Abstract
In the gas-phase, ions of protein complexes typically follow an asymmetric dissociation pathway upon collisional activation, whereby an expelled small monomer takes a disproportionately large amount of the charges from the precursor ion. This phenomenon has been rationalized by assuming that upon activation, a single monomer becomes unfolded, thereby attracting charges to its newly exposed basic residues. Here, we report on the atypical gas-phase dissociation of the therapeutically important, heterodimeric calcium/calmodulin-dependent serine/threonine phosphatase calcineurin, using a combination of tandem mass spectrometry, ion mobility mass spectrometry, and computational modeling. Therefore, a hetero-dimeric calcineurin construct (62 kDa), composed of CNa (44 kDa, a truncation mutant missing the calmodulin binding and auto-inhibitory domains), and CNb (18 kDa), was used. Upon collisional activation, this hetero-dimer follows the commonly observed dissociation behavior, whereby the smaller CNb becomes highly charged and is expelled. Surprisingly, in addition, a second atypical dissociation pathway, whereby the charge partitioning over the two entities is more symmetric is observed. The presence of two gas-phase conformational isomers of calcineurin as revealed by ion mobility mass spectrometry (IM-MS) may explain the co-occurrence of these two dissociation pathways. We reveal the direct relationship between the conformation of the calcineurin precursor ion and its concomitant dissociation pathway and provide insights into the mechanisms underlying this co-occurrence of the typical and atypical fragmentation mechanisms.
Keywords: Calcineurin, Heterodimer protein complex, Native mass spectrometry ion mobility MS, Protein complex dissociation, Asymmetric charge partitioning
Introduction
Protein phosphorylation is a critical process in regulating cellular events such as signal transduction and cell cycle control, and is driven by the interplay between protein kinases and phosphatases. Calcineurin (CN, also known as PP2B) is a member of the serine/threonine phosphatase family. Its activity is regulated by Ca2+ and calmodulin and plays a critical role in many calcium-dependent biological processes, for instance T-cell signaling [1–3]. For its activity, CN interacts with a variety of co-factors, scaffolds, and substrates. Conversely, the phosphatase activity of CN can be inhibited by immunophilin-immunosuppressant agents, such as cyclophilin A and FKBP-FK506 [4–9]. The nuclear factor of activated T-cells (NFAT) is a target for CN. Blocking the phosphatase activity of CN leads to T-cell suppression [5, 7, 10–12].
Full-length CN is a heterodimeric protein (78 kDa) composed of two subunits: a catalytic A subunit (calcineurin A, CNa, 59 kDa) and a regulatory B subunit (calcineurin B, CNb, 19 kDa). Wild-type CNa consists of a globular N-terminal catalytic domain, a helical CNb binding segment, a calmodulin binding segment and an auto-inhibitory peptide. CNb has four EF-hand motifs, typical for calcium binding proteins, where four calcium ions can bind [3, 13–15]. The crystal structure of CN shows that the core structure of CNa has a β-sandwich motif containing the enzyme active site. A long 22-residue helix (~40 Å) of CNa extends from the surface of the catalytic domain accommodating a binding site for CNb [16, 17]. The phosphatase activity of CN is stimulated by both calcium binding to CNb and the calcium-induced binding of calmodulin to CNa [18].
Here we use a native mass spectrometric approach to study the conformational variability of a truncated form of calcineurin (CNt, [62 kDa]), composed of CNa (44 kDa) that misses the calmodulin binding and auto-inhibitory domains, and full length CNb (18 kDa). Collision induced dissociation (CID) is used to activate, unfold and dissociate CNt in a controlled and reproducible manner. The shape/size of the ions is monitored using ion mobility mass spectrometry. During activation, which is thought to be a slow heating process, the accumulated internal energy within the complex becomes distributed and may be followed by subunit ejection. The order of sequential subunit dissociation from a protein complex provides an indication about the structural arrangement within the complex [19–22]. During this dissociation process, the leaving subunit gradually unfolds and attracts a disproportionately high amount of charges relative to its mass. This process leads to a charge dissociation pathway commonly known as “asymmetric charge partitioning” [23, 24].
However, investigating the gas-phase dissociation for CNt we observed a rather unusual dual charge distribution over the formed fragments. In addition to the common asymmetric dissociation pathway, a symmetric charge partitioning mechanism occurred concomitantly. Our data reveal that the dissociation mechanism is determined by the charge state of the precursor complex ion. Lower charge states show a preference for the symmetric charge partitioning pathway, similar to an observation by Pagel et al., showing minimal unfolding of tetrameric transthyretin prior to dissociation, consequently leading to a more evenly distribution of charges over the formed monomer and trimer [25]. Most likely CNt dissociation can follow routes in parallel, due to the presence of two conformational isomers at the start, as shown by IM-MS. The experimentally obtained collisional cross sections (Ω in Å2) of the CNt precursor ion and the fragment ions were compared with values obtained by molecular modeling. From these analyses we propose a model for the activation and dissociation of CNt whereby the more compact structure primarily follows the unusual symmetric charge partitioning pathway. The second conformer, slightly larger, displays the common protein complex dissociation behavior, whereby the small CNb subunit fully unfolds and is expelled with a high number of charges.
Experimental
Materials
Ammonium acetate (7.5 M solution, Sigma-Aldrich, Sigma), imidazole, and 3-nitrobenzoic acid (3-NBA) (for mass spectrometry, Sigma-Aldrich, Fluka, Zwijndrecht, The Netherlands) were used. The experiments were performed using a truncated form of calcineurin (CNt). Here, the CNa subunit contained the phosphatase catalytic domain and CNb binding region, but lacked the calmodulin binding domain and auto-inhibitory sequence; CNb misses amino acids 2–15. Expression and purification was performed as described previously, with only a few exceptions [26, 27]. Bacterial cells were grown at 37 °C to an OD600 of 0.5, and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16–20 h at 18 °C. The CNt complex was purified using a Ni-NTA (nickel-nitrolacetic acid) column (Qiagen), after which the his-tag was removed by thrombin. Next, the protein complex was further purified using a size exclusion and Q-coumn, using a Superdex 200 (GE Healthcare) and Q Sepharose (GE Healthcare) column, respectively.
Sample Preparation
The CNt protein complex (32.1 μM) in 20 mM Tris-HCl buffer including 100 mM NaCl and 1 mM tris(2-carboxyethyl)phosphine (TCEP) at pH 7.9, was buffer exchanged to 150 mM ammonium acetate pH 7.9, using 5 kDa MWCO spin-filter columns (Vivaspin 500; Sartorius Stedim Biotech GmbH, Göttingen, Germany). Mass spectrometric analyses were performed at CNt concentrations between 2 and 5 μM. Final concentration of 10 mM 3-nitro benzoic acid (3-NBA) or imidazole was added to the electrospray solution to enhance or reduce charging of the analytes, respectively. CNt was also sprayed in the presence of 250 μM calcium acetate.
Native (Tandem) MS
One to two μl aliquots of samples were loaded into gold-coated borosilicate capillaries made in house (using a Sutter P-97 puller [Sutter Instruments Co., Novato, CA, USA] and an Edwards scan coat six-sputter-coater [Edwards Laboratories, Milpitas, CA, USA]). (Tandem) mass spectra were acquired on a Q-Tof2 mass spectrometer (Waters Corp., Manchester, UK), modified for the analysis of macromolecular protein complexes [28, 29]. Typically, a capillary voltage of 1250 V was used and a sample cone voltage of 120 V. The source backing pressure was elevated to approximately 6 mbar in order to promote collisional cooling. Xenon was used as the collision gas at a pressure of 2×10−2 mbar. The TOF-MS was operated at a pressure of 1.25×10−6 mbar. Spectra were calibrated using an aqueous cesium iodide solution at a concentration of 25 mg/mL.
Native Ion Mobility MS
IM-MS measurements were performed on the Synapt G1 HDMS (Waters, Manchester, UK). Voltage and pressure settings were similar to those described for the Q-Tof2. The ion mobility cell was filled with nitrogen at a pressure of 0.65 mbar. A ramped wave height of 7–19 V and a wave velocity of 200 m/s were used. The trap and transfer were operated at 12 V. For collision induced dissociation experiments, the trap was operated as a collision cell. Argon was used as the collision gas at a pressure of 3.0×10−2 mbar. The collision (trap) voltage was varied between 12 and 130 V. Calibration of the ion mobility cell, essential to calculate collision cross sections, was performed as described previously using ubiquitin, myoglobin and cytochrome c [30]. Processing of MS data was accomplished using MassLynx 4.1 software (Waters Corp.).
Molecular Modeling
The CCS values of the protein complex structures were calculated using molecular modeling for a modified structure of calcineurin deposited in the Protein Data Bank (PDB: 1AUI) in order to validate the experimental IM-MS data. The quaternary structure was predicted by using the I-TASSER server [31, 32]. The geometries of the CNa and CNb subunits were extracted from the structure of the modified calcineurin using Pymol (Schrödinger, USA). Pymol was also used to predict the possible unfolded structures of the calcineurin and its dissociation products. Unfolding of the proteins was done manually followed by relaxation. To perform the unfolding of the proteins the sculpting function implemented in Pymol was used. After relaxation the coordinates of the proteins were saved in PDB files. Molecular modeling was performed with MOBCAL (using the projection approximation (PA) algorithms), described in detail elsewhere [33]. To calculate theoretical collision cross sections the projection approximation method was used, that in general is shown to be in best agreement with the experimental data of proteins and their complexes [34–36].
Results
Native MS and Tandem MS of Calcineurin
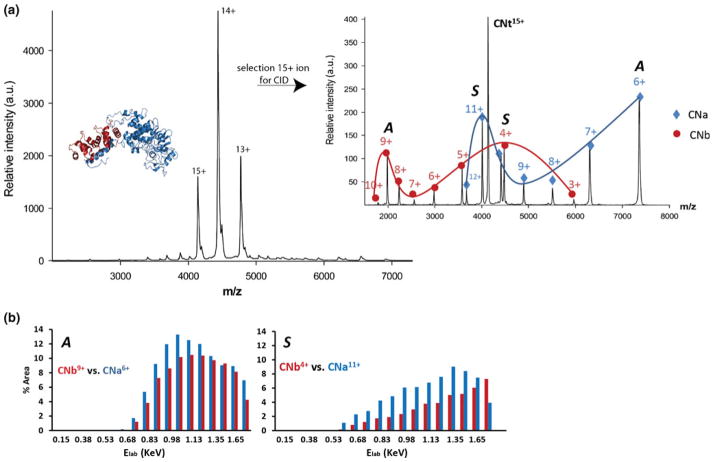
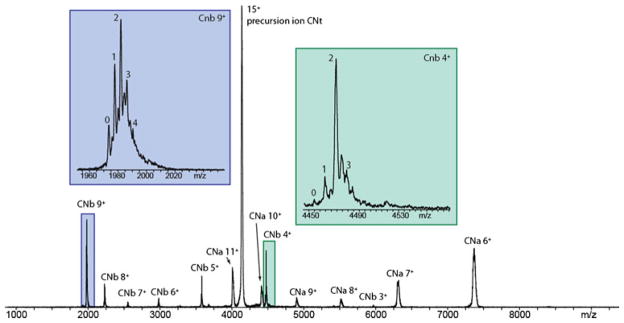
Native nano-ESI on calcineurin (CNt), sprayed from 150 mM aqueous ammonium acetate, pH 7.9, resulted in the detection of the intact hetero-dimer at three distinct charge states; 15+, 14+, and 13+, from which a molecular mass of 62,061±4 Da could be deduced (Figure 1a). To investigate the dissociation behavior of CNt, individual charge states of these precursor ions were selected and subsequently subjected to increasing collisional activation voltages. The acceleration voltage was systematically increased in steps of 5 V. Interestingly, upon dissociation from the CN15+ precursor ion, a dual charge distribution pattern was observed with two distinct maxima in the charge state distributions for both the CNa and CNb fragments, centered around 6+ and 11+ for CNa (Figure 1b, blue line), and 9+ and 4+ for the complementary CNb species (Figure 1b, red line). This indicates that in addition to the common asymmetric charge partitioning pathway (A as represented by the CNa6+ and CNb9+ ion pair), an additional dissociation mechanism occurs, resulting in a more symmetric charge partitioning between both subunits (S as represented by the CNa11+ and CNb4+ ion pair). While for very few systems symmetric dissociation has been observed, CNt is special as in it is a heterodimer. Normally it requires less energy to unfold a smaller protein to a significant extent, in this case CNb. Following this trend the common dissociation mechanism would be asymmetric, whereby a small protein has a high number of charges. For CNt we observe also highly charged CNa product ions generated via a symmetric dissociation route. Clearly, symmetric dissociation reflects the charge partitioning in relation to the protein subunits mass and not the absolute number of charges that each of them acquire after CNt dissociation. In Figure 1c, the relative abundances of the CNa and CNb product ion pairs are plotted versus the ion kinetic energy in the laboratory frame of reference (Elab, equals collision voltage multiplied by ion charge state). Selection and collisional activation of CNt14+ and CNt13+provided very similar results. We observed that the complex dissociates into its individual subunits via both the asymmetric and symmetric pathways. To investigate the possible effect of the precursor ion’s charge on the dissociation behavior, we added imidazole or 3-NBA to the electrospray solution, expanding the envelope to lower charge states with imidazole (12+–10+) and higher charge states with 3-NBA (to 18+–16+)(data not shown). At lower CNt precursor ion charge states the conventional asymmetric charge partitioning diminished and symmetric charge dissociation became more dominant. The higher charge states, on the other hand, showed both pathways in parallel, with initiation of the symmetric dissociation at slightly lower activation energy.
Figure 1.
(a) Native nano-ESI mass spectrum of CNt electrosprayed from 150 mM ammonium acetate, pH 7.9, where the most abundant peaks belong to the 15+, 14+, and 13+ charge states of the CNt hetero-dimer. The inset shows the crystal structure of CNt (CNa subunit in blue, CNb subunit in red). Inset shows a tandem mass spectrum of CNt15+ at an acceleration voltage of 80 V. Blue diamonds and line: charge states of CNa subunit, red filled circles and line: charge states of CNb subunit. (b) Relative abundances (%) of ion pair products formed as a function of the ion kinetic energy in the laboratory frame of reference (Elab, eV). The CNb9+–CNa6+ ion pair (left) represent an asymmetric charge partitioning (A) whereas the symmetric (S) dissociation pathway is illustrated by the CNb4+–CNa11+ ion pair (right)
Unfolding and Dissociation of Calcineurin Monitored by Tandem IM-MS
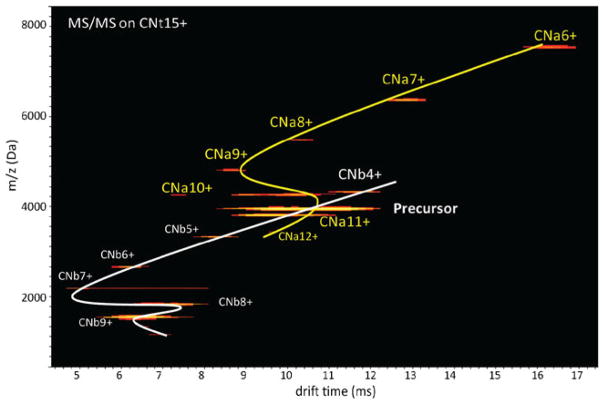
It has been well documented that the dissociation behavior of protein complexes may be linked to their three-dimensional structure [19, 22, 37, 38]. Consequently, the observation of two dissociation mechanisms for CNt may imply the existence of two conformational isomers. Native IM-MS experiments indeed confirmed this, as multiple ion mobility drift times for CNt precursor ions were observed, even at the lowest accelerating voltages. By systematically evaluating the dissociation behavior of various CNt ions we set out to reveal a possible relation between ion structure and charge state partioning behavior. The conformational changes within CNt and expelled CNa and CNb subunits were monitored by IM-MS as a function of acceleration voltage. In Figure 2, drift time profiles of CNt15+, CNa (yellow line), and CNb (white line) product ions are shown generated at an acceleration voltage of 80 V. A close investigation of the contour plot indicates the existence of multiple conformations for the CNt15+ precursor ion, as well as for some of the product ions; most notably for the CNb7+ and CNa10+ ions.
Figure 2.
Two-dimensional drift time profile (m/z versus drift time [ms]) of dissociated CN15+, at an acceleration voltage of 80 V (Elab of 1200 eV). Drift time profiles of the precursor ion and the CNa and CNb product ions are displayed. These drift time profiles were used to derive Ωs for the precursor and product ions
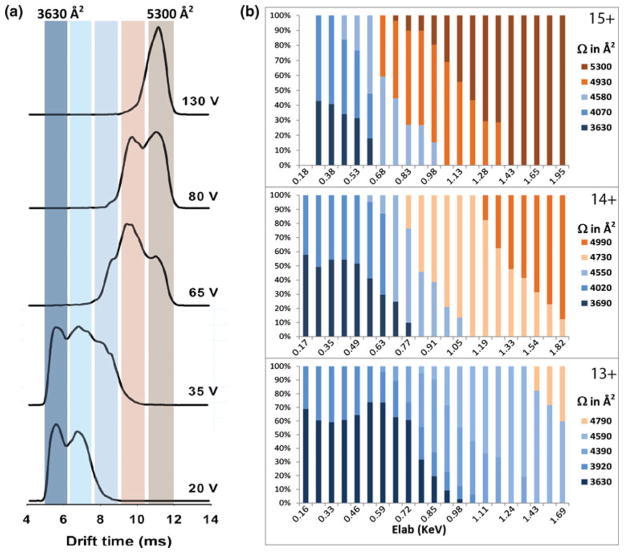
In Figure 3a, IM-MS drift-time profiles of the CNt15+ precursor ions at increasing acceleration voltages are displayed. The two drift-time maxima at 20 V for the CNt15+ ions indicate the existence of two different conformational isomers. As the collision energy is increased, the retention times of these components also increase. We can even observe the existence of multiple components at 35 and 65 V. The calculated Ω values of the CNt15+, CNt14+, and CNt13+ precursor ions are summarized in Figure 3b. Here we plotted the Ω as a function of Elab, to correct for effects of precursor ion charge. These results clearly show that independent of the selected charge state, the precursor ions exhibit more than one conformational isomer under native mass spectrometric conditions. The two major conformational isomers observed at the lowest activation energy have average Ω values of approximately 3,650 and 4,000 Å2, whereby the relative abundance of the larger structure becomes more dominant with increasing charge.
Figure 3.
(a) Zoom in of the IM-MS retention profiles of the CNt15+ precursor ion at an acceleration voltage of 20, 35, 65, 80, and 130 V (Elab: 300–1950 eV). Each maximum is interpreted as a different conformational isomer. These retention time maxima are converted into the Ωs indicated. (b) The relative abundances of the different conformers of the CNt precursor ions (15+, 14+, and 13+) as a function of the increasing Elab (eV). Color bars shown at the right side of the graphs represent different conformational isomers (Ω values, in Å2)
The reported crystal structure of calcineurin (PDB: 1AUI), allows us to calculate a theoretical Ω using the molecular modeling program MOBCAL. Here we used the projection approximation (PA) algorithm, resulting in a value of 3533 Å2. This is in close agreement with the lower experimental Ω value of 3650 Å2. It is known that for some proteins and protein complexes the PA method underestimates the Ω [39]. It is possible to apply a linear correction factor to decrease the deviation between theory and experiment [40–42]. However, it also depends on the system how significant the deviation is [34, 43]. Here we use only the theoretical Ω as a guideline to support our experimental data, whereby we focus on trends in increased or decreased Ω values.
Having calculated a relatively small Ω for CNt, we hypothesize that the larger structure detected, corresponding to a Ω of 4000 Å2, is due to partial unfolding of the CNt hetero-dimer. At this moment, we cannot rule out the possibility that the two CNt conformations originate from a collapsed and native state, instead of a native and unfolded structure. Collapsed protein structures have been observed previously, mainly at higher charge states [35]. As we still observe two distinct conformations at reduced charge states (10+–12+, not shown), collapsing is less likely to occur and the two CNt structures are expected to represent native and partially unfolded states.
Next, we computationally constructed several metastable states of unfolded CNt (using Pymol), and calculated their Ω. Partial unfolding of the CNa subunit, when still bound to CNb, results in a Ω of 4843 Å2. Conversely, partially unfolded CNb, still bound to CNa, increased the Ω to a lesser extent, to 4360 Å2. This reveals that under the conditions used, neither CNa nor CNb completely unfolds in the intact CNt complex. To study if these CNt conformational isomers result in distinct dissociation pathways, we also investigated the dissociation behavior of the lower charge states, whereby we had previously shown that the symmetric charge portioning pathway was dominating. IM-MS performed on CNt10+ precursor ion displayed a Ω of 3440 Å2, within 3 % of the Ω calculated from the crystal structure. Considering the fact that at lower charge states the smaller native-like CNt structure is more abundant, we take this as an argument that the more native conformer of CNt prefers to dissociate via the less common symmetric charge partitioning mechanism.
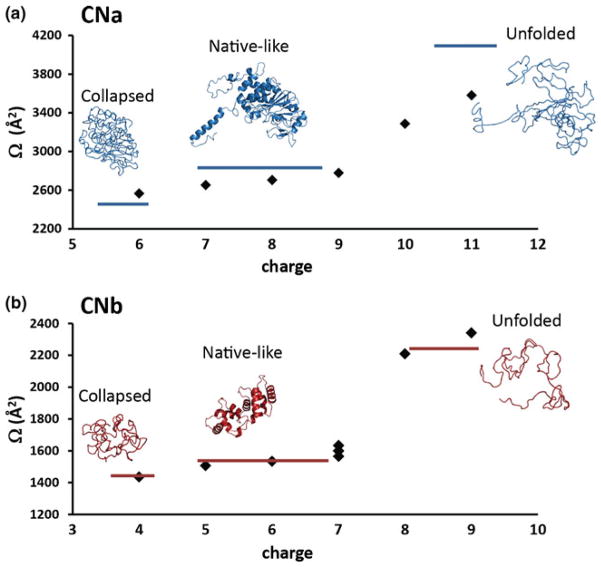
In Figure 4 the experimentally determined Ωs of all the fragment ions, formed by CID on the CNt15+ precursor ions are summarized. The theoretically calculated Ωs of the collapsed and native–like and unfolded CNa and CNb, are indicated as blue and red bars. CNa and CNb were extracted individually from the published crystal structure to calculate their native Ω using the molecular modeling program MOBCAL. Subsequently unfolded structures of the extracted native-like structures were generated using Pymol. The structures of both monomers were unfolded till a certain extent (at those positions most likely to lose their three dimensional conformation first, e.g., extended helices, flexible loops). Afterwards, these new structures were equilibrated in such a way to generate the lowest energy conformations for the particular protein states of CNa and CNb.
Figure 4.
Experimental Ωs (diamonds) and Ωs calculated from structural models of the individual CNa (a) and CNb (b) product ions as a function of their charge. The product ions were formed by performing CID on the CNt15+ precursor ions at a collision voltage of 80 V (Elab 1200 eV). The modeled structures of the collapsed, native-like and fully unfolded product ions are displayed along with their modeled Ωs (blue and red bars)
The experimental Ωs of the CNa and CNb product ions display a similar behavior with increasing charge. In both cases, the collapsed and native-like protein structures are favored when they are formed at lower charge states. The more compact structure of CNa (experimental value 2565 Å2 versus 2413 Å2 calculated) may be due to the collapse of the helix that extends from CNa, involved in CNb binding, in the gas-phase. In the intact CNt complex a helix is extending from the surface of CNa that interacts with CNb. Upon CNb removal the helix becomes an exposed and flexible part of the structure that can easily change its position towards the body of CNa. In case of CNb, that can undergo less dramatic structural rearrangements, a slightly collapsed isoform was modeled (1,427 Å2) which is in good agreement with the experimental value (1,435 Å2) as given in Figure 5b. The larger (more unfolded) conformations are observed at higher charge states. Interestingly, our data indicate that the CNb7+ charge state represents a distinct transition state between the native-like structures at lower charge states and unfolded conformations at higher charge states (Figure 4b).
Figure 5.
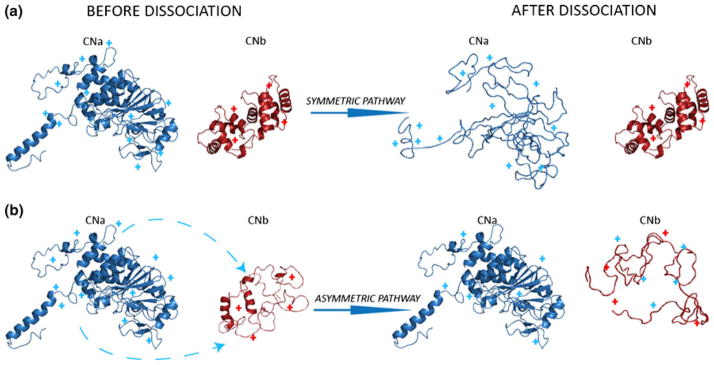
Mechanistic scheme representing the collision induced charge partitioning and dissociation pathways of the CNt heterodimer. Blue and red represent CNa and CNb subunits, respectively. For clarity, their structures are represented as individual components also prior to dissociation. (a) Symmetric charge partitioning pathway. Preceding activation CNa and CNb both remain in their native structure. During dissociation CNa partially unfolds while the structure of CNb is largely unaffected. In this scenario, there is insignificant charge relocation and thus after dissociation the charges are separated equally according to the mass of the individual subunits. (b) Asymmetric dissociation pathway. CNa remains largely in its native structure, while CNb adopts an extended conformation whereby charges are relocated from the CNa to the CNb subunit. After collisional activation CNb is fully unfolded and highly charged. The CNa fragment retains its more native like conformation
Beside the Ωs observed at 80 V acceleration voltage displayed in Figure 5, we monitored the Ωs of the product ions as a function of accelerating voltage used for CID In supplementary Figure 1 this is shown for two series of concomitant product ions, representing the symmetric and asymmetric dissociation pathway, respectively. The CNa11+/CNb4+ concomitant ion pair follows the symmetric charge-partitioning route, clearly, upon dissociation, the larger subunit CNa becomes significantly unfolded, while almost no structural change is observed for CNb. The opposite is observed for the CNa6+/CNb9+ ion pair, formed via the asymmetric charge dissociation pathway, whereby the CNa structure collapses slightly and the CNb becomes largely unfolded.
Discussion and Conclusions
The progression of mass spectrometry in the structural biology field is clearly demonstrated by the increasing number of applications, whereby the technique is used to probe inherent details on protein complex composition, conformation and topology [22, 34, 35, 38, 40, 43]. Especially, tandem mass spectrometry and ion mobility mass spectrometry are used more and more. Over the years, it became apparent that independent of the system studied, protein complexes seemed to display a generic gas-phase dissociation pathway. The assembly is energetically activated, and prior to the release of a single subunit, this subunit (partly) unfolds and attracts a disproportionately large amount of charges of the precursor ion, leaving a modestly charged stripped protein(complex) [23, 37, 44–46]. Although, the majority of all studied protein complexes follow this common trend, more recently it has been demonstrated that this dissociation behavior is not fully generic [29, 47–49]. Charge partitioning is dependent on the ion’s charge state, the conformational flexibility and location of a subunit in a larger protein complex, and interaction area with the other complex components [50, 51]. Williams and coworkers have performed detailed studies to decipher the origin of charge partitioning [45, 52, 53]. For example, ↦;-lactalbumin homodimers, of which both monomers are constrained with intramolecular cross-links primarily dissociate via the symmetric charge partitioning pathway, while the asymmetric route is favored when the monomers are not cross-linked. Clearly, for this homodimer the origin of asymmetric charge partitioning is due to the unfolding event of a single ↦;-lactalbumin monomer just before the actual dissociation event occurs [45]. However, it is still of particular interest to characterize in more detail what drives certain protein complexes to dissociate via a different route.
Here, we studied CNt, which represents an unusual protein complex system, as it can dissociate via both the symmetric and asymmetric pathway. CNt is a heterodimer, composed of a large CNa (44 kDa) and small CNb (18 kDa) subunit. Therefore, after collisional activation, the formation of highly charged CNa product ions was unexpected. However, upon dissociation of CNt, both CNa and CNb fragments ions generated were centered around two different maxima. We found this to be the result of two conformational CNt isomers that are slightly different from each other, a compact native like structure and a slightly unfolded CNt complex. Our tandem and IM-MS data indicate that the compact structure follows the unusual symmetric dissociation pathway, while the extended CNt conformation dissociates via the typical asymmetric charge partitioning route (Figure 5).
Here we postulate two different hypotheses for this unusual behavior of CNt. First, the different CNt conformations observed might correspond to known physiologic conformations of calcineurin that depend on Ca2+ binding to CNb. As mentioned previously, CNb contains four calcium binding sites (EF-hand motifs), two with high affinity and two with moderate affinity. Heterogeneity in binding of Ca2+ to these sites may induce different conformational states of CNb [54]. At the low level of Ca2+ used in the native MS buffer (no additional Ca2+ was added to the spray solution), it is likely that there is incomplete Ca2+ binding. Partially unfolded CNb, due to insufficient Ca2+ binding would be consistent with a larger cross-section and dissociation by the asymmetric route. The more compact and stabilized form of CNb might then, in turn, favor the symmetric dissociation pathway. In the calcineurin crystal structure, CNb is in a compact form, which is stabilized by Ca2+ binding to all four EF-hands and by interaction of all four EF-hands with the CNb-binding helix of CNa. There are two ways that a fraction of CNt might be stabilized in this conformation. First, at the low level of Ca2+ that is still present in the electrospray buffer, the structural Ca2+ binding sites in the C-terminal half of CNb will be occupied by Ca2+, but at least a fraction of the lower-affinity Ca2+ binding sites in the first two EF-hands will be unoccupied [18]. Second, since the CNt dimer lacks a CNa regulatory domain that would compete with the N-terminal part of CNb [54], an appreciable fraction of CNb might assume the conformation seen in the crystal structure, even in the absence of bound Ca2+. In general, our mass spectrometry data confirm this hypothesis, as we do observe differences in Ca2+ binding for the various CNb ions formed after CNt dissociation. The CNb ions formed via the symmetric route occupy a higher number of Ca2+ ions compared with the high charged CNb ions formed via the usual asymmetric dissociation pathway (Figure 6). However, none of the ions show the full occupancy of the four EF-hand motifs by Ca2+, instead we observe that native like CNb ions primarily have two Ca2+ ions bound while for the highly charged unfolded CNb ions also a significant number of ions shows the binding of only one or even no Ca2+ at all. The addition of extra calcium (as an acetate salt) to the spray solution dramatically decreased the quality of our spectra, in terms of ion intensity and resolution. Furthermore, it did not result in the full occupancy of all four sites on CNb.
Figure 6.
Calcium occupancy on the various CNb forms formed after collision induced dissociation of CNt 15+ ion. The symmetric charge partitioning route results in the formation of CNb4+ (purple box), whereby CNb is expected to retain a more native conformation. On the contrary, the CNb9+ ions (green box), originating from the asymmetric dissociation pathway, are more unfolded. In line with this hypothesis the number of calcium ions bound to CNb is significantly lower for these highly charged CNb ions. The intermediate peaks not assigned in both insets originate from Na+ adducts
Our second explanation is more focused on CNa instead of CNb. Within intact CNt an obvious characteristic of CNa is its extending helix. This non-globular protein conformation could be a possible explanation for the symmetric charge partitioning pathway to occur upon CNt dissociation. Normally, the smallest subunit unfolds and thereby increases its surface area; simultaneously the unfolding subunit gathers the majority of the ion’s charges. Under native like conditions the CNa adopts a more extended structure, due to the helix pointing away from the rest of the body of the protein, whereby more chargeable residues become accessible for protonation. Upon CNt activation and the disruption of the noncovalent interactions between CNa and CNb, charge migration occurs towards the direction of CNa that then further unfolds, eventually leading to its dissociation via the symmetric pathway. Interestingly, the number of charges are evenly shared among CNa and CNb, related to their mass, after symmetric charge partitioning. The maxima of the charge state distributions for both fragment ions lies between 4000 and 4500m/z. Still, these “relatively” highly charged CNa ions are not fully unfolded, as parts of this protein still possesses its secondary and tertiary structure. The charge state distribution of fully unfolded CNa, obtained by analyzing CNt under denaturing conditions (data not shown), is centered around 1900m/z, corresponding to an average charge state of the ion that is significantly higher (23+)as for partially unfolded CNa.
Conversely, in the slightly extended CNt structure, already some unfolding in CNb occurred and the protein complex ion will dissociate via the lowest-energy route. In this particular example, it means that the small CNb subunit fully unfolds and dissociation of CNt follows the typical asymmetric route. Here, charge division is based on the accessible surface area, whereby CNb acquires the most.
In conclusion, although it is well established that non-covalent protein complexes dissociate via the prevalent asymmetric charge portioning mechanism expelling a highly charged smaller sub-unit, it is clear, also from the current data, that the structure of individual protein complexes may influences the dissociation behavior. By combining native tandem mass spectrometry, ion mobility mass spectrometry, and computational modeling, we reveal that for calcineurin the distinction between symmetric and asymmetric charge partitioning relies on the structure/conformation of the initial precursor ions. We propose two different explanations for this behavior. Although our data provide some evidence for heterogeneity in Ca2+ binding resulting in different CNb structures, we cannot exclude our second hypothesis. Here, we state that this unusual behavior likely originates from the distinctive structure of calcineurin, wherein a 22-residue helix domain of CNa is strongly involved in CNb binding. We hypothesize that this module plays an initiating role in the dissociation via the symmetric charge partitioning pathway. When more IMMS and computational modeling data become available on a variety of protein complexes, we will be able to better understand the dissociation behavior of the these complexes under tandem MS conditions, with the ultimate goal to use this sort of information for structural elucidation purposes.
Supplementary Material
Acknowledgments
The authors thank Kristina Lorenzen for helpful discussions and Arjan Barendregt and Rebecca Rose for critical reading of the manuscript and for technical support. This work was supported by The Netherlands Organization for Scientific Research (NWO) (VENI 700.58.402, EvD). B.K., I.M.B., E.vD., and A.J.R.H. thank The Netherlands Proteomics Center, embedded in the Netherlands Genomics Initiative for financial support. B.K. was supported by the Center for Biomedical Genetics.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-012-0441-8) contains supplementary material, which is available to authorized users.
References
- 1.Rumi-Masante J, Rusinga FI, Lester TE, Dunlap TB, Williams TD, Dunker AK, Weis DD, Creamer TP. Structural basis for activation of calcineurin by calmodulin. J Mol Biol. 2011;50(2):307–317. doi: 10.1016/j.jmb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80 (4):1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 3.Wang JH, Desai R. A brain protein and its effect on the CA2+-and protein modulator-activated cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1976;72(3):926–932. doi: 10.1016/s0006-291x(76)80220-3. [DOI] [PubMed] [Google Scholar]
- 4.Stoddard BL, Flick KE. Calcineurin-immunosuppressor complexes. Curr Opin Struct Biol. 1996;6(6):770–775. doi: 10.1016/s0959-440x(96)80006-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66(4):807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 6.O’Keefe SJ, Tamura J, Kincaid RL, Tocci MJ, O’Neill EA. FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature. 1992;357(6380):692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- 7.Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357(6380):695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21(2):91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke H, Huai Q. Structures of calcineurin and its complexes with immunophilins-immunosuppressants. Biochem Biophys Res Commun. 2003;311(4):1095–1102. doi: 10.1016/s0006-291x(03)01537-7. [DOI] [PubMed] [Google Scholar]
- 10.Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365(6444):352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 11.Mattila PS, Ullman KS, Fiering S, Emmel EA, McCutcheon M, Crabtree GR, Herzenberg LA. The actions of cyclosporin A and FK506 suggest a novel step in the activation of T lymphocytes. EMBO J. 1990;9(13):4425–4433. doi: 10.1002/j.1460-2075.1990.tb07893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConnell JL, Wadzinski BE. Targeting protein serine/threonine phosphatases for drug development. Mol Pharmacol. 2009;75(6):1249–1261. doi: 10.1124/mol.108.053140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klee CB, Crouch TH, Krinks MH. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc Natl Acad Sci U S A. 1979;76(12):6270–6273. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitken A, Klee CB, Cohen P. The structure of the B subunit of calcineurin. Eur J Biochem. 1984;139(3):663–671. doi: 10.1111/j.1432-1033.1984.tb08055.x. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe Y, Perrino BA, Chang BH, Soderling TR. Identification in the calcineurin A subunit of the domain that binds the regulatory B subunit. J Biol Chem. 1995;270(1):456–460. [PubMed] [Google Scholar]
- 16.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82(3):507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 17.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378(6557):641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 18.Stemmer PM, Klee CB. Dual calcium ion regulation of calcineurin by calmodulin and calcineurin B. Biochemistry. 1994;33(22):6859–6866. doi: 10.1021/bi00188a015. [DOI] [PubMed] [Google Scholar]
- 19.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem Rev. 2007;107(8):3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 20.Benesch JLP, Ruotolo BT, Sobott F, Wildgoose J, Gilbert A, Bateman R, Robinson CV. Quadrupole-time-of-flight mass spectrometer modified for higher-energy dissociation reduces protein assemblies to peptide fragments. Anal Chem. 2009;81(3):1270–1274. doi: 10.1021/ac801950u. [DOI] [PubMed] [Google Scholar]
- 21.van Duijn E, Simmons DA, van den Heuvel RH, Bakkes PJ, van Heerikhuizen H, Heeren RM, Robinson CV, van der Vies SM, Heck AJ. Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J Am Chem Soc. 2006;128(14):4694–702. doi: 10.1021/ja056756l. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinkovic D, Murata T, Bernal RA, Stock D, Robinson CV. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334(6054):380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Light-Wahl KJ, Schwartz BL, Smith RD. Observation of the noncovalent quaternary association of proteins by electrospray ionization mass spectrometry. J Am Chem Soc. 1994;116:5271–5278. doi: 10.1016/1044-0305(94)85034-8. [DOI] [PubMed] [Google Scholar]
- 24.Sciuto SV, Liu J, Konermann L. An electrostatic charge partitioning model for the dissociation of protein complexes in the gas phase. J Am Soc Mass Spectrom. 2011;22:1679–1689. doi: 10.1007/s13361-011-0205-x. [DOI] [PubMed] [Google Scholar]
- 25.Pagel K, Hyung SJ, Ruotolo BT, Robinson CV. Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal Chem. 2010;82(12):5363–5372. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: portrait of a low-affinity signalling interaction. J Mol Biol. 2007;369(5):1296–1306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 27.Mondragon A, Griffith EC, Sun L, Xiong F, Armstrong C, Liu JO. Overexpression and purification of human calcineurin alpha from Escherichia coli and assessment of catalytic functions of residues surrounding the binuclear metal center. Biochemistry. 1997;36(16):4934–4942. doi: 10.1021/bi9631935. [DOI] [PubMed] [Google Scholar]
- 28.Tahallah N, Pinkse M, Maier CS, Heck AJ. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun Mass Spectrom. 2001;15(8):596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- 29.van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Heck AJ. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal Chem. 2006;78(21):7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 30.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3(7):1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Zhang T, Chen K, Shen S, Ruan J, Kurgan L. Sequence based residue depth prediction using evolutionary information and predicted secondary structure. BMC Bioinforma. 2008;9:388. doi: 10.1186/1471-2105-9-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shvartsburg A, Jarrold M. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett. 1996;261(1/2):86–91. [Google Scholar]
- 34.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39(5):1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 35.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310(5754):1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 36.Smith DP, Knapman TW, Campuzano I, Malham RW, Berryman JT, Radford SE, Ashcroft AE. Deciphering drift time measurements from traveling wave ion mobility spectrometry-mass spectrometry studies. Eur J Mass Spectrom. 2009;15(2):113–130. doi: 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- 37.Benesch JL. Collisional activation of protein complexes: picking up the pieces. J Am Soc Mass Spectrom. 2009;20(3):341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Lorenzen K, Vannini A, Cramer P, Heck AJ. Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure. 2007;15(10):1237–1245. doi: 10.1016/j.str.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun Mass Spectrom. 2008;22(20):3297–3304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- 40.Benesch JL, Ruotolo BT. Mass spectrometry: come of age for structural and dynamical biology. Curr Opin Struct Biol. 2011;21(5):641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge state-dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics. J Am Chem Soc. 2012;134(7):3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 42.Bleiholder C, Wyttenbach T, Bowers MT. A novel projection approximation algorithm for the fast and accurate computation of molecular collision cross sections (I) Method. Int J Mass Spectrom. 2011;308:1–10. [Google Scholar]
- 43.Uetrecht C, Heck AJ. Modern biomolecular mass spectrometry and its role in studying virus structure, dynamics, and assembly. Angew Chem. 2011;33:6247–6251. doi: 10.1002/anie.201008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beardsley RL, Jones CM, Galhena AS, Wysocki VH. Non-covalent protein tetramers and pentamers with “n” charges yield monomers with n/4 and n/5 charges. Anal Chem. 2009;81(4):1347–1356. doi: 10.1021/ac801883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jurchen JC, Williams ER. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J Am Chem Soc. 2003;125(9):2817–2826. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madsen JA, Brodbelt JS. Asymmetric charge partitioning upon dissociation of DNA duplexes. J Am Soc Mass Spectrom. 2010;21(7):1144–1150. doi: 10.1016/j.jasms.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones CM, Beardsley RL, Galhena AS, Dagan S, Cheng G, Wysocki VH. Symmetrical gas-phase dissociation of noncovalent protein complexes via surface collisions. J Am Chem Soc. 2006;128(47):15044–15045. doi: 10.1021/ja064586m. [DOI] [PubMed] [Google Scholar]
- 48.Wysocki VH, Jones CM, Galhena AS, Blackwell AE. Surface-induced dissociation shows potential to be more informative than collision-induced dissociation for structural studies of large systems. J Am Soc Mass Spectrom. 2008;19(7):903–913. doi: 10.1016/j.jasms.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wanasundara SN, Thachuk M. Theoretical investigations of the dissociation of charged protein complexes in the gas phase. J Am Soc Mass Spectrom. 2007;18(12):2242–2253. doi: 10.1016/j.jasms.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 50.Kang Y, Douglas DJ. Gas-phase ions of human hemoglobin A, F, and S. J Am Soc Mass Spectrom. 2011;22(7):1187–1196. doi: 10.1007/s13361-011-0138-4. [DOI] [PubMed] [Google Scholar]
- 51.Lorenzen K, van Duijn E. Native mass spectrometry as a tool in structural biology. Curr Protoc Prot Sci. 2010;17(Unit17.12) doi: 10.1002/0471140864.ps1712s62. [DOI] [PubMed] [Google Scholar]
- 52.Jurchen JC, Garcia DE, Williams ER. Gas-phase dissociation pathways of multiply charged peptide clusters. J Am Chem Soc. 2003;14 (12):1373–1386. doi: 10.1016/j.jasms.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurchen JC, Garcia DE, Williams ER. Further studies on the origins of asymmetric charge partitioning in protein homodimers. J Am Soc Mass Spectrom. 2004;15(10):1408–1415. doi: 10.1016/j.jasms.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang SA, Klee CB. Low affinity Ca2+-binding sites of calcineurin B mediate conformational changes in calcineurin A. Biochemistry. 2000;39 (51):16147–16154. doi: 10.1021/bi001321q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.