Abstract
Genetic studies have provided valuable insight into the pathological mechanisms underlying Parkinson’s disease (PD). The elucidation of the genetic components to what was once largely considered a non-genetic disease has given rise to a multitude of cell and animal models enabling the dissection of molecular pathways involved in disease etiology. Here, we review advances obtained from models of dominant mutations in α-synuclein and LRRK2 as well as recessive PINK1, Parkin and DJ-1 mutations. Recent genome-wide association studies have implicated genetic variability at two of these loci, α-synuclein and LRRK2, as significant risk factors for developing sporadic PD. This, coupled to the established role of mitochondrial impairment in both familial and sporadic PD highlights the likelihood of common mechanisms fundamental to the etiology of both.
Keywords: α-synuclein, LRRK2, Parkin, PINK1, DJ-1, PARIS
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that profoundly affects movement. Cardinal symptoms of PD (bradykinesia, resting tremor, rigidity and postural instability) manifest due to the relatively selective loss of nigrostriatal dopamine (DA) neurons that are important for regulating motor function (reviewed in (22)). The onset of sporadic PD is noticeably related to aging, with a sharp rise in incidence seen after the age of 60 (26). Rare ‘early-onset’ PD, which accounts for about 5–10% of all cases, occurs before the age of 50 and is usually monogenic in origin. PD is prevalent in approximately 1% of the population over the age of 60 with some cross-ethnic studies indicating a lower incidence in some African and Asian populations, although these reported differences may be the result of variability in case ascertainment related to screening methods, diagnostic criteria and response rates as well as disparity in disease survival rates (26). While much of the etiology of PD remains unclear, significant advances in understanding the mechanisms of disease pathogenesis have been made in the last two decades with the identification of distinct genetic loci at which pathogenic mutations are associated with Parkinsonism (Table 1). Mendelian loci and the high-risk glucocerebrocidase (GBA) locus together account for between approximately 10–40% of disease risk depending on the population under study (49) and this will likely increase as more risk loci are identified. Recent genome-wide association studies (GWAS) have been fruitful in identifying loci at which common genetic variants increase risk of developing apparently sporadic disease and these studies have confirmed the role of α-synuclein, leucine rich repeat kinase-2 (LRRK2) and microtubule-associated protein tau (MAPT) loci as contributing to disease risk. Looking towards the future, GWAS and exome sequencing will be key to identifying novel PD-associated loci, as exemplified by the recent report of common variants at the histocompatibility complex class IA (HLA), bone marrow stromal cell antigen (BST1) and PARK16 loci as risk factors for PD (47; 109; 118).
Table 1.
Genes and loci associated with Parkinsonism
| PARK locus | Gene | Map position | Clinical phenotype | Pathology |
|---|---|---|---|---|
| PARK1/4 | SNCA | 4q21 | Parkinsonism with common dementia | Lewy bodies |
| PARK2 | Parkin | 6q25-q27 | Early-onset, slowly progressing Parkinsonism | Lewy bodies rarely |
| PARK3 | Unknown | 2p13 | Late-onset Parkinsonism | Lewy bodies |
| PARK5 | UCHL1 | 4p14 | Late-onset Parkinsonism | Unknown |
| PARK6 | PINK1 | 1p35-p36 | Early-onset, slowly progressing Parkinsonism | One case exhibiting Lewy bodies |
| PARK7 | DJ-1 | 1p36 | Early-onset Parkinsonism | Unknown |
| PARK8 | LRRK2 | 12q12 | Late-onset Parkinsonism | Usually Lewy bodies |
| PARK9 | ATP13A2 | 1p36 | Early-onset Parkinsonism with Kufor-Rakeb syndrome | Unknown |
| PARK10 | Unknown | 1p32 | Unclear | Unknown |
| PARK11 | GIGYF2 | 2q36-q37 | Late-onset Parkinsonism | Unknown |
| PARK12 | Unknown | Xq | Unclear | Unknown |
| PARK13 | Omi/HTRA2 | 2p13 | Unclear | Unknown |
| PARK14 | PLA2G6 | 22q13.1 | Parkinsonism with additional features | Lewy bodies |
| PARK15 | FBX07 | 22q12-q13 | Early-onset Parkinsonism | Unknown |
| PARK16? | Unknown | 1q32 | Late-onset Parkinsonism | Unknown |
| FTDP-17 | MAPT | 17q21.1 | Dementia, sometimes Parkinsonism | Neurofibrillary Tangles |
| SCA2 | Ataxin 2 | 12q24.1 | Usually ataxia, sometimes Parkinsonism | Unknown |
| SCA3 | Ataxin 3 | 14q21 | Usually ataxia, sometimes Parkinsonism | Unknown |
| Gaucher’s locus | GBA | 1q21 | Late-onset Parkinsonism | Lewy bodies |
Many pathogenic mutations produce variable pathological and clinical phenotypes that are distinct from typical PD (Table 1). This is consistent with the likely existence of different pathological mechanisms that all lead to the manifestation of clinical Parkinsonism. For example, mutations at spinocerebellar ataxia type (SCA) 2 and 3 loci are associated with L-Dopa-responsive Parkinsonism (65; 133). The presence of expanded polyglutamine repeats in SCA loci of affected individuals is suggestive of a pathogenic process distinct from that caused by mutations in other PD-associated genes. Alternatively, it is conceivable that disparate pathogenic ‘triggers’ arising from different pathogenic mutations could converge on one or more common signaling pathways central to the loss of DA neurons leading to the manifestation of Parkinsonism.
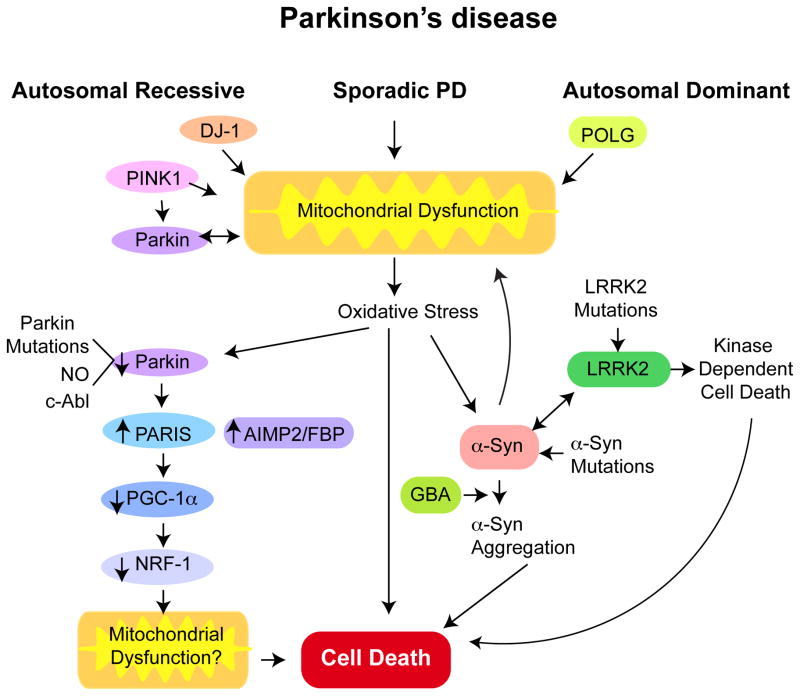
A comprehensive review of all loci associated with PD are found elsewhere (50; 78). Here, we focus on five genes conclusively linked to PD pathogenesis: α-synuclein, LRRK2, PINK1, Parkin and DJ-1. We describe progress made on identifying their normal biological functions, the consequences of pathogenic mutations gleaned from genetic models and highlight recent studies that shed light on the possible interplay between several of these genes in disease development (Figure 2).
Figure 2. Potential pathogenic pathways linking the genetic and sporadic causes of PD.
A key feature of both sporadic and genetic causes of PD is mitochondrial dysfunction. Mutations in the autosomal recessive genes PINK1, Parkin and DJ-1 may directly cause mitochondrial dysfunction. PINK1 may act upstream of Parkin. PINK1 and Parkin may regulate mitochondria mitophagy. Parkin may directly regulate mitochondrial biogenesis through PARIS, which is a transcriptional repressor of the PGC-1α, the master regulator of mitochondrial biogenesis. Dominant mutations in POLG, the catalytic subunit of mitochondrial DNA polymerase causes Parkinsonism in some families. Dominant mutations in LRRK2 and α-synuclein (α-syn) cause PD. Oxidative stress including nitrosative stress and c-Abl phosphorylation of Parkin leads to its inactivation in sporadic PD and subsequent accumulation of substrates that are degraded by the ubiquitin proteasome system, such as AIMP2, FBP1 and PARIS. PARIS may be the key pathogenic Parkin substrate as knockdown of PARIS in an adult conditional knockout of Parkin completely rescues neurodegeneration of DA neurons. α-Synuclein aggregation is a key step in DA neuron degeneration in PD. Oxidative and nitrosative (NO) stress can accelerate α-synuclein and aggregated α-synuclein can damage mitochondria setting in motion a feed forward mechanism. GBA mutations also seem to accelerate α-synuclein aggregation and LRRK2 and α-synuclein interact at some level in the pathogenesis of PD. LRRK2 mutations lead to DA neurodegeneration that is kinase dependent.
α-Synuclein
α-synuclein is a small (140 kDa) protein encoded by the α-synuclein gene, designated SNCA for synuclein, alpha (non A4 component of amyloid precursor) gene and is one of three synuclein family members (α, β, γ) identified in humans (60; 74; 102). Synucleins appear to be exclusive to vertebrates as invertebrates and single-celled organisms do not possess a homolog. Following the identification of the 209G>A (Ala53Thr) pathogenic SNCA mutation in the Contursi kindred (115), two further PD-associated SNCA missense mutations: 188G>A (Glu46Lys) (154) and 88G>C (Ala30Pro) (75) were found (Figure 1). Numerous studies have also established a link between familial PD and duplications or triplications in the SNCA gene (95). Instructively, gene triplication leads to earlier onset and faster progression of disease than duplication indicating that disease severity is dependent on α-synuclein expression levels. These studies have been supported by cell and animal models in which expression of wild type or mutant α-synuclein results in neuronal toxicity (90). Additionally, a link between sporadic PD and SNCA gene expression is supported by evidence of reduced epigenetic silencing of SNCA (63) and SNCA promoter region polymorphisms that might increase α-synuclein expression (104). Studies consistently indicate a relationship between α-synuclein expression levels and the appearance of PD leading to the widely held hypothesis that a gain-of-function by α-synuclein underlies pathogenesis in PD.
Figure 1. PD gene product domains and pathogenic mutations.
Domains are arranged from N- to C- termini. For LRRK2: Ank (Ankyrin-like repeats); LRR (leucine-rich repeats); ROC (Ras of complex GTPase domain); COR (C-terminal of ROC); Kinase and WD40. PINK1: MTS (mitochondrial targeting sequence); TM (putative transmembrane domain); and serine/threonine kinase. For Parkin: UBL (ubiquitin-like) and two RING domains separated by an IBR (in-between RING) domain. α-synuclein has a number of imperfect KTKEGV repeat sequences (white stripes) in the N-terminal region and central NAC (non-amyloid component) region. DJ-1 is a single domain protein. Numbers under the protein indicate domain boundaries. Mutations that segregate with PD are annotated at their approximate position along the protein’s length.
α-Synuclein plays a role in neurotransmitter release
α-Synuclein is expressed at high levels within the vertebrate brain, localized principally to synaptic terminals where it associates with vesicles and the plasma membrane (58). In vitro, α-synuclein appears to be natively unfolded in solution although it is likely that to function in cells, α-synuclein adopts defined conformations, and alpha-helical rich conformations have been observed following the association of α-synuclein with membranes (32). The normal function of α-synuclein remains unclear, although emerging evidence points towards a possible role in neurotransmitter release (93). Specifically, recent in vivo evidence demonstrates that α-synuclein binds to and promotes assembly of SNARE complexes that are required for fusion of vesicles to the pre-synaptic membrane (10; 13). Although knock-out of α-synuclein alone or α-synuclein and β-synuclein together has minimal effects on neuronal function (12), triple knock-out mice lacking all synucleins display deficits in SNARE complex assembly and develop accelerated age-associated motor impairments as well as early-onset mortality, but do not exhibit neurodegeneration (10; 45). Neurotransmition is also perturbed by increased α-synuclein expression in the range predicted for gene multiplication (93). Mechanistically, studies of transfected hippocampal cultures indicate that synaptic vesicle recycling is impaired following α-synuclein overexpression since there is a reduction in synaptic vesicle density at the active zone and in the size of the synaptic vesicle recycling pool compared to control neurons. It is currently unclear whether these effects are due to a gain of function in α-synuclein, or from loss of normal biological function.
α-Synuclein aggregation plays a role in the pathogenesis of PD
In the PD brain, α-synuclein is usually detected in Lewy bodies and Lewy neurites in neurons of the brainstem and cortex, and aggregates of α-synuclein are a major protein component of these structures (77). α-Synuclein exhibits a high propensity to aggregate in vitro and the presence of α-synuclein aggregates in Lewy bodies has led to the hypothesis that aggregation may be important to α-synuclein toxicity (Figure 2) (16). Indeed, mutation or multiplication of α-synuclein generates a protein with increased tendency to develop β-sheet-rich structure and to polymerize into oligomers and higher-order fibrillar aggregates in cells, transgenic animals expressing mutant α-synuclein and human brain (76; 77; 89; 96; 114) (Figure 2). Since A53T and A30P mutations preferentially promote the formation of α-synuclein oligomers and PD patients sometimes lack Lewy bodies, oligomeric α-synuclein may be the primary toxic species in PD pathogenesis (17; 33). Studies in transgenic α-synuclein mice that exhibit only prefibrillar oligomers indicate that α-synuclein is toxic in the oligomeric form. However, fibrils of α-synuclein do play a role in toxicity as the formation of α-synuclein fibrils is required for significant neurodegeneration (25).
Several biological and environmental factors may contribute to formation of α-synuclein oligomers and fibrils. α-synuclein oligomers are predominantly found in membrane-rich fractions and studies suggest that membrane-bound α-synuclein has a higher tendency to aggregate and may seed formation of soluble oligomers elevated in PD (76; 114). The preferential loss of DA neurons in PD may be related to a stabilizing effect of DA (or its oxidation by-products) on protofibrillar α-synuclein aggregates that promote toxicity (18). This affect may be mediated through a direct impact of DA on α-synuclein conformation, which occurs in cultured neurons (96). Aging may contribute to α-synuclein toxicity via the age-related accumulation of oxidative and nitrative α-synuclein modifications, which promote aggregation (Figure 2) (38; 77). Exposure to compounds such as rotenone and paraquat might also promote α-synuclein aggregation (84; 115). Amyloid β peptide (Aβ), which is thought to underlie the proteinopathy of Alzheimer’s disease promotes α-synuclein aggregation via the formation of heterologous Aβ and α-synuclein oligomers. Mice with enhanced levels of α-synuclein and Aβ exhibit augmented neuronal impairment over mice with increased α-synuclein alone (86; 125). Moreover, MAPT promotes the formation of α-synuclein oligomers and fibrils and there is augmented neuronal impairment in mice expressing both MAPT and α-synuclein (39). Thus, the interaction between MAPT, Aβ and α-synuclein might be relevant to patients with combined Alzheimer’s and PD pathology. Mutations in GBA is also strongly associated with the development of PD and other Lewy body disorders (129). The mechanism underlying this association is not known, but may be due to alterations in lipid metabolism or autophagy and lysosomal function.
α-Synuclein exhibits prion-like pathology
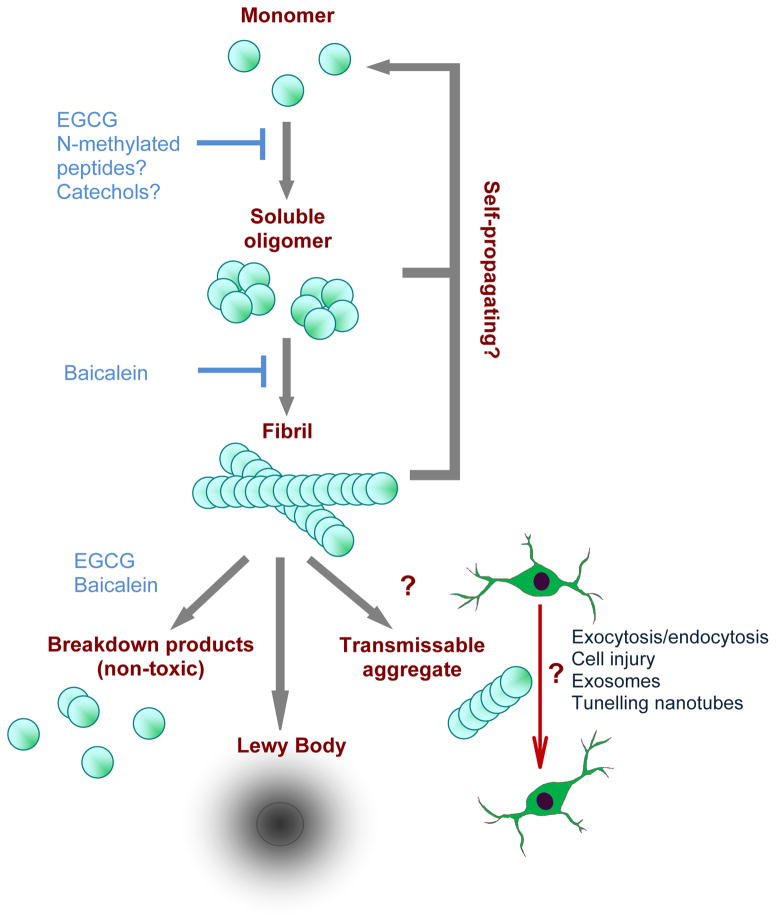
An interesting feature of Lewy body pathology is that it my spread in the brain of PD patients from the lower brainstem to the midbrain and then the cerebral hemispheres (9). One crucial question facing the field is whether the formation of these inclusions are brain region autonomous or conversely, spread from one region to another in a prion-like manner. In favor of the latter hypothesis, embryonic or fetal DA neuron grafts transplanted into the striatum of PD patients develop α-synuclein positive Lewy bodies after only 11–16 years (71; 79). This astounding observation, coupled to reports that α-synuclein is directly transmitted from neurons overexpressing the protein to adjacent healthy embryonic stem cells in culture or in transgenic animals (27), supports the interpretation that α-synuclein aggregates are transferred between neurons. Additionally, misfolded α-synuclein aggregates, akin to the pathogenic prion protein conformer (PrPSc), serve as a template to promote additional misfolding and aggregation of native protein (32; 61). Hence, under certain conditions transmissible α-synuclein aggregates may self-propagate from neuron to neuron, and from one brain region to the next. One puzzling aspect of this theory is how, in transmitting from one neuron to its neighbor, α-synuclein aggregates are able to pass through two cell membrane barriers. It is possible that cell-cell transfer of small transmissible aggregates may occur via exocytosis/endocytosis, cell injury, exosomes or tunneling nanotubes. It will be important to disrupt these processes in a targeted fashion to understand whether they truly underlie the spread of α-synuclein in vivo. If cell-cell transfer of α-synuclein aggregates is relevant to PD etiology, determining the underlying mechanisms will be crucial to understanding disease development and identifying potential strategies to block aggregate transmission (Figure 3).
Figure 3. α-Synuclein aggregation and pathophysiology.
α-Synuclein natively unfolded monomers assemble to form β-sheet-rich soluble oligomers, which further aggregate to form mature fibrils. Aggregates at both stages are thought to act as templates that seed further α-synuclein fibrillogenesis in a feed-forward cycle. Fibrils can deposit into Lewy bodies or might break down into small transmissible aggregates via incomplete degradation that are able to transmit between cells, facilitating the spread of α-synuclein aggregation and toxicity in a prion-like manner. The mechanism of transmission from one neuron to another is unknown. Prevention of oligomer and fibril formation and the disaggregation of mature fibrils to non-toxic breakdown products are therapeutic goals and several compounds, including EGCG, baicalein, N-methylated peptides and catechol-based compounds have been put forward based on their effectiveness in vitro.
Neuroprotection against α-Synuclein toxicity
Protection against toxic α-synuclein aggregation is an attractive therapeutic strategy and several studies have focused on a number of small molecules that interact with α-synuclein and inhibit its aggregation in vitro (7; 30; 46; 144). One approach is to design agents that halt the aggregation process or alternatively, accelerate aggregation into insoluble inclusions with a low toxic profile. A range of molecules that affect α-synuclein amyloidogenesis exist, including antioxidants, β-synuclein, and small N-methylated peptides that bind to and block key regions required for aggregation (46; 83). The flavonoid baicalein blocks α-synuclein fibril formation and disaggregates pre-formed mature fibrils (55; 144). For any potential aggregation inhibitor, it will be important to understand the underlying mechanism. The polyphenol (−)-epigallocatechin (EGCG) potently inhibits α-synuclein fibrillogenesis and, crucially, shows potential to arrest aggregation at a very early step as well as reduce the toxicity of pre-formed aggregates (7; 30). EGCG binds to natively unstructured α-synuclein monomers preventing their conversion to β-sheet-rich structures and therefore blocks the assembly of toxic oligomers and fibrils. Pre-formed α-synuclein aggregates are also bound by EGCG, leading to fibril remodeling, and breakdown to smaller amorphous aggregates that exhibit reduced cellular toxicity. Whether EGCG binding to α-synuclein is protective against fibrillogenesis and pathology in vivo, is not known. One potential complication stems from the ability of EGCG to bind to α-synuclein and stimulate the formation of ‘off-pathway’ oligomers at a relatively fast rate compared to spontaneous α-synuclein fibrillogenesis. Hopefully, EGCG binding and EGCG-mediated oligomerization in vivo will not perturb the normal biological function of α-synuclein and thereby promote neuronal dysfunction.
SIRT2 inhibitors may affect α-synuclein aggregation since an increase in α-synuclein inclusion size was observed in primary neuronal cultures transduced with A53T α-synuclein (46; 97). Intriguingly, loss of tyrosine hydroxylase (TH)-positive neurons in brains of flies expressing α-synuclein targeted to neurons was rescued by feeding flies with SIRT2 inhibitors without affecting levels of transgenic α-synuclein expression. The protective mechanism by which SIRT2 inhibitor treatment prevents α-synuclein related toxicity is unknown although the observed increase in α-synuclein inclusion size and consequent decrease in overall inclusion surface area may lower the interaction of aggregates with cellular macromolecules thereby reducing their toxicity.
Leucine-rich repeat kinase 2 (LRRK2)
LRRK2 is a large multidomain protein with GTP-regulated serine/threonine kinase activity (134). LRRK2 belongs to the ROCO protein family which all possess tandem ROC (Ras of Complex)-COR (C-terminal of ROC) domains. The catalytic core of LRRK2 consists sequentially of ROC, COR and kinase domains and is flanked by upstream ankyrin and leucine-rich repeats and a downstream WD40 domain that likely mediate protein-protein and/or protein-membrane interactions (Figure 1). The kinase domain of LRRK2 and its homolog LRRK1 are most similar in sequence to the receptor-interacting protein kinase and death-domain containing interleukin receptor-associated kinase families (19). Several mutations that cluster in the central GTPase/kinase region of LRRK2 (Y1699C, I2020T R1441C and R1441G) segregate with disease in large families worldwide with autosomal dominant PD (98; 146). A number of other mutations have been identified, but whether they segregate with disease is not clear.
The G2019S kinase domain mutation is the most common LRRK2 mutation segregating with familial PD (21). The mutation is also present in a multitude of apparently sporadic cases (21). G2019S carriers have similar disease risk and progression irrespective of whether they are heterozygous or homozygous for the mutation strongly supporting its dominant nature. Penetrance of the G2019S mutation is age-related reaching approximately 80% in carriers 80 years of age (51). Hence, penetrance is incomplete even at advanced ages and a number of affected individuals remain neurologically unimpaired. Additional studies revealed a third pathogenic mutation at amino residue 1441 (R1441H), and genome-wide association studies independently performed on European and Japanese populations both identified common genetic variants at the LRRK2 locus as significant risk factors for sporadic PD (21).
LRRK2 pathology
One salient feature of pathogenic LRRK2 mutations is that they typically result in clinical phenotypes that resemble late-onset sporadic PD even though Lewy body pathology is sometimes absent or lacking (33; 85). This disconnect between clinical manifestations of PD and the presence of Lewy bodies is similarly found with disease-causing mutations in Parkin (91; 122) and supports the contention that these inclusions are not necessary for the neurodegeneration and clinical phenotypes of PD, and may instead be a consequence of the disease. Mutations outside of the catalytic domains do not appear to segregate with PD, implicating the GTPase and kinase activities of LRRK2 as central to disease development. This does not negate the importance of other LRRK2 regions in disease.
New evidence shows that mutations in the enzymatic core may impact the interaction of 14-3-3 proteins with the N-terminus of LRRK2, slightly upstream of its leucine-rich repeat domain. 14-3-3 proteins are important modulators of cell signaling and several members of this protein family can bind to phosphorylated Ser910 and Ser935 on LRRK2 (94). Importantly, numerous mutations in the Roc, Cor or kinase domains of LRRK2 impair 14-3-3 binding, and the decreased phosphorylation of Ser910 and Ser935 observed following most of these mutations might explain the reduced binding. The possibility of a common pathogenic effect caused by different LRRK2 mutations is exciting but this is dampened by the finding that the G2019S mutation does not appear to decrease 14-3-3 binding (94), hence LRRK2 mutations may cause pathology via different mechanisms.
LRRK2 kinase activity requires functional GTP binding (135) although a reciprocal dependence of GTPase function on kinase activity is not supported by in vitro studies (135). LRRK2 exists primarily as a dimer in vivo (19) and LRRK2 kinase activity may be dependent on dimer formation since LRRK2 monomers or oligomers, in contrast to LRRK2 dimers, appear to be catalytically inactive (112). Consistent with a possible role for LRRK2 dimerization in PD, several pathogenic mutations result in an increased proportion of LRRK2 dimers relative to total protein, hence disruption of dimer formation may be an important therapeutic goal (112).
LRRK2 substrates
Studies on cells and primary neuronal cultures showed that LRRK2 toxicity requires intact kinase activity (Figure 2) (44; 119). This combined with evidence that pathogenic LRRK2 variants (including the common G2019S mutation) often increase autophosphorylation or kinase activity towards pseudosubstrates has generated substantial interest in identifying authentic physiological substrates of LRRK2. To date, a number of candidate LRRK2 substrates that exhibit increased phosphorylation in the presence of PD-linked LRRK2 variants have been put forward from in vitro studies. These include members of the ERM (ezrin/radixin/moesin) protein family, which cross-link the cytoskeleton and plasma membrane (99), members of the MAPK kinase (40) and Ste20 serine/threonine kinase families (139) and eukaryotic translation initiation factor 4E-binding protein (4E-BP) (57). Additional evidence exists to suggest that 4E-BP might be an in vivo LRRK2 substrate, since manipulating expression of the Drosophila LRRK2 ortholog (dLRRK) led to an increase in 4E-BP phosphorylation in dLRRK transgenic flies and decreased phosphorylation in flies with no detectable dLRRK expression (57). 4E-BP phosphorylation disrupts its binding to eukaryotic translation initiation factor 4E (eIF4E) thus freeing eIF4E for its role in mRNA cap-dependent protein translation, suggesting that targets of 4E-BP translational regulation may be important in PD pathogenesis. A link between phosphorylated (inactive) 4E-BP and neurodegeneration is supported by the ability of overexpressed 4E-BP to suppress DA neuron pathology in flies with dLRRK mutations that increase its kinase activity. Similarly, increased 4E-BP activity arising from treatment with rapamycin or loss of dLRRK suppressed degeneration of DA neurons in PINK1 and Parkin mutants (121). Recent data from dietary restriction studies in flies suggest a possible role for 4E-BP activity in regulating mitochondrial function that could explain its protective effects following loss of PINK1 and Parkin and might be relevant to PD pathogenesis. 4E-BP is a target of the nutrient sensing protein mammalian target of rapamycin (mTOR) and in flies, upregulation of 4E-BP following dietary restriction is central to the life span extension caused by reduced feeding. More importantly, the effects of 4E-BP upregulation were mediated by enhanced mitochondrial activity, specifically due to the translational upregulation of nuclear-encoded mitochondrial gene expression (145). Hence, the positive impact of 4E-BP overexpression on mitochondrial activity may negate the otherwise deleterious effects of PINK1 or Parkin deficiencies. Mitochondrial dysfunction is consistently implicated in PD etiology and it is tempting to speculate that aberrant 4E-BP phosphorylation caused by elevated LRRK2 kinase activity might perturb 4E-BP activity under certain stress conditions that could impact mitochondrial function and possibly neuronal viability. This, of course, hinges on a number of conditions, not least the validity of 4E-BP as an authentic LRRK2 substrate, which is currently a matter of debate (73). 4E-BP phosphorylation levels were unaffected by induced or transiently transfected LRRK2 expression in HEK-293 cells, even when the G2019S or R1441C pathological variants were introduced (73). In constrast, 4E-BP was extensively phosphorylated under the same conditions by another kinase, MAPK14, raising doubt over whether 4E-BP is truly a direct substrate for LRRK2 kinase activity.
LRRK2 kinase activity is required neurodegeneration
Besides the need to identify bona fide LRRK2 substrates, it will also be crucial to determine the efficacy of LRRK2 kinase inhibitors as neuroprotective agents in PD. Indeed, a recent screen identified a number of compounds that reduce or completely block LRRK2 kinase activity in vitro (75). Interestingly, the Raf kinase inhibitors, GW5074 and sorafenib, protect against LRRK2 toxicity in cortical neurons whereas ZM336372 which inhibits Raf kinase but not LRRK2 failed to prevent LRRK2 toxicity indicating that the effects were due to impaired LRRK2 kinase activity (75). Similarly, the cyclin-dependent kinase and GSK-3β inhibitor indirubin-3′-monooxime blocked LRRK2 kinase activity and prevented LRRK2-related neuronal toxicity while its functional analog indirubin that has no effect on LRRK2 activity did not, supporting the contention that these effects were mediated via LRRK2 inhibition. Two of these compounds, GW5074 and indirubin-3′-monooxime ameliorate the loss of DA neurons following intrastriatal HSV amplicon-mediated delivery of G2019S-LRRK2 in mice (75). The ability of these compounds to prevent LRRK2 toxicity in vivo warrants additional investigation to develop selective and potent LRRK2 inhibitors and to assess their potential therapeutic benefits in humans.
LRRK2 possibly regulates protein translation
A novel twist in the potential function of LRRK2 comes from recent evidence that it may interact with the microRNA pathway to regulate translation (35). In flies, pathogenic human LRRK2 or dLRRK mutations lead to elevated levels of the cell-cycle control proteins E2F-1 and DP, which occur due to reduced translational repression by let-7 and miR-184*, respectively (35). In light of the finding that E2F-1 is aberrantly expressed in the brains of PD patients and that inappropriate expression of cell-division proteins in post-mitotic neurons may lead to cell death (54), dysregulated translation may represent an important pathogenic output of mutant LRRK2. One caveat to this interpretation is that the effects on E2F-1 and DP protein levels were seen in flies raised and aged at elevated temperatures (effectively under heat shock conditions), creating the possibility that the pathogenic effects of LRRK2 on protein translation may only be seen only under explicit stress conditions. Nonetheless, the interaction of LRRK2 with the microRNA pathway appears to be kinase-dependent and the suppression of this pathway caused by pathogenic LRRK2 with increased kinase activity further underscores the importance of kinase activity to LRRK2 toxicity and potential therapy.
Do LRRK2 and α-synuclein act in the same pathogenic pathway?
Several lines of evidence implicate a possible interplay of LRRK2 and α-synuclein in PD pathology (Figure 2). Neuropathologic examination of brains from PD patients with LRRK2 mutations indicates that Lewy bodies are a consistent finding (108). LRRK2 also co-localizes, in part, with α-synuclein during the early stages of aggregation in the brainstem of patients with PD or dementia with Lewy bodies (1) and LRRK2 immunoreactivity is occasionally detected in LBs (53). LRRK2 may modulates the neuropathological progression in PD-linked A53T α-synuclein transgenic mice (80). Neither overexpression nor ablation of LRRK2 expression in this study led to any observed neurodegeneration in mice up to 20 months of age, whereas co-overexpression of WT or G2019S-LRRK2 accelerated the progression of A53T α-synuclein-mediated neuropathological changes and led to earlier α-synuclein aggregation and abnormal accumulation in striatal neurons. Surprisingly, co-overexpression of LRRK2 lacking a kinase domain also exacerbated α-synuclein-mediated neurodegeneration suggesting that kinase activity is not central to their interaction. Importantly, knock-out of LRRK2 reduced the accumulation and aggregation of A53T α-synuclein and delayed its associated pathology (80) supporting a genetic interaction between α-synuclein and LRRK2. Investigation into possible mechanisms underlying this interaction revealed that disruptions in ER/Golgi trafficking occurred to a greater extent in double transgenic mice than those transgenic for mutant α-synuclein or LRRK2 alone and were mitigated in LRRK2 knock-out and A53T α-synuclein transgenic mice. Taken together, these studies suggest that LRRK2 might promote α-synuclein expression, aggregation and toxicity. One notable feature of LRRK2 is that expression studies in humans and rodents indicate low levels of LRRK2 expression in the substantia nigra (where DA neurons are selectively lost in PD) compared to other brain regions, including the striatum (52). If pathogenic LRRK2 promotes α-synuclein aggregation and commences a cascade of self-propagating aggregation that can transmit from cell to cell, then one interesting possibility to consider is that α-synuclein aggregation may begin outside of substantia nigra DA neurons, for example in the striatum, and eventually transfer to these neurons where it is capable of inducing toxicity.
Complicating a possible relationship between α-synuclein and LRRK2 is the fact that α-synuclein positive Lewy bodies are sometimes absent from the brains of patients with LRRK2-linked PD (33; 85). This is somewhat inconsistent with a role for LRRK2 in promoting α-synuclein aggregation although not entirely since non-fibrillar aggregates could be the predominant species in these cases. Also, multiple investigators have noted a lack of apparent α-synuclein accumulation or aggregation in the brains of LRRK2 knockout mice (3; 80), their appears to be a dramatic age-dependent accumulation and aggregation of α-synuclein in the kidneys of one line of LRRK2 knockout mice (124). Accompanying the accumulation of α-synuclein are defects in the autophagy-lysosomal pathway and apoptotic cell death (124). This finding suggests a possible role for LRRK2 in regulating protein degradation and is at odds with a positive correlation between LRRK2 expression and α-synuclein aggregation. Hence, the precise relationship between LRRK2 and α-synuclein remains somewhat obscure and will require further investigation to unravel.
PINK1 and Parkin in autosomal recessive Parkinsonism
Loss-of-function mutations in PINK1 (PTEN-induced putative kinase 1) or Parkin genes are associated with familial PD (28). Studies suggest that PINK1 and Parkin function in a mitochondrial quality control pathway that goes awry in PD, leading to cellular dysfunction and death (Figure 2) (136). Considering that mitochondrial dysfunction has an established role in sporadic PD pathogenesis, an understanding of dysregulated mitochondria turnover following loss of PINK1/Parkin function and how this contributes to neurodegeneration might provide insight into both familial and sporadic PD.
PINK1 is a mitochondrial associated protein kinase
PINK1 is a protein kinase found in the cytosol and mitochondria, where its kinase domain is thought to face the cytosol (142). PINK1 kinase activity is reduced by a number of PD-associated mutations, although other pathogenic mutations exist outside the kinase domain raising the question as to whether loss of kinase activity is required for PD pathogenesis.
Parkin is an ubiquitin E3 ligase that is inactivated in PD
Parkin is an E3 ubiquitin ligase, which tags proteins for proteasomal degradation and is localized primarily to the cytoplasm (24). PD-linked Parkin mutations give rise to early-onset Parkinsonism often with the absence of Lewy bodies and account for a large proportion of all familial PD cases (24). However, almost as many patients with Parkin mutations have Lewy bodies compared to patients that do not. Most Parkin mutations impact its E3 ligase activity or interactions with E2 enzymes such as UbcH7 and UbcH8 leading to a loss of Parkin function (116; 140). Moreover, Parkin is inactivated by nitrosative, dopaminergic and oxidative stress in sporadic PD indicating that Parkin dysfunction plays a role in autosomal recessive parkinsonism due to Parkin mutations and in sporadic PD (24). Thus, accumulation of Parkin substrates is thought to contribute to DA neurodegeneration due to Parkin inactivation in the pathogenesis of PD. Interestingly the nonreceptor tyrosine kinase, c-Abl phosphorylates tyrosine 143 of Parkin and seems to play a predominant role inactivating Parkin in sporadic PD (Figure 3)(69). Since c-Abl inhibitors and KO of c-Abl maintain Parkin in a catalytically active state, c-Abl inhibition may be an attractive therapy for the treatment of PD (Figure 3).
PARIS is required for neurodegeneration in PD due to Parkin inactivation
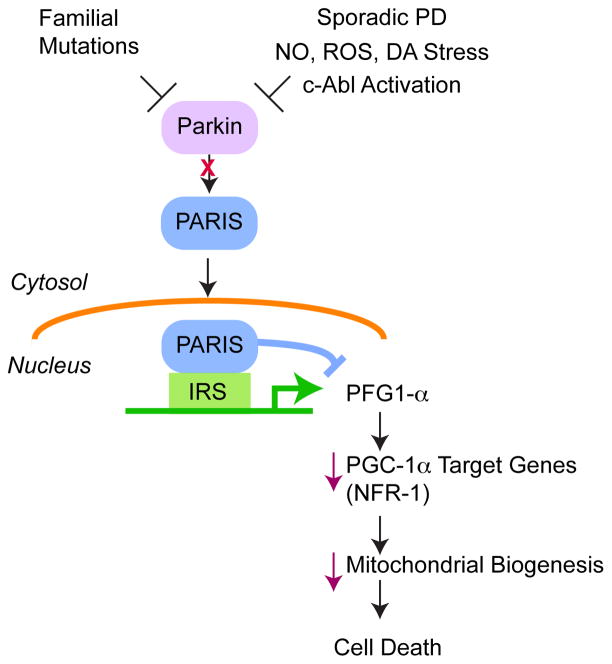
It has been difficult to reconcile a common biochemical pathway for Parkin in PD, in part, due to its ability to mono- and poly-ubiquitinate as well as ubiquitinate proteins with both lysine-48 and lysine-63 chains, as well as a diverse array of substrates (24). A new Parkin Interacting Substrate, PARIS, provides a molecular mechanism for neurodegeneration due to Parkin inactivation in PD (Figure 4) (117). Parkin regulates the levels of PARIS via the ubiquitin proteasome system (UPS). PARIS accumulates in PD brain and models of Parkin inactivation. Peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1α (PGC-1α) expression is potently downregulated by PARIS through transcriptional repression leading to progressive loss of DA neurons that is PARIS dependent in conditional knockout of Parkin in adult mice. Thus, this Parkin-PARIS- PGC-1α pathway contributes to neurodegeneration of DA neurons in PD (117). Other substrates such as the aminoacyl-tRNA synthetase interacting multifunctional protein type 2 (AIMP2) also known as p38/JTV-1 (20; 70) and far upstream element-binding protein 1 (FBP-1) (68) may also play a role in DA neuron degeneration due to Parkin inactivation (Figure 2), but PARIS upregulation is necessary and sufficient to cause DA neuron degeneration in models of Parkin inactivation.
Figure 4. Parkin controls the expression of PGC-1α through PARIS.
PARIS is a key pathogenic substrate of Parkin that accumulates in models of Parkin inactivation and in patients with Parkin mutations and PD patients due to nitrosative (NO), reactive oxygen species (ROS) and dopamine (DA) stress as well as c-Abl phosphorylation. PARIS is a transcriptional repressor that selectively down regulates PGC1-α by binding to insulin response sequences (IRS) in the PGC1-α promoter leading to downregulation of NRF-1, which transcriptionally controls mitochondrial biogenesis leading to DA neuron degeneration.
Mitochondrial abnormalities in PINK1 and Parkin deficient animals
Insight into the interaction between PINK1 and Parkin and pathological consequences of their PD-linked mutations have come from cell and animal models, especially those involving Drosophila melanogaster (14; 28; 43; 100). Complete loss of PINK1 or Parkin function in flies leads to remarkably similar phenotypes such as locomotor impairment, apoptotic muscle degeneration, reduced life span, the appearance of mitochondria with a swollen, rarified appearance and significant (although subtle) loss of DA neurons (43; 101). A contribution of oxidative stress to PD pathology is supported by the observation that flies with PINK1 or Parkin deficits have reduced resistance to exogenous oxidative stressors and moreover, the neuronal death observed in these mutants are prevented by antioxidant overexpression (132; 137). One speculative cause of endogenous oxidative stress in these flies is elevated mitochondrial ROS generation caused by respiratory chain dysfunction in impaired mitochondria.
Importantly, Parkin overexpression in PINK1 deficient flies results in a complete rescue of these phenotypes, whereas enhancing PINK1 expression did not prevent these phenotypes in Parkin mutant flies (14; 100). This genetic interaction is also supported by cell studies in which abnormal mitochondrial morphology and depolarization seen in HeLa cells with endogenous PINK1 deficits are rescued by overexpressing wild type but not PD-linked mutant Parkin (31; 100). Collectively, these studies indicate that Parkin and PINK1 act in a common genetic pathway in which PINK1 is likely to be upstream (28).
PINK1 and Parkin might regulate mitochondrial autophagy
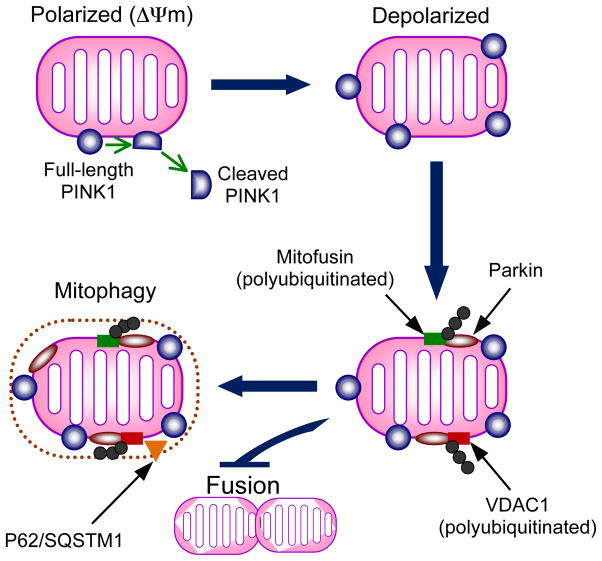
Recent evidence suggests that overexpressed Parkin might be recruited selectively to damaged mitochondria (which have become depolarized due to loss of membrane potential) to promote autophagy in a PINK1-dependent manner (Figure 5) (130). A proposed mechanism for PINK1 selective signaling of dysfunctional mitochondria centers around the voltage-dependent proteolysis of full-length endogenous PINK1 (63 kDa) in healthy mitochondria to a shorter 52 kDa form that is not retained by mitochondria and therefore maintains low levels of PINK1 on healthy (polarized) mitochondria (92). Loss of membrane potential inhibits PINK1 cleavage, leading to its accumulation on the outer mitochondrial membrane (92). Once recruited to the mitochondrion, ubiquitination of potential substrates such as VDAC1 (voltage-dependent anion channel 1) (36) and mitofusin (34; 103; 147) may be instrumental in identifying and segregating damaged mitochondria for degradation in a P62-mediated process (147). Importantly, pathogenic mutations in PINK1 or Parkin appear to perturb Parkin recruitment, substrate ubiqutination and mitophagy (36; 37) thus implicating deficits in Parkin/PINK1-dependent mitochondrial turnover in PD pathology.
Figure 5. Interaction of PINK1 and Parkin in regulating mitochondrial turnover.
The model proposed is based on recent evidence supporting PINK1-dependent recruitment of Parkin to mitochondria and subsequent Parkin substrate polyubiquitination to promote autophagy. On polarized mitochondria (mitochondrial membrane potential intact), PINK1 is cleaved into a short ~52 kDa fragment, which is released into the cytosol. This cleavage is voltage-dependent and impaired by membrane depolarization in dysfunctional mitochondria. Retention of PINK1 at the mitochondrial membrane leads to recruitment of Parkin by unknown mechanisms. Once localized to mitochondria, Parkin polyubiquitylates mitofusin and VDAC1 (voltage-dependent anion channel) and VDAC1 is required for Parkin-mediated mitochondrial clearance. Polyubiqutination of mitofusin may inactivate it and prevent fusion of the dysfunctional mitochondrion with the pool of healthy mitochondria. The autophagic adaptor protein P62/SQSTM1 is recruited to mitochondria and is required for mitochondrial clearance.
Although these studies further our understanding of potential mechanisms that underlie PD, several issues will need to be addressed to better understand the role of Parkin recruitment to mitochondria by PINK1. First, while most studies consistently show that overexpressed wild type Parkin is recruited to mitochondria in a PINK1-dependent process, it may be asked whether the same is true of endogenous Parkin and how PD-linked mutations in endogenous Parkin affect its recruitment. No study to date has shown a role for endogenous Parkin in mitochondrial turnover. Moreover, no study has shown that PINK1/Parkin regulate mitochondria in DA neurons. Also, if PINK1 expression is required for Parkin recruitment and elimination of dysfunctional mitochondria, how is it that (i) the pathological phenotypes resulting from the loss of PINK1 in cells or flies are completely rescued by overexpression of Parkin (31; 100) and (ii) overexpressing Parkin in human fibroblasts from a PINK1 nonsense mutation carrier leads to detectable levels of Parkin in the mitochondrial fraction suggestive of mitochondrial recruitment (106)? Does Parkin have additional roles besides maintaining mitochondrial integrity that allow it to rescue PINK1-deficient animals? Also, what is the precise nature of the PINK1/Parkin interaction at damaged mitochondria? Does PINK1/Parkin actually regulate mitochondrial autophagy (mitophagy) since studies to date have only examined protein markers of mitochondria, which can simply change expression patterns during fission and fusion. It would be important to show that mitophagy of mitochondria is actually regulated by PINK1/Parkin by electron microscopy and quantitative assessment of mitochondrial DNA content. Parkin phosphorylation by PINK1 has been observed (67; 113), but not consistently (131)and Parkin does not appear to ubiquitinate PINK1 or promote its proteasomal degradation (131)leaving a direct functional interaction of Parkin and PINK1 unresolved. If endogenous PINK1 and Parkin are ultimately shown to regulate mitophagy in neurons, PARIS is likely to be part of homeostatic mechanism that increases mitochondrial size and number through it regulation PGC-1α since PGC-1α is a major transcriptional regulator of mitochondrial biogenesis (120) when Parkin decreases the number of mitochondria through mitophagy in response to mitochondrial damage.
DJ-1
Homozygous loss-of-function mutations in DJ-1 result in early-onset PD (8). DJ-1, originally identified as an oncogene, is neuroprotective under oxidative stress conditions. DJ-1 deficient mice exhibit hypersensitivity to oxidative stress and exacerbated DA neuron loss following treatment with MPTP, an oxidative stress-inducing compound (66). Even under normal conditions, these mice display elevated mitochondrial H2O2 and decreased aconitase activity, a sensitive marker of oxidative stress. In addition to these biochemical phenotypes, DJ-1-deficient mice display DA reuptake dysfunction (41) although they lack any overt neurodegeneration (4; 66) suggesting that loss of DJ-1 alone is insufficient to induce cell death and environmental risk factors may be important, at least in mice.
DJ-1 plays a role in maintaining mitochondrial integrity
In human dopaminergic neuroblastoma cells, DJ-1 knock-down leads to mitochondrial depolarization and fragmentation suggesting that DJ-1 is important to maintaining integrity and function of the mitochondrial pool (123). These phenotypes are caused by oxidative stress since antioxidant treatment can prevent them (123). Oxidant-induced modification of DJ-1 at a conserved cysteine residue causes its relocalization to mitochondria, a step that appears to be necessary for its neuroprotection (11). Similarly, a Drosophila DJ-1 ortholog, DJ-1b, protects against oxidative stress and modification of the analogous cysteine residue in fly DJ-1b is required for this function (88). Loss of both orthologs, DJ-1a and DJ-1b, in flies leads to increased cell death following treatment with the ROS-inducing compounds paraquat and rotenone (87). These effects are primarily due to the loss of DJ-1b (87). Double knock-out flies exhibit reduced life span, locomotor deficits and impaired whole body mitochondrial respiration indicative of mitochondrial dysfunction (48) further supporting a role for DJ-1 in maintaining mitochondrial function.
DJ-1 is a redox sensitive chaperone with pleiotropic activities
The mechanism by which DJ-1 protects mitochondria and mitigates cell death induced by oxidative stress occurs, in part, through directly scavenge H2O2, since DJ-1 has peroxiredoxin-like peroxidase activity (4). DJ-1 has other functions, which likely contribute to its protective role. DJ-1 functions as a redox-sensitive RNA-binding protein and it regulates redox-dependent kinase signaling pathways (64; 126). Under oxidative stress conditions, DJ-1 can upregulate synthesis of the antioxidant glutathione (143), and DJ-1 stabilizes Nrf2, a critical regulator of antioxidant gene transcription (15), possibly rendering DJ-1 deficient individuals susceptible to attack from reactive oxygen species. Additional studies have indicated that DJ-1 might modulate the AKT pathway in response to oxidative stress, conferring neuronal protection through pro-survival signaling (2; 126) although work in flies does not support a genetic interaction between DJ-1 and PI3K/AKT signaling under normal conditions.
Considering the substantial evidence that DJ-1, PINK1 and Parkin share similar function in preserving mitochondrial integrity, a possible role for DJ-1 in the PINK1/Parkin pathway has been hypothesized but is largely unfavored by current findings. All three proteins were reported to form a complex that promotes ubiquitination of Parkin substrates in cells and human brain lysate (138), although there is conflicting evidence from size exclusion chromatography in human DA cell cultures, which suggest that none of the proteins form a complex (123). Genetic interaction studies in flies reveal that overexpression of DJ-1a can rescue the pathological muscle phenotypes associated with loss of PINK1, but not Parkin mutant phenotypes. Additionally, neither Parkin nor PINK1 overexpression rescues DJ-1a/DJ-1b double knock-out fly phenotypes and are, surprisingly, lethal in this background (48). In mammalian cells, loss of PINK1 leads to mitochondrial fragmentation that is rescued by Parkin but not DJ-1. Since overexpression of Parkin can rescue phenotypes associated with DJ-1 deficits, it could be hypothesized that DJ-1 acts upstream of PINK1 and Parkin. However, the observation that increased DJ-1 expression protects against rotenone-induced impaired mitochondrial connectivity with or without endogenous PINK1 expression leads to the conclusion that DJ-1 does not act in the same pathway as PINK1/Parkin (123). Given the significant overlap between phenotypes arising from loss of each of these proteins, it is likely that they act in parallel pathways with a common effect. If PINK1 and Parkin act to maintain a healthy mitochondrial pool by triggering autophagy of dysfunctional mitochondria, does DJ-1 have similar effects?
DJ-1 silencing is associated with both upregulated (59; 107; 123; 128) and downregulated (42; 72) markers of autophagy and there is no clear evidence surrounding any direct effects of DJ-1 on mitochondrial autophagy. Hence, additional studies are needed to probe the molecular underpinnings of DJ-1 protection against mitochondrial dysfunction and cell death and to examine the relationship between DJ-1, PINK1 and Parkin in the context of PD pathogenesis. One common thread is the established importance of these three proteins in protecting against oxidative stress. DJ-1 clearly protects cells against oxidative stress and elevated oxidative stress is observed in PINK1 and Parkin mutant Drosophila. In these flies, prominent phenotypes are observed in flight muscle, testes and DA neurons that have a high metabolic rate and therefore a relatively high susceptibility to oxidative stress via reactive oxygen species generation. Hence, strict mitochondria quality control (i.e. elimination of dysfunctional mitochondria by autophagy) may be necessary in tissues with high energy demand to combat the consequences of oxidative stress on cellular function, and DJ-1, PINK1 and Parkin may be crucial to this regulation.
Further evidence for mitochondrial dysfunction in PD
Considerable evidence from genetic studies on PINK1, Parkin and DJ-1 mutants described above supports the importance of mitochondrial dysfunction and oxidative stress in the pathology that results in Parkinsonian phenotypes (23; 110). Mitochondrial impairment is also thought to be central to the development of sporadic PD, triggered by environmental factors in combination with certain susceptibility gene variants and normal age-related loss of mitochondrial function (Figure 2).
Mitochondrial complex I inhibition and PD
Exposure to certain environmental toxins may be an important risk factor for PD (6). In the early 1980s, it was discovered that the injection of narcotics contaminated with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) induced acute Parkinsonism in a number of individuals and led to the selective loss of nigral DA neurons (6). Animal models have since shown that MPTP toxicity is associated with its conversion to the mitochondrial complex I inhibitor MPP+ by monoamine oxidase B followed by selective uptake into DA neurons (62). Certain environmental chemicals such as the herbicide paraquat and pesticide rotenone, are also selective complex I inhibitors, and epidemiological studies consistently point towards a significant increase in PD risk following occupational exposure to these toxins (26; 62). Defects in complex I activity are consistently observed in the substantia nigra of patients with PD (111) suggesting that mitochondrial dysfunction is central to PD pathogenesis. Over half of the proteins encoded by mitochondrial DNA assemble into complex I and certain mitochondrial DNA polymorphisms and haplotypes appear to reduce the risk for developing PD (5; 105), including a specific SNP that results in a threonine to alanine change in NADH3 of complex 1 which strongly associates with this protective effect (127).
Whether any mitochondrial or nuclear-encoded genetic variants for complex I exist that significantly increase the relative risk of developing PD is unclear although continued GWAS should help in determining this. Dominant mutations in POLG, the catalytic subunit of mitochondrial DNA polymerase, co-segregate with a multi-system disorder that includes Parkinsonism in some families (81) and result in the accumulation of mitochondrial DNA deletions in patient tissue. Two follow-up studies further implicate POLG polyglutamine tract variants in idiopathic PD (29; 82) although these results were not replicated in another study (56) indicating that POLG1 is likely to contribute substantially to the etiology of sporadic PD only in certain populations.
PGC-1α dysfunction in PD
Bioenergetic genes, including nuclear-encoded electron transport chain genes that are under control of PGC-1α are underexpressed in PD patients (141). Activation of PGC-1α in cellular models leads to an increase in nuclear-encoded subunits of electron transport chain complexes and blocks neuronal death induced by α-synuclein or rotenone. Moreover, PGC-1α is potently regulated by the Parkin substrate, PARIS (117). Thus, mitochondrial impairment is a putative disease mechanism underlying both familial and sporadic PD, and as such, genetic studies into mitochondrial dysfunction should provide valuable insight into both forms of the disease.
Conclusion
Our understanding of PD pathogenesis has been greatly advanced by genetic studies of disease-causing mutations in cell and animal models. Studies of genes linked to autosomal recessive PD clearly implicate mitochondrial dysfunction and protein mishandling in the pathological progression that leads to neuronal death and clinical PD. Most recently, insight into a putative relationship between PINK1 and Parkin in regulating mitochondrial elimination by autophagy and the regulation of the major transcriptional regulator of mitochondrial biogenesis, PGC-1α, through the Parkin substrate PARIS further emphasizes the importance of impaired mitochondrial function in PD etiology. The role of LRRK2 in PD pathogenesis awaits further clarification, although initial studies indicate an importance of kinase activity in LRRK2 toxicity, which suggests that kinase inhibitors may be relevant to the treatment of PD. As the mechanisms linking genetic mutations to the manifestation of PD become clearer, it is anticipated that our prospects for developing disease-modifying therapies will be greatly improved.
Summary Points.
Evidence points towards a role for α-synuclein in mediating the assembly of SNARE complexes that are required for fusion of synaptic vesicles to the pre-synaptic membrane. Pathogenic mutations or multiplication of α-synuclein along with other factors such as age-related oxidative stress, Aβ, GBA and DA might promote the aggregation of α-synuclein into toxic oligomers and fibrils that are the main component of Lewy bodies. Mechanisms of α-synuclein oligomer toxicity may include formation of membranous pores that permeabilize cellular membranes and self-propagated aggregation that can transmit between cells.
Several LRRK2 mutations cause late-onset PD reminiscent of sporadic disease. LRRK2 kinase activity is required for its toxic effects on neurons, and since certain pathogenic mutations increase LRRK2 kinase activity, this may be important in disease pathogenesis related to these mutations. Consistent with this hypothesis, inhibition of LRRK2 kinase activity can prevent LRRK2 toxicity in vivo.
PINK1 and Parkin are linked with autosomal recessive forms of PD and may act in the same pathway to control mitochondrial turnover. Collected evidence supports a model in which accumulation of PINK1 at damaged mitochondria leads to the recruitment of overexpressed Parkin, which somehow potentially promotes autophagic clearance of damaged mitochondria through a mechanism that likely involves poly-ubiqutination of substrates such as mitofusin and VDAC1.
The identification of PARIS as pathogenic Parkin substrate that accumulates in models of Parkin inactivation leading to DA neurodegeneration through transcriptional repression of PGC-1α. Conditional KO of Parkin in adult animals leads to loss of DA neurons that is PARIS dependent
The identification of common variants at α-synuclein and LRRK2 loci as significant risk factors for developing sporadic PD coupled to the established contribution of mitochondrial dysfunction to sporadic and familial PD indicates that both disease forms share common etiology and emphasize the importance of genetic studies to understanding the molecular underpinnings of the disease.
Future Issues.
Identifying physiological substrates for LRRK2 and PINK1 connected to the phenotypes manifested in genetic PD models is crucial. This will likely reveal key pathways mechanistic to disease and indicate novel therapeutic targets.
Understanding the relationship between PINK1, Parkin, PGC-1α, and PARIS in the pathogenesis of PD and the relationship to mitochondrial biogenesis and mitophagy.
Determining how α-synuclein oligomers and fibrils contribute to neuronal cell death, and the potential importance of oligomeric membrane pores and α-synuclein aggregate prion-like behavior is clearly important to understanding PD and related synucleinopathies.
Better understanding of the molecular relationships between PD-linked gene products will help identify key triggers and pathways in disease caused by different genetic mutations.
Acknowledgments
This work was supported by NS38377, NS048206, NS051764 and the Bachmann Strauss Dystonia and Parkinson’s Disease Foundation. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Abbreviations/Acronyms
- PD
Parkinson’s disease
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- Aβ
Amyloid β peptide
- LRRK2
Leucine-rich repeat kinase 2
- 4E-BP
eukaryotic translation initiation factor 4E-binding protein
- PINK1
PTEN-induced putative kinase 1
- VDAC1
Voltage-dependent anion channel 1
Key Terms/Definitions
- Monogenic disease
Disease caused by the inheritance of a single modified gene
- Lewy body
Cytoplasmic proteinaceous inclusions found in neurons in several neurodegenerative diseases, including PD. Lewy bodies are composed of numerous proteins, including α-synuclein
- Genome-wide association studies
A study of genetic variation across the entire human genome that is designed to identify genetic associations with observable traits or the presence or absence of a disease or condition
- Oxidative stress
Disequilibrium between the generation of reactive oxygen species and their detoxification by endogenous antioxidants leading to macromolecular oxidative damage
- Mitochondrial membrane potential
Voltage gradient across the inner mitochondrial membrane arising from the movement of protons from the matrix to the intermembrane space coupled to flow of electrons through the electron transport chain. Dysfunction of mitochondrial respiration may result in loss of this gradient, and membrane depolarization
Contributor Information
Ian Martin, Email: imarti11@jhmi.edu.
Valina L. Dawson, Email: vdawson@jhmi.edu.
Ted M. Dawson, Email: tdawson@jhmi.edu.
Literature Cited
- 1.Alegre-Abarrategui J, Ansorge O, Esiri M, Wade-Martins R. LRRK2 is a component of granular alpha-synuclein pathology in the brainstem of Parkinson’s disease. Neuropathol Appl Neurobiol. 2008;34:272–83. doi: 10.1111/j.1365-2990.2007.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleyasin H, Rousseaux MW, Marcogliese PC, Hewitt SJ, Irrcher I, et al. DJ-1 protects the nigrostriatal axis from the neurotoxin MPTP by modulation of the AKT pathway. Proc Natl Acad Sci U S A. 2010;107:3186–91. doi: 10.1073/pnas.0914876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J Neurosci. 2009;29:15846–50. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A. 2007;104:14807–12. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Autere J, Moilanen JS, Finnila S, Soininen H, Mannermaa A, et al. Mitochondrial DNA polymorphisms as risk factors for Parkinson’s disease and Parkinson’s disease dementia. Hum Genet. 2004;115:29–35. doi: 10.1007/s00439-004-1123-9. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee R, Starkov AA, Beal MF, Thomas B. Mitochondrial dysfunction in the limelight of Parkinson’s disease pathogenesis. Biochim Biophys Acta. 2009;1792:651–63. doi: 10.1016/j.bbadis.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieschke J, Russ J, Friedrich RP, Ehrnhoefer DE, Wobst H, et al. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc Natl Acad Sci U S A. 2010;107:7710–5. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 9.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 10.Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–7. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–8. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra S, Fornai F, Kwon HB, Yazdani U, Atasoy D, et al. Double-knockout mice for alpha- and beta-synucleins: effect on synaptic functions. Proc Natl Acad Sci U S A. 2004;101:14966–71. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra S, Gallardo G, Fernandez-Chacon R, Schluter OM, Sudhof TC. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123:383–96. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 14.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 15.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103:15091–6. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–20. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 17.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc Natl Acad Sci U S A. 2000;97:571–6. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–9. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 19.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci. 2010;11:791–7. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–37. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 21.Dachsel JC, Farrer MJ. LRRK2 and Parkinson disease. Arch Neurol. 2010;67:542–7. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
- 22.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 23.Dawson T, Mandir A, Lee M. Animal models of PD: pieces of the same puzzle? Neuron. 2002;35:219–22. doi: 10.1016/s0896-6273(02)00780-8. [DOI] [PubMed] [Google Scholar]
- 24.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord. 2010;25(Suppl 1):S32–9. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66:646–61. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5:525–35. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- 27.Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106:13010–5. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr Opin Neurobiol. 2007;17:331–7. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Eerola J, Luoma PT, Peuralinna T, Scholz S, Paisan-Ruiz C, et al. POLG1 polyglutamine tract variants associated with Parkinson’s disease. Neurosci Lett. 2010;477:1–5. doi: 10.1016/j.neulet.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15:558–66. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 31.Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–8. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A. 2009;106:5645–50. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaig C, Marti MJ, Ezquerra M, Rey MJ, Cardozo A, Tolosa E. G2019S LRRK2 mutation causing Parkinson’s disease without Lewy bodies. J Neurol Neurosurg Psychiatry. 2007;78:626–8. doi: 10.1136/jnnp.2006.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–41. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 37.Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–8. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 38.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, et al. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–9. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 39.Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, et al. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300:636–40. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 40.Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–68. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- 41.Goldberg MS, Pisani A, Haburcak M, Vortherms TA, Kitada T, et al. Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial Parkinsonism-linked gene DJ-1. Neuron. 2005;45:489–96. doi: 10.1016/j.neuron.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Polo R, Niso-Santano M, Moran JM, Ortiz-Ortiz MA, Bravo-San Pedro JM, et al. Silencing DJ-1 reveals its contribution in paraquat-induced autophagy. J Neurochem. 2009;109:889–98. doi: 10.1111/j.1471-4159.2009.06020.x. [DOI] [PubMed] [Google Scholar]
- 43.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–41. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Greten-Harrison B, Polydoro M, Morimoto-Tomita M, Diao L, Williams AM, et al. alphabetagamma-Synuclein triple knockout mice reveal age-dependent neuronal dysfunction. Proc Natl Acad Sci U S A. 2010;107:19573–8. doi: 10.1073/pnas.1005005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta A, Dawson VL, Dawson TM. What causes cell death in Parkinson’s disease? Ann Neurol. 2008;64(Suppl 2):S3–15. doi: 10.1002/ana.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42:781–5. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–52. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–6. doi: 10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardy J, Lewis P, Revesz T, Lees A, Paisan-Ruiz C. The genetics of Parkinson’s syndromes: a critical review. Curr Opin Genet Dev. 2009;19:254–65. doi: 10.1016/j.gde.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 51.Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7:583–90. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higashi S, Moore DJ, Colebrooke RE, Biskup S, Dawson VL, et al. Expression and localization of Parkinson’s disease-associated leucine-rich repeat kinase 2 in the mouse brain. J Neurochem. 2007;100:368–81. doi: 10.1111/j.1471-4159.2006.04246.x. [DOI] [PubMed] [Google Scholar]
- 53.Higashi S, Moore DJ, Yamamoto R, Minegishi M, Sato K, et al. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. J Neuropathol Exp Neurol. 2009;68:994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:3585–90. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong DP, Fink AL, Uversky VN. Structural characteristics of alpha-synuclein oligomers stabilized by the flavonoid baicalein. J Mol Biol. 2008;383:214–23. doi: 10.1016/j.jmb.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudson G, Tiangyou W, Stutt A, Eccles M, Robinson L, et al. No association between common POLG1 variants and sporadic idiopathic Parkinson’s disease. Mov Disord. 2009;24:1092–4. doi: 10.1002/mds.22310. [DOI] [PubMed] [Google Scholar]
- 57.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–43. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irizarry MC, Kim TW, McNamara M, Tanzi RE, George JM, et al. Characterization of the precursor protein of the non-A beta component of senile plaques (NACP) in the human central nervous system. J Neuropathol Exp Neurol. 1996;55:889–95. doi: 10.1097/00005072-199608000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Irrcher I, Aleyasin H, Seifert EL, Hewitt SJ, Chhabra S, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19:3734–46. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 60.Jakes R, Spillantini MG, Goedert M. Identification of two distinct synucleins from human brain. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- 61.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A. 2008;105:19666–71. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Javitch JA, D’Amato RJ, Strittmatter SM, Snyder SH. Parkinsonism-inducing neurotoxin, N-methyl-4-phenyl-1,2,3,6 -tetrahydropyridine: uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc Natl Acad Sci U S A. 1985;82:2173–7. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jowaed A, Schmitt I, Kaut O, Wullner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci. 2010;30:6355–9. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kahle PJ, Waak J, Gasser T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic Biol Med. 2009;47:1354–61. doi: 10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Kim JM, Hong S, Kim GP, Choi YJ, Kim YK, et al. Importance of low-range CAG expansion and CAA interruption in SCA2 Parkinsonism. Arch Neurol. 2007;64:1510–8. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- 66.Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A. 2005;102:5215–20. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y, Park J, Kim S, Song S, Kwon SK, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–80. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 68.Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J Biol Chem. 2006;281:16193–6. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- 69.Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–6. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–78. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14:504–6. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 72.Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One. 2010;5:e9367. doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, et al. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lavedan C, Leroy E, Dehejia A, Buchholtz S, Dutra A, et al. Identification, localization and characterization of the human gamma-synuclein gene. Hum Genet. 1998;103:106–12. doi: 10.1007/s004390050792. [DOI] [PubMed] [Google Scholar]
- 75.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–8. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 77.Lee VM, Trojanowski JQ. Mechanisms of Parkinson’s disease linked to pathological alpha-synuclein: new targets for drug discovery. Neuron. 2006;52:33–8. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 78.Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 79.Li JY, Englund E, Holton JL, Soulet D, Hagell P, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 80.Lin X, Parisiadou L, Gu XL, Wang L, Shim H, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–27. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–82. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 82.Luoma PT, Eerola J, Ahola S, Hakonen AH, Hellstrom O, et al. Mitochondrial DNA polymerase gamma variants in idiopathic sporadic Parkinson disease. Neurology. 2007;69:1152–9. doi: 10.1212/01.wnl.0000276955.23735.eb. [DOI] [PubMed] [Google Scholar]
- 83.Madine J, Doig AJ, Middleton DA. Design of an N-methylated peptide inhibitor of alpha-synuclein aggregation guided by solid-state NMR. J Am Chem Soc. 2008;130:7873–81. doi: 10.1021/ja075356q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manning-Bog AB, McCormack AL, Li J, Uversky VN, Fink AL, Di Monte DA. The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: paraquat and alpha-synuclein. J Biol Chem. 2002;277:1641–4. doi: 10.1074/jbc.C100560200. [DOI] [PubMed] [Google Scholar]
- 85.Marti-Masso JF, Ruiz-Martinez J, Bolano MJ, Ruiz I, Gorostidi A, et al. Neuropathology of Parkinson’s disease with the R1441G mutation in LRRK2. Mov Disord. 2009;24:1998–2001. doi: 10.1002/mds.22677. [DOI] [PubMed] [Google Scholar]
- 86.Masliah E, Rockenstein E, Veinbergs I, Sagara Y, Mallory M, et al. beta-amyloid peptides enhance alpha-synuclein accumulation and neuronal deficits in a transgenic mouse model linking Alzheimer’s disease and Parkinson’s disease. Proc Natl Acad Sci U S A. 2001;98:12245–50. doi: 10.1073/pnas.211412398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr Biol. 2005;15:1572–7. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 88.Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc Natl Acad Sci U S A. 2006;103:12517–22. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller DW, Hague SM, Clarimon J, Baptista M, Gwinn-Hardy K, et al. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62:1835–8. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 90.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 91.Mori H, Kondo T, Yokochi M, Matsumine H, Nakagawa-Hattori Y, et al. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51:890–2. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- 92.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, et al. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum Mutat. 2010;31:763–80. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Outeiro TF, Klucken J, Bercury K, Tetzlaff J, Putcha P, et al. Dopamine-induced conformational changes in alpha-synuclein. PLoS One. 2009;4:e6906. doi: 10.1371/journal.pone.0006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 98.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 99.Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–80. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park J, Lee SB, Lee S, Kim Y, Song S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 101.Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–94. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 102.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 103.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Puschmann A, Wszolek ZK, Farrer M, Gustafson L, Widner H, Nilsson C. Alpha-synuclein multiplications with parkinsonism, dementia or progressive myoclonus? Parkinsonism Relat Disord. 2009;15:390–2. doi: 10.1016/j.parkreldis.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 105.Pyle A, Foltynie T, Tiangyou W, Lambert C, Keers SM, et al. Mitochondrial DNA haplogroup cluster UKJT reduces the risk of PD. Ann Neurol. 2005;57:564–7. doi: 10.1002/ana.20417. [DOI] [PubMed] [Google Scholar]
- 106.Rakovic A, Grunewald A, Seibler P, Ramirez A, Kock N, et al. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet. 2010;19:3124–37. doi: 10.1093/hmg/ddq215. [DOI] [PubMed] [Google Scholar]
- 107.Ren H, Fu K, Mu C, Li B, Wang D, Wang G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett. 2010;297:101–8. doi: 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 108.Ross OA, Toft M, Whittle AJ, Johnson JL, Papapetropoulos S, et al. Lrrk2 and Lewy body disease. Ann Neurol. 2006;59:388–93. doi: 10.1002/ana.20731. [DOI] [PubMed] [Google Scholar]
- 109.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 110.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 111.Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–7. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 112.Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J Biol Chem. 2009;284:36346–56. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–63. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sharon R, Bar-Joseph I, Frosch MP, Walsh DM, Hamilton JA, Selkoe DJ. The formation of highly soluble oligomers of alpha-synuclein is regulated by fatty acids and enhanced in Parkinson’s disease. Neuron. 2003;37:583–95. doi: 10.1016/s0896-6273(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 115.Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, et al. An in vitro model of Parkinson’s disease: linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. J Neurosci. 2002;22:7006–15. doi: 10.1523/JNEUROSCI.22-16-07006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 117.Shin J-H, Ko HS, Kang H, Lee Y, Lee YI, et al. PARIS (ZNF746) Represses PGC-1α Contributing to Neurodegeneration in Parkinson’s Disease. Cell. 2011 doi: 10.1016/j.cell.2011.02.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–3. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 120.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 121.Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–35. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, et al. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44:437–41. doi: 10.1212/wnl.44.3_part_1.437. [DOI] [PubMed] [Google Scholar]
- 123.Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, et al. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci U S A. 2010;107:9879–84. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsigelny IF, Crews L, Desplats P, Shaked GM, Sharikov Y, et al. Mechanisms of hybrid oligomer formation in the pathogenesis of combined Alzheimer’s and Parkinson’s diseases. PLoS One. 2008;3:e3135. doi: 10.1371/journal.pone.0003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105:10244–9. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–11. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vasseur S, Afzal S, Tardivel-Lacombe J, Park DS, Iovanna JL, Mak TW. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc Natl Acad Sci U S A. 2009;106:1111–6. doi: 10.1073/pnas.0812745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Velayati A, Yu WH, Sidransky E. The role of glucocerebrosidase mutations in Parkinson disease and Lewy body disorders. Curr Neurol Neurosci Rep. 2010;10:190–8. doi: 10.1007/s11910-010-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vives-Bauza C, Przedborski S. PINK1 points Parkin to mitochondria. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12068. [DOI] [PubMed] [Google Scholar]
- 131.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang D, Qian L, Xiong H, Liu J, Neckameyer WS, et al. Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc Natl Acad Sci U S A. 2006;103:13520–5. doi: 10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang JL, Xiao B, Cui XX, Guo JF, Lei LF, et al. Analysis of SCA2 and SCA3/MJD repeats in Parkinson’s disease in mainland China: genetic, clinical, and positron emission tomography findings. Mov Disord. 2009;24:2007–11. doi: 10.1002/mds.22727. [DOI] [PubMed] [Google Scholar]
- 134.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.West AB, Moore DJ, Choi C, Andrabi SA, Li X, et al. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–32. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 136.Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 137.Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2005;102:8024–9. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiong H, Wang D, Chen L, Choo YS, Ma H, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–60. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zach S, Felk S, Gillardon F. Signal transduction protein array analysis links LRRK2 to Ste20 kinases and PKC zeta that modulate neuronal plasticity. PLoS One. 2010;5:e13191. doi: 10.1371/journal.pone.0013191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–9. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou C, Huang Y, Shao Y, May J, Prou D, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–7. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem. 2005;280:43150–8. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 144.Zhu M, Rajamani S, Kaylor J, Han S, Zhou F, Fink AL. The flavonoid baicalein inhibits fibrillation of alpha-synuclein and disaggregates existing fibrils. J Biol Chem. 2004;279:26846–57. doi: 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- 145.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–60. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 147.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–23. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]