Abstract
Our long-term goal is the design of a human L-asparaginase (hASNase3) variant, suitable for use in cancer therapy without the immunogenicity problems associated with the currently used bacterial enzymes. Asparaginases catalyze the hydrolysis of the amino acid asparagine to aspartate and ammonia. The key property allowing for the depletion of blood asparagine by bacterial asparaginases is their low micromolar KM value. In contrast, human enzymes have a millimolar KM for asparagine. Towards the goal of engineering an hASNase3 variant with micromolar KM, we conducted a structure/function analysis of the conserved catalytic threonine triad of this human enzyme. As a member of the N-terminal nucleophile (Ntn) family, to become enzymatically active, hASNase3 must undergo autocleavage between residues Gly167 and Thr168. To determine the individual contribution of each of the three conserved active site threonines (threonine triad, Thr168, Thr186, Thr219) for the enzyme-activating autocleavage and asparaginase reactions, we prepared the T168S, T186V and T219A/V mutants. These mutants were tested for their ability to cleave and to catalyze asparagine hydrolysis, in addition to being examined structurally. We also elucidated the first Ntn plant-type asparaginase structure in the covalent intermediate state. Our studies indicate that while not all the triad threonines are required for the cleavage reaction, all are essential for the asparaginase activity. The increased understanding of hASNase3 function resulting from these studies reveals the key regions that govern cleavage and the asparaginase reaction, which may inform the design of variants that attain a low KM for asparagine.
Keywords: Asparaginase, Enzyme mechanism, Mutagenesis, X-ray crystallography, Cancer biology
INTRODUCTION
L-Asparaginases catalyze the hydrolysis of the amino acid L-asparagine (Asn) to L-aspartate (Asp) and ammonia. Enzymes with asparaginase activity are classified based on sequence and structural homology into two families: bacterial, also called type I and type II, and plant-type asparaginases, also called type III [1-3]. For several decades, the bacterial type II enzymes have been studied extensively because of their utility in treating acute lymphoblastic leukemia (ALL) [4-8]. Unlike most normal cells, ALL cells are unable to synthetize the non-essential amino acid Asn due to loss of asparagine synthetase expression [9]. Consequently, their survival becomes highly dependent on the uptake of Asn from blood. In the treatment of ALL with asparaginases, the hydrolysis of blood Asn deprives the leukemic cells of this amino acid, which ultimately leads to cell death. Unfortunately, the clinically used bacterial asparaginases often trigger an immune response leading to enzyme inactivation and various immunogenic side effects. To overcome this immunogenicity problem, we propose the use of human asparaginases instead of bacterial enzymes.
The human genome encodes at least three enzymes capable of hydrolyzing Asn. The first, belonging to the bacterial-type family, is called 60 kDa lysophospholipase due to its ability to hydrolyze lysophospholipids in addition to Asn [10]. The asparaginase activity is present in its N-terminal domain that is homologous to the Escherichia coli type I and II asparaginases. A second – and probably the most-studied human enzyme with asparaginase activity – is lysosomal aspartylglucosaminidase (AGA), a plant-type enzyme whose main function is to remove carbohydrate groups linked to Asn [11, 12]. Finally, the third enzyme – and the focus of this study – is the human L-asparaginase [13, 14]. Like AGA, it is also a plant-type asparaginase, which we refer to as hASNase3 due to its high structural homology with the E. coli type III asparaginase.
Plant-type asparaginases such as hASNase3 are members of the N-terminal nucleophile (Ntn) hydrolase family [15, 16]. Enzymes in this family are synthesized as a single polypeptide precursor, devoid of enzymatic activity. These enzymes must undergo autoproteolytic cleavage to form two subunits, α and β, to attain an enzymatically active state. Previous mechanistic studies on Ntn enzymes were conducted on the human [17] and bacterial versions of AGA [18], with some studies on the E. coli ASNase3 [19]. These studies allowed the authors to propose a mechanism where three active site threonine residues participate in substrate hydrolysis [17]. A novelty of this proposed mechanism is the role played by the β-subunit's N-terminal amino group of the catalytic threonine. The amino group of this threonine functions to activate its hydroxyl group, thus rationalizing the need for Ntn enzymes to undergo cleavage in order to attain enzymatic activity (prior to cleavage, this amino group is involved in a peptide bond with the preceding residue).
Surprisingly, hASNase3 purifies uncleaved (i.e. enzymatically inactive), indicating a very slow autocleavage rate [20, 21]. In contrast, AGA and other studied bacterial ASNase3 enzymes are purified as fully cleaved enzymes (i.e. enzymatically active). For the Flavobacterium glycosylasparaginase, the aspartic acid preceding the scissile bond was proposed to act as the base activating the following threonine to initiate cleavage [22]. In contrast, a glycine precedes the analogues threonine in hASNase3, indicating that a different base must be compensating. Previously, we reported the ability of the free amino acid glycine to promote the cleavage reaction of hASNase3 [21], which occurs between Gly167 (becoming the end of the α-subunit) and Thr168 (becoming the beginning of the β-subunit). This finding is consistent with a conclusion voiced by Michalska et al [19] that the mechanism of autocleavage is likely to be different between different classes of Ntn enzymes. Two additional active site threonine residues, Thr186 and Thr219, are conserved within this enzyme family and together build a catalytic triad. Mutagenesis of analogous threonines have been reported for human AGA [8, 17], but the fact that autocleavage remained efficient for many of the mutants suggest fundamental differences to hASNase3. Therefore, while the basic hydrolytic mechanism of Ntn enzymes has been largely elucidated (and would presumably apply to hASNase3, but would need to be confirmed), questions remain regarding the role of the catalytic triad to the essential autocleavage reaction.
As previously discussed, replacing the therapeutically used bacterial asparaginases with a less-immunogenic human enzyme would have clinical advantages. A prerequisite for the clinical use of asparaginases is a low micromolar KM value for the substrate Asn (serum Asn concentration ~50 μM [23]). The wild type version of hASNase3 has a KM value in the millimolar range, making it unsuitable as a replacement for the bacterial asparaginases. To inform the design of hASNase3 variants with low Asn KM, and for engineering variants that autocleave efficiently without the need for glycine activation, we initiated a structure/function study of this enzyme. In this work we focused on the threonine catalytic triad, dissecting the role of each threonine for both the enzyme-activating cleavage reaction and for the asparaginase reaction. We also report, to our knowledge for the first time, an Ntn plant-type asparaginase structure in the covalent intermediate state – a state after nucleophilic attack on the substrate Asn side chain and before release of the product Asp. Together, these structural and biochemical experiments reveal important details on critical residues involved in the hASNase3 cleavage self-activation and catalytic activity, and suggest an important functional role for the highly conserved N-terminal segment situated next to the active site (His8-Gly9-Gly10), which we refer to as the HGG loop.
RESULTS AND DISCUSSION
Individual role of threonines 168, 186 and 219 for hASNase3 autocleavage
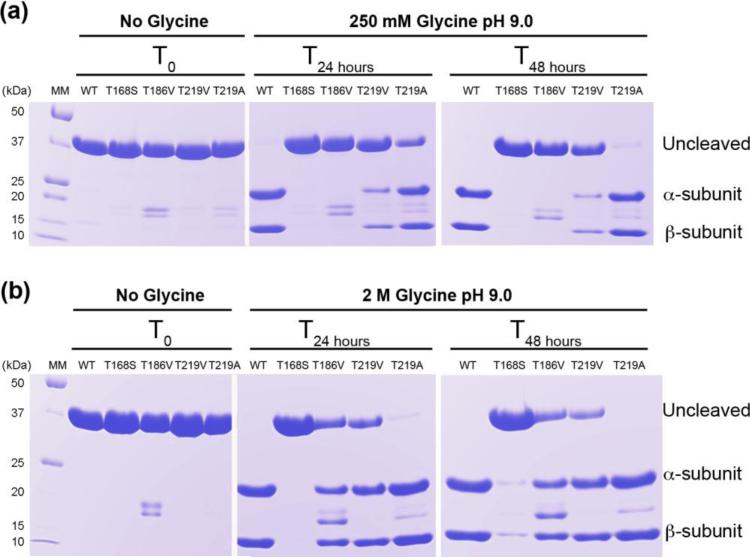
We recently reported that the free amino acid glycine selectively acts to promote the autocleavage reaction of hASNase3 in a temperature, time and concentration-dependent manner [21]. We took advantage of this discovery to evaluate the capacity of four hASNase3 mutants to undergo the autocleavage reaction in order to identify which of the active site threonines is critical for enzyme activation. Wild type (WT) hASNase3 is expressed and purified as a single polypeptide (uncleaved inactive form) that migrates as a single band of ~37 kDa on SDS-PAGE denaturing gel (Figure 1a and b). A 24-hour incubation at room temperature in 250 mM glycine solution pH 9.0 is sufficient to promote full cleavage of the WT enzyme (active form). In the context of the cleaved enzyme, the two lower molecular weight polypeptides at ~20 and ~12 kDa are referred to as the α- and β-subunits, respectively (Figure 1). To probe the role of the three conserved threonines (168, 186 and 219 according to hASNase3 numbering) regarding self-activation, we generated the following mutations: T168S, T186V, T219A and T219V (the rationale for each mutation is discussed below).
FIGURE 1.
Glycine activation of WT hASNase3 and catalytic triad threonine mutants. (a) WT hASNase3 and mutants were incubated for 2 days at 25 °C in storage buffer containing 250 mM glycine pH 9.0. Enzyme samples prior to incubation are labeled under No Glycine and T0 and migrate as a ~37 kDa band on the gel, corresponding to the uncleaved (inactive) form of the enzyme. After 24 h incubation, the WT hASNase3 migrates as two lower molecular bands corresponding to the α- and β-subunits and is fully activated. The T219A mutant is partially and almost fully activated after 24 h and 48h incubation, respectively, while the T219V mutant is only partially cleaved after 48 h. The T168S and T186V mutants remain in their uncleaved inactive form. (b) Same as (a) but enzymes were incubated in a 2.0 M glycine pH 9.0 solution. In this condition, the WT hASNase3 is fully cleaved after 24 h incubation. The T219A mutant shows a faster cleavage rate and is almost fully activated after 24 h incubation. The T168S mutant remains essentially uncleaved while the T186V and T219V mutants are partially cleaved.
All mutants expressed comparably to the WT enzyme and the purified mutant enzymes exhibited the expected single band at ~37 kDa on a SDS-PAGE gel corresponding to the uncleaved state (Figure 1). After a 250 mM glycine treatment, the T168S and T186V mutants showed no cleavage even after 48 hours incubation (Figure 1a). In contrast, the T219V mutant displayed partial cleavage (bands at ~20 and ~12 kDa) while a complete, albeit slow cleavage was observed for the T219A mutant after 48 hours (note that the additional faint bands at ~16 and 18 kDa are interpreted as contaminants, based on the fact that they do not increase in intensity as cleavage progresses).
As glycine can accelerate the autocleavage reaction in a concentration dependent manner, we increased the amount of glycine during incubation from 250 mM to 2.0 M in order to confirm that the Thr168 and Thr186 residues are indeed essential for the cleavage reaction. Despite the presence of 2.0 M glycine, the T168S mutant remained predominantly uncleaved, showcasing the essential role of Thr168 for the cleavage reaction (Figure 1b). In contrast, the increase in glycine concentration promoted partial cleavage of the T186V mutant. This result suggests an important but not essential role for Thr186 in the cleavage step. Recapitulating the observations made in the 250 mM glycine experiments, the Thr219 mutants behaved somewhat differently to each other, with the T219V mutant still remaining only partially cleaved even after 48 hours, despite the presence of 2.0 M glycine. In contrast, the T219A mutant was fully cleaved already after 24 hours. This supports the conclusion that Thr219 is not essential for the cleavage reaction, but the different behavior between the T219V and T219A mutants suggests a cleavage-inhibiting effect in the case of valine at that position.
Based on these results, we rank the members of the threonine triad by decreasing importance for the cleavage self-activating reaction as follows: Thr168>>Thr186>Thr219. In order to better understand the role and mechanism of these threonines during the process of self-activation, we solved the three dimensional structures of each mutant. Additionally, we solved a high-resolution wild type hASNase3 structure that was used for comparison with the mutant structures.
High resolution wild type hASNase3 structure
To better understand structural changes between WT and mutant enzymes, in this report we make use of a new 1.5 Å resolution structure of uncleaved WT hASNase3. Data collection and refinement statistics are listed in Table 1. In previous work [20, 21] we were able to elucidate several hASNase3 structures of cleaved and/or uncleaved enzyme in the apo state or in complex with product and/or glycine. However, the highest resolution structure obtained prior to this work was 1.75 Å (PDB ID: 4GDV [20]) corresponding to the partially cleaved hASNase3 in complex with aspartate. It has to be mentioned that despite extending the resolution to 1.5 Å for the new WT uncleaved hASNase3 structure reported here, we still could not observe traceable electron density for the region spanning residues 153 to 165, which appears to be too flexible even in the uncleaved state (in other words, despite being uncleaved, only Leu166 and Gly167 could be modeled prior to the would-be cleavage site with Thr168). We use this high-resolution WT hASNase3 structure for all the comparisons with the threonine mutant structures discussed below.
Table 1.
Data collection and refinement statistics
| Mutant | WT-uncleaved | T168S | T186V | T219A | T219V | Covalent- intermediate |
|---|---|---|---|---|---|---|
| PDB codes | 4O0C | 4O0D | 4O0E | 4O0F | 4O0G | 4O0H |
| Data collection statistics | ||||||
| X-ray source and detector | LS-CAT ID-G | SERCAT ID-22 | LS-CAT ID-D | Rigaku RU-200 | LS-CAT ID-D | Rigaku RU-200 |
| MARCCD 300 | MARCCD 300 | MARCCD 300 | R-AXIS IV ++ | MARCCD 300 | R-AXIS IV ++ | |
| Wavelength (A) | 0.97856 | 1.00000 | 1.07820 | 1.5418 | 1.07820 | 1.5418 |
| Temperature (K) | 100 | 100 | 100 | 93 | 100 | 93 |
| Resolutiona, b (A) | 1.50 (a30-4.46; b1.59- 1.50) | 1.95 (a30-8.70; b2.00- 1.95) | 1.71 (a30-5.06;b1.81- 1.71) | 1.92 (a30-5.67; b2.04- 1.92) | 2.10 (a30-6.16; b2.22- 2.10) | 1.97 (a30-5.82; b2.09- 1.97) |
| Number of Reflections | ||||||
| Observed | 752752 | 214997 | 330581 | 126398 | 125411 | 129948 |
| Unique | 92426 | 36456 | 62537 | 43516 | 34954 | 38615 |
| Completeness (%) | 99.0 (b94.3) | 82.6 (b71.4) | 98.1 (b93.1) | 96.3 (b87.6) | 97.9 (b96.0) | 91.9 (b80.5) |
| Rsym (%) | 9.2 (a4.2; b34.8) | 10.6 (a6.5; b52.3) | 6.2 (a3.4; b55.9) | 6.0 (a3.0; b37.5) | 9.8 (a3.2; b55.9) | 9.7 (a5.0; b43.5) |
| AverageI/σ(I) | 13.2 (b2.86) | 11.7 (b3.18) | 14.41 (b1.65) | 11.78 (b1.86) | 11.54 (b2.79) | 9.16 (b2.05) |
| Space group | P65 | P65 | P65 | P65 | P65 | P65 |
| Unit cell (A): a=b, c | 59.2, 297.8 | 59.7, 299.7 | 59.3, 298.3 | 59.4, 299.2 | 59.4, 306.8 | 59.6, 298.5 |
| Refinement statistics | ||||||
| Refinement program | REFMAC5 | REFMAC5 | REFMAC5 | REFMAC5 | REFMAC5 | REFMAC5 |
| Twinning fraction | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Rcryst (%) | 15.4 (b21.0) | 17.8 (b22.0) | 17.5 (b18.8) | 16.7 (b19.4) | 16.8 (b20.1) | 16.8 (b17.8) |
| Rfee (%) | 20.5 (b30.5) | 23.4 (b30.9) | 22.5 (b30.8) | 21.5 (b28.1) | 23.6 (b34.1) | 22.6 (b24.0) |
| Resolution range (A) | 30-1.5 | 30-1.95 | 30-1.71 | 30-1.92 | 30-2.10 | 30-1.97 |
| Protein molecules per a.u. | 2 | 2 | 2 | 2 | 2 | 2 |
| Number of atoms: | ||||||
| Protein (protA, protB) | 2146, 2121 | 2185, 2175 | 2168, 2169 | 2164, 2140 | 2120, 2122 | 2173, 2173 |
| Water molecules | 339 | 216 | 177 | 155 | 152 | 193 |
| Na+ | 5 | 2 | 2 | 4 | 2 | 2 |
| Cl- | 2 | - | - | - | - | - |
| Glycine | - | 4 x 5 | - | 4 x 5 | - | - |
| DMSO | 2 x 4 | - | - | - | - | - |
| R.m.s. deviation from ideal: | ||||||
| Bond length (A) | 0.012 | 0.012 | 0.011 | 0.012 | 0.014 | 0.012 |
| Bond angles (°) | 1.559 | 1.591 | 1.603 | 1.741 | 1.766 | 1.748 |
| Average B-factors (A2) | ||||||
| Protein (protA, protB) | 16.1, 16.0 | 19.8, 19.8 | 26.7, 26.6 | 26.6, 26.8 | 28.7, 29.2 | 25.5, 25.6 |
| Water molecules | 20.5 | 19.7 | 26.7 | 23.9 | 25.7 | 23.9 |
| Na+ | 18.2 | 14.4 | 27.9 | 26.1 | 27.9 | 27.7 |
| Cl- | 25.6 | - | - | - | - | - |
| Glycine | - | 23.5 | - | 28.6 | - | - |
| DMSO | 23.7 | - | - | - | - | - |
| Ramachandran plot (%): | ||||||
| most favored regions | 90.8 | 89.4 | 88.7 | 89.3 | 88.6 | 89.7 |
| additionally allowed regions | 8.8 | 10.0 | 10.7 | 9.9 | 11.0 | 9.7 |
| generously allowed/disallowed regions | 0.4 | 0.6 | 0.6 | 0.8 | 0.4 | 0.6 |
Low and
High (last) resolution shells in parentheses
Comparison of WT and T168S hASNase3: role of Thr168
A recent analysis of serine and cysteine proteases clarifies that these catalytic residues cannot be replaced by threonine due to the steric constraints imposed by the threonine γ-methyl group [24]. However, these constraints are eliminated in Ntn enzymes, explaining the compatibility of threonine (though Ntn enzymes also use serine and cysteine). In the case of the proteasome β-subunit, the first member of “Thr proteases”, replacing the threonine by a serine reduces the reaction rate [25]. This was explained by the γ-methyl group of the threonine favoring the reactive threonine rotamer [24]. Likewise, for the Flavobacterium glycosylasparaginase, an enzyme very similar to hASNase3, mutation of the catalytic threonine to either cysteine or serine resulted in reduced but measurable enzymatic activity [26]. Different to our observation with hASNase3, where replacing the catalytic threonine by a serine largely abolishes autoproteolysis (Figure 1), the analogous mutation in the bacterial enzyme only reduces autoproteolysis by about 50% [26]. To understand the importance of threonine at position 168 for intramolecular cleavage of hASNase3 we determined the structure of the T168S variant at 1.95 Å (Table 1). The T168S mutant as well as all three other variants discussed here crystallized at the same condition as the WT enzyme, i.e. 2.2-2.5 M sodium malonate pH 7.0. Moreover, crystals adopted the same space group and unit cell dimensions, contained two molecules (homodimer) in the asymmetric unit and also presented the same twinning characteristics. As such, we can exclude crystallization conditions or crystal contact artifacts as the cause of any observed differences between the structures.
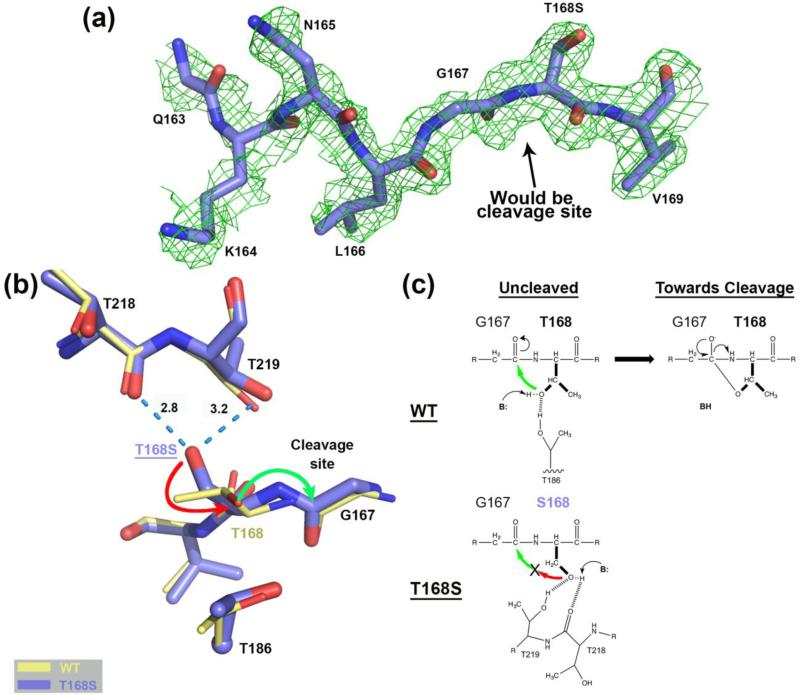
The rationale behind mutating Thr168 to serine instead of the isosteric valine is that lack of a hydroxyl group would by necessity prevent the cleavage reaction. Due to the hydroxyl functionality in serine, one would have predicted that this residue would be able to substitute for threonine at this position. Yet, as revealed by the autoproteolysis experiments discussed earlier, the T168S mutant remains essentially uncleaved, even in the presence of glycine. In fact, in the structure of the T168S mutant we could model more residues N-terminal to what would have been the cleavage site than in any other of our previous uncleaved hASNase3 structures (Figure 2a).
FIGURE 2.
Comparison of the hASNase3 T168S mutant to the WT uncleaved high-resolution structure. (a) Simulated annealing omit map (2Fo-Fc) from CNS contoured at 1σ around residues Gln163 to Val169 of the T168S structure Protomer A. (b) WT hASNase3 uncleaved structure (thin yellow sticks) and the T168S mutant (thick blue sticks) are superimposed and a close-up of the cleavage site is presented. Serine at position 168 interacts with the carbonyl of Thr218 and the side chain of Thr219 (dashed blue lines with distances in Angstroms). The side chain of Ser168 would have to rotate 180° (red arrow) to be able to proceed to nucleophilic attack on Gly167 carbonyl (green arrow) and break the peptide bond. Thr186 side chain is also represented with its α-carbon displayed as a sphere. (c) On top, proposed scheme showing the WT hASNase3 nucleophilic attack from Thr168 side chain on the Gly167 carbonyl with B: representing the base responsible for additional Thr168 activation. At the bottom, the same reaction within the T168S mutant is hampered by Ser168 side chain being maintained by interactions with threonines 218 and 219 and thus oriented in a catalytically non-competent conformation.
The WT and T168S hASNase3 structures were superimposed (root mean square deviation (rmsd) of 0.32 Å for 540 Cαatoms) and a close-up of their active site is represented in Figure 2b. The major difference occurs in the orientation of the side chain of the residue at position 168. In the WT structure, the Thr168 hydroxyl shares a hydrogen atom with the hydroxyl of Thr186 (Figure 2b and c; see also Figure 3). This interaction acts to orient the Thr168 hydroxyl group in the direction of Gly167. This interaction is crucial for the first step in the peptide-bond cleavage reaction, as once activated by a base, Thr168 can then initiate a nucleophilic attack on the Gly167 carbonyl moiety (green arrow, Figure 2b). In contrast, in the T168S structure, the hydroxyl of Ser168 is orientated in an opposite direction, being at hydrogen bond distance to the Thr218 carbonyl group and the hydroxyl of Thr219. These interactions strongly stabilize the Ser168 side chain away from Gly167. Hence the lack of a γ-methyl group in serine allows for the stabilization of an inactive rotamer, resulting in this variant being essentially unable to cleave itself (Figure 2b and c).
FIGURE 3.
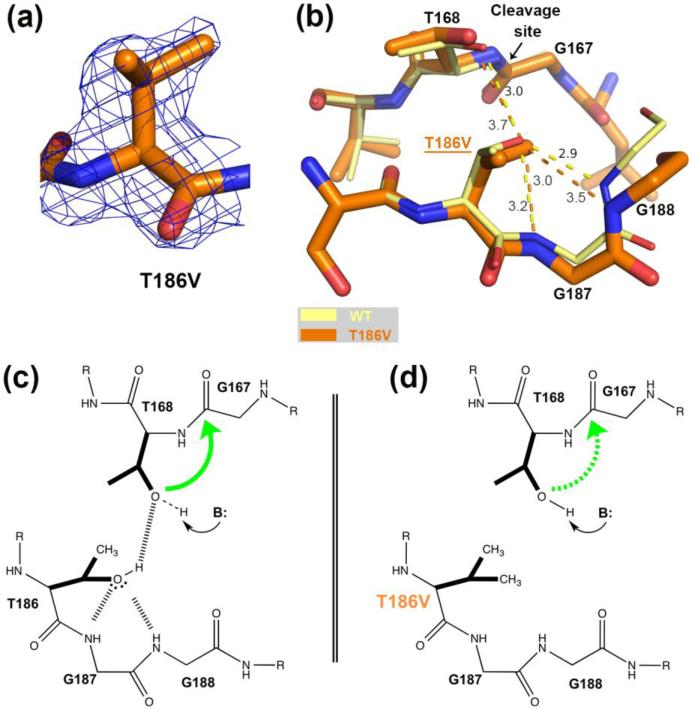
Comparison of the hASNase3 T186V mutant to the WT uncleaved high-resolution structure. (a) Fo-Fc omit map contoured at 2.2σ around valine at position 186. Residue 186 from Protomer A was removed from the model that then underwent several rounds of refinement to eliminate model bias. (b) WT hASNase3 uncleaved structure (thin yellow sticks) and the T186V mutant (thick orange sticks) are superimposed and a close-up of the cleavage site is presented. In the WT enzyme, the hydroxyl group of Thr186 hydroxyl is oriented by the main chain NH atoms of highly conserved glycines 187 and 188 (3.0 Å and 2.9 Å respectively) and by Thr168 (3.0 Å). Dashed lines represent hydrogen bonding interactions with distances given in Angstroms. When the isosteric valine replaces the threonine at position 186, the side chain maintains the same orientation but the distance to Thr168 increases to 3.7 Å in order to avoid steric clashes from their respective side chains. (c) Scheme proposing a cleavage mechanism for WT hASNase3 in which Thr186 builds a hydrogen bond donor interaction to the Thr168 hydroxyl and hydrogen bond acceptor interactions to the Gly187/188 NH atoms. As a result of accepting a proton from Thr186, the Thr168 hydroxyl becomes more likely to be activated by a base (B:). The green arrow represents the nucleophilic attack from Thr168 hydroxyl on Gly167 carbonyl. (d) In the T186V mutant, valine at position 186 is unable to participate in hydrogen bond interactions. As a result, Thr168 activation is more difficult to attain (dashed green arrow).
Comparison of WT and T186V hASNase3: role of Thr186
Incubation of the T186V mutant with glycine showed that this mutant retains the ability to cleave, albeit at a slow rate. Moreover, to compensate for the lack of the Thr186 hydroxyl group, the cleavage is dependent on a high amount of base (i.e. glycine) to activate Thr168. This indicates that while Thr186 is not crucial for activation it is still very important. We determined the structure of the T186V mutant to better understand the role of this residue (Figure 3a).
The T186V hASNase3 structure was solved in the uncleaved state at a resolution of 1.71 Å (Table 1) and the overlay with the WT structure (rmsd of 0.25 Å for 482 Cαatoms) reveals nearly identical structures (Figure 3b). Based on these structures, and as previously noted for homologous bacterial enzymes [3, 27], in Figure 3c we propose a cleavage mechanism for the WT situation in which the Thr186 hydroxyl accepts two hydrogen bonds from the conserved Gly187/Gly188 loop (3.0 Å and 2.9 Å, respectively). As a result of these interactions, the Thr186 hydroxyl is oriented to act as a hydrogen bond donor for Thr168. The formation of the hydrogen bond between Thr186 (as donor) and Thr168 (as acceptor) is expected to make the hydroxyl's proton of Thr168 more labile; by Thr168 accepting a hydrogen bond from Thr186, the electron density on the oxygen atom of Thr168 is reduced and this will lower the pKa for this hydroxyl group. In other words, the anion is stabilized relative to the hydroxyl state and the anion is more nucleophilic. Being now further activated by a base (e.g. glycine, as proposed by Su et al. [21]), the Thr168 Oγ atom can proceed to attack the Gly167 carbonyl, thus initiating enzyme cleavage (green arrow, Figure 3c).
As noted above, the function of the Gly187/Gly188 NH groups is to orient the hydroxyl of Thr186. Despite lacking a hydroxyl group, this orientation is maintained for the valine side chain in the T186V mutant (3.2 Å and 3.5 Å away from the Gly187/Gly188 NH groups, respectively). However, the distance of the valine to the hydroxyl of Thr168 increases to 3.7 Å from 3.0 Å in the case of a threonine at position 186 (Figure 3b and d). This difference in distance is due to adjustment in position of both Thr168 and Val186, which is done to minimize the unfavorable close interaction between their respective side chains. In conclusion, the T186V mutant structure supports the proposed role for the hydroxyl of Thr186 as an activator of the Thr168 hydroxyl group.
Comparison of WT and T219A/V hASNase3: role of Thr219
The hASNase3 glycine incubation experiments have revealed that the T219A variant can be relatively easily activated compared to the other threonine mutants examined, but at a slower rate than the WT enzyme (Figure 1). Aware of the fact that modifying an amino acid with a smaller one could mislead interpretation of the role of Thr219, we also replaced the threonine with its isosteric amino acid valine to generate the T219V variant. Indeed, as discussed previously, we observed a different behavior with this mutant, where it is barely cleaved even after a 48-hour incubation in a solution of 2.0 M glycine pH 9.0 (Figure 1).
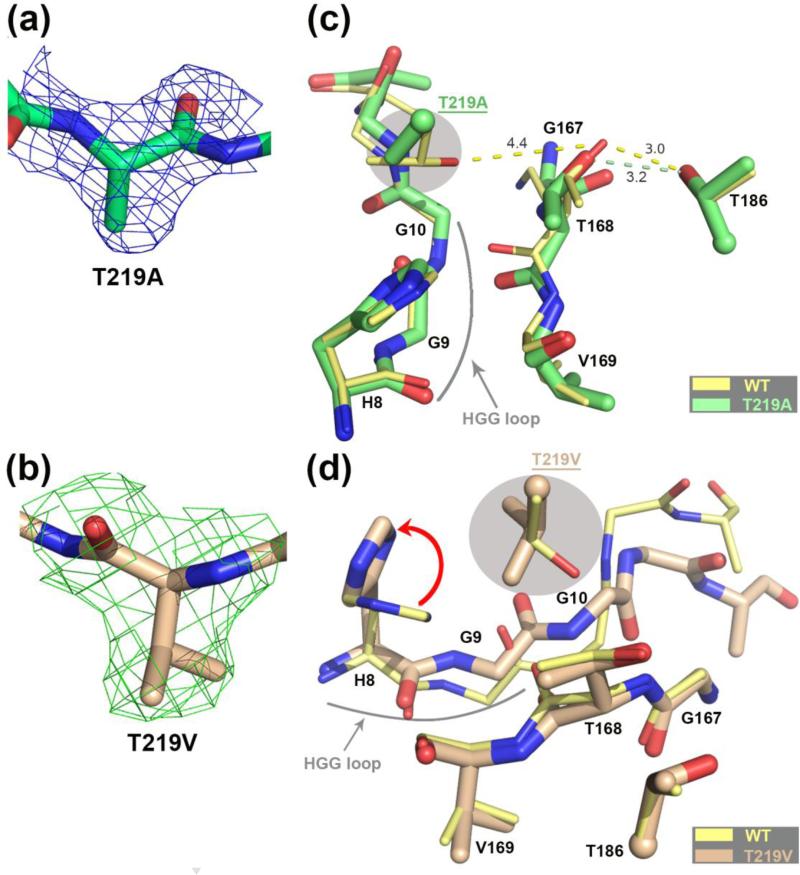
To attain more insight into the function of Thr219, we solved the structures of the hASNase3 T219A and T219V mutants in their uncleaved state at a resolution of 1.95 Å and 2.1 Å, respectively (Table 1 and Figure 4a and b). Both structures were aligned and compared to the uncleaved WT hASNase3 structure (T219A/WT: rmsd of 0.30 Å for 531 Cα atoms and T219V/WT: rmsd of 0.32 Å for 511 Cα atoms). The low rmsd values demonstrate that the overall structure of the mutants is mostly identical to that of the wild type enzyme. A close-up of the area around Thr219 is presented in Figure 4 c and d. Interestingly, even when focusing on the mutation site, we could not detect any notable structural difference between the WT enzyme and the T219A variant (Figure 4c), which is consistent with the observation that the cleavage rate of this mutant is only slightly slower than the WT enzyme (Figure 1).
FIGURE 4.
Comparison of the hASNase3 T219A/V mutants to the WT uncleaved high-resolution structure. (a) Fo-Fc omit map contoured at 2.2σ around alanine at position 219. Residue 219 from Protomer A was removed from the model that then underwent several rounds of refinement to eliminate model bias. (b) Fo-Fc omit map contoured at 2.2σ around valine at position 219. Residue 219 from Protomer A was removed from the model that then underwent several rounds of refinement to eliminate model bias. (c) WT hASNase3 uncleaved structure (thin yellow sticks) and the T219A mutant (thick green sticks) are superimposed and a close-up around the cleavage region is presented. Side chains of residues 186 and 219 (highlighted in grey) are presented with the α-carbon displayed as a sphere. Distances between residue 168 and residues 186 and 219 are in Angstroms and represented in dashed lines. The highly conserved HGG loop is also represented. (d) WT hASNase3 uncleaved structure (thin yellow sticks) and the T219V mutant (thick tan sticks) are superimposed and a close-up around the cleavage region is presented. The valine side chain at position 219 (highlighted in grey) rotates ~135° compared to the original threonine, forcing the side chain of His8 to rotate 90° to avoid steric clashes (red arrow). In this conformation the Gly9 carbonyl also has to rotate ~60° facing the opposite side of Thr168 resulting in a change in conformation of the residues following Gly9 (see also Figure 5d).
In contrast, at the mutation site, the T219V mutant displays structural changes explaining why cleavage is more adversely affected by this mutation (Figure 4d). As noticed with T219A, it seems that the residue at position 219 doesn't directly affect the positioning or orientation of the Thr168 side chain. However, it appears to be influencing the movement of the neighboring glycine-rich HGG loop instead. The HGG loop is a conserved sequence at the N-terminal region of the enzyme that changes conformation between the cleaved and uncleaved states. Our study was one of the first to report a change in the Gly9 carbonyl orientation when the enzyme is cleaved: in the uncleaved state the Gly9 carbonyl interacts with the side chain of His8, whereas in the cleaved state the carbonyl of Gly9 flips to the opposite side [20, 28]. In fact, we exploit this structural feature to distinguish the cleaved from uncleaved state.
In the T219V mutant structure we observe, for the first time in an hASNase3 structure, a reorientation of His8 (Figure 4d, red arrow). Moreover, the valine introduced at position 219 adopts a different orientation (~135° rotation) compared to the position of the original threonine. In this conformation, Val219 faces the His8 imidazole group, which compensates by rotating to avoid a steric clash. In the T219V variant structure, we observe a not-seen before conformation of Gly9 whose carbonyl rotates ~60° in the opposite direction of Thr168 compared to uncleaved hASNase3 WT. In such a conformation and because of the geometrical restraints, the following glycines 10 and 11 have to adopt a conformation not seen previously. The interplay between the HGG-loop conformation and the nature of the residue at position 219 reveals that it is not a coincidence that the HGG motif is strictly conserved among the plant-type asparaginases, and the T219V structure adds to the mounting evidence indicating an indirect role for the HGG loop in the cleavage reaction.
Implication of threonines 168, 186 and 219 for enzymatic activity of hASNase3
We evaluated each member of the threonine triad for its role in the asparaginase activity (i.e. Asn hydrolysis). In addition, for WT hASNase3 we also determined the isoaspartyl aminopeptidase activity using β-L-Asp-LPhe as substrate. Since uncleaved enzymes are by default devoid of enzymatic activity, we first activated the wild type and mutant enzymes by incubating with glycine (see Materials and Methods for more details). The kinetic parameters measured for wild type hASNase3 are presented in Table 2A. Our measured KM values are in the range of those determined previously by Cantor et al [16]. However, we observe a slightly slower substrate hydrolysis rate that could be due to the different enzymatic assay used or the method used to approximate the active (i.e. cleaved) enzyme concentration.
Table 2.
Kinetic characterization of WT hASNase3
| A. Kinetic parameters of WT hASNase3a | |||||
|---|---|---|---|---|---|
| Asn | P-L-Asp-L-Phe | ||||
| Km (mM) | kcat (S-1) | kcat/KM (mM-1s-1) | Km (mM) | kcat (S-1) | kcat/KM (mM-1s-1) |
| 2.09 (± 0.16) | 3.19 (± 0.07) | 1.52 (± 0.15) | 0.12 (± 0.02) | 0.80 (± 0.06) | 6.66 (± 1.61) |
| B. Enzyme concentrations ([E]) used to determine asparaginase activity | ||||
|---|---|---|---|---|
| % Cleavageb | Activityc | [E]d no correction | [E]e with correction | |
| WT | 100 | Yes | 75 nM Enzyme | 75 nM Enzyme |
| T186V | 45 | Not detected | 400 nM Enzyme | 180 nM Enzyme |
| T219A | 100 | Not detected | 310 nM Enzyme | 310 nM Enzyme |
| T219V | 50 | Not detected | 310 nM Enzyme | 155 nM Enzyme |
Reactions were conducted at 37 °C, pH 7.5 and in triplicate.
Determined by gel densitometry analysis
Measured with [ASN] = 20 mM
Total [E] in cuvette
Approximated cleaved [E] in cuvette
The T168S mutant was unable to be cleaved and thus, no activity assays were performed on this variant. Interestingly, even though all other variants were partially (T186V, T219V) or completely cleaved (T219A), we were not able to detect any asparaginase activity under the experimental conditions (Table 2B). To confirm the lack of residual asparaginase activity of the mutant enzymes, we employed much higher enzyme concentrations relative to that used when assaying the wild type, and very high substrate concentrations. Despite this, no asparaginase activity was detected for the mutants, indicating that all three threonines are required to catalyze the hydrolysis reaction.
Insight into hASNase3 Asn hydrolysis reaction through the covalent intermediate structure
We were successful in trapping wild type hASNase3 in the covalent intermediate state by performing a very short soak with the substrate Asn, and solved its structure at a resolution of 1.97 Å (Table 1 and Figure 5a). The covalent intermediate represents a state after nucleophilic attack by the Thr168 side chain on the carbonyl group of the Asn substrate side chain, and the expulsion of the amino group. To our knowledge, this is the first elucidated Ntn plant-type (i.e. type III) asparaginase structure in this state (note that a covalent intermediate structure was previously reported for the type I/II asparaginases [29, 30], but these enzymes are different from the type III enzymes discussed here).
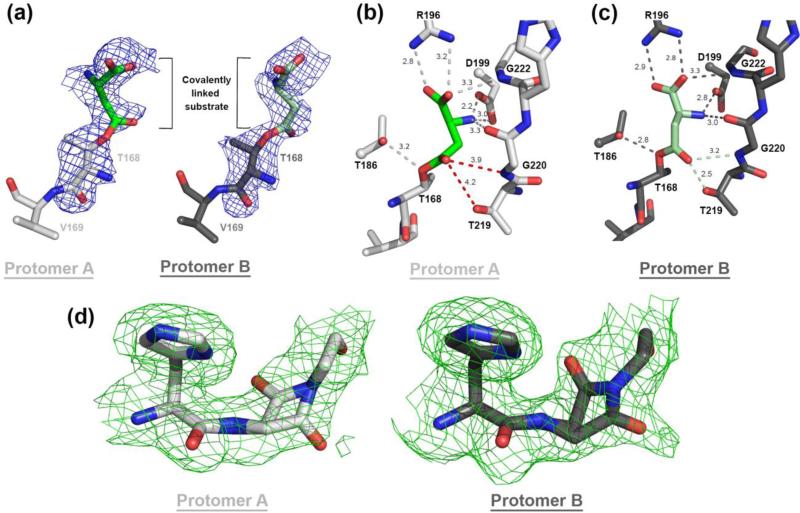
FIGURE 5.
The WT hASNase3 structure in the covalent intermediate state. (a) Fo – Fc omit map (contour level of 2.2σ) surrounding the covalently linked substrate to Thr168. The substrate covalently linked to Thr168 conformation is defined in unbiased electron density that was calculated before the Thr168-substrate was built in both protomers. (b) Close-up of the WT hASNase3 active site protomer A structure (grey). Dashed lines indicate interactions between covalently linked substrate (green) and hASNase3 surrounding residues (distances in Angstroms). The red dashed lines show the lack of interaction with the oxyanion hole formed by Thr219 and Gly220. (c) Same as (b) but from protomer B showing that the interactions between the covalent substrate and Thr219-Gly220 are maintained (green dashed lines). Thus, the state observed in protomer B more accurately demonstrates the role of these residues in stabilizing the negatively charged tetrahedral intermediate. (d) Fo-Fc omit map contoured at 2.2σ around His8-Gly9 region. Residues His8 and Gly9 were removed from the model that then underwent several rounds of refinement to eliminate model bias. We had to model the Gly9 carbonyl as 50% in the “cleaved” and “uncleaved” conformations.
A close-up view of the active site of the covalent intermediate structure is presented in Figure 5b and c. Both protomers of the crystallographic asymmetric unit display the covalent intermediate state and have a nearly identical structure (rmsd of 0.40 Å over 278 Cαatoms) with limited differences at the active site. Interestingly, the covalently linked substrate adopts a slightly different orientation of the β–carbon between protomers A and B but maintains the same overall interactions with the active site residues (Figure 5b and c). The covalently linked substrate's α-carboxylic group makes a bidentate interaction with the Arg196 side chain and with the Gly222 NH atom. The α-amino group is in close contact with the Asp199 side chain and the Gly220 carbonyl.
With regard to interactions with the threonine triad, Thr168 is covalently connected with the substrate. Thr186 is at 2.8 / 3.2 Å (depending on protomer) distance to the oxygen atom of Thr168 that is linked to the substrate. The interaction between Thr219 and the substrate (side-chain carbonyl group) diverges largely between the two protomers, being at a 2.5 Å distance in protomer B but extending to 4.2 Å in protomer A. Similarly, the distance between the NH group of Gly220 and the substrate side-chain carbonyl group differs from 3.2 Å in protomer B to 3.9 Å in protomer A. Thr219 and Gly220 have been reported by us and others to be part of the oxyanion hole that has been proposed to stabilize the negatively charged tetrahedral intermediate [17, 20, 28].
To obtain a more detailed understanding of how Asn hydrolysis is catalyzed by hASNase3, we compared this new covalent intermediate state to the substrate/product bound state as previously observed in the structure of the fully cleaved hASNase3 – Asp complex (PDB: 4GDW [20]; Figure 6). We chose protomer B of the covalent intermediate structure for this comparison since the intermediate – Thr219/Gly220 distances are shorter, and would therefore more likely represent the state immediately following the formation of the reaction intermediate.
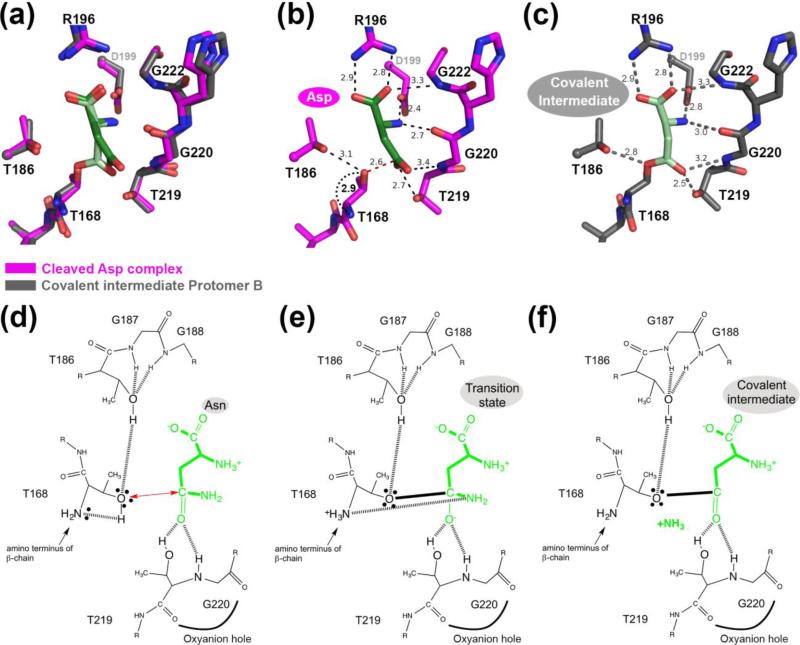
FIGURE 6.
Comparison of fully cleaved WT hASNase3 in complex with Asp and in the covalent intermediate state. (a) Fully cleaved WT hASNase3 structure in complex with Asp (PDB ID 4GDW; enzyme in magenta, Asp in dark green) and the WT covalent intermediate hASNase3 protomer B structure (enzyme in grey and covalently linked substrate in lighter green) are superimposed and a closeup of the active site is presented. (b) WT hASNase3 active site from the fully cleaved Asp complex structure. Black dashed lines indicate interactions between Asp (dark green) and hASNase3. The red dashed line shows the distance between the Thr168 hydroxyl group and the Asp β-carboxylate carbon atom (2.6 Å). The curved dotted black line indicates the distance between the freed amino group from Thr168 activating its hydroxyl for nucleophilic attack on the substrate side chain (all distances are in Angstroms). (c) WT hASNase3 active site from the covalent intermediate protomer B structure. Grey dashed lines indicate interactions between the covalently linked substrate (light green) and hASNase3. (d) hASNase3 asparaginase mechanism. After enzyme cleavage and binding of Asn, the freed amino group of Thr168 activates its side chain, allowing for nucleophilic attack on the substrate side chain (red arrow). This reaction is facilitated by the interaction between Thr186 and Thr168 side chains. (e) The resulting negatively charged tetrahedral intermediate is stabilized by the oxyanion hole composed of Thr219 and Gly220 in the transition state. (f) After release of ammonia, the being-hydrolyzed amino acid remains covalently linked to Thr168 in a state we refer to as the covalent intermediate.
The overlay of WT hASNase3 covalent intermediate protomer B and the cleaved hASNase3 Asp complex reveals an overall identical structure with an rmsd of 0.27 Å over 552 Cαatoms. Figure 6 (panels d, e and f) proposes a schematic of the hASNase3 asparaginase reaction from the initiating nucleophilic attack to the formation of the covalent intermediate with distances reported in Figure 6 panels b and c. The processing of Asn to Asp and ammonia is thought to begin with the nucleophilic attack by the activated side chain of Thr168 on the substrate's side chain carbonyl group. Similar to the role it plays in the cleavage reaction (as discussed above), the hydroxyl of Thr186 acts as a hydrogen bond donor for Thr168. This is a vital interaction that increases the nucleophilicity of Thr168. Additionally activating Thr168 is its own α-amino group. Unlike the pre-cleavage state where the amino group of Thr168 makes a peptide bond to Gly167, in the post-cleavage state it is a free amino group. As such, it acts as a base to activate the Thr168 hydroxyl (distance between α-amino and hydroxyl of Thr168 2.9 Å, Figure 6b and d), which then proceeds to attack the substrate's side chain (distance of Thr168 hydroxyl and Asn Cβ-atom 2.6 Å, Figure 6b). Upon nucleophilic attack, the negative charge that develops on the Asn side chain O atom in the transition state is stabilized by the oxyanion hole formed by Thr219 and Gly220 (Figure 6e). After release of ammonia, the being-hydrolyzed amino acid remains covalently linked to Thr168 (Figure 6c & f) until an activated water molecule attacks the ester carbon atom and liberates the product (see mechanism previously proposed by us [20] for the rest of the reaction).
Interestingly, in the covalent intermediate structure, we were required to model the Gly9 carbonyl as 50% in conformations referred to as “cleaved” and “uncleaved” positions (Figure 5d). Hence, the HGG loop seems not to only play a role in the cleavage reaction but also for substrate hydrolysis. This observation further supports the importance of this highly conserved region.
The behavior of the T186V and T219A/V mutants regarding the asparaginase reaction becomes now somewhat clearer. We mentioned earlier that a valine at position 186 impedes hASNase3 cleavage because this residue is unable to activate Thr168 for autocleavage. Additionally, the cleaved T186V enzyme is devoid of asparaginase activity. This suggests that for promoting the attack on the Asn side chain by the Thr168 hydroxyl group, Thr186 must work in concert with the Thr168's own α-amino group. That is, for nucleophilic attack on the side chain of the substrate asparagine, Thr168 requires activation both by its own amino group and by the hydroxyl of Thr186. Concerning T219A, this mutant can be easily cleaved but yet does not attain asparaginase activity (Figure 1, Table 2B). The covalent intermediate structure suggests that Thr219 has a critical role in stabilizing the tetrahedral intermediate (Figure 6c). Additionally, the structure of the covalent intermediate reveals flexibility in the HGG loop, suggesting that the conformation of this region may be also playing a part in catalyzing asparagine hydrolysis.
Mutants of hASNase3 generated to date based on the increased understanding of its function have failed to provide an enzyme with a lower Km value. For example, replacing the histidine of the HGG loop with an alanine with the goal of promoting flexibility and the cleaved state resulted in an unstable enzyme. Likewise, several mutations of Ile189, situated in the proximity of the substrate, did not result in an enzyme with the desired low Km property. These initial mutagenesis efforts suggest a need to combine several amino acid changes to attain the goal of a stable enzyme with low Km.
CONCLUSIONS
In summary, the experiments conducted on hASNase3 provide structure/function evidence for the role of the catalytic triad composed of three conserved threonines in type III asparaginases. We found that correct positioning of the Thr168 hydroxyl group is critical for autocleavage, and hence even when conservatively replaced by a serine, no active enzyme is made. Thr186 is important but not essential for autocleavage, yet is crucial for hASNase3 activity. On the other hand, Thr219 is not essential for autocleavage, but like the other two threonines of the triad, is required for asparaginase activity. Studies of the Thr219 mutants suggest that flexibility of the HGG loop influences both enzyme cleavage and its asparaginase activity. Therefore, the engineering of this enzyme in order to lower its Asn KM value for use as a replacement of bacterial enzyme in cancer therapy would have to take into consideration both the importance of the threonine triad as well as the implication of the HGG loop to promote the cleaved and enzymatically-competent conformation. While such a mutated hASNase3 variant may also be immunogenic, it is predicted to be less so than the currently used bacterial enzymes.
Materials and Methods
Cloning, Expression and Purification of human ASNase3
Human ASNase3 was cloned and purified as previously described in Nomme et al. [20]. All point mutations were inserted using the QuikChange® Site-Directed Mutagenesis Kit from Agilent Technologies and confirmed by sequencing.
Briefly, for expression and purification, E. coli BL21(DE3) C41 carrying the hASNase3 plasmid (modified pET14b to include a His-SUMO tag) were grown at 37 °C in 2YT media until reaching an optical density at 600 nm of 0.6-0.8. Expression was induced using 0.5 mM isopropyl β-D-1-thiogalactopyranoside and the temperature was reduced to 18 °C overnight. After lysis by sonication, the enzyme was purified on a metal affinity column charged with nickel. The His-SUMO tag was then cleaved in solution by SUMO protease, and followed by a last step of gel filtration. The hASNase3 containing fractions were then concentrated to 38-53 mg/ml, aliquoted, and stored at −80 °C.
Enzyme activation by glycine
Wild type hASNase3 and variants were activated by incubating for 4 days in a 2.0 M glycine pH 9.0 solution at room temperature (RT). Glycine was then washed out by dialysis overnight at RT against a solution containing 25 mM Tris/HCl pH 7.5, 200 mM NaCl and 2 mM DTT.
Kinetic Assay
The catalytic activity of hASNase3 was determined using a spectroscopic NADH-dependent enzyme-coupled assay [31, 32]. All measurements were taken in triplicate at 37 °C in a buffer containing 100 mM Tris/HCl pH 7.5, 0.4 mM α-ketoglutarate and 0.4 mM NADH with 75 nM hASNase3 wild type. To determine the KM and kcat values, we varied L-asparagine/β-L-Asp-L-Phe concentration and kept the enzyme concentration fixed. Using SigmaPlot the Michaelis–Menten equation was fit to the data (SigmaPlot 2000/Enzyme Kinetics Module 1.0, from Systat Software, Inc., San Jose California USA, www.sigmaplot.com).
Crystallization of hASNase3
For crystallization setups, 2 μL of hASNase3 at 35-40 mg/ml (in 25 mM Tris/HCl pH 7.5, 200 mM NaCl, 2 mM DTT) were mixed with 1 μL of 0.6 M sodium malonate, pH 7.0 on a glass cover slip, and left to undergo vapor diffusion using the hanging drop method at 20 °C over a reservoir of 2.2-2.5 M sodium malonate, pH 7.0. As described in a previous study by us [20], for crystals grown in malonate, we observed precipitant molecules occupying the substrate binding site. To solve this precipitant-competition issue, all crystals were first transferred to a 75% saturated NaCl, 25 mM MES/NaOH pH 6.5 solution (wash solution) for 15 min in order to wash the malonate molecules out from the active site. We also showed that a short soak (2-5 min) in a 2-3 M glycine, pH 7.5 solution (cleavage solution) was normally enough to complete full cleavage of the wild type protein in the crystal [20]. To obtain the high resolution wild type hASNase3 structure crystals were transferred into the cleavage solution for 2 min followed by 5 min in the wash solution in order to wash out the glycine from the previous step and finally into a 75% saturated NaCl, 25 mM MES/NaOH pH 6.5, 20% glycerol solution (cryo solution) supplemented with 62.5 mM β-Asp-Phe for 2 min for cryoprotection. Unexpectedly, the enzyme remained uncleaved and no substrate was observed in the active site. The T168S mutant crystals were transferred to the cleavage solution and then moved to the cryo solution for 1 min. The T186V structure was obtained by transferring crystals into the cryo solution containing 62.5 mM β-Asp-Phe for 1 min. However, no substrate was observed in the active site. The T219A mutant crystals were transferred to the cleavage solution for 2 days followed by a 1 min soak in the cryo solution. The T219V mutant crystal was moved to the cryo solution for 1 min for cryoprotection. Finally, to obtain the wild type protein in the covalent intermediate state, crystals were first transferred to the cleavage solution for 2 min. Glycine was then washed out by transferring the crystals back into the wash solution. A last transfer to a 250 mM asparagine, 75% saturated NaCl, 25 mM MES/NaOH pH 6.5, 20% glycerol solution for 2 min was used as source of asparagine and as the cryoprotectant. Longer soaks resulted in an electron density at the active site corresponding to the product aspartate (note that we cannot distinguish by electron density between asparagine or aspartate, but interpret the resulting density in crystals that were soaked a longer time with asparagine as coming from the product aspartate).
Data collection and structure solution of hASNase3
Diffraction data for the hASNase3 wild type covalent intermediate and the T219A mutant were collected using the in-house X-ray source (Rigaku RU- 200 rotating anode) with an R-Axis IV++ image plate detector while data for the uncleaved wild type hASNase3, T168S, T186V and T219V mutants were collected at the Advanced Photon Source (APS) located at Argonne National Laboratory, using the SERCAT 22-ID and LS-CAT beamlines (Table 1). Data were processed using XDS [33].
All crystals were perfectly twinned, with the true space group being P65 (apparent space group, P6522), and contained two copies of hASNase3 in the asymmetric unit. The data sets were analyzed by phenix.xtriage [34] and depending on the data set, the usual parameters that indicate the presence of twinning (e.g. Wilson ratio <I^2>/<I>^2; L test; H test) were clear about the presence of twinning or less so. However, the R vs R analysis consistently indicated twinning. This analysis suggests that our crystals contain pseudo rotational symmetry (PRS). PRS has been shown to mask the presence of twinning by partially compensating for its effect [35]. An excellent treatise on this topic is discussed in the paper by Lebedev et. al. [35] Supplementary Table S1 presents the twinning statistics for all data sets computed for the entire resolution range plus for the highest resolution shell. At high resolution the masking effects of PRS on the twinning statistics are diminished.
The structure was solved by molecular replacement (Molrep [36] CCP4) using the human ASNase3 as starting model (PDB entry 4GDV), refined with Refmac5 [37] and model built using Coot [38]. All structures had residue Asp119 of both protomers in the generously allowed/disallowed regions of the Ramachandran plot; the correctness of the modeling was verified with omit maps. We are aware of the short distance (2.2 Å) between the N-atom of the covalent intermediate and the carboxylate of Asp199 in protomer A. Refinement using Refmac5 and phenix both yielded the same short distance, and the model is consistent with omit maps. Structural figures were prepared using the PyMOL Molecular Graphics System, Version 1.6.0 Schrödinger, LLC.
Supplementary Material
Highlights.
Structures of human L-asparaginase (hASNase3) T168S, T186V, T219A/V mutants and wild type covalent intermediate are presented.
Thr168 is crucial for the activating autocleavage step and all triad threonines are required for asparaginase activity.
The conserved HGG loop plays a critical role in autocleavage, as well as in asparagine hydrolysis.
Improved understanding of hASNase3 function can inform the design of variants suitable as replacement to the immunogenic bacterial asparaginases currently used in cancer therapy.
Acknowledgments:
We thank Dr. Amanda Schalk for helpful discussions. We also thank the staff at SER-CAT and LS-CAT for assistance in data collection. We are grateful for the expert advice of Dr. Peter Zwart regarding the twinning analysis.
This work was supported by a National Institutes of Health grant R21 CA155424.
Abbreviations
- hASNase3
human L-asparaginase 3
- Ntn
N-terminal nucleophile
- Asn
L-asparagine
- Asp
L-aspartate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB accession numbers: 4O0C, 4O0D, 4O0E, 4O0F, 4O0G, 4O0H.
References
- 1.Borek D, Jaskolski M. Sequence analysis of enzymes with asparaginase activity. Acta Biochim Pol. 2001;48:893–902. [PubMed] [Google Scholar]
- 2.Michalska K, Bujacz G, Jaskolski M. Crystal structure of plant asparaginase. J Mol Biol. 2006;360:105–16. doi: 10.1016/j.jmb.2006.04.066. [DOI] [PubMed] [Google Scholar]
- 3.Michalska K, Jaskolski M. Structural aspects of L-asparaginases, their friends and relations. Acta Biochim Pol. 2006;53:627–40. [PubMed] [Google Scholar]
- 4.Stecher AL, de Deus PM, Polikarpov I, Abrahao-Neto J. Stability of L-asparaginase: an enzyme used in leukemia treatment. Pharm Acta Helv. 1999;74:1–9. doi: 10.1016/s0031-6865(99)00009-6. [DOI] [PubMed] [Google Scholar]
- 5.Duval M, Suciu S, Ferster A, Rialland X, Nelken B, Lutz P, Benoit Y, Robert A, Manel AM, Vilmer E, Otten J, Philippe N. Comparison of Escherichia coli-asparaginase with Erwinia-asparaginase in the treatment of childhood lymphoid malignancies: results of a randomized European Organisation for Research and Treatment of Cancer-Children's Leukemia Group phase 3 trial. Blood. 2002;99:2734–9. doi: 10.1182/blood.v99.8.2734. [DOI] [PubMed] [Google Scholar]
- 6.Asselin BL, Ryan D, Frantz CN, Bernal SD, Leavitt P, Sallan SE, Cohen HJ. In vitro and in vivo killing of acute lymphoblastic leukemia cells by L-asparaginase. Cancer Res. 1989;49:4363–8. [PubMed] [Google Scholar]
- 7.Ertel IJ, Nesbit ME, Hammond D, Weiner J, Sather H. Effective dose of L-asparaginase for induction of remission in previously treated children with acute lymphocytic leukemia: a report from Childrens Cancer Study Group. Cancer Res. 1979;39:3893–6. [PubMed] [Google Scholar]
- 8.Swain AL, Jaskolski M, Housset D, Rao JK, Wlodawer A. Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy. Proc Natl Acad Sci U S A. 1993;90:1474–8. doi: 10.1073/pnas.90.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broome JD. Studies on the mechanism of tumor inhibition by L-asparaginase. Effects of the enzyme on asparagine levels in the blood, normal tissues, and 6C3HED lymphomas of mice: differences in asparagine formation and utilization in asparaginase-sensitive and -resistant lymphoma cells. J Exp Med. 1968;127:1055–72. doi: 10.1084/jem.127.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugimoto H, Odani S, Yamashita S. Cloning and expression of cDNA encoding rat liver 60-kDa lysophospholipase containing an asparaginase-like region and ankyrin repeat. J Biol Chem. 1998;273:12536–42. doi: 10.1074/jbc.273.20.12536. [DOI] [PubMed] [Google Scholar]
- 11.Oinonen C, Tikkanen R, Rouvinen J, Peltonen L. Three-dimensional structure of human lysosomal aspartylglucosaminidase. Nat Struct Biol. 1995;2:1102–8. doi: 10.1038/nsb1295-1102. [DOI] [PubMed] [Google Scholar]
- 12.Saito S, Ohno K, Sugawara K, Suzuki T, Togawa T, Sakuraba H. Structural basis of aspartylglucosaminuria. Biochem Biophys Res Commun. 2008;377:1168–72. doi: 10.1016/j.bbrc.2008.10.142. [DOI] [PubMed] [Google Scholar]
- 13.Bush LA, Herr JC, Wolkowicz M, Sherman NE, Shore A, Flickinger CJ. A novel asparaginase-like protein is a sperm autoantigen in rats. Mol Reprod Dev. 2002;62:233–47. doi: 10.1002/mrd.10092. [DOI] [PubMed] [Google Scholar]
- 14.Evtimova V, Zeillinger R, Kaul S, Weidle UH. Identification of CRASH, a gene deregulated in gynecological tumors. Int J Oncol. 2004;24:33–41. [PubMed] [Google Scholar]
- 15.Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–9. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 16.Cantor JR, Stone EM, Chantranupong L, Georgiou G. The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with beta-aspartyl peptidase activity. Biochemistry. 2009;48:11026–31. doi: 10.1021/bi901397h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tikkanen R, Riikonen A, Oinonen C, Rouvinen R, Peltonen L. Functional analyses of active site residues of human lysosomal aspartylglucosaminidase: implications for catalytic mechanism and autocatalytic activation. EMBO J. 1996;15:2954–60. [PMC free article] [PubMed] [Google Scholar]
- 18.Guan C, Liu Y, Shao Y, Cui T, Liao W, Ewel A, Whitaker R, Paulus H. Characterization and functional analysis of the cis-autoproteolysis active center of glycosylasparaginase. J Biol Chem. 1998;273:9695–702. doi: 10.1074/jbc.273.16.9695. [DOI] [PubMed] [Google Scholar]
- 19.Michalska K, Hernandez-Santoyo A, Jaskolski M. The mechanism of autocatalytic activation of plant-type L-asparaginases. J Biol Chem. 2008;283:13388–97. doi: 10.1074/jbc.M800746200. [DOI] [PubMed] [Google Scholar]
- 20.Nomme J, Su Y, Konrad M, Lavie A. Structures of apo and product-bound human L-asparaginase: insights into the mechanism of autoproteolysis and substrate hydrolysis. Biochemistry. 2012;51:6816–26. doi: 10.1021/bi300870g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y, Karamitros CS, Nomme J, McSorley T, Konrad M, Lavie A. Free glycine accelerates the autoproteolytic activation of human asparaginase. Chem Biol. 2013;20:533–40. doi: 10.1016/j.chembiol.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian X, Guan C, Guo HC. A dual role for an aspartic acid in glycosylasparaginase autoproteolysis. Structure. 2003;11:997–1003. doi: 10.1016/s0969-2126(03)00150-3. [DOI] [PubMed] [Google Scholar]
- 23.Cooney DA, Capizzi RL, Handschumacher RE. Evaluation of L-asparagine metabolism in animals and man. Cancer Res. 1970;30:929–35. [PubMed] [Google Scholar]
- 24.Buller AR, Townsend CA. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc Natl Acad Sci U S A. 2013;110:E653–61. doi: 10.1073/pnas.1221050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisselev AF, Songyang Z, Goldberg AL. Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J Biol Chem. 2000;275:14831–7. doi: 10.1074/jbc.275.20.14831. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Guan C, Aronson NN., Jr. Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase. J Biol Chem. 1998;273:9688–94. doi: 10.1074/jbc.273.16.9688. [DOI] [PubMed] [Google Scholar]
- 27.Michalska K, Brzezinski K, Jaskolski M. Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate. J Biol Chem. 2005;280:28484–91. doi: 10.1074/jbc.M504501200. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Cantor JR, Yogesha SD, Yang S, Chantranupong L, Liu JQ, Agnello G, Georgiou G, Stone EM, Zhang Y. Uncoupling intramolecular processing and substrate hydrolysis in the N-terminal nucleophile hydrolase hASRGL1 by circular permutation. ACS Chem Biol. 2012;7:1840–7. doi: 10.1021/cb300232n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun MK, Nourse A, White SW, Rock CO, Heath RJ. Crystal structure and allosteric regulation of the cytoplasmic Escherichia coli L-asparaginase I. J Mol Biol. 2007;369:794–811. doi: 10.1016/j.jmb.2007.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palm GJ, Lubkowski J, Derst C, Schleper S, Rohm KH, Wlodawer A. A covalently bound catalytic intermediate in Escherichia coli asparaginase: crystal structure of a Thr-89-Val mutant. FEBS Lett. 1996;390:211–6. doi: 10.1016/0014-5793(96)00660-6. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez CA, Cai X, Elozory A, Liu C, Panetta JC, Jeha S, Molinelli AR, Relling MV. High-throughput asparaginase activity assay in serum of children with leukemia. Int J Clin Exp Med. 2013;6:478–87. [PMC free article] [PubMed] [Google Scholar]
- 32.Hejazi M, Piotukh K, Mattow J, Deutzmann R, Volkmer-Engert R, Lockau W. Isoaspartyl dipeptidase activity of plant-type asparaginases. Biochem J. 2002;364:129–36. doi: 10.1042/bj3640129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebedev AA, Vagin AA, Murshudov GN. Intensity statistics in twinned crystals with examples from the PDB. Acta Crystallogr D Biol Crystallogr. 2006;62:83–95. doi: 10.1107/S0907444905036759. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.