Abstract
The recent explosion of interest in microRNAs (miRNAs) in the nervous system has recently expanded to the investigation of their role in neurodegeneration. These studies have begun to reveal the influence of miRNAs on neuronal survival and the accumulation of toxic proteins associated with neurodegeneration as well as providing clues as to how these toxic proteins can influence miRNA expression.
In recent years, several classes of small regulatory RNAs have been identified in a variety of tissues in many species. One such class of small RNAs are microRNAs (miRNAs), 18–25 nucleotide long RNAs that are generated by a series of cleavage events from long, polymerase II-transcribed RNA1. miRNAs act to guide the RNA-induced silencing complex (RISC) to mRNAs that have a target sequence that is complementary to the miRNA (Figure 1A). The interaction between miRNA and target need not be completely complementary: the most important pairing involves nucleotides 2–7 of the miRNA, the so-called ‘seed sequence’. With some rare exceptions2, an mRNA targeted by RISC will be translationally silenced or will be destabilized and degraded3. In either case, the outcome is a decrease in protein production, with consequences for biological function that depend upon the mRNA targeted. In the nervous system these include effects on neurogenesis4–6, dendritic outgrowth7, 8 and dendritic spine formation9, 10.
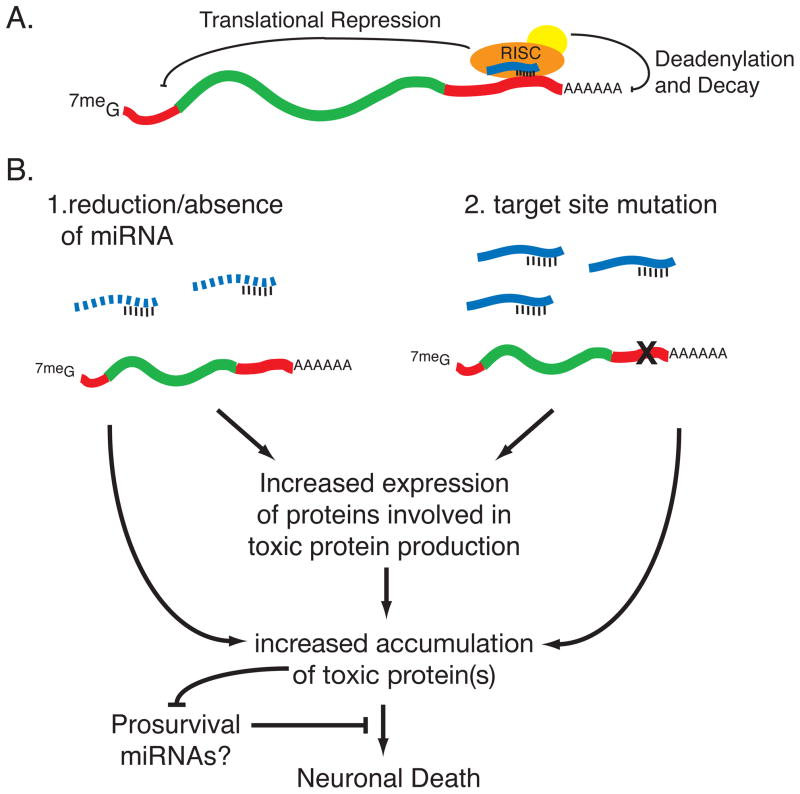
Figure 1.
Messenger RNA repression by microRNA and its affect on neuredegeneration. a| MicroRNAs (blue) bind their target mRNAs through sequences in the 3′ UTR. MicroRNAs require only a short span of sequence complementarity at the 5′ end of the miRNA to guide the RNA-induced silencing complex (RISC) to the mRNA, which promotes either translational repression or mRNA decay. Although many mechanisms for RNA silencing have been proposed, the prevailing view in the field is that RISC-mediated translational control occurs at the step of translation initiation. RISC-induced mRNA decay is thought to occur by deadenylation of the poly(A) tail followed by mRNA destruction. b| Proposed mechanisms by which miRNAs could influence neurodegeneration. Alterations in miRNA function could result from changes in miRNA expression through genetic or epigenetic changes, resulting in either the reduction or absence of the miRNA. Alternatively, mutation of a miRNA binding site in the 3′-UTR of a target mRNA can disrupt miRNA-mediated repression. miRNAs have been shown to regulate proteins involved in the production of toxic proteins as well as toxic proteins themselves. Thus, reductions on miRNA activity may lead to the increased accumulation of toxic proteins which in turn could cause neuronal death or affect the expression of as of yet unidentified prosurvival miRNAs. miR-9 may be an example of a prosurvival miRNA through its interaction with REST/CoREST34.
Alterations in the tuning of protein production can have serious consequences and is linked to many neurodegenerative diseases. For example, one extra copy of the normal α-synuclein gene was sufficient to cause Parkinson’s disease (PD) in one family11. Similarly, duplication of the gene encoding the amyloid precursor protein (APP)12, 13 or mutations in its regulatory region that increase its transcriptional activity14, 15 can lead to early onset Alzheimer’s disease (AD) or increased risk of AD, respectively. These examples highlight what is thought to be a central mechanism in neurodegenerative diseases: increased accumulation of toxic protein, leading to neuronal dysfunction.
miRNAs could modulate the accumulation of these toxic proteins by regulating the mRNA encoding the toxic protein itself or by regulating the mRNAs encoding proteins that modulate expression of the disease causing protein (Figure 1B). Furthermore, miRNAs might contribute to the pathogenesis of neurodegenerative disease downstream of the accumulation of toxic proteins by altering the expression of proteins that promote or inhibit cell survival. In this article, we discuss our current understanding of the contributions of miRNAs to neurodegenerative disease (Table 1) and consider how recent advances in technology may be implemented to advance the field.
Table 1.
Human microRNAs linked to neurodegenerative disease
| microRNA | Mode of Action | Targets | Ref. |
|---|---|---|---|
| miR-19, miR-101, miR-130 | Supresses ATAXIN1 accumulation | ATAXIN1 | 33 |
| miR-9/miR-9* | Supresses negative interaction between Huntingtin and REST/CoREST | REST and CoREST | 34 |
| miR-29a/b | Supresses accumulation of toxic Aβ peptide | BACE1 | 26 |
| miR-133b | Dopamine neuron specification and survival (?) | PITX3 | 16 |
| miR-433 | Indirectly supresses expression of α-synuclein | FGF20 | 28 |
| miR-659 | Represses GRN expression | GRN | 25 |
miRNAs support neuronal survival
Neural cell death is the defining feature of all neurodegenerative diseases and the underlying cause of many functional deficits. Understanding the pathways that promote and prevent cell death in the nervous system is therefore essential for an understanding of disease pathology and to devise effective treatment strategies. Neuronal survival is supported by a variety of proteins, including those that provide trophic support such as brain-derived neurotrophic factor (BDNF), glial-derived neurotrophic factor (GDNF), and nerve growth factor (NGF). Pro-survival proteins such as BCL-2 and BCL-xL, also act to inhibit cell death programs. miRNAs might regulate neuronal survival by inhibiting negative regulators of these pro-survival factors, or might regulate the pro-survival proteins themselves in response to survival cues such as neuronal activity.
A blunt, but useful, approach to define the roles of miRNAs in any function, including neuronal survival, is to disable the miRNA biogenesis pathway. In particular, these studies have been used to investigate the role of miRNAs in neuronal survival during development. One of the first studies to investigate the role of miRNAs in neuronal survival utilized a cerebellar Purkinje cell-specific knockout of Dicer, an enzyme essential for the generation of miRNA16. This resulted in the depletion of all mature miRNAs in these cells and was associated with a progressive neurodegenerative phenotype characterized by ataxia (loss of motor control) and Purkinje cell degeneration17.
In another study, Dicer was inactivated under the control of the dopamine receptor 1-Cre (DR1-Cre) driver which caused the gene to be deleted in the DR1-expressing neurons of the striatum18. The Dicer mutants generated with DR1-Cre had significantly lower brain masses than their wild-type littermates, a phenotype that was suggestive of neurodegeneration. Surprisingly there were signs of reactive gliosis – a condition associated with neuronal cell death – but no clear sign of degeneration in the adult DR1-Cre mutants. The authors of the study therefore suggested that the decreased brain mass might have resulted from a combination of neuronal death during development and hypertrophy of Dicer-null neurons.
Similar observations were made in Dicer mutants generated using the Calmodulin kinase II-Cre (CamKII-Cre) driver, which inactivates Dicer in several regions, including the forebrain19. These mice had substantially smaller brains than control mice, which was shown to be due in part to increased cell death in the early postnatal period. The complexity of the dendritic architecture of CA1 hippocampal neurons was also dramatically reduced in the CamKII-Cre knockouts, although it is not clear whether this was a degenerative or developmental effect. For both DR1-Cre and CamKII-Cre Dicer ablations, developmental neuronal death seemed to contribute to the gross reduction in brain mass, consistent with miRNAs playing a significant role in neuronal development. As is the case with most developmental processes, neuronal number is determined by balancing the levels of cell division and programmed cell death. Therefore, it is difficult to say whether the gross phenotypes observed in the DR-1-Cre and CamKII-Cre Dicer mutants are due to cell death, the absence of cell proliferation, or both. Deletion of Dicer in mature mouse olfactory neurons had little effect on the survival or function of these neurons, although ablation of Dicer in immature olfactory neurons results in failed differentiation of the neurons20. Deletion of Dicer in the developing cortex of mice using the Emx1-Cre line5 results in the ablation of Dicer from embryonic day (E)9.5, which corresponds to the initiation of cortical neurogenesis. As in the olfactory system, cortical progenitor pools seem unaffected by the removal of miRNAs. Furthermore, the first wave of neurogenesis seems to occur normally; however, by E12.5 massive numbers of apoptotic cells were observed in the neuronal layer of the developing cortex.
Although problems associated with the long half-lives of miRNAs and Dicer made these experiments complicated, these results seems to suggest that miRNA are not required in the neural progenitors, but are required for the specification and survival of some types of mature neurons. Whether key individual miRNA/target pairs or broad regulation of the neuronal proteome is the critical factor for neuronal survival in these knockout models remains to be determined. Together these studies have established a critical role for miRNAs in the survival of neurons, but the precise mechanism by which miRNA promote survival will require further research.
A direct connection between a pro-survival gene and a miRNA in a neurodegenerative disease was made by the discovery of a common single nucleotide polymorphism (SNP) in the 3′ UTR of the progranulin (GRN) gene. GRN is a secreted protein with anti-apototic properties that have been described outside the CNS21. Significantly, mutations in GRN are linked to familial forms of frontotemporal lobe demntia (FTLD)22–24. This 3′ UTR SNP was found to be associated with a sub-type of frontotemporal lobe dementia 25 and enhances the ability of the human-specific miR-659 to bind to and regulate the translation of GRN mRNA. The authors showed that GRN protein is reduced in tissue from patients with the disease-associated SNP. Using quantitative reverse transcription polymerase chain reaction (qRT-PCR), they found that miR-659 is expressed in the brain. However, miR-659’s relative abundance in the tissue affected in FTLD is unknown, making the relevance of this particular miRNA in the pathogenesis of FTLD currently unclear. Nevertheless, this study may provide the basis for further fruitful inquiry into the role of miRNA regulation in FTLD.
miRNAs alter protein accumulation
A common theme among many neurodegenerative conditions is the accumulation of proteins that are toxic to neurons. There are many points in the pathways leading to production of toxic proteins at which miRNAs could conceivably act and at which changes in miRNA activity might be important. Most directly, there might be a loss of direct miRNA regulation of a toxic protein’s mRNA. But perhaps equally as importantly, there could be a loss of miRNA-mediated regulation of proteins involved in the production or degradation of toxic proteins.
A recent study profiled miRNAs in AD patients and age-matched controls and found a small number of miRNAs with modestly altered expression levels26. Armed with this data, the authors used in silico predictions of miRNA targets to determine which miRNAs to pursue (Box 1). Among the down-regulated miRNAs, miR-29a/b were shown to be capable of regulating the beta-site APP cleaving enzyme 1 (BACE1) 3′ UTR in a luciferase reporter assay. BACE1 has a central role in producing the toxic Aβ peptide, the principal component of the plaques that characterize AD. Indeed, the authors demonstrated a significant correlation between lower expression of miR-29a/b and higher expression of BACE1 in brain tissue. miR-29a/b is thought to be ubiquitously expressed in neurons and astroglia, suggesting that the specific reduction in its expression is not a secondary consequence of the death of specific neuronal populations. Transfection of miR-29a/b in HEK293 cells expressing APP – the precursor from which Aβ peptide is formed – significantly reduced the production of the Aβ peptide. This suggests that miR-29a/b is capable of significantly modulating BACE1 and consequently the production of the toxic Aβ peptide. Currently there is no evidence to indicate that genetic polymorphisms in miR-29a/b or the 3′ UTRs of APP or BACE1 contribute to AD, suggesting that the miR-29a/b-BACE1 interaction may not be causative in familial forms of AD27.
Box 1. miRNA Target Prediction Programs.
A miRNA requires very few base pair interactions to effectively silence a target mRNA. The most critical region for interaction is the ‘seed sequence’ (nucleotides 2–7 of the miRNA). Since a sequence of this length will occur with high frequency in the genome by chance alone, predicting functional miRNA target sites, even within the constraint of 3′ UTRs, is challenging. Most target prediction programs identify hundreds of potential targets for any given miRNA. This may lead to user bias since it is likely that there will be a gene of interest within the list of hundreds of predicted targets. This is particularly true for heavily studied genes with long, well annotated 3′ UTRs. Most successful target prediction alogrithims rely on evolutionary conservation to identify 6-mer, or longer, seed region homologies that may indicate functional homology. This obviously precludes clade-specific target sites that may have important functional significance. Two recent studies of miRNA mutants were used to investigate the impact of the loss of a single miRNA on proteome-wide protein level41, 42. These two studies agreed that the most comprehensive and accurate prediction programs are currently TargetScan and PicTar. Diana-microT has also been evaluated with favorable outcome. Despite the relatively good performance of TargetScan and PicTar, two-thirds of their predicted targets were not affected by the absence of the miRNA41 (see REF 43 for a review).
Another mechanism by which miRNAs could affect protein accumulation in neurodegenerative disease is through polymorphisms in 3′ UTRs that could either add or eliminate miRNA target sites in mRNAs. For example, a recent study of risk factors for PD identified a point mutation in the 3′ UTR of the fibroblast growth factor 20 (FGF20) gene that disrupts a miR-433 binding site28. FGFs have been shown to regulate α-synuclein in vitro29 and increases in α-synuclein expression can act as a causative agent in the development of PD30. The 3′UTR polymorphism can increase the expression of FGF20 and alter the expression of α-synuclein in cell culture. One major caveat of these findings is that the low level of miR-433 detected in the brain may mean it is unlikely that this specific interaction has relevance to PD pathology. However, it is possible that miR-433 is expressed in a minor cell population with relevance for PD. To address this possibility, future studies could use in situ hybridization to determine if dopamine neurons or neighboring cells are enriched in miR-433 expression. This could lending credence to the hypothesis that a miRNA with a low expression level could effect α-synuclein expression though FGF signaling in disease-relevant cell types. However, it is also worth noting that a recent study was unable to reproduce any association between PD and FGF2031.
One of the early observations that suggested that miRNAs might be involved in neurodegenerative disease was made in a Drosophila model of spinal cerebellar ataxia32. In this study, neurodegeneration caused by overexpression of a poly-glutamine expanded human ataxin in the Drosophila eye was enhanced by a heterozygous mutation in Dicer1. Conversely, overexpression of the bantam miRNA suppressed ataxin-induced neurodegeneration, although the mechanism by which the suppression occured remains unclear. Overexpression of poly-glutamine expanded human ataxin is also toxic in HEK293 cells; in this model, as in the fly eye, ataxin toxicity was enhanced by knockdown of Dicer suggesting that the enhancement is a generalizable phenomenon. Although this study did not produce evidence that miRNAs directly regulate ataxin expression, a related study showed that a variety of miRNAs were capable of regulating the human Ataxin mRNA in HEK293 cells33. Importantly, the authors showed that these miRNAs are expressed in Purkinje cells, one of the main target cell types of the disease. Either the knockdown of these miRNAs or the deletion of the miRNA binding sites in the 3′ UTR of the Ataxin mRNA accentuated Ataxin-induced toxicity in HEK293 cells. Experiments in Purkinje cells and in vivo will be required to further determine the importance of these miRNA-Ataxin interactions in the brain.
miRNAs downstream of toxic proteins
Although the accumulation of toxic proteins is thought to be the cause of many neurodegenerative conditions, the mechanism by which the toxic proteins cause cell death remains controversial. One possible mechanism by which they might do this is by interfering with miRNA-mediated regulation of pro-survival proteins.
Profiling of miRNA expression in tissue from patients with Huntington’s Disease (HD) demonstrated significant decreases in miR-9/9* expression as disease progressed34.. The authors showed that alterations in miR-9 and miR-9* could affect the expression of the RE-1 silencing transcription factor (REST) and its co-repressor protein, CoREST. This was significant because upregulation of the REST repressor complex can have deleterious effects on neuronal gene expression, repressing pro-survival genes like BDNF that may contribute to HD pathogenesis35. These results, together with those of other recently published HD studies suggest that a reciprocal transcription-translation circuit exists through which REST can in turn repress miR-9 transcription. The activity of REST is increased by mutant Huntingtin protein35, the protein that causes HD. Previous studies have shown that REST binds to DNA upstream of neuronally expressed miRNAs including the three genomic loci from which miR-9 is transcribed, as well as miR-124 and miR-13236.
The interaction between REST and the upstream regions of miRNAs was confirmed by another group studying the impact of REST-mediated transcription in the context of HD37. These authors also investigated the expression of miRNA in post-mortem HD brain. Although many of the conclusions of these two studies were similar, there was little similarity between findings regarding the expression of miRNAs in post-mortem HD brain. For instance, REF 37 reported a significant decline in miR-132 expression in post-mortem HD cortex, whereas REF 34 noted a significant increase in miR-132 expression in late stage HD brain. This may be a consequence of differing methods of measuring miRNA expression levels, the noted heterogeneity in gene expression profiles in HD brains, or may indicate that the small sample size of each study is insufficient to power analysis of the data.
Profiling post-mortem tissue also identified reduced miR-133b in the midbrain of PD patients16. One trivial explanation for the reduction of mR-133b in PD patients is that it is enriched in the dopamine neurons that are lost during disease progression. This seems to be the case as miR-133b is enriched in the midbrain and is depleted in mouse models that are deficient in dopamine neurons, suggesting that loss of miR-133b is downstream of the accumulation of toxic protein and dopamine neuron death. However the authors of this study also uncovered a developmental feedback loop through which PITX3, a transcription factor that has a key role in dopamine neuron development, regulates miR-133b transcription and miR-133b in turn represses PITX3 synthesis. Whether this regulatory loop contributes to the survival of midbrain dopamine neurons, or whether miR-133b regulates other important factors in dopamine neurons remains to be determined.
Future Efforts
Exploiting miRNA biology to understand and treat neurologic disease is a novel and exciting opportunity. How will our understanding of miRNA in neurodegeneration intersect with therapeutics? miRNA-based interventions that enhance the endogenous neuroregenerative or neuroprotective capacity of the CNS are of course attractive. However, at the current time, targeting specific miRNAs in order to directly treat neurodegenerative diseases faces many challenges. The first challenge is understanding the breadth of the regulation of proteins by miRNAs. A single miRNA may regulate the expression of a few proteins or a large network of proteins. The cellular feedback loops and regulation of miRNA expression are not yet known. There is also a paucity of validated miRNA targets and there are difficulties in delivering of miRNA reagents to the brain. However, these are resolvable challenges. The delivery of oligonucleotides, ribozymes, siRNA and mRNA using viral and non-viral methods for gene therapy is currently the focus of extensive efforts in medical research. Although effective delivery to neural tissues has yet to be realized, significant advances have been achieved through nanotechnology38. Using oligonucleotide-based miRNA therapy has the advantage of being transient, whereas viral or transgenic modification presents a variety of risks including viral-induced inflammation and oncogenesis.
Bearing in mind these challenges, there may nevertheless come a time when regulation of the expression or activity of miRNAs may be possible and have a clear therapeutic benefit. One possible strategy would be to enhance the expression miRNAs that target toxic proteins by providing either synthetic miRNA or virally expressed miRNA. Inhibiting miRNA-mediated repression of a neuroprotective mRNA may also represent an important therapeutic approach. Using miRNA in an ex vivo setting to expand cultures or to push stem cells to adopt appropriate fates also holds promise for models of neurodegenerative disease or replacement therapies. All of these approaches will require significant advances in delivery technology in order to be successful In addition, a precise understanding of miRNA-target relationships, including the cells in which each are expressed, is an absolute requirement for therapies to be effective. In order to arrive at this point there must be improvements in how we investigate miRNA expression and function.
One such improvement would be the use of cell-type specific miRNA expression profiles. Although there is clear value in expression profiling of tissue from disease patients, knowing where and when the miRNAs are expressed has important implications not only for disease mechanisms, but also for possible therapeutic intervention. Recent advancements in in situ hybridization techniques may rapidly improve what has been a largely ignored caveat to miRNA profiling studies39. Similarly, very few profiling experiments have begun to consider miRNA expression in neurons versus glia. This is an obvious and important question when considering the mechanism of miRNA action. Currently, our understanding of miRNA action suggests that they are stoichiometric inhibitors of mRNA translation. Therefore, only the most abundant miRNAs in a given tissue or cell type should be considered relevant to the biology of the tissue or cell. This further emphasizes the importance of localization of miRNAs to specific cell types in the identification of miRNAs that are likely to have relevance in neurodegenerative conditions.
Unbiased methodologies for miRNA target identification will also be an important advance. In silico methods for target identification are constantly improving (Box 1). Although already much improved, experimental methods are still emerging and will be essential tools for understanding how changes in miRNA expression will affect the proteome of a cell. A recent study may point the way toward resolving this question40. By using high-throughput sequencing coupled with cross-linking immunoprecipitation (HITS-CLIP) of RNA that is bound by RISC, the authors were able to identify both the miRNAs that are expressed in the brain and their likely mRNA targets. This represents an important leap forward in experimental miRNA target identification but still does not resolve questions of cell-type specific miRNA expression. It is likely that these technical advancements will yield significant details that will push forward our understanding of the neural miRNA system in health and disease.
Acknowledgments
We apologize to authors whose papers were not discussed here due to the short format. This work was funded by USPHS DA00266. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–9. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–9. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Pietri Tonelli D, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–21. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. Embo J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–16. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 11.Chartier-Harlin MC, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–9. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 12.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–6. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 13.Sleegers K, et al. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–83. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 14.Theuns J, et al. Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am J Hum Genet. 2006;78:936–46. doi: 10.1086/504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brouwers N, et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer’s disease. Brain. 2006;129:2984–91. doi: 10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–8. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuellar TL, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–9. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–30. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi PS, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. Bioessays. 2009 doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 22.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 23.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 24.Gass J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 25.Rademakers R, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–42. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettens K, et al. APP and BACE1 miRNA genetic variability has no major role in risk for Alzheimer disease. Hum Mutat. 2009;30:1207–13. doi: 10.1002/humu.21027. [DOI] [PubMed] [Google Scholar]
- 28.Wang G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rideout HJ, Dietrich P, Savalle M, Dauer WT, Stefanis L. Regulation of alpha-synuclein by bFGF in cultured ventral midbrain dopaminergic neurons. J Neurochem. 2003;84:803–13. doi: 10.1046/j.1471-4159.2003.01574.x. [DOI] [PubMed] [Google Scholar]
- 30.Skovronsky DM, Lee VM, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol. 2006;1:151–70. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 31.Wider C, et al. FGF20 and Parkinson’s disease: no evidence of association or pathogenicity via alpha-synuclein expression. Mov Disord. 2009;24:455–9. doi: 10.1002/mds.22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–63. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y, et al. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci. 2008;11:1137–9. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 36.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson R, et al. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–45. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Midoux P, Pichon C, Yaouanc JJ, Jaffres PA. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br J Pharmacol. 2009;157:166–78. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pena JT, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–41. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009 doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]