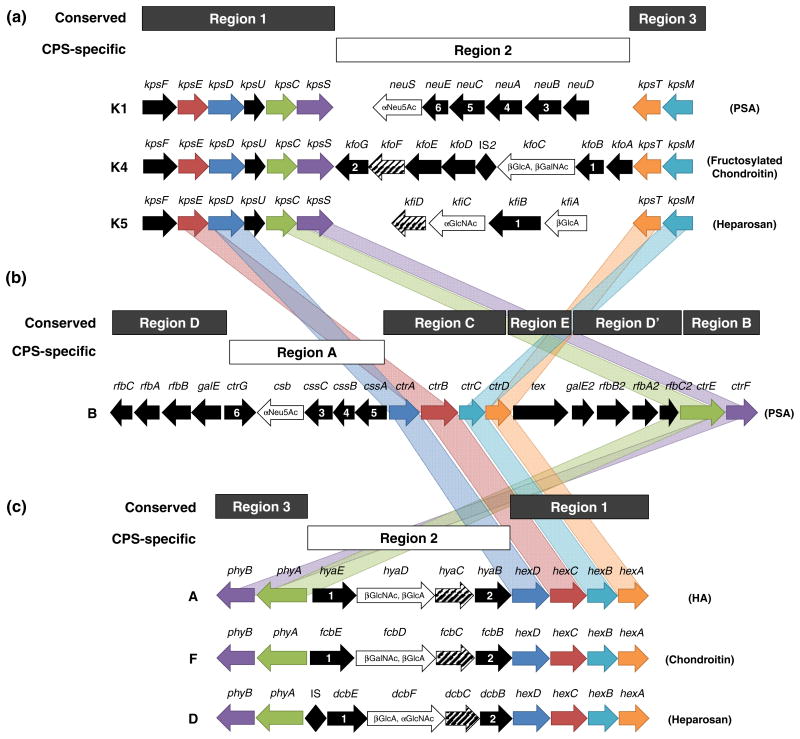
The gene cluster encoding biosynthesis and transport of Group 2 capsules, including the capsule types of primary interest in this review, is depicted in Fig. 4a. Regions 1 (kpsFEDUCS) and 3 (kpsMT) genes are conserved in Group 2 capsular E. coli and encode the enzymes and transport proteins responsible for initiation of chain elongation and translocation to the cell surface, while Region 2 genes encode the glycosyltransferases and other enzymes responsible for biosynthesis of the K-antigen-specific CPS. In comparison to Groups 1, 3, and 4, expression of Group 2 CPS is subject to thermoregulation. Promoters upstream of Region 1 and Region 3 are sufficient for transcription of all genes within the CPS cluster at temperatures near the optimum of 37°C, but no Region 1 or 3 transcripts are detectable at temperatures below 20°C (Cieslewicz & Vimr, 1996; Simpson et al., 1996; Stevens et al., 1997; Whitfield & Roberts, 1999; Rowe et al., 2000; Xue et al., 2009). Specifically, kpsMT of Region 3 encodes the ABC transporter responsible for translocation of the fully synthesized CPS across the inner membrane (Reizer et al., 1992). The transporter consists of multiple protein products, where two units of KpsM function as the inner membrane spanning domain and two units of KpsT serve as the nucleotide-binding domain (Pavelka et al., 1994; Pigeon & Silver, 1994; Steenbergen & Vimr, 2008). Region 3 is organized into a single transcriptional unit such that the start codon of kpsT overlaps the stop codon of kpsM by two base-pairs, and it has been suggested that the two proteins are translationally coupled to facilitate their interaction at the inner membrane (Smith et al., 1990; Pavelka et al., 1991). Upon binding of ATP, KpsT undergoes a conformational change that is conveyed to KpsM, which then experiences a change in conformation to enable transport of CPS using the energy gained from ATP hydrolysis (Bliss et al., 1996).
Fig. 4.
CPS gene loci in Gram-negative bacteria expressing ABC-transporter dependent CPS assembly pathways. (a) Gene loci encoding enzymes and transport proteins required for assembly of Group 2 E. coli K-antigens K1 (polysialic acid), K4 (chondroitin), and K5 (heparosan). Genes encoded by Regions 1 and 3 are well-conserved within Group 2 E. coli strains and encode enzymes required for CPS translocation across the cell wall, while Region 2 encodes CPS-specific glycosyltransferases and other biosynthetic enzymes. (b) Gene locus encoding proteins required for assembly of N. meningitidis serogroup B CPS. Region A encodes CPS-specific biosynthetic enzymes and varies between serogroups, while Regions B-E are highly conserved in all N. meningitidis serogroups. Regions B and C encode CPS translocation proteins homologous to genes in Regions 1 and 3 of Group 2 E. coli (homologous genes connected with gray bands), Regions D and D′ encode LPS assembly genes, and Region E has no known function. (c) Gene loci encoding P. multocida type A, D, and F CPS biosynthetic and transport proteins. Region 1 and 3 encode translocation genes whose functions are relatively well conserved in P. multocida, and Region 2 encodes CPS biosynthetic enzymes unique to the serotype specified. Homologous inter-species transport genes are color-coded and connected by bands of matching colors. Genes encoding glycosyltransferases are illustrated as white arrows with black outline and glycosyltransferase activity denoted within (bifunctional glycosyltransferases are labeled as found in nature, with N-terminal domain displayed at 5′ end of gene and C-terminal domain displayed at 3′ end of gene). UDP-glucose dehydrogenase is frequently encoded in CPS-specific biosynthetic clusters and is indicated here with diagonal lines. Other genes with known and putative homologs are designated with matching numbers. Note: genes and operons are not drawn to scale.
Region 1 encodes genes implicated in biosynthesis of the poly-KDO linker, as well as in translocation initiation and transport through the periplasm. The genes kpsED encode two proteins that receive the CPS from KpsMT and transport it to the outer membrane (Wunder et al., 1994; Rosenow et al., 1995). Functional deletions of kpsED lead to accumulation of CPS in the periplasm, which supports the role of KpsED in CPS translocation (Silver et al., 1988; Bronner et al., 1993; Pazzani et al., 1993). KpsE has been described as an adaptor protein that spans the periplasm to guide CPS from the ABC-transporter toward KpsD, a channel allowing CPS passage through the outer membrane (Rosenow et al., 1995). The gene kpsF encodes d-arabinose 5-phosphate isomerase, a homotetramer (Meredith & Woodard, 2006) that interconverts d-ribulose 5-phosphate and d-arabinose 5-phosphate with higher turnover toward d-arabinose 5-phosphate. The adjacent gene kpsU encodes CMP-KDO synthetase, a dimer (Jelakovic et al., 1996) that converts d-arabinose 5-phosphate provided by KpsF to nucleotide-activated CMP-KDO (Rosenow et al., 1995). The absolute roles of the cytosolic proteins encoded by kpsC and kpsS are not entirely elucidated. Group 2 strains with deletions of kpsC (ΔkpsC) or kpsS (ΔkpsS) accumulate high molecular weight polysaccharide intracellularly, implicating these two proteins in control of polymer length as well as in translocation initiation (Larue et al., 2011; Willis et al., 2013). The accumulating cytosolic polysaccharide inside ΔkpsC and ΔkpsS K1 strains was recently found to be non-lipidated (Willis et al., 2013) despite conflicting past reports (Frosch & Müller, 1993; Tzeng et al., 2005), suggesting that either KpsC or KpsS might catalyze the transfer of CPS to the glycolipid anchor. Given that most genes required for CPS biosynthesis and transport are typically located within the CPS biosynthetic gene cluster, it is also possible that either KpsC or KpsS is a β-KDO-polymerase catalyzing the biosynthesis of the poly-β-KDO-linker (Willis et al., 2013). Further studies are required to determine the roles played by KpsC and KpsS in biosynthesis and translocation initiation.
Common to Region 2 are genes encoding the glycosyltransferases required for assembly of the K-antigen-specific CPS structure. Glycosyltransferases in Group 2 E. coli are processive and catalyze the addition of high-energy nucleotide-activated sugar monomers to the nonreducing end of the growing CPS. It should be noted that identical CPSs in disparate bacteria, such as P. multocida, are biosynthesized by non-processive enzymes (DeAngelis et al., 2003). Studies have shown that Group 2 glycosyltransferases colocalize to the cytoplasmic side of the inner membrane with other proteins from the CPS gene cluster and that the proteins form a hierarchical transenvelope hetero-oligomeric complex to efficiently couple CPS assembly and transport (Rigg et al., 1998). Also often encoded in Region 2 with the glycosyltransferases are enzymes that biosynthesize CPS precursors but that do not actively participate in chain elongation (Cimini et al., 2012). In certain instances, enzymes within the CPS biosynthetic gene cluster are predicted to duplicate the function of enzymes encoded elsewhere in the chromosome (Muñoz et al., 1998). However, sequence divergence between the copies suggests that there could be an advantage conferred by the extra copy. Spatial co-localization of such duplicated enzymes with the CPS biosynthetic complex might ensure higher local concentrations of CPS precursors. Finally, CPS clusters possess genes with unknown functions that do not appear necessary for CPS production (Krahulec et al., 2005), while other encoded proteins lacking detectable catalytic activity have been shown to associate with the biosynthetic complex and increase biosynthetic productivity, possibly by lending structural integrity to the biosynthetic complex or by fostering protein-protein interactions (Hodson et al., 2000).
CPS biosynthesis in N. meningitidis is not as well characterized as in the more genetically tractable microbe, E. coli. However, the genomes of representative strains from all known serogroups have been sequenced, and an ABC-transporter dependent capsule assembly pathway with homology to Group 2 and 3 E. coli strains is conserved among all N. meningitidis capsular strains (Harrison et al., 2013). Unique to each serogroup, of course, are CPS biosynthetic enzymes for serotype-specific polysaccharide production. Despite the homology of many CPS transport genes between N. meningitidis and E. coli, the two distinct CPS loci exhibit limited synteny. Six regions known as A-D, D′, and E exist within the CPS gene locus of N. meningitidis, occurring in the order D-A-C-E-D′-B (Fig. 4b). CPS-specific biosynthetic genes are encoded within Region A, and CPS transport proteins are encoded within Regions C and B. Protein sequence alignments have been used to identify CPS transport proteins in N. meningitidis, and a new gene nomenclature has recently been proposed to ensure consistent descriptions between strains, where the names of all N. meningitidis CPS transport genes begin with “ctr” to denote capsule transport (Harrison et al., 2013). The four genes encoding the transmembrane complex ctrA, ctrB, ctrC, and ctrD are adjacent to each other within Region C of N. meningitidis, which contrasts the organization of the E. coli homologs kpsD, kpsE, kpsM, and kpsT, respectively. It seems intuitive that the proteins required for CPS translocation across the cell wall, including the ABC transporter proteins (KpsM/CtrC and KpsT/CtrD), the periplasm spanning adaptor protein (KpsE/CtrA), and the outer membrane protein (KpsD/CtrB), would be encoded within a single operon as in N. meningitidis. However, it is likely that ancient genomic rearrangements have led to the separation of kpsED and kpsMT between two independent transcripts in E. coli. N. meningitidis genes ctrE (formerly lipA) and ctrF (formerly lipB) are encoded in Region B and are homologs of E. coli genes kpsC and kpsS, respectively. Despite the recent demonstration that ΔkpsC and ΔkpsS E. coli K1 strains accumulate non-lipidated CPS (Willis et al., 2013), mutations in N. meningitidis genes ctrE and ctrF have been shown to lead to intracellular accumulation of lipidated CPS (Tzeng et al., 2005). This disparity could represent a slight variation between species in an otherwise highly similar transport system, where lipidation and translocation initiation events are decoupled in N. meningitidis but intertwined in E. coli. Further research differentiating between these steps and assigning biochemical functions to CtrE and CtrF will help resolve this discrepancy. Region D and D′ encode duplicates of genes required for biosynthesis of N. meningitidis LPS (Hammerschmidt et al., 1994), while the function of Region E is unknown.
P. multocida also harbors a CPS gene locus that shares significant homology with Group 2 E. coli and N. meningitidis CPS gene loci (Fig. 4c). The topology of P. multocida type A, D, and F gene clusters more closely resembles that of E. coli, where a central CPS-specific region is flanked by two regions, Region 1 and 3, coding for translocation proteins. Specifically, Region 1 encodes the transport genes hexA, hexB, hexC, and hexD that are homologous to E. coli kpsT, kpsM, kpsE, and kpsD, respectively, while Region 3 encodes phyA and phyB, homologs to kpsC and kpsS genes predicted to lipidate CPS and initiate translocation (Chung et al., 1998). The genes in both regions are highly conserved among members of all five serogroups (A, B, D, E, and F), but P. multocida type B and E CPS loci exhibit slight rearrangements in gene order (Boyce et al., 2010). Region 3 gene lipA (homologous to phyA) is instead located between Regions 1 and 2, whereas lipB (homologous to phyB) maintains synteny with other serogroups due to its preserved location on the opposite side of Region 2 (Boyce et al., 2000a). For serogroups A and B, Region 1 genes hexA-D are known as cexA-D. CPS-specific biosynthesis in all serogroups is guided by the variable enzymes encoded in Region 2, including the synthases responsible for CPS polymerization. Experimental validation of the putative functions of most P. multocida Region 1 and 3 proteins is lacking, but a ΔhexA mutant strain was acapsular, supporting the role of HexA in CPS transport through the cell wall (Chung et al., 2001).
Biosynthesis
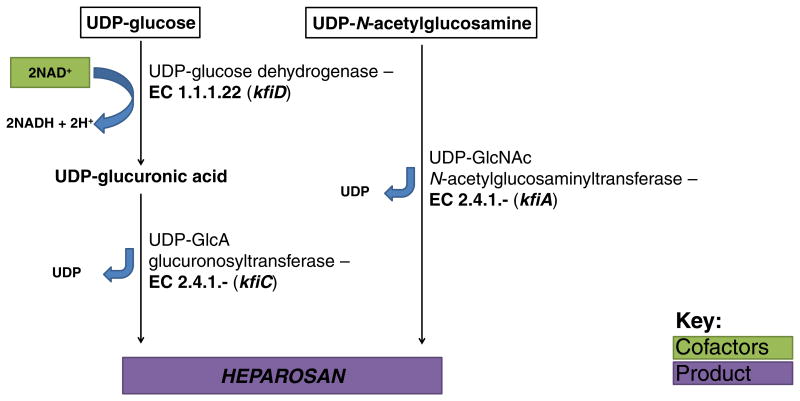
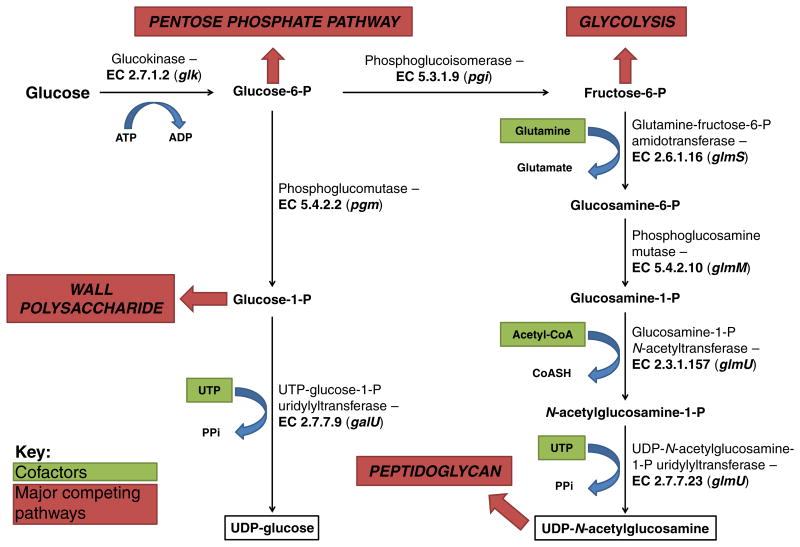
Several model E. coli strains possessing K1, K4, and K5 capsules have been sequenced, and their amino/nucleotide sugar metabolism is well conserved with only slight genetic and metabolic differences (Chen et al., 2006; Shuting Lu et al., 2011; Cress et al., 2013a, b, c). The conserved biosynthetic steps for these E. coli CPSs are representative of many bacteria and are shown alongside major competing metabolic pathways in Fig. 5, with CPS intermediates boxed in black. Two activated UDP-sugar intermediates are required for biosynthesis of the GAG-like CPSs, while only one of these two precursors is required for biosynthesis of PSA, a non-GAG CPS. The cytosolic reactions constituting these two intermediate pathways act as sinks on upper glycolysis and can be considered as two distinct modules, represented by the two branches in Fig. 5.
Fig. 5.
Central biosynthetic pathway for CPS precursor production in E. coli. This metabolic model is representative of early CPS biosynthesis for many bacteria, including those of interest in this review.
In the first module, glucose-6-phosphate (G6P) is converted to UDP-glucose by sequential action of two enzymes, while UDP-glucose is further converted to UDP-GlcA (the immediate GAG precursor) in GAG-producing K4 and K5 strains. Phosphoglucomutase (encoded by pgm) isomerizes G6P to glucose-1-phosphate (G1P), which is then converted by the UTP:G1P uridylyltransferase galU to UDP-glucose through transfer of a uridylyl group from UTP, releasing a pyrophosphate (PPi). The second module consists of four enzymes catalyzing the formation of UDP-N-acetylglucosamine (UDP-GlcNAc) from fructose-6-phosphate (F6P). F6P-amidotransferase encoded by glmS transfers an amine group from glutamine to F6P to form glucosamine-6-phosphate, which is further isomerized to glucosamine-1-phosphate by phosphoglucosamine mutase (glmM). An acetyl group is then transferred to glucosamine-1-phosphate to form N-acetylglucosamine-1-phosphate, a reaction catalyzed by glucosamine-1-phosphate N-acetyltransferase (glmU). The gene glmU encodes a bifunctional enzyme that subsequently transfers an uridylyl group from UTP to N-acetylglucosamine-1-phosphate, forming UDP-GlcNAc and releasing PPi. Specific biosynthesis of CPS from these intermediates and transport out of the cell in these model strains will be described in greater detail within. It is important to note here that these CPSs have been found in other species (Table 1), and it is expected that the rapid increase in microbial genome sequencing projects will continue to reveal disparate bacteria sharing related capsular gene loci. Since HA capsules are not known to exist in E. coli, the genetic and biosynthetic description will be presented later.
Polysialic acid
PSA is not a GAG, but like GAGs it is an acidic, linear polysaccharide found in vertebrate tissues. PSA is composed of repeating sialic acid (Neu5Ac) monomers, where the glycosidic linkage configuration is organism-dependent and commonly found as either α-2,8 or α-2,9 linkages or a combination of the two. In E. coli K1, N. meningitidis serogroup B, M. nonliquefaciens, and M. haemolytica A2, PSA possesses the mammalian-like [→8) Neu5Ac (2→]n structure seen in Fig. 3. Other strains possess immunogenic PSA capsules due to the non-animal glycosidic linkages. For instance, sialic acid units in E. coli K92 CPS are α-2,9 linked, and in N. meningitidis serogroup C are alternating α-2,8 and α-2,9 linked.
Polysialic acid biosynthesis
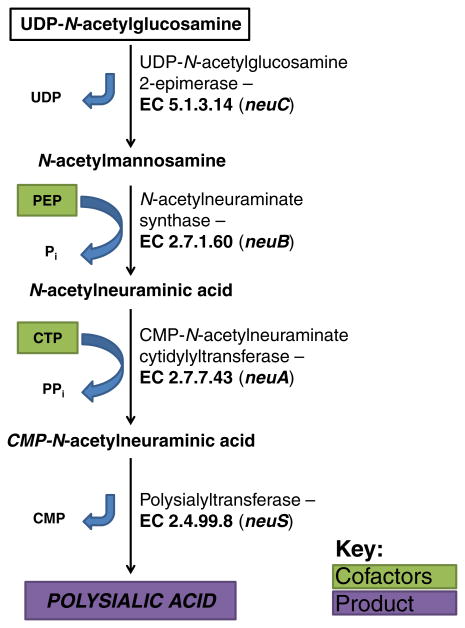
E. coli strains expressing K1 CPS share Region 1 and 3 of the CPS gene cluster with other Group 2 capsular bacteria (Fig. 4a) (Roberts, 1996). Region 2 consists of 6 genes specific for biosynthesis of K1 PSA. As depicted in Fig. 6, the first committed step of PSA biosynthesis is catalyzed by UDP-GlcNAc 2-epimerase (encoded by neuC), which epimerizes UDP-GlcNAc to N-acetylmannosamine (ManNAc) (Vann et al., 2004). NeuNAc synthase (neuB) then catalyzes the condensation of ManNAc and phosphoenolpyruvate (PEP) to NeuNAc and inorganic phosphate (Pi), where three carbons from PEP extend the monosaccharide from six to nine carbons (Annunziato et al., 1995). CMP-NeuNAc cytidyltransferase (neuA) utilizes a single molecule of CTP to activate NeuNAc with the transfer of CMP, thereby releasing pyrophosphate (PPi) (Silver et al., 1988). The processive sialic acid polymerase, polysialyltransferase, encoded by neuS sequentially adds NeuNAc to the nonreducing end of the nascent PSA chain (Silver et al., 1988). Although it has been suggested that neuD encodes a Neu5Ac O-acetyltransferase due to the presence of a hexapeptide repeat motif characteristic of an acyltransferase superfamily (Vimr & Steenbergen, 2006), this possibility is unlikely since K1 CPS is not O-acetylated in many strains encoding neuD. Another study demonstrated heterodimerization between NeuD and NeuB, suggesting that NeuD plays a stabilization role during chain elongation (Daines & Silver, 2000). The exact function of NeuE is unknown, and further efforts will be required to understand its role in polymerization initiation (Reid & Cuthbertson, 2012).
Fig. 6.
Biosynthetic pathway for PSA production in E. coli K1.
Many K1 CPS strains have been found to possess PSA that has been O-acetylated at the C7 or C9 hydroxyl group (Orskov et al., 1979). These are chemical modifications that increase resistance to desiccation and reduce biofilm formation, while coincidentally increasing immunogenicity (Mordhorst et al., 2009). However, the O-acetylation is a dynamic phenotype that varies within a population and appears to be controlled by a stochastic “on-off” switching mechanism at the genetic level (King et al., 2007). It was determined that this phase variation (also known as form variation) only occurs in K1 strains that have been lysogenized by a K1-specific lambdoid bacteriophage and thus possess a chromosomal accretion element known as CUS-3, a remnant of the infection. The neuO gene, encoding the K1 O-acetyltransferase responsible for the chemical modification of PSA, is encoded within CUS-3. The 5′ end of neuO is subject to slip strand DNA mispairing in which a 7 nucleotide repeat sequence is gained or lost at the 5′ end, leading to a frameshift and corrupted translation. By this mechanism, individuals in a CUS-3-harboring K1 population randomly partition between acetylation-on and acetylation-off variants, a phenomenon that presumably confers a population-level evolutionary advantage where the likelihood of persistence increases in adverse environmental pressures (Deszo et al., 2005). Incredibly, the O-acetyltransferase catalytic efficiency has also been shown to increase linearly with the number of tandem, in-frame repeats that manifest as tandem heptapeptide repeats and form a disordered N-terminal domain (Schulz et al., 2011). The function of the disordered region remains unknown. PSA purified from K1 strains lacking the CUS-3 region was invariably lacking O-acetylation (Deszo et al., 2005).
N. meningitidis serogroup B CPS is identical to K1 CPS, although the genes encoding the biosynthetic enzymes are organized differently and share only 30-40% amino acid sequence identity. N. meningitidis serogroup B Region A encodes cssABC (formerly synABC or siaABC), csb (formerly synD or siaD), and ctrG (formerly NMB0065) (Fig. 4b). The cssA gene encodes an UDP-GlcNAc 2-epimerase that shares 32% identity with NeuC from K1 (Murkin et al., 2004), while cssB codes for a CMP-Neu5Ac cytidylyltransferase with 34% identity to K1 NeuA (Edwards & Frosch, 1992; Ganguli et al., 1994). The cssC gene shares 37% identity with its K1 homolog NeuB, a Neu5Ac synthase (Vimr et al., 1989; Ganguli et al., 1994). The csb gene codes for a polysialyltransferase with 33% identity to K1 NeuS (Frosch et al., 1991). Finally, the gene products of ctrG and K1 NeuE share 27% identity. The role of CtrG in polymerization initiation is not entirely elucidated, but is appears to play a similar role as NeuE in coupling CPS biosynthesis with traversal through the cell wall (Hobb et al., 2010). Thus, the pathways biosynthesizing polysialic acid in these two Gram-negative organisms are metabolically and functionally identical. Furthermore, protein homology demonstrates high functional conservation of the ATP-dependent transport pathways. Biosynthesis of α-2,8-PSA capsules in other species, including M. haemolytica A2 (Adlam et al., 1987) and M. nonliquefaciens (Devi et al., 1991), has not been studied in depth.
Polysialic acid in evasion of immune system
Compared to other CPSs, K1 E. coli strains are particularly non-immunogenic due to the fact that the structure of CPS mimics the substance in the host (Brooks et al., 1980). The chemical structure of K1 polysaccharide is identical to the PSA on the embryonic form of NCAM, which is related to the organization of the neural tissue (Finne et al., 1983). Therefore, they are relatively more virulent because the immune response towards K1 is usually non-existent due to the mistaken recognition of the encapsulated bacteria as “self”, letting them pass protective barriers. Interestingly, this only applies to a certain host age range. The K1 organism's carriage rates are 22-42% among infant and children without sex differentiation while the highest among women aged 16-31 years old (Sarff et al., 1975). Some studies also indicate that K1 strains are poor activators for initiating the alternative complement pathway of immune response (Bortolussi et al., 1979). The anti-complementary effect is due to PSA's increasing the binding of inhibitor B1H to C3b, preventing formation of C3 convertase and blocking activation of the complement cascade (Harber et al., 1986; Leying et al., 1990). The failure to accumulate C3b on the cell surface effectively prevents phagocytosis and, thus, enhances the virulence of E. coli K1 strain.
E. coli K1 strains frequently cause infections of the urinary tract (Johnson, 1991), which according to Wiles, is one of the most common sites associated with human disease (Wiles et al., 2008). Moreover, K1 is also mainly responsible for causing acute pyelonephritis (Kaijser, 1973) since they can be easily found among the bacterial strains isolated from clinical specimens during acute pyelonephritis (Hanson et al., 1977). In addition, the K1 antigen is also found on strains of the extraintestinal pathogenic E. coli (ExPEC) (Wiles et al., 2008). K1 E. coli are generally thought to be the second most common cause of human neonatal meningitis (next to group B streptococci) and approximately 80% of American and European strains implicated in the disease have PSA capsules (Orskov & Orskov, 1992). Some studies have also suggested that bacterial survival within brain microvascular endothelial cells was enhanced by K1 CPS (Pluschke et al., 1983; Kim et al., 2003; Scholl et al., 2005).
PSA is also the CPS of two serogroups of N. meningitidis, a common causative agent of meningitis in children and adults. Early work on the CPS of N. meningitidis serogroup B found that mutants deficient in capsule production lost all pathogenicity in mice (Masson et al., 1982). Several studies have demonstrated that the CPS aids in the resistance of the bacterial cells to the innate immune system (Jarvis & Vedros, 1987; Spinosa et al., 2007). Similar to other CPSs, the PSA capsule of N. meningitidis has been found to hinder adhesion and invasion (Spinosa et al., 2007), and it is unable to activate the complement pathway (Jarvis & Vedros, 1987). This is most likely a result of the capsule masking immunogenic adhesins and invasins on the surface of the bacterial cell. However, it has also been shown that the CPS is vital for the survival of the bacterium in the bloodstream (Jarvis & Vedros, 1987) and important for survival inside human cells (Spinosa et al., 2007). In the bloodstream, the CPS allows N. meningitidis to evade uptake and degradation by macrophages (Jarvis & Vedros, 1987), while intracellularly the encapsulated bacterial cells are resistant to antimicrobial peptides, which act by binding bacterial membranes and increasing their permeability (Spinosa et al., 2007).
Several studies have shown the ability to successfully produce antibodies protective against N. meningitidis serogroup B by immunization with a vaccine containing N-propionyl and de-N-acetylated sialic acid derivatives (Pon et al., 1997; Granoff et al., 1998; Moe et al., 2009). Importantly, the antibodies were shown to be unreactive to human PSA. More specifically, vaccines containing de-N-acetylated sialic acid derivatives were shown to possess the ability to protect against N. meningitidis in multiple ways, including complement-dependent bactericidal activity and passive protection in infant mice (Moe et al., 2009). Because the vaccines show protection against N. meningitidis serogroup B, but not purified human PSA, it is possible that some amount of de-N-acetylated sialic acid is present in the CPS of N. meningitidis serogroup B. It has been proposed that de-N-acetylated sialic acid elicits a T-cell dependent immune response due to its zwitterionic nature, characterized by the presence of both positively-charged free amino groups at de-N-acetylated positions and negatively charged carboxyl groups along the polymer backbone (Moe et al., 2009). In contrast to the more common negatively charged or neutral CPSs, zwitterionic CPSs are known to be T-dependent antigens, which are bound, processed, and presented by major histocompatibility complex class II (MHCII) to stimulate helper T-cells through what is known as the MHCII endocytic pathway (Cobb et al., 2004; Surana & Kasper, 2012). These remarkable findings suggest that enzymatic de-N-acetylation of other acidic CPSs such as heparosan, chondroitin, and hyaluronan could represent a strategy for eliciting natural immune response to pathogenic infection.
Chondroitin
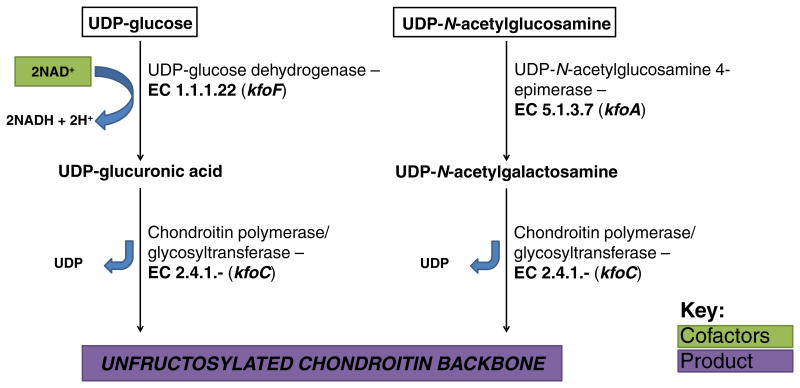
CPS produced by strains of K4 E. coli and P. multocida type F is structurally related to the GAG CS, which is involved in a range of molecular interactions in humans as previously described. As loss of CS from cartilage in humans leads to osteoarthritis, nutritional supplementation is a common treatment strategy, making CS a pharmaceutically and nutraceutically valuable product (Wildi et al., 2011). CS is currently harvested from animal sources (cow, pig, shark, fish, and bird cartilage) (Huskisson, 2008), but there is a growing interest in moving to sustainable microbial production platforms to minimize contamination and to control product consistency. One such production strategy involves harvesting a CS precursor from cultures of E. coli strains biosynthesizing a K4 capsule (Zanfardino et al., 2010; Restaino et al., 2011, 2012; Schiraldi et al., 2011, 2012; Trilli et al., 2012). This biotechnological relevance has provoked increased interest in improving biosynthesis of K4 CPS by manipulating E. coli metabolism (Restaino et al., 2012; Cimini et al., 2013; Cress et al., 2013b; Wu et al., 2013). K4 CPS is similar in structure to unsulfated CS, with the exception of an acid-labile, bisecting β-fructofuranose attached to C3 of the GlcA residue (Fig. 3). Alternatively, the P. multocida type F CPS is a linear polysaccharide identical to unsulfated chondroitin (DeAngelis et al., 2002). Commercial interest in this GAG has increased knowledge regarding its biosynthesis in microbes.
Chondroitin biosynthesis
E. coli strains expressing the K4 capsule share Regions 1 and 3 of the Group 2 CPS biosynthetic cluster, but the biosynthetic enzymes unique to K4 CPS are encoded by Region 2 and presumably form a biosynthetic complex at the cytosolic side of the inner cell membrane (Fig. 4a). Biosynthesis of K4 CPS precursors can be segmented into two distinct modules drawing from upper glycolysis through the intracellular pool of UDP-glucose and UDP-GlcNAc (Fig. 7). In the first module, UDP-glucose is converted to UDP-GlcA by UDP-glucose dehydrogenase (UGDH) encoded by kfoF and associated with the K4 CPS biosynthetic enzyme complex (Ninomiya et al., 2002). Although it is not uncommon for multiple copies of UDP-glucose dehydrogenase to exist in E. coli genomes, the existence of two other copies in the genome of the model K4 strain U1-41 (Cress et al., 2013b) suggests that kfoF has evolved to perform a distinct physiological role. Since UDP-glucose is a key metabolite in many pathways, it is plausible that the association of the kfoF-encoded copy of UDP-glucose dehydrogenase with the capsular biosynthetic enzyme complex serves to spatially constrain the chemical reaction—conversion of UDP-glucose to UDP-GlcA near the K4 CPS glycosyltransferase on the inner cell membrane would increase the local concentration of UDP-GlcA and effectively channel valuable UDP-glucose toward production of K4 CPS without significant loss to other cellular reactions. In the second module, UDP-GlcNAc 4-epimerase encoded by kfoA catalyzes the formation of UDP-GalNAc (Ninomiya et al., 2002). The two activated sugar precursors from each module are sequentially added to the nonreducing end of the growing polysaccharide chain by chondroitin polymerase (encoded by kfoC), a bifunctional glycosyltransferase catalyzing the transfer of both GlcA and GalNAc and release of two molecules of UDP per disaccharide extension (Ninomiya et al., 2002). The crystal structure of KfoC has been obtained in the presence of UDP-activated precursors, indicating the existence of two active sites for addition of UDP-GalNAc by the N-terminal domain and UDP-GlcA by the C-terminal domain (Osawa et al., 2009).
Fig. 7.
Biosynthetic pathway for production of K4 CPS, chondroitin, in E. coli K4.
Several studies have suggested that K4 chondroitin is fructosylated subsequent to polymerization of the backbone (Lidholt & Fjelstad, 1997), but the enzyme responsible for fructosylation has remained unreported until recently. Initial searches for the fructosyltransferase focused on proteins in Region 2 of the K4 biosynthetic gene cluster that were predicted to possess glycosyltransferase motifs. One such enzyme encoded by kfoG possessed a putative glycosyltransferase domain; however, disruption/deletion of the gene did not prevent fructosylation (Krahulec et al., 2005). A series of recent patents purport that deletion of kfoE results in production of unfructosylated chondroitin, suggesting that kfoE encodes a fructosyltransferase (Trilli et al., 2012). The enzyme has not been characterized in its purified form, however, and biochemical characterization will be required to assign fructosyltransferase activity to kfoE. Several genes in the K4 CPS cluster, including kfoB, kfoD, and kfoG, have unknown function. However, kfoB shares 38% identity with a gene in E. coli K5 (kfiB) that is believed to be structurally important in K5 CPS biosynthesis. This might suggest a stabilizing role for KfoB that is critical for assembling or colocalizing biosynthetic enzymes near the transenvelope CPS assembly complex, but further work will be required to determine the precise functions of these proteins.
The unfructosylated, unsulfated chondroitin backbone constituting the P. multocida type F CPS is biosynthesized by genes in Region 2 of the P. multocida CPS locus. The region consists of four genes, fcbB, fcbC, fcbD, and fcbE, only one of which has been characterized. The chondroitin synthase known as PmCS is encoded by fcbD and is homologous to E.coli K4 kfoC (DeAngelis et al., 2002). PmCS is also a bifunctional polymerase possessing two distinct glycosyltransferase domains that sequentially add GlcA and GalNAc residues to the growing polysaccharide chain (DeAngelis & Padgett-McCue, 2000). Similar to KfoC, the N-terminal domain of PmCS possesses GalNAc-transferase activity, and the C-terminus possesses GlcA-transferase activity (Osawa et al., 2009; Otto et al., 2012). While deletion of kfoE in E. coli K4 has been reported to abrogate chondroitin fructosylation, it would be interesting to determine if heterologous expression of kfoE in P. multocida type F complements expression of fructosylated CPS. P. multocida genes fcbB, fcbC, and fcbE are homlogous to E. coli K4 genes kfoG, kfoF, and kfoB, respectively, but the biological functions have not been studied (Townsend et al., 2001).
Chondroitin in evasion of immune system
The K4 antigen is not implicated in human disease as frequently as K1 and K5 antigens, but K4 strains have been associated with human and animal infection. K4 E. coli strain U1-41 is a uropathogen (Rodriguez et al., 1988), and the K4 capsule has been found on strains causing diarrhea in humans (Orskov et al., 1985). EHEC K4 strains have also been isolated in calves (Moxley & Francis, 1986; Stordeur et al., 2000). The unfructosylated chondroitin backbone is identical to the mammalian CS precursor, but unlike the K1 and K5 antigens, the K4 antigen host-related bacterial polymer is substituted with an additional sugar residue not present in the mature, sulfated GAG. The β-linked fructose on K4 CPS is an antigenic determinant that imparts immunogenicity to the E. coli K4 capsule and is responsible for a conformational change resulting in significantly higher viscosity compared to defructosylated K4 CPS (Rodriguez et al., 1988). Therefore, K4 CPS may be easily recognized by anti-K4 antibodies and induce a complement dependent immune response. However, the fructosyl group is labile and can be easily removed in mild acidic conditions. The conversion of K4 antigen to a non-fructosylated chondroitin results in the non-immunogenicity of K4 CPS (Rodriguez et al., 1988; Jann & Jann, 1997). Thus, it has been suggested that such lability enables dynamic physiological conditions in the host, such as low pH in certain host tissues, cells, or compartments (Jann & Jann, 1997), to modulate the presence of this immunodominant residue on the K4 capsule. A K4 strain also might benefit outside the host from a more viscous capsule and inside the host from a less viscous, non-immunogenic capsule.
The chondroitin CPS of P. multocida type F is non-immunogenic since it lacks the fructose residue present in E. coli K4 CPS and is thus identical to the animal precursor to CS. It has been shown that treatment of P. multocida type F with chondroitinase, a lyase that is known to cleave unsulfated chondroitin, results in increased phagocytosis by swine neutrophils (Rimler et al., 1995), presumably through elimination of the capsule.
Heparosan
Pharmaceutical grade heparin has traditionally been derived from porcine or bovine mucosal tissues, but a contamination crisis leading to several hundred deaths worldwide and causing severe allergic reactions in many patients prompted exploration of microbial heparosan production followed by enzymatic conversion to bioengineered heparin with purity and biological activity meeting United States Pharmacopeia (USP) standards (Wang et al., 2011). E. coli K5 strains have been shown to serve as a viable production platform due to the similarity between K5 CPS and the mammalian precursor for heparin (Wang et al., 2010, 2011; Ly et al., 2011). Recent work has also demonstrated the ability of an engineered heparosan-producing E. coli BL21 strain to naturally secrete the polysaccharide (Zhang et al., 2012), presumably utilizing transport proteins encoded by Region 1 and 3 genes from its endogenous Group 2 capsular export system (Andreishcheva & Vann, 2006). This work demonstrated the feasibility of producing heparosan in well-characterized production strains, albeit at much lower concentrations than wild-type K5 strains. Efforts in this area have contributed to the understanding of heparosan biosynthesis.
Heparosan biosynthesis
Similar to K4 CPS, the biosynthesis of K5 CPS can be partitioned into two modules each producing the requisite activated sugar precursor for heparosan polymerization. The monomeric constituents of K5 CPS are GlcA and GlcNAc, which draw carbon from G6P and F6P in the form of two activated sugar precursors, UDP-GlcA and UDP-GlcNAc, respectively. UDP-GlcA biosynthesis in K5 is identical to that in K4 with the exception of the UDP-glucose dehydrogenase associated with the capsular biosynthetic complex. Conversion of UDP-glucose to UDP-GlcA in the first module is catalyzed by an UDP-glucose dehydrogenase encoded by kfiD from Region 2 in the K5 capsule gene cluster (Fig. 4a) (Sieberth et al., 1995). The kfiD gene encodes the third copy of UDP-glucose dehydrogenase in the genome of the model strain Bi 8337-41 (Cress et al., 2013c) and the probiotic strain Nissle 1917 (Cress et al., 2013a), and biochemical studies provide evidence for its association with the K5 CPS biosynthetic complex also comprised of proteins encoded by kfiA, kfiB, and kfiC (Rigg et al., 1998). While kfiB is thought to perform a membrane-localization and complex-stabilizing or scaffolding role rather than a catalytic one (Hodson et al., 2000; Zhang et al., 2012), kfiA and kfiC are glycosyltransferases that alternatively elongate the heparosan chain at the nonreducing end (Petit et al., 1995). The final reaction of the first module is catalyzed by kfiC-encoded UDP-GlcA glucuronosyltransferase, which attaches a GlcA residue to the nascent K5 polysaccharide and releases UDP. The second module in K5 CPS biosynthesis encompasses the reactions converting F6P to UDP-GlcNAc as shown in Fig. 8, and the final reaction involves transfer of GlcNAc from UDP-GlcNAc to the growing polysaccharide by kfiA-encoded UDP-GlcNAc N-acetylglucosaminyltransferase, a step that frees an additional UDP.
Fig. 8.
Biosynthetic pathway for production of K5 CPS, heparosan, in E. coli K5.
Relatively little is known about most of the proteins encoded in Region 2 of P. multocida type D (dcbB, dcbC, dcbE). An exception is the well-studied heparosan synthase PmHS1 encoded by dcbF, also known as hssA (Kane et al., 2006). This bifunctional synthase possesses two fused glycosyltransferase domains, where a segment of the N-terminal β-1,4-glucuronosyltransferase domain is homologous to E. coli KfiC, and a section of the C-terminal α-1,3-N-acetylglucosaminyltransferase domain is homologous to KfiA (Kane et al., 2006; Otto et al., 2012). Drawing analogy with other CPS biosynthetic gene clusters, the putative UDP-glucose dehydrogenase encoded by dcbC likely supplies UDP-GlcA to PmHS1 for chain elongation. Finally dcbE encodes a homolog of E. coli kfiB, and dcbB is a putative glycosyltransferase sharing homology with kfoG. Surprisingly another heparosan synthase (PmHS2) with similar glycosyltransferase domain organization and ∼70% identity to PmHS1 is located outside of the CPS gene locus in the GAG-producing P. multocida type A, D, and F strains (DeAngelis & White, 2004). The biological role of PmHS2 has not been characterized in vivo, but it has been speculated that PmHS2 and the HA and chondroitin synthases in type A and F strains, respectively, might be differentially regulated by environmental conditions to allow variation in the type of GAG displayed on the cell surface (DeAngelis & White, 2004). Owing to their broad substrate specificities compared to PmHS1, purified PmHS2 and KfiA have been used to synthesize novel unnatural polysaccharides from analogs of native UDP-sugar donors, and PmHS2 has also been used to prepare heparosan for the chemoenzymatic production of heparin (Liu et al., 2010; Li et al., 2013).
Heparosan in evasion of immune system
The structure of E. coli K5 CPS is identical to that of heparosan, which is the first polymeric intermediate during the biosynthesis of heparin in the host (Navia et al., 1983). In addition, the mature heparin chain also contains some unsulfated and unepimerized [→4) β-D-GlcA (1→4) α-D-GlcNAc (1→]n repeats identical to the K5 disaccharide (Vann et al., 1981). Thus, the mimicry of the K5 CPS structure makes it non-immunogenic. Since traditional CPS typing relied on antigenicity, the capsule of E. coli K5 was originally classified as K-non-typeable (Jann & Jann, 1987). In part due to the difficulty of identifying the K5 antigen and the frequency or extent in different infections, more effective methods of typing were developed, including one by Gupta et al. that utilized a specific phage to type K5 in various infections (Kaijser & Jodal, 1984).
The capsules of E. coli K5 strains are most commonly found in urinary tract infections (UTI) (Sandberg et al., 1988) such as pyelonephritis, cystitis, and asymptomatic bacteriuria. Although K5 CPS has been found to be a common E. coli capsule antigen in UTI infections, it is also prevalent in strains causing sepsis (Kaijser & Jodal, 1984). In one of the studies using specific K5 phage typing, K5 was found in the 17.1% strains in the case of sepsis followed by 12.4% strains in the case of asymptomatic bacteriuria (Kaijser & Jodal, 1984). Moreover, the K5 capsule has also been shown to promote the persistence of E. coli in the rat large intestinal microflora (Hérias et al., 1997). The same study indicated that the enhanced intestinal colonization of K5 encapsulated strains allowed higher cell densities to be reached and secondarily led to increased translocation through the intestinal mucosa to mesenteric lymph nodes. Translocation is thought to be a normal physiological process regulating immunity to gut bacteria (Wells et al., 1988), but it can also reach the blood stream causing sepsis and meningitis (Lambert-Zechovsky et al., 1992). In the tissues, the capsule might further protect the organisms from phagocytosis and complement-mediated killing (Hérias et al., 1997).
The heparosan CPS of the animal pathogen P. multocida type D has also been demonstrated to be an important virulence factor for the organism. Acapsular variants of toxigenic P. multocida type D lose their virulence in a murine model, and lesions caused by acapsular strains are less severe than their encapsulated counterpart (Jacques et al., 1993).
Hyaluronan
The unsulfated GAG known as HA exists in a wide-range of mammalian tissues and as the primary constituent of the CPS of several microbes, including the well-studied bacteria S. pyogenes type A and C and P. multocida type A (other species possessing HA capsules are listed in Table 1). Although the monosaccharide constituents of this acidic polysaccharide are identical to heparosan, the glycosidic linkages are distinct and lead to a slightly different repeating disaccharide structure. HA is noted as one of the most hygroscopic molecules to exist in nature, and it is estimated that hydrated HA contains a 1000-fold mass of water compared to its own weight (Laurent & Fraser, 1992). The high biocompatibility of HA stems from its natural presence in many mammalian tissues, and it is thus utilized in cosmetic and medical applications (Cimini et al., 2012), including tissue engineering (Allison & Grande-Allen, 2006). HA has also long been known as a regulator of cancer progression, and a high-molecular-mass HA unique to naked mole rats was recently shown to mediate resistance to cancer, an unusual but famous property contributing to the species' extraordinary longevity (Tian et al., 2013). Thus, commercial HA production is appealing for its use in a wide range of applications. Industrial preparation of HA was originally achieved by extraction of HA from animal sources such as rooster combs, but there is interest in moving towards more sustainable production practices such as microbial fermentation (Boeriu et al., 2013). Since the existence of a bacterial HA synthase was first documented in pathogenic group A S. pyogenes (DeAngelis et al., 1993a, b), HA production has been evaluated in nonpathogenic Streptococci and recombinant strains (Yu & Stephanopoulos, 2008; Yu et al., 2008; Liu et al., 2011), and now microbial HA has been fully commercialized (DeAngelis, 2012).
Hyaluronan biosynthesis
Microbial production of HA has primarily been studied in streptococcal species and P. multocida type A. Although not all enzymes catalyzing biosynthetic steps toward HA have been definitively proven, biosynthetic models have been proposed based on known CPS genes in other organisms (Liu et al., 2011; Cimini et al., 2012; Boeriu et al., 2013). HA and heparosan share the same precursors (UDP-GlcA and UDP-GlcNAc), so the metabolic models for these CPSs are likely analogous. The similarity of early biosynthetic steps for nucleotide activated sugar precursors across a wide range of bacteria also supports the assertion that biosynthetic steps are conserved between heparosan and HA strains. Since UDP-glucose and UDP-GlcNAc are important building blocks for many microbial glycans and other glycoconjugates, it would be surprising to see significant inter-species variation in the anabolism of these critical components. The obvious distinction between heparosan and HA biosynthetic pathways are the glycosyltransferases that polymerize the distinct CPSs. HA synthases from Streptococci and P. multocida are bifunctional enzymes that alternatively add GlcA and GlcNAc to the nascent polymer (DeAngelis et al., 1993b; Kumari & Weigel, 1997; DeAngelis et al., 1998; Ward et al., 2001). As opposed to heparosan synthases that form alternating α- and β-linkages between monosaccharides, however, HA synthases only form β-linkages. A striking feature of HA synthases from different species is that they polymerize in opposite directions; Class I HA synthases of Streptococci catalyze monosaccharide addition at the reducing end of the chain, while Class II HA synthases in P. multocida elongate the chain by addition to the non-reducing end (DeAngelis, 2012).
As discussed in previous sections, Regions 1 and 3 of the P. multocida CPS gene cluster are presumably responsible for CPS transport through an ABC-transporter dependent pathway. Genes hyaB, hyaC, hyaD, and hyaE in Region 2 of the P. multocida type A CPS locus are involved in HA biosynthesis (Chung et al., 1998). Putative functions have been ascribed to P. multocida hyaB, hyaC, and hyaE based upon their homology to kfoG, kfiD/kfoF, and kfiB/kfoB, respectively, where HyaC is an UDP-glucose dehydrogenase providing UDP-GlcA to HA synthase, and HyaE could serve as a scaffold or structural component. In fact, a P. multocida type A ΔhyaE mutant was shown to be acapsular, suggesting that HyaE is critical for proper translocation of the HA CPS (Crouch et al., 2012). In another study, an acapsular P. multocida type A mutant was generated by deletion of hyaB, which implicates HyaB as another key protein in CPS translocation (Steen et al., 2010). The hyaD gene encodes the HA synthase PmHAS, an enzyme surprisingly sharing high identity with the chondroitin synthase PmCS from P. multocida type F (DeAngelis et al., 1998). Like most other GAG synthases, PmHAS possesses two independent glycosyltransferase domains that functions in a non-processive manner. Specifically, the N-terminus of PmHAS possesses a GlcNAc-transferase domain, while the C-terminus possesses a GlcA-transferase domain (Jing & DeAngelis, 2000; DeAngelis et al., 2003).
Details regarding HA assembly in Gram-positive Streptococci are lacking as well, but the HA biosynthetic operon has been cloned from four species, including Streptococcus pyogenes, Streptococcus uberis, Streptococcus equisimilis, and Streptococcus zooepidemicus subsp. equi. This operon possesses hasA (HA synthase) and hasB (UDP-glucose dehydrogenase; homologous to hyaC in P. multocida type A) in all four species (Crater & van de Rijn, 1995). In all but S. uberis, hasC (UTP-glucose-1-P uridylyltransferase) is encoded downstream of hasB. In S. uberis, hasC is located elsewhere in the genome (Ward et al., 2001). The CPS operon in S. equisimilis and S. zooepidemicus also possesses hasD, a gene encoding the bifunctional glucosamine-1-P N-acetyltransferase/UDP-GlcNAc-1-P uridylyltransferase with homology to glmU from Gram-negative bacteria. Finally, S. zooepidemicus subsp. equi has an additional gene in the operon known as hasE, which encodes phophoglucoisomerase (Widner et al., 2005). Phosphoglucoisomerase is essential for many normal cellular processes, but with respect to CPS biogenesis, it could be important for balancing the intracellular abundance of the two UDP-sugar precursors to HA (Chen et al., 2009; Prasad et al., 2010).
Genes encoded in the CPS operons of different species might lead to more subtle variations in HA biogenesis through the expression of dedicated enzymes that increase the availability of UDP-sugar precursors to meet the demands of CPS production. It is noteworthy that the presence of such supplementary enzymes is inconsistent between species producing identical CPS, as exemplified by the extra copy of HasD in the CPS locus of S. equisimilis and S. zooepidemicus compared to S. uberis and S. pyogenes (Blank et al., 2008). A possible explanation is that dedicated biosynthetic enzymes might be required in the metabolic background of one species to ensure sufficient intermediate metabolite availability, where, in a different species, sufficient precursor concentration exists without these dedicated enzymes. It cannot be discounted that the genetic difference is random; however, maintenance of function of these duplicate, CPS-dedicated enzymes suggests that they confer an evolutionary advantage.
Hyaluronan in evasion of immune system
P. multocida is an animal pathogen, mainly in avian, bovine, and swine hosts, but can be transmitted to humans through animal bites, particularly from cats and dogs. Multiple studies have been performed that demonstrate the prevalence of the differing serotypes, based on the different CPSs utilized, of P. multocida in healthy and diseased organisms. In one study, 289 strains that were isolated from a variety of both healthy and ill animals (including bovine, small ruminants, buffalo, swine, rabbits, dogs, cats, and poultry) were serotyped based on PCR detection of the capsular biosynthesis genes capA, B, D, E, and F. The study found that the HA capsule of type A P. multocida was the most common amongst the isolates, followed by type D (Ewers et al., 2006). A similar study investigated the capsules found in porcine pneumonia and atrophic rhinitis samples, and likewise found that A was the most common, followed by D (Davies, 2003). A 2012 study used multiplex PCR to determine the capsular genotype of isolates from 121 animals in Malaysia, and found that the capsular genotype was specific for infections in different hosts, with capsular type A predominantly found in avian, rabbit, and porcine samples, capsular type B found mostly in cattle and buffalo samples, capsular type D found mainly in goat samples, and type F capsules found only in a cattle sample (Mohamad et al., 2012). While the organism is most often associated with disease in chickens and pigs, isolates have more recently been studied from chimpanzees and humans. In a study of isolates from the lungs of wild chimpanzees affected by an outbreak of respiratory disease in 2004, researchers found that all of the isolates had the biosynthesis genes required for the production of a HA capsule (Köndgen et al., 2011). Another study, this time on 143 isolates of Pasteurella from humans, again found that type A was the most common serotype identified, and it was predominantly isolated from respiratory tissue, while isolates identified as type B, D, and F were found more commonly than type A in soft tissue infections such as bite wounds (Donnio et al., 2004).
The capsule of P. multocida serotype A is composed of the GAG HA. Several studies have shown that this capsule plays an important role in both virulence and evasion of the host immune system. Studies on the capsule's role in virulence of P. multocida have shown that the HA capsule enhances virulence, and that inhibiting the production of the capsule attenuates the pathogenicity. For instance, one group showed that PBA930, a mutant P. multocida type A strain deficient in capsule export, was attenuated in mice when compared to the wild type strain, and that lethality was restored to wild type levels when the strain was complemented with a plasmid encoding the genes responsible for export (Chung et al., 2001). Another such study showed that invasion of acapsular P. multocida mutant cells into chicken embryo fibroblast cells was decreased 12 to 16-fold when compared to the wild type of the same strain (Al-Haj Ali et al., 2004). The same study also showed that encapsulated strains were more adhesive to the chicken embryo fibroblasts than the acapsular mutants. Interestingly, treatment with hyaluronidase did not affect invasion or adhesion, while trypsin and periodic acid treatments both inhibited invasion and adhesion only in encapsulated strains, indicating that HA may be playing a role in masking the receptors responsible for adhesion and invasion by these cells. Moreover, another study showed that the thickness of the capsule seems to also play a role in the virulence of P. multocida infections in chickens, with increasing capsule thickness correlating positively with virulence, with thinly-capsulated strains being attenuated even at high doses (Borrathybay et al., 2003a).
Several studies have attempted to understand how the HA capsule of P. multocida type A aids the bacteria in evading the immune system of its host. Studies have shown that this capsule aids in evading multiple faucets of the immune system, including phagocytic uptake, phagocytic killing, and the complement system. One such study found that a mutant strain of P. multocida lacking the genes required for capsular export was sensitive to killing in 90% chicken serum, while encapsulated strains and complemented mutant strains were not (Chung et al., 2001). Interestingly, the group had previously determined that the capsule of serotype B P. multocida, which has a non-GAG capsule, composed of mannose, galactose, and arabinose, did not infer resistance to complement in chicken serum (Boyce et al., 2000b). This seems to indicate an important role in capsules composed of GAGs to infer resistance to complement in serum.
In addition to resistance against complement activity in serum, studies have also indicated that the HA capsule plays a role in making the bacteria resistant to phagocytosis by macrophages. A 2004 study showed that immunization of mice with crude capsular extract (serotype A) did not induce the production of antibodies against HA, but did induce antibodies against a 39 kDa protein that is only present in encapsulated strains of P. multocida. The study also used immunoelectron microscopy to show that the 39 kDa protein was localized at the capsule, and that mice that were immunized with the antibodies against this protein were protected from at least 2 serotype A strains of P. multocida (Al-haj Ali et al., 2004). This illustrates that one function of the capsule in promoting virulence is to mask surface antigens on the bacterial cell from the host immune system. Another study examined the capsule's role in virulence by comparing phagocytic uptake of P. multocida cells with and without capsules and found that encapsulated strains were resistant to phagocytosis, and that this resistance was lowered when the capsules were removed enzymatically with hyaluronidase (Poermadjaja & Frost, 2000). Another study found that encapsulated strains of P. multocida type A were less hydrophobic than acapsular strains, which could be responsible for inhibiting interactions between the cells and hydrophobic components on the outer surface of macrophages, and also found that treatment with hyaluronidase or mechanical shearing of the bacterial cells significantly reduced the surface charge of the cells, which could also play a role in interactions with macrophages or other immune system components (Watt et al., 2003). Together, the studies on the influence of the HA capsule of P. multocida serotype A indicate that capsule enhances virulence by evading the host immune system in a number of ways.
S. pyogenes, also commonly known as Group A Streptococcus (GAS), is the causative agent of human streptococcal pharyngitis, rheumatic fever, and soft tissue infections including necrotizing fascitis. The capsule of S. pyogenes is also composed of HA. Many studies have been performed on GAS in order to elicit the importance and role of the HA capsule in virulence. As in P. multocida, the capsule of GAS has been shown to enhance virulence. An interesting 2004 study showed that incidence of mucoid (encapsulated) GAS isolates correlated temporally with incidence of rheumatic fever (Veasy et al., 2004). One group used mutations in the CsrRS regulation system to show that increased capsule production correlated with increased virulence (Engleberg et al., 2001). Another study on GAS looked to determine the extent to which the HA capsule contributed to virulence compared to another important virulence factor in GAS, the M protein. Similar to the capsule of P. multocida type A, this study found that the capsule of GAS played an important role in resistance to phagocytosis in serum (Fillit et al., 1986). Another study showed similar results, finding that encapsulated GAS was able to grow in human blood, while acapsular GAS was not, and that encapsulated GAS was resistant to phagocytosis, while acapsular was not. The same study found that the loss of the HA capsule resulted in a 100-fold loss of virulence in mice (Wessels et al., 1991).
In contrast to the immune-resistance roles in P. multocida, the HA capsule in GAS has been implicated in additional roles in virulence. One study using transmission electron microscopy found that the binding of the GAS capsule to skin epithelial CD-44 receptors induced cytoskeletal rearrangements that allowed the bacterial cells to invade (Cywes & Wessels, 2001). However, an earlier study found that the HA capsule of GAS did not correlate with invasion into skin cells, but did find that the non-encapsulated GAS was significantly less virulent when it invaded skin cells, causing fewer and less severe lesions (Schrager et al., 1996). Another study, aiming to look at the role of both the HA capsule and the M protein in a baboon model, found that acapsular GAS persisted half as long in the throat as encapsulated GAS, and also indicated a role for the capsule in enhancing microbial resistance to antibody-mediated phagocytic killing (Ashbaugh et al., 2000). The studies on the importance of the HA capsule in the virulence of Group A Streptococcus have shown that its role is very similar to that of the HA capsule of P. multocida. Its primary role seems to be to protect the bacterial cell from host immune response, but it also has additional roles in invasion of host cells.
Concluding remarks and future perspectives
Owing to the diversity and abundance of distinct microbial virulence factors and their multifunctional, synergistic properties, strictly decoupling the role of CPSs in pathogenicity from other contributing factors is a daunting and risky task. The wealth of research devoted to bacterial capsules has nevertheless expanded our understanding of the mechanisms utilized by pathogens to persist in host tissues and cavities without provoking severe immune responses. In light of the concerning trend of pathogenic strains evolving and acquiring drug-resistance, however, the scientific community should endeavor to identify vulnerabilities in this first line of defense for capsular pathogens and prioritize studies aimed at reducing virulence through capsule manipulation or interference with capsular biosynthesis. Given the nature of this review, we believe that a degree of speculation about possible studies and therapeutic strategies is warranted in this perspectives section.
Understanding virulence, pathogenicity, and protective roles of capsules
Further studies will help to better understand the role of the capsule in the pathogenicity of bacteria, as exemplified by the potential directions outlined in this section. For instance, the presence or absence of the capsule clearly impacts bacterial virulence, but little is known about the effect of intermediate degrees of encapsulation. Although CPS structure is likely the dominant virulence determinant, the mass of CPS per cell, CPS chain length, capsule density, and capsule thickness might be important contributing factors to persistence against host immune systems. A thick capsule of high-molecular weight, non-immunogenic polysaccharide might be expected to serve as a more efficacious shield of cell surface components than a tenuous coating composed of an identical polysaccharide of shorter chain length. Indeed, it should be noted that strains of the same capsular type will express different quantities or molecular weight of CPS (Hickey et al., 2013), and it is clear that within many pathogenic bacterial species, the more capsule expressed the more virulent they are (Lee et al., 1991; Luong & Lee, 2002). Since there are other genetic differences between such wild-type strains, however, specifically implicating quantitative capsule expression as a virulence factor has not been straightforward. Although studying the impact of variable microbial capsule “coverage” in animal infection models is a difficult prospect, the relationship between degree of coverage and pathogenicity is worth investigating because it could support or obviate the design of drugs that facilitate clearance of encapsulated pathogens through partial or complete removal of non-immunogenic CPSs. If even minimal capsule coverage is sufficient to inhibit immune response, then a more appropriate drug design strategy might target early stages of CPS biosynthesis to preclude capsule formation entirely.
The increasing prevalence of synthetic biology tools could help resolve some intriguing questions about immune response with respect to varying levels of encapsulation. Genetic mutations in past CPS virulence studies have consistently been static, where the presence or absence of capsule is set prior to inoculation by deleting or heterologously expressing capsule biosynthesis genes. One can envision engineering virulent wild-type strains with dynamic transcriptional regulatory circuits capable of modulating CPS production in response to environmental cues (Khalil & Collins, 2010; Chang et al., 2012a), or even in an inducer-free, oscillatory manner that takes advantage of mutually repressible genes (Elowitz & Leibler, 2000; Danino et al., 2010). Similarly, common metabolic engineering strategies such as overexpressing capsule biosynthesis genes or downregulating and deleting genes in competing metabolic pathways could be implemented to create strains producing different quantities of CPS (Yu & Stephanopoulos, 2008; Yu et al., 2008; Zhang et al., 2012). After inoculating model animals with these engineered strains, it is conceivable that the effect on immunogenicity of transient variation in CPS coverage could be monitored to understand critical coverage levels and to measure rates of capsule assembly and degradation in vivo. In any such studies, it will likely be important to consider that differential capsule production might not correlate with capsule coverage. For instance, if it can be assumed that total mass of CPS attached to the outside of a bacterium is intrinsically limited by surface area of the outer leaflet or number of available attachment sites, then where would excess CPS localize after this saturation point is reached? In strains engineered to overexpress capsular biosynthetic machinery, it might be expected that excess CPS would accumulate extracellularly without leading to a corresponding augmentation of capsule thickness or density and without affecting immunogenicity. Alternatively, excess CPS might remain loosely associated with the cell through noncovalent interactions with the capsule, a scenario that could lead to capsule thickening and attenuated host immune response. Related questions that remain to be fully resolved are the following: can bacteria sense change in the quantity of CPS attached to the outer membrane, and can such information actuate a change in CPS production levels? Alterations in bacterial metabolism upon capsule perturbation could also be examined using metabolomic, genomic, and proteomic studies to elucidate native capsule regulatory mechanisms and other unknown players in capsule assembly.
In contrast to modulating CPS production in vivo, an alternative method for studying the relationship between capsule coverage and immunogenicity would be to subject capsular bacteria to CPS-specific lyases or glycosidases that would remove the capsule post-colonization. For instance, since carbohydrate-cleaving enzymes have been shown to degrade bacterial capsules in vitro (Rimler et al., 1995), it is conceivable that an infected animal could be treated with these purified enzymes to strip the invading pathogen of its capsule. Alternatively, pathogenic microbes could be engineered to secrete lyases or glycosidases to degrade their own CPS in situ, effectively exposing cell surface antigenic determinants to the host immune system on command from an external signal. In a related example, the wild-type E. coli K5 strain naturally produces and secretes a heparosan lyase that depolymerizes its own capsule (Legoux et al., 1996). Engineering the secretion of enzymes that are not naturally secreted can be challenging, but recent successes in this area suggest that diverse types of recombinant proteins can be engineered for secretion in Gram-negative bacteria like E. coli or Gram-positive bacteria like Bacillus subtilis or other bacilli by fusing secretion peptide tags to one end of the protein. Interested readers are directed to comprehensive reviews on the subject (Simonen & Palva, 1993; Tjalsma et al., 2004; Mergulhao et al., 2005). Prior to such studies, however, an additional question that should be addressed is whether treatment with a lyase alters the immunogenicity of CPS. The lyase from K5-specific coliphage creates an unnatural double bond in the external-facing GlcA residue through a β-elimination mechanism (Hänfling et al., 1996), which could presumably serve as an antigenic “flag” on the outer surface of the glycocalyx; in contrast, treatment of K1 capsule with a sialidase (the cognate polysialic acid glycosidase, or carbohydrate hydrolyzing enzyme) would preserve the natural polysaccharide terminus without introduction of an unsaturated bond. Thus, it is important to consider unintended immunogenic consequences of the class of depolymerizing enzyme utilized. To decouple the role of CPS structure from other virulence factors, similar inducible circuits could be designed to switch from expression of wild-type CPS machinery to expression of biosynthetic enzymes for production of a different CPS structure, a scenario in which the wild-type capsule would be progressively supplanted by a capsule with dissimilar physical properties and antigens. In this manner, the confounding variables stemming from different virulence factors between strains could be minimized when studying immune response against distinct CPS structures.
Another valuable approach would be to develop genome-scale metabolic reconstructions of capsular bacteria and study, in silico, environmental or media conditions leading to differential CPS production level. Constraints-based metabolic modeling techniques such as flux balance analysis (Orth et al., 2010) and metabolite essentiality analysis (MEA) (Kim et al., 2007) excel at identifying non-intuitive strategies for modulating bacterial metabolism toward a specific phenotype and have already been used to study pathogenic bacteria and predict validated antibiotics (Shen et al., 2010). By individually constraining production of all metabolites to zero, one can probe the capacity of a metabolic network to sustain a cellular phenotype (such as maintaining biomass production/flux, the objective function of the optimization algorithm) when certain compounds are removed from metabolism. Metabolites that are found to be essential for the phenotype of interest can be derivatized to compete with the natural metabolite and hinder phenotype manifestation. In this manner, a novel antibiotic was rationally designed and shown to out-perform an existing therapeutic for Vibrio vulnificus infection (Kim et al., 2011). MEA could be applied in a similar manner to genome-scale models of capsular pathogens while using CPS production as the objective function. As minimal subnetworks required to sustain the capsule are discovered in simulations, gene deletions could be used to validate predictions in vivo. Analogs of essential CPS metabolites could then be screened for CPS inhibition.
Although countless experiments could be contemplated, the implication is clear: there is still much to learn about the role of capsules in microbial pathogenesis, and creative strategies capitalizing on new genetic and computational tools should be devised to probe the limits of the immune system in detecting and clearing exposed pathogens and to predict molecules capable of interfering with capsule biosynthesis and transport. Exploration of the gap between outright killing of pathogenic bacteria and simply exposing them to the immune system could lead to development of therapeutic strategies for treating infection by multi-drug resistant strains, and it could also minimize society's contribution to the alarming spread of antibiotic resistance.
Potential therapeutic approaches
Therapeutic strategies that replace or supplement antibiotic treatment by capitalizing on capsule susceptibility will likely depend upon the type of CPS expressed by the invading pathogen, thus development of rapid and inexpensive diagnostic tools to guide treatment options will be a critical component of any therapeutic strategy. Serological characterization of surface antigens is hindered by the inability to generate, with high specificity, antibodies against important CPS structures that are identical to human glycans. Although MLST analyses and PCR based techniques guided by conserved capsule flanking regions are the current gold standard for molecular typing, these methods are limited due to the requirement of equipment that is not ubiquitous. Furthermore, molecular typing could lead to false positives due to expectations of CPS production that might not manifest due to the presence of mutations or additional CPS modifying enzymes located outside of the assayed gene loci. Rapid CPS diagnostics should be cheap, deployable to any location in the world, and should exhibit very high specificity for the target molecule to limit false positives.
One technology that satisfies these constraints is known as a molecular beacon, or a nucleic acid aptamer that has been engineered to emit a fluorometric or colorimetric signal upon binding of target molecules (Raj & van Oudenaarden, 2009). Aptamers are nucleic acid oligomers that have been selected from large libraries of random sequences due to their affinity and selectivity for a target molecule (Voigt, 2006). Highly specific aptamer affinity to a target molecule is easily attained by performing negative selection screens against structurally related molecules to remove cross-reacting oligonucleotides from the candidate aptamer pool. Aptamers with dissociation constants in the low nanomolar range have been generated against carbohydrates (Sun et al., 2010), and even recently against the PSA from the capsule of E. coli K1 (Cho et al., 2013). Target binding is associated with conformation change in the oligonucleotide; hence molecular beacons can be readily designed with covalently-bound fluorophore and quencher molecules that remain in close proximity in the unbound state to mask fluorescence but separate enough to unveil the fluorescent moiety when bound to the target (Tombelli et al., 2005). Addition of molecular beacon to a sample containing the cognate CPS would enable fluorescence but would require a high intensity light source for excitation and detection. Alternatively, aptamer sequences have been fused with ribozymes to, upon binding of target molecule, actuate production of a colored compound by reaction with other reagents in solution (Vinkenborg et al., 2011; Tang et al., 2012). Diagnosis of CPS structure with molecular beacons would be rapid, cheap, and easily performed by any healthcare worker.
Subsequent to diagnosis of the predominant CPSs, targeted therapeutic strategies can be implemented. One strategy for treatment of intestinal infections might involve ingestion of a probiotic strain that has been previously characterized to exclude or inhibit pathogens (Fig. 9a) (Lebeer et al., 2010). The term “probiotics” describes microorganisms that confer health benefits to the host, particularly with respect to human health (Marco et al., 2006). For instance, the probiotic strain E. coli Nissle 1917 has been shown to outcompete the encapsulated uropathogenic E. coli (UPEC) strain CFT073 in several growth conditions, including growth in urine (Hancock et al., 2010), and it exhibits bactericidal activity against many other pathogenic microbes as well (Storm et al., 2011). Prophylactic administration of Nissle 1917 in a porcine model was even shown to abolish secretory diarrhea upon challenge with an ETEC strain (Schroeder et al., 2006). In addition to the ability of Nissle 1917 to outcompete pathogens, it has been shown in another experiment using a porcine model to persist in the gut for up to a month after inoculation (Barth et al., 2009). Nissle 1917 is of particular interest in this review because of its non-immunogenic heparosan capsule, which might confer an advantage over other probiotics in outcompeting extraintestinal pathogens in, for example, the urinary tract. Furthermore, the ease of genetic manipulation of E. coli compared to other bacteria could make Nissle 1917 a model probiotic target for more complicated engineering strategies. The most common probiotics in use today are, however, members of the Lactobacillus and Bifidobacterium generas, which are utilized primarily in the GI tract. In addition, previous work has demonstrated the feasibility of engineering probiotic bacteria for vaccine delivery, as exemplified by a study in which nasal administration of Lactococcus lactis, recombinantly expressing pneumonococcal protective protein on its surface, induced protection against Streptococcus pneumoniae infection in young mice (Vintiñi et al., 2010). A related but speculative therapeutic strategy would be engineering probiotic strains to detect pathogenic signals and secrete pathogen-specific CPS-depolymerizing enzymes, effectively presenting the surface of the pathogen to the host immune system in a strategy that might complement any other positive effects already conferred by the probiotic strain (Fig. 9c). This strategy would require engineering a sensor capable of controlling expression and secretion of a CPS-degrading enzyme. One candidate is known as a riboregulator, an RNA sequence capable of binding a ligand through an aptamer domain, which induces a conformational change sequestering an antisense domain and allowing expression of the cognate mRNA. Riboregulators have been used to control translation of proteins in response to various ligands as discussed elsewhere (Khalil & Collins 2010); thus, it is expected that incorporation of an RNA aptamer domain capable of binding a specific CPS could be incorporated into a riboregulator to control expression of a secretion-tagged, CPS-depolymerizing lyase or glycosidase. An alternative to engineering enzyme-secreting probiotic strains would be to simply purify recombinantly expressed CPS-cleaving enzymes from fermentations of genetically tractable, secretion-capable production strains and to administer the purified enzyme to the patient. In one study illustrating the potential of this strategy, intraperitoneal injection of an anti-K1 glycosidase (endosialidase E from a bacteriophage) to rat pups with previously induced blood-borne E. coli K1 infection significantly reduced mortality and also showed prophylactic efficacy (Mushtaq et al., 2004). The glycosidase treatment was shown not reduce the viability of the pathogen, but rather to expose the bacteria to complement-mediated killing; bacteremia typically subsided within 24 h post injection and prevented death in most cases. Many studies have also demonstrated the promise of alginate lyase therapeutics for degrading Pseudomonas aeruginosa biofilms in the airways of cystic fibrosis patients, including a recent investigation of alginate lyase-PEG conjugates for degradation of P. aeruginosa biofilms on abiotic surfaces (Lamppa et al., 2011).
Fig. 9.
Strategies to combat capsule-bearing pathogens. Probiotic or engineered strains are designated in blue, and pathogenic strains are colored red with or without an orange capsule. (a) Ingestion of wild-type probiotic or engineered strains that outcompete pathogenic bacteria. (b) Small molecule inhibitors of CPS biosynthetic enzymes (upper) or CPS translocation proteins (lower). Triangles represent small-molecule inhibitors; purple circular (“Pac-Man”) symbol represents polysaccharide glycosyltransferase. (c) Treatment with probiotic bacteria engineered to secrete lyases or glycosidases (drawn as scissors) with activity against pathogen's CPS (upper) or treatment with purified enzyme (lower). (d) Treatment with natural or engineered bacteriophage to lyse bacteria bearing specific CPS type.
Phage therapy is another approach that should be considered for its potential to combat pathogenic bacteria possessing non-immunogenic capsules (Fig. 9d). Interest in the use of bacteriophages, or viruses that infect bacteria, declined in the West after the advent of antibiotics, but the alarming trend of drug-resistant bacterial infections has provoked reconsideration throughout the last decade of phage therapy as a strategy to replace or potentiate antibiotic treatment. Despite the decreasing investigation of phage therapy in Europe and the Americas after the commercialization of antibiotics, much work devoted to this topic was continued in states of the former Soviet Union and Poland throughout the twentieth century, including the use of bacteriophage to treat human diseases (O'Flaherty et al., 2009). An excellent review details the successful application of phage therapy for a wide range of infections and addresses the safety of phages from a medical standpoint (Sulakvelidze et al., 2001). A practical example of phage treatment includes the U.S. Food and Drug Administration's approval in 2006 of the use of a phage cocktail to prevent adulteration of ready-to-eat meat products by Listeria monocytogenes (Shuren, 2006). Furthermore, promising recent work has demonstrated the ability to engineer bacteriophages like T7 to degrade infectious biofilms (Azeredo & Sutherland, 2008) by expressing CPS-specific (Scholl et al., 2005) or EPS-specific (Lu & Collins, 2007) depolymerizing enzymes on their surface or tail spike, or by expressing these enzymes intracellularly during infection for subsequent release and targeting of neighboring bacterial cells after lysis. One strategy that phages have naturally evolved is the ability to “dig” for cell surface receptors that have been masked by cell surface biomolecules, such as CPS, by depolymerizing the coating. This is exemplified by K1 and K5 specific coliphages (Stummeyer et al., 2004; Thompson et al., 2010) that express evolved tail spike proteins capable of CPS-specific degradation. To date, various bacteriophages have been shown to possess CPS-degrading enzymes specific for heparosan (Thompson et al., 2010), polysialic acid (Stummeyer et al., 2004), and hyaluronan (Baker et al., 2002), but no bacteriophages are known to possess chondroitin lyases. However, bacterial enzymes have been successfully expressed by bacteriophage, so it could be valuable to explore bacteriophage surface or tail spike expression of chondroitinase to target and depolymerize the chondroitin capsules of certain pathogenic P. multocida and E. coli strains. Taken together, these studies not only highlight the potential to utilize highly specific phages as antibacterial agents against encapsulated bacteria, but they suggest that it should be possible to engineer bacteriophages to specifically target and lyse pathogenic microbes possessing non-immunogenic capsules.
Finally, small molecule therapeutics could be used to interfere with CPS biosynthesis, CPS translocation, or even CPS attachment to the outer leaflet of the outer membrane (Fig. 9b). In a recent publication, Seed et al. developed a high-throughput screening method to search large chemical libraries for capsule inhibitors (Goller & Seed, 2010). Using a CPS-dependent bacteriophage (that requires surface-exposed capsule for adsorption and subsequent ejection of phage nucleic acid into the host) as an indicator for presence of intact capsule, bacteria that had been incubated with library compounds were evaluated for capsule production. One compound, dubbed “C7”, inhibited capsule formation in UTI89, a UPEC K1 strain. In a fascinating turn of events, C7 also inhibited capsule formation in UPEC K5 strain DS17. The finding that assembly of two different CPS structures was inhibited by the same compound, coupled with data showing insignificant accumulation of intracellular or extracellular CPS, it was hypothesized that C7 inhibits an early stage of CPS biogenesis in all Group 2 capsular E. coli. This study exemplifies the approaches that could lead to incredibly valuable therapeutics against infectious pathogens. Very recently, another study demonstrated the use of glycomimetics, rationally designed to block export of an E. coli CPS by “plugging” Wza, a pore-like translocon protein in the Wzy-dependent transport pathway (Kong et al., 2013). It is our hope that aforementioned strategies will contribute to a better understanding of capsule biosynthesis in a metabolic context and inspire discussion leading to CPS-targeting therapeutic and prophylactic solutions to a host of serious diseases caused by masquerading microbial pathogens.
Acknowledgments
We would like to thank all of our coworkers and collaborators, as well as the funding agencies that make this work possible. B. F. Cress, J. A. Englaender, and W. He are partially supported by graduate fellowships from Rennselaer Polytechnic Institute. Research in the Linhardt laboratory on the subject of this review is supported by NIH grants HL62244, HL094463, and HL096972.
References
- Adlam C, Knights JM, Mugridge A, Williams JM, Lindon JC. Production of colominic acid by Pasteurella haemolytica serotype A2 organisms. FEMS Microbiol Lett. 1987;42:23–25. [Google Scholar]
- Al-haj Ali H, Sawada T, Hatakeyama H, Katayama Y, Ohtsuki N, Itoh O. Invasion of chicken embryo fibroblast cells by avian Pasteurella multocida. Vet Microbiol. 2004;104:55–62. doi: 10.1016/j.vetmic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Al-haj Ali H, Sawada T, Hatakeyama H, Ohtsuki N, Itoh O. Characterization of a 39kDa capsular protein of avian Pasteurella multocida using monoclonal antibodies. Vet Microbiol. 2004;100:43–53. doi: 10.1016/j.vetmic.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Allison DD, Grande-Allen KJ. Review. Hyaluronan: a powerful tissue engineering tool. Tissue Eng. 2006;12:2131–2140. doi: 10.1089/ten.2006.12.2131. [DOI] [PubMed] [Google Scholar]
- Almeida A, Albuquerque P, Araujo R, Ribeiro N, Tavares F. Detection and discrimination of common bovine mastitis-causing streptococci. Vet Microbiol. 2013;164:370–377. doi: 10.1016/j.vetmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Almeida RA, Oliver SP. Antiphagocytic effect of the capsule of Streptococcus uberis. Zbl Vet Med B. 1993;40:707–714. doi: 10.1111/j.1439-0450.1993.tb00195.x. [DOI] [PubMed] [Google Scholar]
- Andreishcheva EN, Vann WF. Escherichia coli BL21(DE3) chromosome contains a group II capsular gene cluster. Gene. 2006;384:113–119. doi: 10.1016/j.gene.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Annunziato PW, Wright LF, Vann WF, Silver RP. Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1995;177:312–319. doi: 10.1128/jb.177.2.312-319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai T, Timoney JF, Kuwamoto Y, Fujita Y, Wada R, Inoue T. In vivo pathogenicity and resistance to phagocytosis of Streptococcus equi strains with different levels of capsule expression. Vet Microbiol. 1999;67:277–286. doi: 10.1016/s0378-1135(99)00051-6. [DOI] [PubMed] [Google Scholar]
- Ashbaugh CD, Moser TJ, Shearer MH, White GL, Kennedy RC, Wessels MR. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell Microbiol. 2000;2:283–292. doi: 10.1046/j.1462-5822.2000.00050.x. [DOI] [PubMed] [Google Scholar]
- Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol. 2010;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- Azeredo J, Sutherland IW. The use of phages for the removal of infectious biofilms. Curr Pharm Biotechno. 2008;9:261–266. doi: 10.2174/138920108785161604. [DOI] [PubMed] [Google Scholar]
- Baker J, Dong S, Pritchard D. The hyaluronan lyase of Streptococcus pyogenes bacteriophage H4489A. Biochem J. 2002;365:317–322. doi: 10.1042/BJ20020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett B, Ebah L, Roberts IS. Genomic structure of capsular determinants. Curr Top Microbiol. 2002;264:137–155. [PubMed] [Google Scholar]
- Barth S, Duncker S, Hempe J, Breves G, Baljer G, Bauerfeind R. Escherichia coli Nissle 1917 for probiotic use in piglets: evidence for intestinal colonization. J Appl Microbiol. 2009;107:1697–1710. doi: 10.1111/j.1365-2672.2009.04361.x. [DOI] [PubMed] [Google Scholar]
- Bendaoud M, Vinogradov E, Balashova NV, Kadouri DE, Kachlany SC, Kaplan JB. Broad-spectrum biofilm inhibition by Kingella kingae exopolysaccharide. J Bacteriol. 2011;193:3879–3886. doi: 10.1128/JB.00311-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter-Suermann D, Goergen I, Frosch M. Monoclonal antibodies to weak immunogenic Escherichia coli and meningococcal capsular polysaccharides. In: Lark DL, editor. Protein-Carbohydrate Interactions in Biological Systems: The Molecular Biology of Microbial Pathogenicity. Academic Press; London: 1986. pp. 395–396. [Google Scholar]
- Blank LM, Hugenholtz P, Nielsen LK. Evolution of the hyaluronic acid synthesis (has) operon in Streptococcus zooepidemicus and other pathogenic streptococci. J Mol Evol. 2008;67:13–22. doi: 10.1007/s00239-008-9117-1. [DOI] [PubMed] [Google Scholar]
- Bliss JM, Garon CF, Silver RP. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology. 1996;6:445–452. doi: 10.1093/glycob/6.4.445. [DOI] [PubMed] [Google Scholar]
- Boeriu CG, Springer J, Kooy FK, van den Broek LAM, Eggink G. Production Methods for Hyaluronan. Int J Carbohyd Chem. 2013;2013:1–14. [Google Scholar]
- Born J, Klaus J, Assmann KJ, Lindahl U, Berden JH. N-Acetylated domains in heparin sulfates revealed by a monoclonal antibody against the Escherichia coli K5 capsular polysaccharide. Distribution of the cognate epitope in normal human kidney and transplant kidney with chronic vascular rejection. J Biol Chem. 1996;271:22802–22809. doi: 10.1074/jbc.271.37.22802. [DOI] [PubMed] [Google Scholar]
- Borrathybay E, Sawada T, Kataoka Y, Okiyama E, Kawamoto E, Amao H. Capsule thickness and amounts of a 39kDa capsular protein of avian Pasteurella multocida type A strains correlate with their pathogenicity for chickens. Vet Microbiol. 2003a;97:215–227. doi: 10.1016/j.vetmic.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Borrathybay E, Sawada T, Kataoka Y, Ohtsu N, Takagi M, Nakamura S, Kawamoto E. A 39kDa protein mediates adhesion of avian Pasteurella multocida to chicken embryo fibroblast cells. Vet Microbiol. 2003b;97:229–243. doi: 10.1016/j.vetmic.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Bortolussi R, Ferrieri P, Björkstén B, Quie PG. Capsular K1 polysaccharide of Escherichia coli: relationship to virulence in newborn rats and resistance to phagocytosis. Infect Immun. 1979;25:293–298. doi: 10.1128/iai.25.1.293-298.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøvre K, Bryn K, Closs O, Hagen N, Frøholm LO. Surface polysaccharide of Moraxella non-liquefaciens identical to Neisseria meningitidis group B capsular polysaccharide. A chemical and immunological investigation. NIPH Ann. 1983;6:65–73. [PubMed] [Google Scholar]
- Boyce JD, Chung JY, Adler B. Genetic organisation of the capsule biosynthetic locus of Pasteurella multocida M1404 (B:2) Vet Microbiol. 2000a;72:121–134. doi: 10.1016/s0378-1135(99)00193-5. [DOI] [PubMed] [Google Scholar]
- Boyce JD, Chung JY, Adler B. Pasteurella multocida capsule: composition, function and genetics. J Biotechnol. 2000b;83:153–160. doi: 10.1016/s0168-1656(00)00309-6. [DOI] [PubMed] [Google Scholar]
- Boyce JD, Harper M, Wilkie IW, Adler B. Pasteurella. In: Gyles CL, Prescott J, Songer G, Thoen CO, editors. Pathogenesis of Bacterial Infections. Fourth. Wiley-Blackwell; 2010. pp. 325–346. [Google Scholar]
- Bronner D, Sieberth V, Pazzani C, Smith A, Boulnois G, Roberts I, Jann B, Jann K. Synthesis of the K5 (group II) capsular polysaccharide in transport-deficient recombinant Escherichia coli. FEMS Microbiol Lett. 1993;113:279–284. doi: 10.1111/j.1574-6968.1993.tb06527.x. [DOI] [PubMed] [Google Scholar]
- Brooks HJ, O'Grady F, McSherry MA, Cattell WR. Uropathogenic properties of Escherichia coli in recurrent urinary-tract infection. J Med Microbiol. 1980;13:57–68. doi: 10.1099/00222615-13-1-57. [DOI] [PubMed] [Google Scholar]
- Bushell SR, Mainprize IL, Wear MA, Lou H, Whitfield C, Naismith JH. Wzi is an Outer Membrane Lectin that Underpins Group 1 Capsule Assembly in Escherichia coli. Structure. 2013;21:844–853. doi: 10.1016/j.str.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byarugaba DK, Minga UM, Gwakisa PS, Katunguka-Rwakishaya E, Bisgaard M, Olsen JE. Virulence characterization of Avibacterium paragallinarum isolates from Uganda. Avian Pathol. 2007;36:35–42. doi: 10.1080/03079450601102947. [DOI] [PubMed] [Google Scholar]
- Calvinho LF, Almeida RA, Oliver SP. Potential virulence factors of Streptococcus dysgalactiae associated with bovine mastitis. Vet Microbiol. 1998;61:93–110. doi: 10.1016/s0378-1135(98)00172-2. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Edit. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotech. 2012a;23:679–688. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RCC, Chiu K, Ho YS, So KF. Modulation of neuroimmune responses on glia in the central nervous system: implication in therapeutic intervention against neuroinflammation. Cell Mol Immunol. 2009;6:317–326. doi: 10.1038/cmi.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Yang B, Zhao X, Linhardt RJ. Analysis of glycosaminoglycan-derived disaccharides by capillary electrophoresis using laser-induced fluorescence detection. Anal Biochem. 2012b;427:91–98. doi: 10.1016/j.ab.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. P Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Marcellin E, Hung J, Nielsen LK. Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J Biol Chem. 2009;284:18007–18014. doi: 10.1074/jbc.M109.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Lee BR, Cho BK, Kim JH, Kim BG. In vitro selection of sialic acid specific RNA aptamer and its application to the rapid sensing of sialic acid modified sugars. Biotechnol Bioeng. 2013;110:905–913. doi: 10.1002/bit.24737. [DOI] [PubMed] [Google Scholar]
- Chung JY, Zhang Y, Adler B. The capsule biosynthetic locus of Pasteurella multocida A:1. FEMS Microbiol Lett. 1998;166:289–296. doi: 10.1111/j.1574-6968.1998.tb13903.x. [DOI] [PubMed] [Google Scholar]
- Chung JY, Wilkie I, Boyce JD, Townsend KM, Frost AJ, Ghoddusi M, Adler B. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect Immun. 2001;69:2487–2492. doi: 10.1128/IAI.69.4.2487-2492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslewicz M, Vimr E. Thermoregulation of kpsF, the first region 1 gene in the kps locus for polysialic acid biosynthesis in Escherichia coli K1. J Bacteriol. 1996;178:3212–3220. doi: 10.1128/jb.178.11.3212-3220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, De Rosa M, Schiraldi C. Production of glucuronic acid-based polysaccharides by microbial fermentation for biomedical applications. Biotechnol J. 2012;7:237–250. doi: 10.1002/biot.201100242. [DOI] [PubMed] [Google Scholar]
- Cimini D, De Rosa M, Carlino E, Ruggiero A, Schiraldi C. Homologous overexpression of rfaH in E. coli K4 improves the production of chondroitin-like capsular polysaccharide. Microb Cell Fact. 2013;12:1–12. doi: 10.1186/1475-2859-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BA, Wang Q, Tzianabos AO, Kasper DL. Polysaccharide processing and presentation by the MHCII pathway. Cell. 2004;117:677–687. doi: 10.016/j.cell.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Crater DL, van de Rijn I. Hyaluronic acid synthesis operon (has) expression in group A streptococci. J Biol Chem. 1995;270:18452–18458. doi: 10.1074/jbc.270.31.18452. [DOI] [PubMed] [Google Scholar]
- Cress BF, Linhardt RJ, Koffas MAG. Draft Genome Sequence of Escherichia coli Strain Nissle 1917 (Serovar O6:K5:H1) Genome Announc. 2013a;1:1–2. doi: 10.1128/genomeA.00047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress BF, Greene ZR, Linhardt RJ, Koffas MAG. Draft Genome Sequence of Escherichia coli Strain ATCC 23502 (Serovar O5:K4:H4) Genome Announc. 2013b;1:1–2. doi: 10.1128/genomeA.00046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress BF, Greene ZR, Linhardt RJ, Koffas MAG. Draft Genome Sequence of Escherichia coli Strain ATCC 23506 (Serovar O10:K5:H4) Genome Announc. 2013c;1:1–2. doi: 10.1128/genomeA.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross AS. The biologic significance of bacterial encapsulation. Curr Top Microbiol. 1990;150:87–95. doi: 10.1007/978-3-642-74694-9_5. [DOI] [PubMed] [Google Scholar]
- Crouch CF, LaFleur R, Ramage C, Reddick D, Murray J, Donachie W, Francis MJ. Cross protection of a Mannheimia haemolytica A1 Lkt-/Pasteurella multocida ΔhyaE bovine respiratory disease vaccine against experimental challenge with Mannheimia haemolytica A6 in calves. Vaccine. 2012;30:2320–2328. doi: 10.1016/j.vaccine.2012.01.063. [DOI] [PubMed] [Google Scholar]
- Cunningham RK, Söderström TO, Gillman CF, Van Oss CJ. Phagocytosis as a surface phenomenon. V. Contact angles and phagocytosis of rough and smooth strains of Salmonella typhimurium, and the influence of specific antiserum. Immunol Commun. 1975;4:429–442. doi: 10.3109/08820137509057331. [DOI] [PubMed] [Google Scholar]
- Cywes C, Wessels MR. Group A Streptococcus tissue invasion by CD44-mediated cell signalling. Nature. 2001;414:648–652. doi: 10.1038/414648a. [DOI] [PubMed] [Google Scholar]
- Daines DA, Silver RP. Evidence for multimerization of neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J Bacteriol. 2000;182:5267–5270. doi: 10.1128/jb.182.18.5267-5270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Kasper DL. Novel tools for modulating immune responses in the host-polysaccharides from the capsule of commensal bacteria. In: Alt FW, editor. Advances in Immunology. Vol. 106. Elsevier Inc.; 2010. pp. 61–91. [DOI] [PubMed] [Google Scholar]
- Davies RL. Characterization and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J Med Microbiol. 2003;52:59–67. doi: 10.1099/jmm.0.05019-0. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Papaconstantinou J, Weigel PH. Isolation of a Streptococcus pyogenes gene locus that directs hyaluronan biosynthesis in acapsular mutants and in heterologous bacteria. J Biol Chem. 1993a;268:14568–14571. [PubMed] [Google Scholar]
- DeAngelis PL, Papaconstantinou J, Weigel PH. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem. 1993b;268:19181–19184. [PubMed] [Google Scholar]
- DeAngelis PL, Jing W, Drake RR, Achyuthan AM. Identification and molecular cloning of a unique hyaluronan synthase from Pasteurella multocida. J Biol Chem. 1998;273:8454–8458. doi: 10.1074/jbc.273.14.8454. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pathogens, and algal viruses. Cell Mol Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL, Padgett-McCue AJ. Identification and molecular cloning of a chondroitin synthase from Pasteurella multocida type F. J Biol Chem. 2000;275:24124–24129. doi: 10.1074/jbc.M003385200. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat Rec. 2002a;268:317–326. doi: 10.1002/ar.10163. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL. Microbial glycosaminoglycan glycosyltransferases. Glycobiology. 2002b;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, White CL. Identification and molecular cloning of a heparosan synthase from Pasteurella multocida type D. J Biol Chem. 2002;277:7209–7213. doi: 10.1074/jbc.M112130200. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Gunay NS, Toida T, Mao W, Linhardt RJ. Identification of the capsular polysaccharides of Type D and F Pasteurella multocida as unmodified heparin and chondroitin, respectively. Carbohyd Res. 2002;337:1547–1552. doi: 10.1016/s0008-6215(02)00219-7. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Oatman LC, Gay DF. Rapid chemoenzymatic synthesis of monodisperse hyaluronan oligosaccharides with immobilized enzyme reactors. J Biol Chem. 2003;278:35199–35203. doi: 10.1074/jbc.M306431200. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, White CL. Identification of a distinct, cryptic heparosan synthase from Pasteurella multocida types A, D, and F. J Bacteriol. 2004;186:8529–8532. doi: 10.1128/JB.186.24.8529-8532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL. Glycosaminoglycan polysaccharide biosynthesis and production: today and tomorrow. Appl Microbiol Biot. 2012;94:295–305. doi: 10.1007/s00253-011-3801-6. [DOI] [PubMed] [Google Scholar]
- Deszo EL, Steenbergen SM, Freedberg DI, Vimr ER. Escherichia coli K1 polysialic acid O-acetyltransferase gene, neuO, and the mechanism of capsule form variation involving a mobile contingency locus. P Natl Acad Sci USA. 2005;102:5564–5569. doi: 10.1073/pnas.0407428102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SJ, Schneerson R, Egan W, Vann WF, Robbins JB, Shiloach J. Identity between polysaccharide antigens of Moraxella nonliquefaciens, group B Neisseria meningitidis, and Escherichia coli K1 (non-O acetylated) Infect Immun. 1991;59:732–736. doi: 10.1128/iai.59.2.732-736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnio PY, Lerestif-Gautier AL, Avril JL. Characterization of Pasteurella spp. Strains isolated from human infections. J Comp Pathol. 2004;130:137–142. doi: 10.1016/j.jcpa.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Dougherty BA, van de Rijn I. Molecular characterization of a locus required for hyaluronic acid capsule production in group A streptococci. J Exp Med. 1992;175:1291–1299. doi: 10.1084/jem.175.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso LM, Bono JL, Keen JE. Molecular serotyping of Escherichia coli O26:H11. Appl Environ Microb. 2005;71:4941–4944. doi: 10.1128/AEM.71.8.4941-4944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MS, Kasper DL, Jennings HJ, Baker CJ, Nicholson-Weller A. Capsular sialic acid prevents activation of the alternative complement pathway by type III, group B streptococci. J Immunol. 1982;128:1278–1283. [PubMed] [Google Scholar]
- Edwards U, Frosch M. Sequence and functional analysis of the cloned Neisseria meningitidis CMP-NeuNAc synthetase. FEMS Microbiol Lett. 1992;75:161–166. doi: 10.1016/0378-1097(92)90397-7. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- Erbel PJA, Barr K, Gao N, Gerwig GJ, Rick PD, Gardner KH. Identification and biosynthesis of cyclic enterobacterial common antigen in Escherichia coli. J Bacteriol. 2003;185:1995–2004. doi: 10.1128/JB.185.6.1995-2004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD. Proteoglycans and Glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Second. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- Esko JD, Doering TL, Raetz CR. Eubacteria and Archaea. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Second. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- Ewers C, Lübke-Becker A, Bethe A, Kiebling S, Filter M, Wieler LH. Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet Microbiol. 2006;114:304–317. doi: 10.1016/j.vetmic.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Fillit HM, McCarty M, Blake M. Induction of Antibodies to Hyaluronic Acid by Immunization of Rabbits with Encapsulated Streptococci. J Exp Med. 1986;164:762–776. doi: 10.1084/jem.164.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke A, Bronner D, Nikolaev AV, Jann B, Jann K. Biosynthesis of the Escherichia coli K5 polysaccharide, a representative of group II capsular polysaccharides: polymerization in vitro and characterization of the product. J Bacteriol. 1991;173:4088–4094. doi: 10.1128/jb.173.13.4088-4094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finne J. Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. J Biol Chem. 1982;257:11966–11970. [PubMed] [Google Scholar]
- Finne J, Leinonen M, Mäkelä PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. Human disease isolates of serotype M4 and M22 Group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio. 2012;3:e00413–12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregolino E, Ivanova R, Lanzetta R, Molinaro A, Parrilli M, Paunova-Krasteva T, Stoitsova SR, De Castro C. Occurrence and structure of cyclic Enterobacterial Common Antigen in Escherichia coli O157:H- Carbohyd Res. 2012;363:29–32. doi: 10.1016/j.carres.2012.09.017. [DOI] [PubMed] [Google Scholar]
- Frosch M, Müller A. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol Microbiol. 1993;8:483–493. doi: 10.1111/j.1365-2958.1993.tb01592.x. [DOI] [PubMed] [Google Scholar]
- Frosch M, Görgen I, Boulnois GJ, Timmis KN, Bitter-Suermann D. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. P Natl Acad Sci USA. 1985;82:1194–1198. doi: 10.1073/pnas.82.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- Gallagher JT. The extended family of proteoglycans: social residents of the pericellular zone. Curr Opin Cell Biol. 1989;1:1201–1218. doi: 10.1016/s0955-0674(89)80072-9. [DOI] [PubMed] [Google Scholar]
- Ganguli S, Zapata G, Wallis T, Reid C, Boulnois G, Vann WF, Roberts IS. Molecular cloning and analysis of genes for sialic acid synthesis in Neisseria meningitidis group B and purification of the meningococcal CMP-NeuNAc synthetase enzyme. J Bacteriol. 1994;176:4583–4589. doi: 10.1128/jb.176.15.4583-4589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glode MP, Robbins JB, Liu TY, Gotschlich EC, Orskov I, Orskov F. Cross-antigenicity and immunogenicity between capsular polysaccharides of group C Neisseria meningitides and of Escherichia coli K92. J Infect Dis. 1977;135:94–104. doi: 10.1093/infdis/135.1.94. [DOI] [PubMed] [Google Scholar]
- Goller CC, Seed PC. High-throughput identification of chemical inhibitors of E. coli Group 2 capsule biogenesis as anti-virulence agents. PLoS ONE. 2010;5:e11642. doi: 10.1371/journal.pone.0011642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granoff DM, Bartoloni A, Ricci S, et al. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J Immunol. 1998;160:5028–5036. [PubMed] [Google Scholar]
- Hafez M, Hayes K, Goldrick M, Warhurst G, Grencis R, Roberts IS. The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect Immun. 2009;77:2995–3003. doi: 10.1128/IAI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt S, Birkholz C, Zähringer U, Robertson BD, van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- Hancock V, Vejborg RM, Klemm P. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics. 2010;284:437–454. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
- Hänfling P, Shashkov AS, Jann B, Jann K. Analysis of the enzymatic cleavage (beta elimination) of the capsular K5 polysaccharide of Escherichia coli by the K5-specific coliphage: reexamination. J Bacteriol. 1996;178:4747–4750. doi: 10.1128/jb.178.15.4747-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BR, Neely MN. Coordinate regulation of Gram-positive cell surface components. Curr Opin Microbiol. 2012;15:204–210. doi: 10.1016/j.mib.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson LA, Ahlstedt S, Fasth A, Jodal U, Kaijser B, Larsson P, Lindberg U, Olling S, Sohl-Åkerlund A, Svanborg-Edén C. Antigens of Escherichia coli, human immune response, and the pathogenesis of urinary tract infections. J Infect Dis. 1977;136(Suppl):S144–S149. doi: 10.1093/infdis/136.supplement.s144. [DOI] [PubMed] [Google Scholar]
- Harber MJ, Topley N, Asscher AW. Virulence factors of urinary pathogens. Clin Sci. 1986;70:531–538. doi: 10.1042/cs0700531. [DOI] [PubMed] [Google Scholar]
- Harrison OB, Claus H, Jiang Y, et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis. 2013;19:566–573. doi: 10.3201/eid1904.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, Torii K, Hasegawa T, Ohta M. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol. 2004;42:186–192. doi: 10.1128/JCM.42.1.186-192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger M, Avery OT. The soluble specific substance of pneumococcus. J Exp Med. 1923;38:73–79. doi: 10.1084/jem.38.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérias MV, Midtvedt T, Hanson LA, Wold AE. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect Immun. 1997;65:531–536. doi: 10.1128/iai.65.2.531-536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey AM, Bhaskar U, Linhardt RJ, Dordick JS. Effect of eliminase gene (elmA) deletion on heparosan production and shedding in Escherichia coli K5. J Biotechnol. 2013;165:175–177. doi: 10.1016/j.jbiotec.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobb RI, Tzeng YL, Choudhury BP, Carlson RW, Stephens DS. Requirement of NMB0065 for connecting assembly and export of sialic acid capsular polysaccharides in Neisseria meningitidis. Microbes Infect. 2010;12:476–487. doi: 10.1016/j.micinf.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson N, Griffiths G, Cook N, Pourhossein M, Gottfridson E, Lind T, Lidholt K, Roberts IS. Identification that KfiA, a protein essential for the biosynthesis of the Escherichia coli K5 capsular polysaccharide, is an alpha-UDP-GlcNAc glycosyltransferase. The formation of a membrane-associated K5 biosynthetic complex requires KfiA, KfiB, and KfiC. J Biol Chem. 2000;275:27311–27315. doi: 10.1074/jbc.M004426200. [DOI] [PubMed] [Google Scholar]
- Höök M, Kjellén L, Johansson S. Cell-surface glycosaminoglycans. Annu Rev Biochem. 1984;53:847–869. doi: 10.1146/annurev.bi.53.070184.004215. [DOI] [PubMed] [Google Scholar]
- Horwitz MA, Silverstein SC. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J Clin Invest. 1980;65:82–94. doi: 10.1172/JCI109663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huskisson E. Glucosamine and Chondroitin for Osteoarthritis. J Int Med Res. 2008;36:1161–1179. doi: 10.1177/147323000803600602. [DOI] [PubMed] [Google Scholar]
- Jacques M, Kobisch M, Bélanger M, Dugal F. Virulence of capsulated and noncapsulated isolates of Pasteurella multocida and their adherence to porcine respiratory tract cells and mucus. Infect Immun. 1993;61:4785–4792. doi: 10.1128/iai.61.11.4785-4792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jann B, Jann K. Structure and biosynthesis of the capsular antigens of Escherichia coli. Curr Top Microbiol. 1990;150:19–42. doi: 10.1007/978-3-642-74694-9_2. [DOI] [PubMed] [Google Scholar]
- Jann K, Jann B. Polysaccharide antigens of Escherichia coli. Rev Infect Dis. 1987;9(Suppl 5):S517–S526. doi: 10.1093/clinids/9.supplement_5.s517. [DOI] [PubMed] [Google Scholar]
- Jann K, Jann B. Capsules of Escherichia coli. In: Sussman M, editor. Escherichia Coli: Mechanisms of Virulence. Cambridge University Press; Cambridge CB2 2RU, United Kingdom: 1997. pp. 113–143. [Google Scholar]
- Jann K, Jann B. Bacterial polysaccharides related to mammalian structures. In: Dumitriu S, editor. Polysaccharides: Structural Diversity and Functional Versatility. Marcel Dekker; New York, NY: 1998. pp. 211–236. [Google Scholar]
- Jarvis GA, Vedros NA. Sialic acid of group B Neisseria meningitidis regulates alternative complement pathway activation. Infect Immun. 1987;55:174–180. doi: 10.1128/iai.55.1.174-180.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelakovic S, Jann K, Schulz GE. The three-dimensional structure of capsule-specific CMP: 2-keto-3-deoxy-manno-octonic acid synthetase from Escherichia coli. FEBS Lett. 1996;391:157–161. doi: 10.1016/0014-5793(96)00724-7. [DOI] [PubMed] [Google Scholar]
- Jennings HJ, Roy R, Michon F. Determinant specificities of the groups B and C polysaccharides of Neisseria meningitidis. J Immunol. 1985;134:2651–2657. [PubMed] [Google Scholar]
- Jensen A, Kilian M. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol. 2012;50:113–126. doi: 10.1128/JCM.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Li J, Han F, Duan G, Lu X, Gu Y, Yu W. Antibiofilm activity of an exopolysaccharide from marine bacterium Vibrio sp. QY101. PLoS ONE. 2011;6:e18514. doi: 10.1371/journal.pone.0018514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing W, DeAngelis PL. Dissection of the two transferase activities of the Pasteurella multocida hyaluronan synthase: two active sites exist in one polypeptide. Glycobiology. 2000;10:883–889. doi: 10.1093/glycob/10.9.883. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Nickerson KG, Liao J, Grossbard L, Osserman EF, Glickman E, Chess L, Robbins JB, Schneerson R, Yang YH. A human monoclonal macroglobulin with specificity for alpha(2----8)-linked poly-N-acetyl neuraminic acid, the capsular polysaccharide of group B meningococci and Escherichia coli K1, which crossreacts with polynucleotides and with denatured DNA. J Exp Med. 1986;164:642–654. doi: 10.1084/jem.164.2.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaijser B. Immunology of Escherichia coli: K antigen and its relation to urinary-tract infection. J Infect Dis. 1973;127:670–677. doi: 10.1093/infdis/127.6.670. [DOI] [PubMed] [Google Scholar]
- Kaijser B, Jodal U. Escherichia coli K5 antigen in relation to various infections and in healthy individuals. J Clin Microbiol. 1984;19:264–266. doi: 10.1128/jcm.19.2.264-266.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaijser B, Hanson LA, Jodal U, Lidin-Janson G, Robbins JB. Frequency of E. coli K antigens in urinary-tract infections in children. Lancet. 1977;1:663–666. doi: 10.1016/s0140-6736(77)92111-0. [DOI] [PubMed] [Google Scholar]
- Kamhi E, Joo EJ, Dordick JS, Linhardt RJ. Glycosaminoglycans in infectious disease. Biol Rev Camb Philos. 2013;88:928–943. doi: 10.1111/brv.12034. [DOI] [PubMed] [Google Scholar]
- Kane TA, White CL, DeAngelis PL. Functional characterization of PmHS1, a Pasteurella multocida heparosan synthase. J Biol Chem. 2006;281:33192–33197. doi: 10.1074/jbc.M606897200. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Kim SY, Jeong H, Kim TY, Kim JJ, Choy HE, Yi KY, Rhee JH, Lee SY. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol Syst Biol. 2011;7:1–15. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Elliott SJ, Di Cello F, Stins MF, Kim KS. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol. 2003;5:245–252. doi: 10.1046/j.1462-5822.2003.t01-1-00271.x. [DOI] [PubMed] [Google Scholar]
- Kim PJ, Lee DY, Kim TY, Lee KH, Jeong H, Lee SY, Park S. Metabolite essentiality elucidates robustness of Escherichia coli metabolism. P Natl Acad Sci USA. 2007;104:13638–13642. doi: 10.1073/pnas.0703262104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Oh S, Kim SH. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem Bioph Res Co. 2009;379:324–329. doi: 10.1016/j.bbrc.2008.12.053. [DOI] [PubMed] [Google Scholar]
- King MR, Steenbergen SM, Vimr ER. Going for baroque at the Escherichia coli K1 cell surface. Trends Microbiol. 2007;15:196–202. doi: 10.1016/j.tim.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Klainer AS, Geis I. Agents of bacterial disease. Medical Department, Harper & Row; 1975. [Google Scholar]
- Köndgen S, Leider M, Lankester F, Bethe A, Lübke-Becker A, Leendertz FH, Ewers C. Pasteurella multocida involved in respiratory disease of wild chimpanzees. PLoS ONE. 2011;6:e24236. doi: 10.1371/journal.pone.0024236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Wang W, Tao J, Wang L, Wang Q, Sabananthan A, Gilbert GL. A molecular-capsular-type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA-cpsB sequencing and wzy- or wzx-specific PCR. J Med Microbiol. 2005;54:351–356. doi: 10.1099/jmm.0.45924-0. [DOI] [PubMed] [Google Scholar]
- Kong L, Harrington L, Li Q, Cheley S, Davis BG, Bayley H. Single-molecule interrogation of a bacterial sugar transporter allows the discovery of an extracellular inhibitor. Nat Chem. 2013;5:651–659. doi: 10.1038/nchem.1695. [DOI] [PubMed] [Google Scholar]
- Korhonen TK, Valtonen MV, Parkkinen J, Väisänen-Rhen V, Finne J, Orskov F, Orskov I, Svenson SB, Mäkelä PH. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahulec J, Krahulcová J, Medová M, Velebny V. Influence of KfoG on capsular polysaccharide structure in Escherichia coli K4 strain. Mol Biotechnol. 2005;30:129–134. doi: 10.1385/MB:30:2:129. [DOI] [PubMed] [Google Scholar]
- Kröncke KD, Orskov I, Orskov F, Jann B, Jann K. Electron microscopic study of coexpression of adhesive protein capsules and polysaccharide capsules in Escherichia coli. Infect Immun. 1990;58:2710–2714. doi: 10.1128/iai.58.8.2710-2714.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuberan B, Linhardt R. Carbohydrate Based Vaccines. Curr Org Chem. 2000;4:653–677. [Google Scholar]
- Kumari K, Weigel PH. Molecular cloning, expression, and characterization of the authentic hyaluronan synthase from group C Streptococcus equisimilis. J Biol Chem. 1997;272:32539–32546. doi: 10.1074/jbc.272.51.32539. [DOI] [PubMed] [Google Scholar]
- Lambert-Zechovsky N, Bingen E, Denamur E, Brahimi N, Brun P, Mathieu H, Elion J. Molecular analysis provides evidence for the endogenous origin of bacteremia and meningitis due to Enterobacter cloacae in an infant. Clin Infect Dis. 1992;15:30–32. doi: 10.1093/clinids/15.1.30. [DOI] [PubMed] [Google Scholar]
- Lamppa JW, Ackerman ME, Lai JI, Scanlon TC, Griswold KE. Genetically engineered alginate lyase-PEG conjugates exhibit enhanced catalytic function and reduced immunoreactivity. PLoS ONE. 2011;6:e17042. doi: 10.1371/journal.pone.0017042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield RC. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue K, Ford RC, Willis LM, Whitfield C. Functional and structural characterization of polysaccharide co-polymerase proteins required for polymer export in ATP-binding cassette transporter-dependent capsule biosynthesis pathways. J Biol Chem. 2011;286:16658–16668. doi: 10.1074/jbc.M111.228221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent TC, Fraser JR. Hyaluronan. FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Banks SD, Li JP. Virulence, immunity, and vaccine related to Streptococcus pneumoniae. Crit Rev Microbiol. 1991;18:89–114. doi: 10.3109/10408419109113510. [DOI] [PubMed] [Google Scholar]
- Legoux R, Lelong P, Jourde C, et al. N-acetyl-heparosan lyase of Escherichia coli K5: gene cloning and expression. J Bacteriol. 1996;178:7260–7264. doi: 10.1128/jb.178.24.7260-7264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leying H, Suerbaum S, Kroll HP, Stahl D, Opferkuch W. The capsular polysaccharide is a major determinant of serum resistance in K-1-positive blood culture isolates of Escherichia coli. Infect Immun. 1990;58:222–227. doi: 10.1128/iai.58.1.222-227.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ly M, Linhardt RJ. Proteoglycan sequence. Mol Biosyst. 2012;8:1613–1625. doi: 10.1039/c2mb25021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu Hai, Thon V, Chen Yi, Muthana MM, Qu J, Hie L, Chen X. Donor substrate promiscuity of the N-acetylglucosaminyltransferase activities of Pasteurella multocida heparosan synthase 2 (PmHS2) and Escherichia coli K5 KfiA. Appl Microbiol Biot. 2013;2:1–8. doi: 10.1007/s00253-013-4947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholt K, Fjelstad M. Biosynthesis of the Escherichia coli K4 capsule polysaccharide. A parallel system for studies of glycosyltransferases in chondroitin formation. J Biol Chem. 1997;272:2682–2687. doi: 10.1074/jbc.272.5.2682. [DOI] [PubMed] [Google Scholar]
- Lifely MR, Nowicka UT, Moreno C. Analysis of the chain length of oligomers and polymers of sialic acid isolated from Neisseria meningitidis group B and C and Escherichia coli K1 and K92. Carbohyd Res. 1986;156:123–135. doi: 10.1016/s0008-6215(00)90104-6. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Galliher PM, Cooney CL. Polysaccharide lyases. Appl Biochem Biotech. 1986;12:135–176. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Toida T. Role of glycosaminoglycans in cellular communication. Accounts Chem Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu Y, Li J, Du G, Chen J. Microbial production of hyaluronic acid: current state, challenges, and perspectives. Microb Cell Fact. 2011;10:1–9. doi: 10.1186/1475-2859-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Xu Y, Chen M, Weïwer M, Zhou X, Bridges AS, DeAngelis PL, Zhang Q, Linhardt RJ, Liu J. Chemoenzymatic design of heparan sulfate oligosaccharides. J Biol Chem. 2010;285:34240–34249. doi: 10.1074/jbc.M110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodinová-Žaadniková R, Tlaskalová-Hogenová H, Sonnenborn U. Local and serum antibody response in full–term and premature infants after artificial colonization of the intestine with E. coli strain Nissle 1917 (Mutaflor®) Pediatr Allergy Immu. 1992;3:43–48. [Google Scholar]
- Loos M. The complement system: activation and control. Curr Top Microbiol. 1985;121:7–18. doi: 10.1007/978-3-642-45604-6_2. [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang X, Zhu Y, Kim KS, Yang J, Jin Q. Complete genome sequence of the neonatal-meningitis-associated Escherichia coli strain CE10. J Bacteriol. 2011;193:7005. doi: 10.1128/JB.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. P Natl Acad Sci USA. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong TT, Lee CY. Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect Immun. 2002;70:3389–3395. doi: 10.1128/IAI.70.7.3389-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly M, Wang Z, Laremore TN, Zhang F, Zhong W, Pu D, Zagorevski DV, Dordick JS, Linhardt RJ. Analysis of E. coli K5 capsular polysaccharide heparosan. Anal Bioanal Chem. 2011;399:737–745. doi: 10.1007/s00216-010-3679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotech. 2006;17:204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Cover image. J Biol Chem. 2009;284(27) doi: 10.1074/jbc.M109.008094. cover. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson L, Holbein BE, Ashton FE. Virulence linked to polysaccharide production in serogroup B Neisseria meningitidis. FEMS Microbiol Lett. 1982;13:187–190. [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Meredith TC, Woodard RW. Characterization of Escherichia coli D-arabinose 5-phosphate isomerase encoded by kpsF: implications for group 2 capsule biosynthesis. Biochem J. 2006;395:427–432. doi: 10.1042/BJ20051828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, Woodard RW. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J Biol Chem. 2007;282:7790–7798. doi: 10.1074/jbc.M611034200. [DOI] [PubMed] [Google Scholar]
- Mergulhao FJM, Summers DK, Monteiro GA. Recombinant protein secretion in Escherichia coli. Biotechnol Adv. 2005;23:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Minshew BH, Jorgensen J, Swanstrum M, Grootes-Reuvecamp GA, Falkow S. Some characteristics of Escherichia coli strains isolated from extraintestinal infections of humans. J Infect Dis. 1978;137:648–654. doi: 10.1093/infdis/137.5.648. [DOI] [PubMed] [Google Scholar]
- Moe GR, Bhandari TS, Flitter BA. Vaccines containing de-N-acetyl sialic acid elicit antibodies protective against Neisseria meningitidis groups B and C. J Immunol. 2009;182:6610–6617. doi: 10.4049/jimmunol.0803677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad SAS, Azmi NA, Mohamed R. 2012 IEEE Symposium on Humanities, Science and Engineering Research. IEEE; 2012. Screening and identification of Pasteurella multocida capsular types among animal cases in Malaysia; pp. 737–742. [Google Scholar]
- Mordhorst IL, Claus H, Ewers C, et al. O-acetyltransferase gene neuO is segregated according to phylogenetic background and contributes to environmental desiccation resistance in Escherichia coli K1. Environ Microbiol. 2009;11:3154–3165. doi: 10.1111/j.1462-2920.2009.02019.x. [DOI] [PubMed] [Google Scholar]
- Moxley RA, Francis DH. Natural and experimental infection with an attaching and effacing strain of Escherichia coli in calves. Infect Immun. 1986;53:339–346. doi: 10.1128/iai.53.2.339-346.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon ER, Kroll JS. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- Muñoz R, García E, López R. Evidence for horizontal transfer from Streptococcus to Escherichia coli of the kfiD gene encoding the K5-specific UDP-glucose dehydrogenase. J Mol Evol. 1998;46:432–436. doi: 10.1007/pl00006322. [DOI] [PubMed] [Google Scholar]
- Murkin AS, Chou WK, Wakarchuk WW, Tanner ME. Identification and mechanism of a bacterial hydrolyzing UDP-N-acetylglucosamine 2-epimerase. Biochemistry-US. 2004;43:14290–14298. doi: 10.1021/bi048606d. [DOI] [PubMed] [Google Scholar]
- Mushtaq N, Redpath MB, Luzio JP, Taylor PW. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Ch. 2004;48:1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia JL, Riesenfeld J, Vann WF, Lindahl U, Rodén L. Assay of N-acetylheparosan deacetylase with a capsular polysaccharide from Escherichia coli K5 as substrate. Anal Biochem. 1983;135:134–140. doi: 10.1016/0003-2697(83)90741-8. [DOI] [PubMed] [Google Scholar]
- Ninomiya T, Sugiura N, Tawada A, Sugimoto K, Watanabe H, Kimata K. Molecular cloning and characterization of chondroitin polymerase from Escherichia coli strain K4. J Biol Chem. 2002;277:21567–21575. doi: 10.1074/jbc.M201719200. [DOI] [PubMed] [Google Scholar]
- Nithya C, Begum MF, Pandian SK. Marine bacterial isolates inhibit biofilm formation and disrupt mature biofilms of Pseudomonas aeruginosa PAO1. Appl Microbiol Biot. 2010;88:341–358. doi: 10.1007/s00253-010-2777-y. [DOI] [PubMed] [Google Scholar]
- O'Flaherty S, Ross RP, Coffey A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev. 2009;33:801–819. doi: 10.1111/j.1574-6976.2009.00176.x. [DOI] [PubMed] [Google Scholar]
- O'Hanlon KA, Catarame TMG, Duffy G, Blair IS, McDowell DA. RAPID detection and quantification of E. coli O157/O26/O111 in minced beef by real-time PCR. J Appl Microbiol. 2004;96:1013–1023. doi: 10.1111/j.1365-2672.2004.02224.x. [DOI] [PubMed] [Google Scholar]
- Orskov F, Orskov I. Escherichia coli serotyping and disease in man and animals. Can J Microbiol. 1992;38:699–704. [PubMed] [Google Scholar]
- Orskov F, Orskov I, Sutton A, Schneerson R, Lin W, Egan W, Hoff GE, Robbins JB. Form variation in Escherichia coli K1: determined by O-acetylation of the capsular polysaccharide. J Exp Med. 1979;149:669–685. doi: 10.1084/jem.149.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orskov I, Orskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orskov I, Birch-Andersen A, Duguid JP, Stenderup J, Orskov F. An adhesive protein capsule of Escherichia coli. Infect Immun. 1985;47:191–200. doi: 10.1128/iai.47.1.191-200.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Sugiura N, Shimada H, Hirooka R, Tsuji A, Shirakawa T, Fukuyama K, Kimura M, Kimata K, Kakuta Y. Crystal structure of chondroitin polymerase from Escherichia coli K4. Biochem Bioph Res Co. 2009;378:10–14. doi: 10.1016/j.bbrc.2008.08.121. [DOI] [PubMed] [Google Scholar]
- Ostblom A, Adlerberth I, Wold AE, Nowrouzian FL. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants' commensal microbiotas. Appl Environ Microb. 2011;77:2303–2308. doi: 10.1128/AEM.02405-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto NJ, Green DE, Masuko S, Mayer A, Tanner ME, Linhardt RJ, DeAngelis PL. Structure/function analysis of Pasteurella multocida heparosan synthases: toward defining enzyme specificity and engineering novel catalysts. J Biol Chem. 2012;287:7203–7212. doi: 10.1074/jbc.M111.311704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka MS, Wright LF, Silver RP. Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991;173:4603–4610. doi: 10.1128/jb.173.15.4603-4610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka MS, Hayes SF, Silver RP. Characterization of KpsT, the ATP-binding component of the ABC-transporter involved with the export of capsular polysialic acid in Escherichia coli K1. J Biol Chem. 1994;269:20149–20158. [PubMed] [Google Scholar]
- Pazzani C, Rosenow C, Boulnois GJ, Bronner D, Jann K, Roberts IS. Molecular analysis of region 1 of the Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J Bacteriol. 1993;175:5978–5983. doi: 10.1128/jb.175.18.5978-5983.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson PK, Wilkinson BJ, Kim Y, Schmeling D, Quie PG. Influence of encapsulation on staphylococcal opsonization and phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1978;19:943–949. doi: 10.1128/iai.19.3.943-949.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Rigg GP, Pazzani C, Smith A, Sieberth V, Stevens M, Boulnois G, Jann K, Roberts IS. Region 2 of the Escherichia coli K5 capsule gene cluster encoding proteins for the biosynthesis of the K5 polysaccharide. Mol Microbiol. 1995;17:611–620. doi: 10.1111/j.1365-2958.1995.mmi_17040611.x. [DOI] [PubMed] [Google Scholar]
- Pigeon RP, Silver RP. Topological and mutational analysis of KpsM, the hydrophobic component of the ABC-transporter involved in the export of polysialic acid in Escherichia coli K1. Mol Microbiol. 1994;14:871–881. doi: 10.1111/j.1365-2958.1994.tb01323.x. [DOI] [PubMed] [Google Scholar]
- Pluschke Gerd, Mayden J, Achtman M, Levine RP. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect Immun. 1983;42:907–913. doi: 10.1128/iai.42.3.907-913.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poermadjaja B, Frost A. Phagocytic uptake and killing of virulent and avirulent strains of Pasteurella multocida of capsular serotype A by chicken macrophages. Vet Microbiol. 2000;72:163–171. doi: 10.1016/s0378-1135(99)00196-0. [DOI] [PubMed] [Google Scholar]
- Pon RA, Lussier M, Yang QL, Jennings HJ. N-Propionylated group B meningococcal polysaccharide mimics a unique bactericidal capsular epitope in group B Neisseria meningitidis. J Exp Med. 1997;185:1929–1938. doi: 10.1084/jem.185.11.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SB, Jayaraman G, Ramachandran KB. Hyaluronic acid production is enhanced by the additional co-expression of UDP-glucose pyrophosphorylase in Lactococcus lactis. Appl Microbiol Biot. 2010;86:273–283. doi: 10.1007/s00253-009-2293-0. [DOI] [PubMed] [Google Scholar]
- Rafiq I, Parthasarathy H, Tremlett C, Freeman LJ, Mullin M. Infective endocarditis caused by Moraxella nonliquefaciens in a percutaneous aortic valve replacement. Cardiovasc Revasc Med. 2011;12:184–186. doi: 10.1016/j.carrev.2010.03.082. [DOI] [PubMed] [Google Scholar]
- Rahn A, Beis K, Naismith JH, Whitfield C. A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol. 2003;185:5882–5890. doi: 10.1128/JB.185.19.5882-5890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Ann Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph AP, Carapetis JR. Group a streptococcal diseases and their global burden. Curr Top Microbiol. 2013;368:1–27. doi: 10.1007/82_2012_280. [DOI] [PubMed] [Google Scholar]
- Reid AN, Cuthbertson L. Biosynthesis of Capsular Polysaccharides and Exopolysaccharides. In: Reid CW, Twine SM, Reid AN, editors. Bacterial Glycomics: Current Research, Technology and Applications. Caister Academic Press; Norfolk, UK: 2012. pp. 27–54. [Google Scholar]
- Reizer J, Reizer A, Saier MH. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1992;1:1326–1332. doi: 10.1002/pro.5560011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restaino OF, Cimini D, De Rosa M, Catapano A, Schiraldi C. High cell density cultivation of Escherichia coli K4 in a microfiltration bioreactor: a step towards improvement of chondroitin precursor production. Microb Cell Fact. 2011;10:1–10. doi: 10.1186/1475-2859-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restaino OF, Di Lauro I, Cimini D, Carlino E, De Rosa M, Schiraldi C. Monosaccharide precursors for boosting chondroitin-like capsular polysaccharide production. Appl Microbiol Biot. 2012;97:1699–1709. doi: 10.1007/s00253-012-4343-2. [DOI] [PubMed] [Google Scholar]
- Rice JA, Carrasco-Medina L, Hodgins DC, Shewen PE. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev. 2007;8:117–128. doi: 10.1017/S1466252307001375. [DOI] [PubMed] [Google Scholar]
- Rigg GP, Barrett B, Roberts IS. The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiol-SGM. 1998;144(Pt 10):2905–2914. doi: 10.1099/00221287-144-10-2905. [DOI] [PubMed] [Google Scholar]
- Rimler RB, Register KB, Magyar T, Ackermann MR. Influence of chondroitinase on indirect hemagglutination titers and phagocytosis of Pasteurella multocida serogroups A, D and F. Vet Microbiol. 1995;47:287–294. doi: 10.1016/0378-1135(95)00127-1. [DOI] [PubMed] [Google Scholar]
- Rimler RB, Rhoades KR. Serogroup F, a new capsule serogroup of Pasteurella multocida. J Clin Microbiol. 1987;25:615–618. doi: 10.1128/jcm.25.4.615-618.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JB, McCracken GH, Gotschlich EC, Orskov F, Orskov I, Hanson LA. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. New Engl J Med. 1974;290:1216–1220. doi: 10.1056/NEJM197405302902202. [DOI] [PubMed] [Google Scholar]
- Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Roberts IS, Mountford R, High N, Bitter-Suermann D, Jann K, Timmis K, Boulnois G. Molecular cloning and analysis of genes for production of K5, K7, K12, and K92 capsular polysaccharides in Escherichia coli. J Bacteriol. 1986;168:1228–1233. doi: 10.1128/jb.168.3.1228-1233.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez ML, Jann B, Jann K. Structure and serological characteristics of the capsular K4 antigen of Escherichia coli O5:K4:H4, a fructose-containing polysaccharide with a chondroitin backbone. Eur J Biochem. 1988;177:117–124. doi: 10.1111/j.1432-1033.1988.tb14351.x. [DOI] [PubMed] [Google Scholar]
- Rosenow C, Roberts IS, Jann K. Isolation from recombinant Escherichia coli and characterization of CMP-Kdo synthetase, involved in the expression of the capsular K5 polysaccharide (K-CKS) FEMS Microbiol Lett. 1995;125:159–164. doi: 10.1111/j.1574-6968.1995.tb07352.x. [DOI] [PubMed] [Google Scholar]
- Rosenow C, Esumeh F, Roberts IS, Jann K. Characterization and localization of the KpsE protein of Escherichia coli K5, which is involved in polysaccharide export. J Bacteriol. 1995;177:1137–1143. doi: 10.1128/jb.177.5.1137-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J Bacteriol. 2000;182:2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Watanabe M, Silver J, Troy FA, Vimr ER. Specific alteration of NCAM-mediated cell adhesion by an endoneuraminidase. J Cell Biol. 1985;101:1842–1849. doi: 10.1083/jcb.101.5.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg T, Kaijser B, Lidin-Janson G, Lincoln K, Orskov F, Orskov I, Stokland E, Svanborg-Edén C. Virulence of Escherichia coli in relation to host factors in women with symptomatic urinary tract infection. J Clin Microbiol. 1988;26:1471–1476. doi: 10.1128/jcm.26.8.1471-1476.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarff LD, McCracken GH, Schiffer MS, Glode MP, Robbins JB, Orskov I, Orskov F. Epidemiology of Escherichia coli K1 in healthy and diseased newborns. Lancet. 1975;1:1099–1104. doi: 10.1016/s0140-6736(75)92496-4. [DOI] [PubMed] [Google Scholar]
- Sawata A, Kume K. Relationships between virulence and morphological or serological properties of variants dissociated from serotype 1 Haemophilus paragallinarum strains. J Clin Microbiol. 1983;18:49–55. doi: 10.1128/jcm.18.1.49-55.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiraldi C, Cimini D, De Rosa M. Production of chondroitin sulfate and chondroitin. Appl Microbiol Biot. 2010;87:1209–1220. doi: 10.1007/s00253-010-2677-1. [DOI] [PubMed] [Google Scholar]
- Schiraldi C, Carcarino IL, Alfano A, Restaino OF, Panariello A, De Rosa M. Purification of chondroitin precursor from Escherichia coli K4 fermentation broth using membrane processing. Biotechnol J. 2011;6:410–419. doi: 10.1002/biot.201000266. [DOI] [PubMed] [Google Scholar]
- Schiraldi C, Alfano A, Cimini D, De Rosa M, Panariello A, Restaino Odile F. Application of a 22L scale membrane bioreactor and cross-flow ultrafiltration to obtain purified chondroitin. Biotechnol Progr. 2012;28:1012–1018. doi: 10.1002/btpr.1566. [DOI] [PubMed] [Google Scholar]
- Scholl D, Adhya S, Merril C. Escherichia coli K1's capsule is a barrier to bacteriophage T7. Appl Environ Microb. 2005;71:4872–4874. doi: 10.1128/AEM.71.8.4872-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Rogers S, Adhya S, Merril CR. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J Virol. 2001;75:2509–2515. doi: 10.1128/JVI.75.6.2509-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager HM, Rheinwald JG, Wessels MR. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Invest. 1996;98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, Duncker S, Barth S, Bauerfeind R, Gruber AD, Deppenmeier S, Breves G. Preventive effects of the probiotic Escherichia coli strain Nissle 1917 on acute secretory diarrhea in a pig model of intestinal infection. Digest Dis Sci. 2006;51:724–731. doi: 10.1007/s10620-006-3198-8. [DOI] [PubMed] [Google Scholar]
- Schulz EC, Bergfeld AK, Ficner R, Mühlenhoff M. Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO. PLoS ONE. 2011;6:e17403. doi: 10.1371/journal.pone.0017403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Liu J, Estiu G, Isin B, Ahn YY, Lee DS, Barabási AL, Kapatral V, Wiest O, Oltvai ZN. Blueprint for antimicrobial hit discovery targeting metabolic networks. P Natl Acad Sci USA. 2010;107:1082–1087. doi: 10.1073/pnas.0909181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuren J. Food additives permitted for direct addition to food for human consumption; bacteriophage preparation. Federal Register. 2006;71:47729–47732. [PubMed] [Google Scholar]
- Sieberth V, Rigg GP, Roberts IS, Jann K. Expression and characterization of UDPGlc dehydrogenase (KfiD), which is encoded in the type-specific region 2 of the Escherichia coli K5 capsule genes. J Bacteriol. 1995;177:4562–4565. doi: 10.1128/jb.177.15.4562-4565.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RP, Aaronson W, Vann WF. The K1 capsular polysaccharide of Escherichia coli. Rev Infect Dis. 1988;10(Suppl 2):S282–S286. doi: 10.1093/cid/10.supplement_2.s282. [DOI] [PubMed] [Google Scholar]
- Silver RP, Finn CW, Vann WF, Aaronson W, Schneerson R, Kretschmer PJ, Garon CF. Molecular cloning of the K1 capsular polysaccharide genes of E. coli. Nature. 1981;289:696–698. doi: 10.1038/289696b0. [DOI] [PubMed] [Google Scholar]
- Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DA, Hammarton TC, Roberts IS. Transcriptional organization and regulation of expression of region 1 of the Escherichia coli K5 capsule gene cluster. J Bacteriol. 1996;178:6466–6474. doi: 10.1128/jb.178.22.6466-6474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Boulnois GJ, Roberts IS. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- Spinosa MR, Progida C, Talà A, Cogli L, Alifano P, Bucci C. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun. 2007;75:3594–3603. doi: 10.1128/IAI.01945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen JA, Steen JA, Harrison P, Seemann T, Wilkie I, Harper M, Adler B, Boyce JD. Fis is essential for capsule production in Pasteurella multocida and regulates expression of other important virulence factors. PLoS Pathog. 2010;6:e1000750. doi: 10.1371/journal.ppat.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen SM, Vimr ER. Biosynthesis of the Escherichia coli K1 group 2 polysialic acid capsule occurs within a protected cytoplasmic compartment. Mol Microbiol. 2008;68:1252–1267. doi: 10.1111/j.1365-2958.2008.06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Clarke BR, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–1012. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- Stevens P, Huang SN, Welch WD, Young LS. Restricted complement activation by Escherichia coli with the K-1 capsular serotype: a possible role in pathogenicity. J Immunol. 1978;121:2174–2180. [PubMed] [Google Scholar]
- Stordeur P, China B, Charlier G, Roels S, Mainil J. Clinical signs, reproduction of attaching/effacing lesions, and enterocyte invasion after oral inoculation of an O118 enterohaemorrhagic Escherichia coli in neonatal calves. Microbes Infect. 2000;2:17–24. doi: 10.1016/S1286-4579(00)00290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm DW, Koff SA, Horvath DJ, Li B, Justice SS. In vitro analysis of the bactericidal activity of Escherichia coli Nissle 1917 against pediatric uropathogens. J Urology. 2011;186:1678–1683. doi: 10.1016/j.juro.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid–degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2004;12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Mikami T. Chondroitin/dermatan sulfate in the central nervous system. Curr Opin Struc Biol. 2007;17:536–545. doi: 10.1016/j.sbi.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG. Bacteriophage therapy. Antimicrob Agents Ch. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Gunzer F, Westendorf AM, Buer J, Scharfe M, Jarek M, Gössling F, Blöcker H, Zeng AP. Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J Biotechnol. 2005;117:147–161. doi: 10.1016/j.jbiotec.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Sun W, Du L, Li M. Aptamer-based carbohydrate recognition. Curr Pharm Design. 2010;16:2269–2278. doi: 10.2174/138161210791792877. [DOI] [PubMed] [Google Scholar]
- Surana NK, Kasper DL. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol Rev. 2012;245:13–26. doi: 10.1111/j.1600-065X.2011.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Liu Y, Ali MM, Kang DK, Zhao W, Li J. Colorimetric and ultrasensitive bioassay based on a dual-amplification system using aptamer and DNAzyme. Anal Chem. 2012;84:4711–4717. doi: 10.1021/ac203274k. [DOI] [PubMed] [Google Scholar]
- Thelin MA, Bartolini B, Axelsson J, Gustafsson R, Tykesson E, Pera E, Oldberg A, Maccarana M, Malmstrom A. Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J. 2013;280:2431–2446. doi: 10.1111/febs.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Pourhossein M, Waterhouse A, Hudson T, Goldrick M, Derrick JP, Roberts IS. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J Biol Chem. 2010;285:23963–23969. doi: 10.1074/jbc.M110.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013 doi: 10.1038/nature12234. http://dx.doi.org/10.1038/nature12234. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- Tjalsma H, Antelmann H, Jongbloed JD, et al. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol R. 2004;68:207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosens Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. 2001;39:924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilli A, Busiello I, Daly S, Bagatin F. Biotechnological production of chondroitin. IPO Patent Number 2012004063 2012
- Troy FA. Polysialylation: from bacteria to brains. Glycobiology. 1992;2:5–23. doi: 10.1093/glycob/2.1.5. [DOI] [PubMed] [Google Scholar]
- Tzeng Y, Datta AK, Strole CA, Lobritz MA, Carlson RW, Stephens DS. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect Immun. 2005;73:1491–1505. doi: 10.1128/IAI.73.3.1491-1505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, Latour-Lambert P, Ghigo JM. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. P Natl Acad Sci USA. 2006;103:12558–12563. doi: 10.1073/pnas.0605399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oss CJ, Gillman CF, Neumann AW. Phagocytic engulfment and cell adhesiveness as cellular surface phenomena. Vol. 2. Marcel Dekker; New York and Basel: 1975. [Google Scholar]
- Vann WF, Schmidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4. A polymer similar to desulfo-heparin. Eur J Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- Vann WF, Daines DA, Murkin AS, Tanner ME, Chaffin DO, Rubens CE, Vionnet J, Silver RP. The NeuC protein of Escherichia coli K1 is a UDP N-acetylglucosamine 2-epimerase. J Bacteriol. 2004;186:706–712. doi: 10.1128/JB.186.3.706-712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126:841–845. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Veasy LG, Tani LY, Daly JA, Korgenski K, Miner L, Bale J, Kaplan EL, Musser JM, Hill HR. Temporal association of the appearance of mucoid strains of Streptococcus pyogenes with a continuing high incidence of rheumatic fever in Utah. Pediatrics. 2004;113:e168–e172. doi: 10.1542/peds.113.3.e168. [DOI] [PubMed] [Google Scholar]
- Vimr ER, Steenbergen SM. Mobile contingency locus controlling Escherichia coli K1 polysialic acid capsule acetylation. Mol Microbiol. 2006;60:828–837. doi: 10.1111/j.1365-2958.2006.05158.x. [DOI] [PubMed] [Google Scholar]
- Vimr ER, Aaronson W, Silver RP. Genetic analysis of chromosomal mutations in the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1989;171:1106–1117. doi: 10.1128/jb.171.2.1106-1117.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat Chem Biol. 2011;7:519–527. doi: 10.1038/nchembio.609. [DOI] [PubMed] [Google Scholar]
- Vintiñi E, Villena J, Alvarez S, Medina M. Administration of a probiotic associated with nasal vaccination with inactivated Lactococcus lactis-PppA induces effective protection against pneumoccocal infection in young mice. Clin Exp Immunol. 2010;159:351–362. doi: 10.1111/j.1365-2249.2009.04056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotech. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- Volpi N. Mass spectrometry characterization of Escherichia coli K4 oligosaccharides from 2-mers to more than 20-mers. Rapid Commun Mass Sp. 2007;21:3459–3466. doi: 10.1002/rcm.3245. [DOI] [PubMed] [Google Scholar]
- Volpi N, Zhang Z, Linhardt RJ. Mass spectrometry for the characterization of unsulfated chondroitin oligosaccharides from 2-mers to 16-mers. Comparison with hyaluronic acid oligomers. Rapid Commun Mass Sp. 2008;22:3526–3530. doi: 10.1002/rcm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ly M, Zhang F, Zhong W, Suen A, Hickey AM, Dordick JS, Linhardt RJ. E. coli K5 fermentation and the preparation of heparosan, a bioengineered heparin precursor. Biotechnol Bioeng. 2010;107:964–973. doi: 10.1002/bit.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang B, Zhang Z, Ly M, Takieddin M, Mousa S, Liu J, Dordick JS, Linhardt RJ. Control of the heparosan N-deacetylation leads to an improved bioengineered heparin. Appl Microbiol Biot. 2011;91:91–99. doi: 10.1007/s00253-011-3231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PN, Field TR, Ditcham WG, Maguin E, Leigh JA. Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect Immun. 2001;69:392–399. doi: 10.1128/IAI.69.1.392-399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt JM, Wade MM, Holman SC, Wilson WW, Keil DE, Pruett SB, Jacques M, Champlin FR. Influence of serotype A capsulation on cell surface physiologic factors in Pasteurella multocida. Colloid Surface B. 2003;28:227–238. [Google Scholar]
- Wei Z, Fu Q, Chen Y, Cong P, Xiao S, Mo D, He Z, Liu X. The capsule of Streptococcus equi ssp. zooepidemicus is a target for attenuation in vaccine development. Vaccine. 2012;30:4670–4675. doi: 10.1016/j.vaccine.2012.04.092. [DOI] [PubMed] [Google Scholar]
- Weintraub A. Immunology of bacterial polysaccharide antigens. Carbohyd Res. 2003;338:2539–2547. doi: 10.1016/j.carres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- Wells CL, Maddaus MA, Simmons RL. Proposed mechanisms for the translocation of intestinal bacteria. Rev Infect Dis. 1988;10:958–979. doi: 10.1093/clinids/10.5.958. [DOI] [PubMed] [Google Scholar]
- Wessels MR, Rubens CE, Benedí VJ, Kasper DL. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. P Natl Acad Sci USA. 1989;86:8983–8987. doi: 10.1073/pnas.86.22.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. P Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- Wibawan IW, Pasaribu FH, Utama IH, Abdulmawjood A, Lämmler C. The role of hyaluronic acid capsular material of Streptococcus equi subsp. zooepidemicus in mediating adherence to HeLa cells and in resisting phagocytosis. Res Vet Sci. 1999;67:131–135. doi: 10.1053/rvsc.1998.0287. [DOI] [PubMed] [Google Scholar]
- Widner B, Behr R, Von Dollen S, Tang M, Heu T, Sloma A, Sternberg D, DeAngelis PL, Weigel PH, Brown S. Hyaluronic acid production in Bacillus subtilis. Appl Environ Microb. 2005;71:3747–3752. doi: 10.1128/AEM.71.7.3747-3752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildi LM, Raynauld JP, Martel-Pelletier J, Beaulieu A, Bessette L, Morin F, Abram F, Dorais M, Pelletier JP. Chondroitin sulphate reduces both cartilage volume loss and bone marrow lesions in knee osteoarthritis patients starting as early as 6 months after initiation of therapy: a randomised, double-blind, placebo-controlled pilot study using MRI. Ann Rheum Dis. 2011;70:982–989. doi: 10.1136/ard.2010.140848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis LM, Stupak J, Richards MR, Lowary TL, Li J, Whitfield C. Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. P Natl Acad Sci USA. 2013;110:7868–7873. doi: 10.1073/pnas.1222317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelstein JA. The role of complement in the host's defense against Streptococcus pneumoniae. Reviews of infectious diseases. 1981;3:289–298. doi: 10.1093/clinids/3.2.289. [DOI] [PubMed] [Google Scholar]
- Wu JR, Chen PY, Shien JH, Shyu CL, Shieh HK, Chang F, Chang PC. Analysis of the biosynthesis genes and chemical components of the capsule of Avibacterium paragallinarum. Vet Microbiol. 2010;145:90–99. doi: 10.1016/j.vetmic.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Wu Q, Yang A, Zou W, Duan Z, Liu J, Chen J, Liu L. Transcriptional engineering of Escherichia coli K4 for fructosylated chondroitin production. Biotechnol Progr. 2013 doi: 10.1002/btpr.1777. http://dx.doi.org/10.1002/btpr.1777. [Epub ahead of print] [DOI] [PubMed]
- Wunder DE, Aaronson W, Hayes SF, Bliss JM, Silver RP. Nucleotide sequence and mutational analysis of the gene encoding KpsD, a periplasmic protein involved in transport of polysialic acid in Escherichia coli K1. J Bacteriol. 1994;176:4025–4033. doi: 10.1128/jb.176.13.4025-4033.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue P, Corbett D, Goldrick M, Naylor C, Roberts IS. Regulation of expression of the region 3 promoter of the Escherichia coli K5 capsule gene cluster involves H-NS, SlyA, and a large 5′ untranslated region. J Bacteriol. 2009;191:1838–1846. doi: 10.1128/JB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Zhu L, Guo H, Mei Li, Li J, Wang PG. Formation of a new O-polysaccharide in Escherichia coli O86 via disruption of a glycosyltransferase gene involved in O-unit assembly. Carbohyd Res. 2006;341:2254–2260. doi: 10.1016/j.carres.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Yu H, Stephanopoulos G. Metabolic engineering of Escherichia coli for biosynthesis of hyaluronic acid. Metab Eng. 2008;10:24–32. doi: 10.1016/j.ymben.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Yu H, Tyo K, Alper H, Klein-Marcuschamer D, Stephanopoulos G. A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnol Bioeng. 2008;101:788–796. doi: 10.1002/bit.21947. [DOI] [PubMed] [Google Scholar]
- Zanfardino A, Restaino OF, Notomista E, Cimini D, Schiraldi C, De Rosa M, De Felice M, Varcamonti M. Isolation of an Escherichia coli K4 kfoC mutant over-producing capsular chondroitin. Microb Cell Fact. 2010;9:1–8. doi: 10.1186/1475-2859-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder JL, Galli SJ. Mast-cell heparin demystified. Nature. 1999;400:714–715. doi: 10.1038/23360. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu L, Teng L, Chen J, Liu J, Li J, Du G, Chen J. Metabolic engineering of Escherichia coli BL21 for biosynthesis of heparosan, a bioengineered heparin precursor. Metab Eng. 2012;14:521–527. doi: 10.1016/j.ymben.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Sun Y, Zhang X, Kong Y, Xie Z, Zhu Y, Zhou E, Jiang S. Evaluation of two experimental infection models for Avibacterium paragallinarum. Vet Microbiol. 2010;141:68–72. doi: 10.1016/j.vetmic.2009.07.034. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang Q, Wen L, Xu J, Shao Z, Chen M, Chen M, Reeves PR, Cao B, Wang L. Development of a multiplex PCR assay for detection and genogrouping of Neisseria meningitidis. J Clin Microbiol. 2012;50:46–51. doi: 10.1128/JCM.00918-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler G, Schmidt G, Orskov I, Orskov F, Falkenhagen U, Naumann G. K-antigen identification, hemolysin production, and hemagglutination types of Escherichia coli O6 strains isolated from patients with urinary tract infections. Zbl Bakt-Int J Med M. 1990;274:372–381. doi: 10.1016/s0934-8840(11)80695-x. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]