Summary
Meiosis expressed gene 1 (Meig1) was originally identified in a search for mammalian genes potentially involved in meiosis. Seven mouse Meig1 transcripts with the same coding region, but different 5′-UTRs, have been identified. These transcripts have different tissue distributions, two are only present in the testis. In the testis, Meig1 is present in germ cells and Sertoli cells. A Meig1 conditional knockout model has been generated. When Meig1 was inactivated globally by crossing with Cmv-Cre transgenic mice, the Meig1-deficient males were sterile due to severe spermiogenic defects, and had no obvious defects in meiosis. To further study its role in individual cell types in the testis, the Meig1flox mice were crossed with Hsp2a-Cre, Prm-Cre, and Amh-Cre mice, in which the Cre recombinase is driven by the heat shock protein 2 (Hsp2a) gene promoter (expressed in spermatocytes), the protamine 1 gene promoter (expressed in post-meiotic spermatids) and the anti-Mullerian hormone (Amh) gene promoter (expressed in Sertoli cells) respectively. Both Meig1 mRNA and protein were undetectable in testis of the Hsp2a-Cre; Meig1flox/flox mice and all the mutant adult males tested were sterile. This phenotype mirrors that of the Cmv-Cre; Meig1flox/flox mice. Even though the total testicular Meig1 mRNA and protein expression levels were dramatically reduced in testis of the Prm-Cre; Meig1flox/flox males, all the mice tested were fertile, and there was no significant difference in sperm count and sperm motility compared with age-matched Meig1flox/flox male mice. Disruption of Meig1 in the Sertoli cells did not affect the MEIG1 protein expression. Amh-Cre; Meig1flox/flox males were fertile, and produced the same amount of spermatozoa as age-matched Meig1flox/flox mice. The testicular histology was also normal. Our results indicate that MEIG1 regulates spermiogenesis through effects in germ cells alone, and that the Meig1 gene must be active during a discrete period in spermatogenesis after which it is dispensable.
Keywords: germ cells, -Meig1, mouse, spermiogenesis, testis
Introduction
Mouse meiosis expressed gene 1 (Meig1) was originally identified in a search for mammalian genes involved in meiotic processes (Don & Wolgemuth, 1992). Two murine Meig1 transcripts, 11a2 and 2a2, were identified previously (11a2 and 2a2 are ID number of the clones identified, 11a2 also called Meig1_v1, and Meig1-002 in Ensembl database; 2a2 also called Meig1_v2, and Meig1-001 in Ensembl database); both contain three exons. The two isoforms share the same open reading frame and 3′-UTR, but differ in their 5′-UTRs. Each has a non-translated exon 1. Meig1_v1 message was found to be expressed specifically in somatic cells in the testis. However, the predominant Meig1_v2 isoform was found to be germ cell-specific. The Meig1_v2 transcript begins to accumulate in testis at days 8–9 of post-natal (pn) development, coinciding with the entry of germ cells into meiosis, and is expressed most abundantly at pn day 14, and at subsequent stages, when spermatocytes enter the pachytene stage. In situ hybridization analysis showed that the Meig1 expression level was low in leptotene cells, and increased as the cells progressed through zygotene and pachytene stages. In addition, Meig1 message was also detected in embryonic ovary after day 15 of gestation when the cells entered the pachytene stage of meiosis 1, but not in adult ovary, suggesting that Meig1 is a meiosis-associated gene (Don et al., 1994; Chen-Moses et al., 1997; Steiner et al., 1999). Recent transcriptional profile studies revealed that Meig1 message is also present in Sertoli cells in foetal gonads, and a Sertoli cell line TTE3 (Tabuchi et al., 2006; Bouma et al., 2007).
Although the molecular weight of MEIG1 is estimated to be 10 kDa based on its amino acid composition, the protein migrates as a 14 kDa band in Western blots, probably because of its basic isoelectric point. However, MEIG1 protein contains multiple consensus sequences for serine and threonine phosphorylation. Therefore, post-translational modifications might also affect MEIG1 mobility in the electrophoretic separation. There is evidence that MEIG1 protein is phosphorylated and can form a dimer in vivo. Furthermore, it has been suggested that the phosphorylated dimer enters the nucleus during the first meiotic prophase and binds to meiotic chromatin (Ever et al., 1999).
Several studies suggested that MEIG1 is essential for spermatogenesis, and is related to ciliary function. Meig1 message is dramatically reduced in heat shock transcription factor 2 (Hsf2) mutant mice, and this may result in impaired spermatogenesis and reduced fertility in Hsf2 mutant mice (Wang et al., 2003). Bioinformatic analysis revealed that Meig1 is most abundantly expressed in tissues rich in highly ciliated cells, such as, testis, lung and olfactory sensory neurons, and is therefore predicted to be important for cilia (McClintock et al., 2008). Our previous study showed that MEIG1 is associated with WD repeats of SPAG16 protein (Zhang et al., 2004), and Spag16 mutant mice lacking the WD repeat region have a severe spermatogenesis defect due to haploinsufficiency, suggesting that MEIG1 might function in the same complex as SPAG16 protein.
To study the function of mouse MEIG1 protein, we further characterized this gene. Instead of two, seven mouse Meig1 transcripts have been identified in our laboratory, all are translated into the same protein (Zhang et al., 2009). These seven transcripts have different tissue distributions. As noted above, two are only present in the testis (Zhang et al., 2009). Even though no known functional domains have been identified, MEIG1 protein sequences were found to be highly conserved among different species (Salzberg et al., 2010).
To further study the role of the Meig1 gene, a Meig1 conditional knockout model has been generated in our laboratory. The Meig1 gene was inactivated globally by crossing the conditional knockout mice with Cmv-Cre transgenic mice. The global Meig1 knockout mice revealed that homozygous mutant mice are viable, but the males are sterile, producing only a few spermatozoa that are morphologically abnormal. Unexpectedly, the Meig1 mutant male mice had no obvious defect in meiosis, as was originally predicted from earlier work by Don and colleagues (Don & Wolgemuth, 1992; Don et al., 1994; Chen-Moses et al., 1997; Steiner et al., 1999). The mice were sterile as a result of impaired spermatogenesis at the stage of elongation and condensation (Zhang et al., 2009). The manchette was dramatically disrupted in the Meig1-deficient spermatids when viewed by transmission electron microscopy. The male infertility phenotype was later confirmed by a conventional Meig1 knockout model generated recently by the Don laboratory (Salzberg et al., 2010).
To investigate MEIG1's role in germ cells and Sertoli cells, the Meig1flox/flox mice were crossed with transgenic mice that express Cre recombinase specifically in these individual cells, and the phenotypes were analysed. Our results demonstrate that MEIG1 plays an essential role in germ cells, but not in Sertoli cells.
Materials and Methods
Generation of cell type-specific Meig1 mutant mice
Amh-Cre (Stock Number: 007915), Hsp2a-Cre (Stock Number: 008870), and Prm-Cre (Stock Number: 003328) mice were purchased from Jackson laboratory (Bar Harbor, ME, USA). These transgenic mice were crossed with Meig1flox/flox mice, and the resulting Cre; Meig1flox/+mice were backcrossed with Meig1flox/flox mice to create the Cre; Meig1flox/flox mice. The detailed breeding strategy used to create conditional Meig1 mutant mice is shown in Supplemental Figures 1, 3 and 5.
Genotyping and determination of tissue/cell type-specific disruption of the Meig1 gene
Genotypes of floxed Meig1 mice were determined by PCR using DNA isolated from tail slips following the same strategy as described previously (Zhang et al., 2009). PCR was also conducted to identify Cre-positive mice using a primer set that can amplify the Cre recombinase gene with the following primers: forward primer: 5′-CAGTTCGATTACGATCAG-3′, and reverse primer: 5′-GACTTAGGCTATCGTAC-3′.
To determine tissue-specific deletion of the floxed Meig1 region, DNA was isolated from the indicated tissues of the Cre-Meig1flox/flox mice, and PCR was conducted using the primer set that flanks the floxed region. Testicular germ cells and somatic cells were also collected from adult Cre-Meig1flox/flox mice, and DNA was isolated for PCR to determine cell type-specific deletion of floxed Meig1 region. Briefly, mixed testicular cells were prepared from adult Cre-Meig1flox/flox mice by enzymatic dissociation with collagenase (1 mg/mL) at 34 °C for 12 min, followed by digestion with trypsin (0.5 mg/mL) for 10 min at 34 °C. The cells were washed with DMEM, and suspended with DMEM (10% FBS), and cultured at 34 °C. Six hours after plating, somatic cells stick to the culture plates, and germ cells are suspended in the medium, thus the two populations were separated (Chang et al., 2011). Beside Sertoli cells, the somatic cell preparation also contains a small number of Leydig cells and peritubular cells.
Northern blot analysis
Total RNA was isolated from mouse testes with Trizol (Life Technologies, Inc., Grand Island, NY, USA). A quantity of 30 μg of these RNAs was separated on a 1% denaturing agarose gel. The RNA was subsequently transferred to a Hybond N+ nylon membrane. The blots were hybridized with the 32P-α-dCTP labelled Meig1 probe generated previously (Zhang et al., 2009). After hybridization, the membranes were exposed to X-ray films.
Western blot analysis
Equal amounts of protein (50 μg/lane) were heated to 95 °C for 10 min in 4 × sample buffer, loaded onto 10% sodium dodecyl sulphate-polyacrylamide gels, electrophoretically separated and transferred to polyvinylidene difluoride membranes. The membranes were blocked, and then incubated with indicated antibodies overnight at 4 °C. After being washed, the blots were incubated with an anti-rabbit, or anti-mouse immunoglobulin conjugated to horseradish peroxidase for 1 h at room temperature. The proteins were detected with Super Signal Pico Chemiluminescent system (Pierce, Rockford, IL, USA).
Assessment of fertility and fecundity
To assess fertility and fecundity, littermate males (3–4 months old) of different genotypes were each placed in cages with two mature wild-type females for at least 2 months. The number of mice achieving a pregnancy and the number of offspring from each mating set or pregnancy were recorded.
Sperm motility assays
Spermatozoa were collected after swimming out from the caudae epididymides into modified Whitten's medium (15 mM Hepes-sodium salt, 1.2 mM MgCl2, 100 mM NaCl, 4.7 mM KCl, 1 mM pyruvic acid and 4.8 mM lactic acid) for 10 min at 37 °C. The solution had a final pH of 7.35. Spermatozoa were counted and diluted to 2 × 106 spermatozoa/mL, and motility was analysed in a 100-μm chamber (Cell Vision-USA, Delvavan, WI) using the IVOS Sperm Analyzer (Hamilton-Thorne Research, Beverly, MA, USA). The parameters that were used for the analysis setup are described in the technical guide for the IVOS system, version 12.2 (mouse setup 1). Sperm populations were analysed as soon as possible after their release from the epididymides. For each sperm sample, a total of 10 fields were analysed (Zhang et al., 2006).
Histology, immunochemistry and transmission electron microscopy
Testes, epididymides and cauda epididymal spermatozoa were prepared for light and transmission electron microscopy using standard methods, as previously described. The same mice that were used to assess fertility were used for histological analysis of the testes (Zhang et al., 2002).
Ethics
Procedures involving animals were conducted under the approval of the Institutional Animal Care and Use Committee of the Virginia Commonwealth University in accordance with the Guide for Care and Use of Laboratory Animals (Protocol #AM10297).
Results
Disruption of the Meig1 gene at the stage of meiosis in germ cells results in a severe spermatogenesis defect
In the testis, MEIG1 is predominantly expressed in the germ cells. To investigate the role of MEIG1 in germ cells, Meig1flox/flox males were crossed with Hsp2a-Cre females so that Meig1 is disrupted in germ cells at the stage of meiosis (Supplemental Figure 1).
Mice were genotyped by PCR and tested to determine if they carried the Hsp2a-Cre transgene (Supplemental Figure 2A). To evaluate tissue-specific deletion of the Meig1 gene, genomic DNA was isolated from the testis, heart and liver from Hsp2a-Cre; Meig1flox/flox mice. PCR was conducted using the primer set flanking the floxed region. The deletion band was observed only in the testis, but not in the heart and liver (Supplemental Figure 2). Testis somatic cells and germ cells were separated, and DNA was isolated from them. PCR results indicated that the deletion band was present only in the germ cells, not in the somatic cells (Supplemental Figure 2).
Northern blot and Western blot results revealed that testicular Meig1 mRNA and protein were undetectable in the Hsp2a-Cre; Meig1flox/flox mice (Fig. 1). Even though all the Hsp2a-Cre; Meig1flox/flox mice were viable and showed no gross abnormalities, and the testis weight was also comparable to age-matched Meig1flox/flox mice, all of the males tested were infertile, and the epididymidal sperm number was significantly lower than that in the Meig1flox/flox mice (Table 1).
Figure 1.
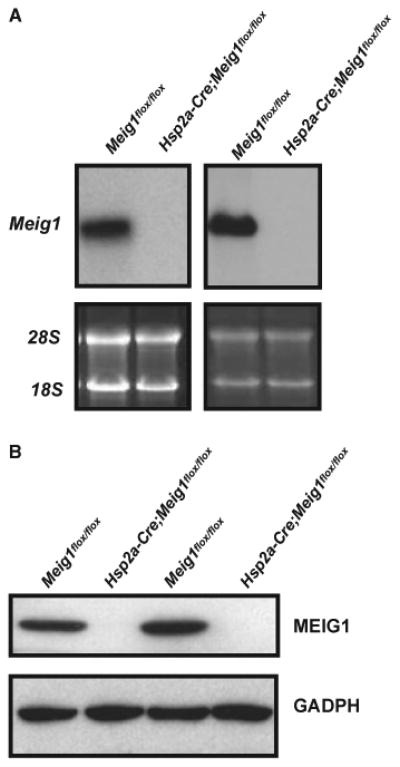
Expression of Meig1 mRNA and protein is underdetectable in the testis of the Hsp2-Cre; Meig1flox/flox mice. Total testicular RNA and protein were extracted from adult Meig1flox/flox and Hsp2a-Cre; Meig1flox/flox mice. Northern blot (A) was conducted using a full-length Meig1 cDNA as the probe; Western blot (B) was performed using an anti-MEIG1 antibody. Notice that there were no detectable Meig1 mRNA and protein in the testis of Hsp2a-Cre; Meig1flox/flox mice.
Table 1. Hsp2a/Cre; Meig1flox/flox males are infertile.
| Mice genotype | Male fertility | Litter size (n = 8)a | Sperm count (106, n = 8)ab | Testis/body weight (mg/g)a | Seminal vesicle/body weight (mg/g)a |
|---|---|---|---|---|---|
| Meig1flox/flox | 8/8 | 9.2 ± 0.7 | 25.68 ± 1.32 | 8.06 ± 0.66 | 6.34 ± 0.56 |
| Hsp2a-Cre; Meig1flox/flox | 8/0 | 0 | 0.051 ± 0.004 | 8.02 ± 0.46 | 6.18 ± 0.39 |
Values are Means ± SEM.
p < 0.01.
Microscopy revealed that spermatogenesis is arrested in the Hsp2a-Cre; Meig1flox/flox mice. Most spermatids failed to develop to the elongating stage [Fig. 2(A)]. Epididymides from Hsp2a-Cre; Meig1flox/flox mice only contained debris and degenerating spermatozoa [Fig. 2(B)]. The remaining epididymidal spermatozoa showed no motility as examined by visual inspection with a light microscope, and most spermatozoa had round or detached heads [Fig. 2(C)].
Figure 2.
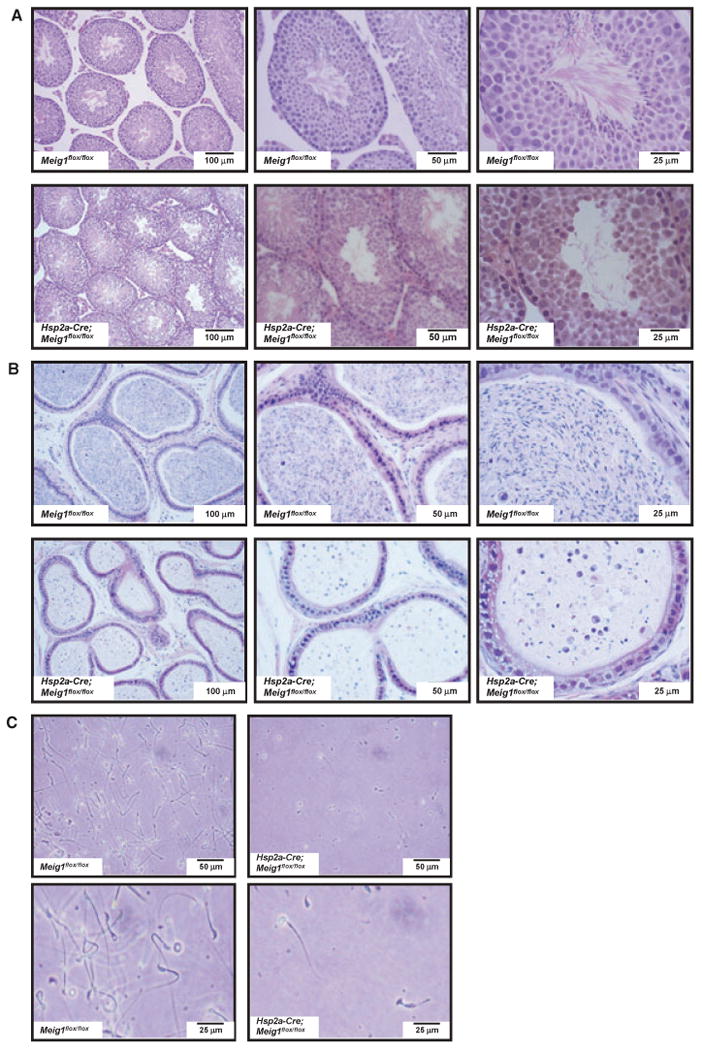
Disruption of Meig1 gene in the germ cells during meiosis stage results in severe spermatogenesis defect. (A) Representative testicular histology from a Meig1flox/flox mouse and a Hsp2a-Cre; Meig1flox/flox mouse. The Meig1flox/flox animal reveals normal architecture of the seminiferous tubules and interstitial tissue, but spermatogenesis is arrested in the Hsp2a-Cre; Meig1flox/flox mouse. (B) Representative epididymides histology from a Meig1flox/flox mouse and a Hsp2a-Cre; Meig1flox/flox mouse. The lumen is filled with mature spermatozoa in the Meig1flox/flox animal, but only cell debris can be seen in the Hsp2a-Cre; Meig1flox/flox animal. (C) Morphology of epididymal spermatozoa collected from a Meig1flox/flox mouse and a Hsp2a-Cre; Meig1flox/flox mouse. All spermatozoa in the Meig1flox/flox animal show normal morphology. However, very few spermatozoa can be observed in the Hsp2a-Cre; Meig1flox/flox animal, and most collected spermatozoa had round or detached heads.
Transmission electron microscopy demonstrated that spermatids in wild-type mice undergo normal developmental processes, including flagellum formation with ‘9 + 2’ axonemes and chromatin condensation associated with normal manchette structure [Fig. 3(A)]. However, in the Hsp2a-Cre; Meig1flox/flox mice, very few flagella were formed, and the sperm axonemes and accessory structures were disorganized [Fig. 3(B)]. In particular, manchette structures in spermatids were either disrupted or absent [Fig. 3(B)a–b], spermatid tails were not formed normally [Fig. 3(B)c] and the redundant nuclear envelope appeared to be rather enlarged [Fig. 3(B)d].
Figure 3.
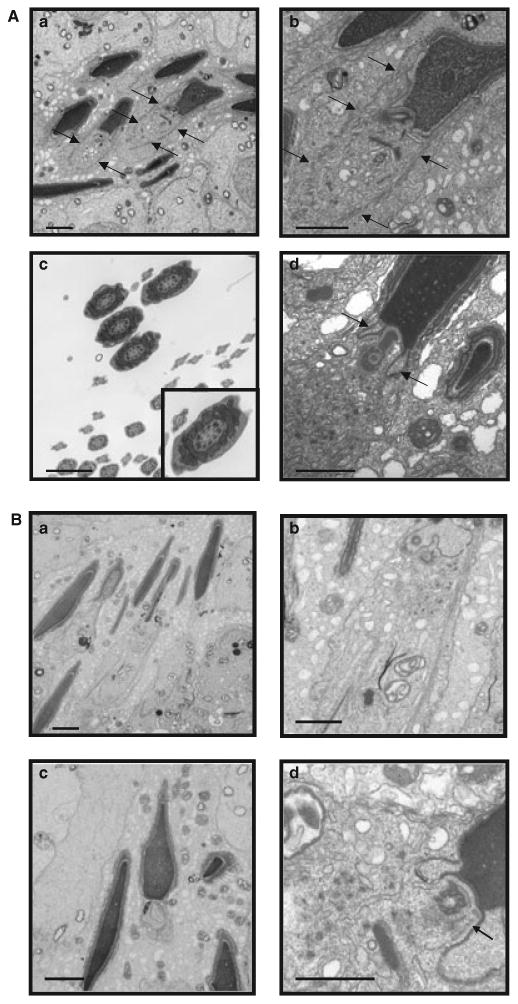
Testicular ultrastructure in adult Meig1flox/flox and Hsp2a-Cre; Meig1flox/flox mice. (A) Representative transmission electronic microscopy images from adult Meig1flox/flox mice. (a) Condensing spermatids with normal manchette structure (arrows) in Meig1flox/flox testes. (b) Higher magnification of a condensing spermatid in (a) with normal manchette structure (arrows). (c) Tail cross-sections with normal ‘9 + 2’ axonemes. (d) Normal condensing spermatid head and redundant nuclear envelope (arrows). Bars = 2 microns. (B) Representative transmission electronic microscopy images from adult Hsp2a-Cre; Meig1flox/flox mice. (a) Condensing spermatids with missing or disorganized manchettes. (b) Higher magnification of a condensing spermatid in (a) with abnormal manchette structure.(c) Spermatid with abnormal tail formation (centre). (d) Spermatid with enlarged redundant nuclear envelope (arrow). Bars = 2 microns.
Disruption of the Meig1 gene post-meiotically does not affect spermatogenesis and sperm motility
Germ cells undergo dramatic morphological changes after meiosis. To determine if disruption of the Meig1 gene post-meiotically affects spermatogenesis, the Meig1flox/flox mice were crossed with Prm-Cre mice to create Prm-Cre; Meig1flox/flox mice (Supplemental Figure 3).
The mice were genotyped following the same strategy as for the Hsp2a-Cre; Meig1flox/flox mice (Supplemental Figure 4A), and similar to the Hsp2a-Cre; Meig1flox/flox mice, the deletion band was observed only in the testis germ cells, but not in other somatic tissues, including the heart, liver, lung and testis somatic cells (Supplemental Figure 4B).
Testicular Meig1 mRNA and protein expression levels were significantly reduced in the Prm-Cre; Meig1flox/flox mice (Fig. 4). However, all the males tested were fertile and sired normal numbers of offspring. Epididymidal sperm counts were normal (Table 2). Testis and epididymides were indistinguishable under light microscope observation compared with the Prm-Cre; Meig1flox/flox and Meig1flox/flox mice. Both showed normal histology (Fig. 5A,B). Sperm morphology was normal [Fig. 5(C)], and there was no reduction in sperm motility in the Prm-Cre; Meig1flox/flox mice [Fig. 5(D)].
Figure 4.
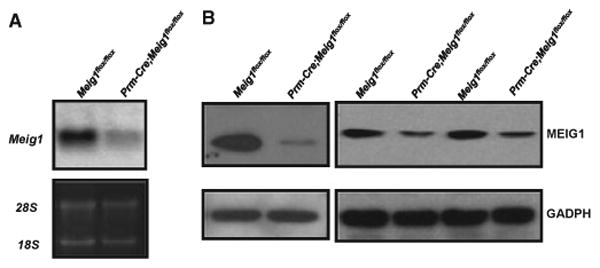
Expression of Meig1 mRNA and protein is significantly reduced in the testis of the Prm-Cre; Meig1flox/flox mice. Total testicular RNA and protein were extracted from adult Meig1flox/flox and Prm-Cre; Meig1flox/flox mice. Northern blot (A) was conducted using a full-length Meig1 cDNA as the probe; Western blot (B) was performed using an anti-MEIG1 antibody. Notice that both Meig1 mRNA and protein expression levels in the Prm-Cre; Meig1flox/flox mice are significantly lower than that in the control Meig1flox/flox mice.
Table 2. Prm-Cre; Meig1flox/flox males demonstrate normal fertility.
| Mice genotype | Male fertility | Litter size (n = 8)a | Sperm count (106, n = 8)a | Testis/body weight (mg/g)a | Seminal vesicle/body weight (mg/g)a |
|---|---|---|---|---|---|
| Meig1flox/flox | 10/10 | 9.1 ± 0.6 | 24.52 ± 1.32 | 8.14 ± 0.76 | 6.27 ± 0.35 |
| Prm-Cre; Meig1flox/flox | 10/10 | 8.9 ± 0.7 | 21.86 ± 1.48 | 8.22 ± 0.46 | 6.38 ± 0.42 |
Values are Means ± SEM.
Figure 5.
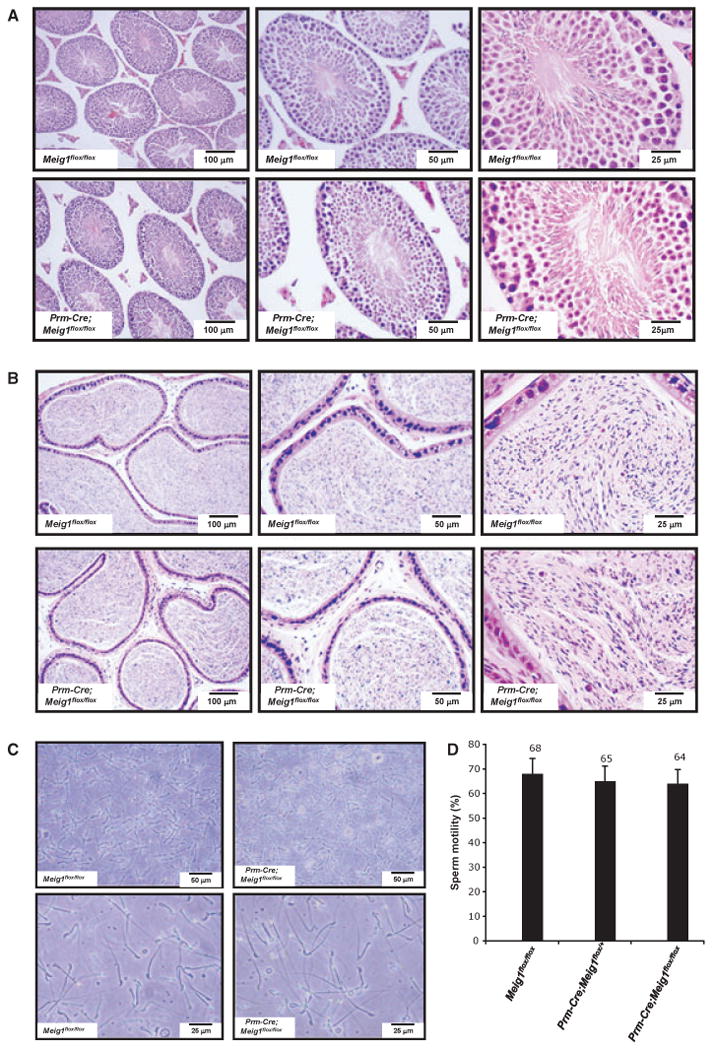
Disruption of Meig1 gene in the germ cells post-meiotically does not result in spermatogenesis defect. (A) Representative testicular histology from a Meig1flox/flox mouse and a Prm-Cre; Meig1flox/flox mouse. Both animals show normal architecture of the seminiferous tubules and interstitial tissue. (B) Representative epididymides histology from a Meig1flox/flox mouse and a Prm-Cre; Meig1flox/flox mouse. The lumen is filled with mature spermatozoa in both animals. (C) Morphology of epididymal spermatozoa collected from a Meig1flox/flox mouse and a Prm-Cre; Meig1flox/flox mouse. (D) All spermatozoa in both groups appear to be normal.
Disruption of the Meig1 gene in Sertoli cells does not affect spermatogenesis
Besides germ cells, Meig1 is expressed in the Sertoli cells in the testis. To investigate whether MEIG1 also regulates spermatogenesis through Sertoli cells, Amh-Cre mice were crossed with the Meig1flox/flox mice so that Meig1 was disrupted only in Sertoli cells (Supplemental Figure 5).
Amh-Cre; Meig1flox/flox mice were identified using the same strategy as described in identifying the Hsp2a-Cre; Meig1flox/flox and Prm-Cre; Meig1flox/flox mice (Supplemental Figure 6A). The deletion band was observed only in the total testis and testis somatic cells, but not in testis germ cells and other somatic tissue, including the heart (Supplemental Figure 6).
Northern blot and Western blot analysis showed that there were no differences in Meig1 mRNA and protein expression between the Amh-Cre; Meig1flox/flox mice and the Meig1flox/flox mice (Fig. 6).
Figure 6.
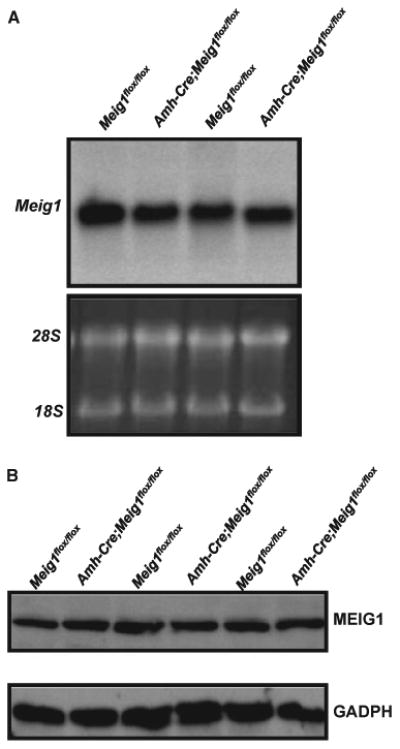
Expression levels of Meig1 mRNA and protein are not changed in the testis of the Amh-Cre; Meig1flox/flox mice. Total testicular RNA and protein were extracted from adult Meig1flox/flox and Amh-Cre; Meig1flox/flox mice. Northern blot (A) was conducted using a full-length Meig1 cDNA as the probe; Western blot (B) was performed using an anti-MEIG1 antibody. Notice that there was no difference in Meig1 mRNA and protein expression between the two groups.
Like the Hsp2a-Cre; Meig1flox/flox mice, all the Amh-Cre; Meig1flox/flox mice were viable and showed no gross abnormalities. The testis weight was also comparable to age-matched Meig1flox/flox mice. All the males tested were fertile, and sired normal number of offspring, and these mice had normal number of spermatozoa collected from epididymides (Table 3).
Table 3. Amh-Cre; Meig1flox/flox males have normal fertility.
| Mice genotype | Male fertility | Litter size (n = 9)a | Sperm count (106, n = 8)a | Testis/body weight (mg/g)a | Seminal vesicle/body weight (mg/g)a |
|---|---|---|---|---|---|
| Meig1flox/flox | 9/9 | 8.9 ± 0.7 | 24.71 ± 2.68 | 7.95 ± 0.52 | 6.20 ± 0.42 |
| Amh-Cre; Meig1flox/flox | 9/9 | 8.6 ± 0.6 | 23.15 ± 2.13 | 8.05 ± 0.68 | 6.08 ± 0.36 |
Values are Means ± SEM.
Amh-Cre; Meig1flox/flox males showed normal testis architecture. All stages of spermatogenesis were present [Fig. 7(A)]. Epididymidal morphology from Amh-Cre, Meig1flox/flox and Meig1flox/flox mice was also comparable. Mature spermatozoa filled the whole cavity [Fig. 7(B)]. The epididymidal spermatozoa showed normal morphology under light microscopy [Fig. 7(C)], and normal motility when estimated by visual inspection with a light microscope.
Figure 7.
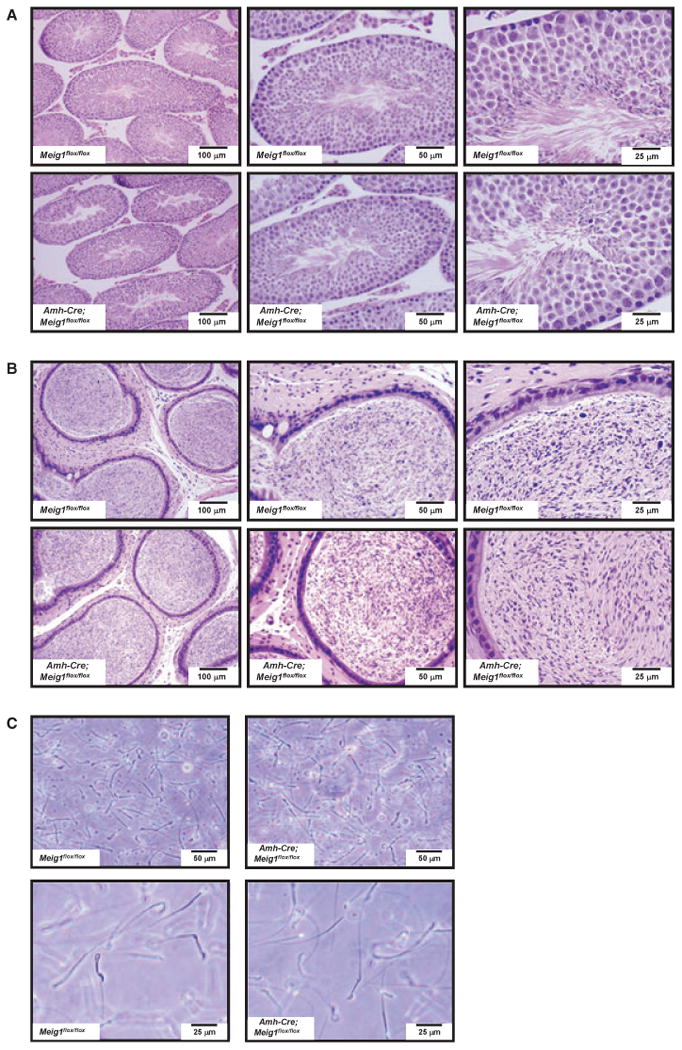
Disruption of Meig1 gene in the Sertoli cells does not affect normal spermatogenesis. (A) Representative testicular histology from a Meig1flox/flox mouse and a Amh-Cre; Meig1flox/flox mouse. Both reveal normal architecture of the seminiferous tubules and interstitial tissue. (B) Representative epididymides histology from a Meig1flox/flox mouse and a Amh-Cre; Meig1flox/flox mouse. The lumen is filled with mature spermatozoa in both mice. (C) Morphology of epididymal spermatozoa collected from a Meig1flox/flox mouse and a Amh-Cre; Meig1flox/flox mouse. All spermatozoa in both groups appear to be normal.
Discussion
Our previous studies and others demonstrated that MEIG1 plays an essential role in spermiogenesis. The mouse Meig1 gene encodes at least seven isoforms that are present in multiple tissues (Zhang et al., 2009). Some isoforms are expressed only in the testis (Zhang et al., 2009). The testis contains both germ cells and somatic cells, and the somatic cells also play a role in regulating spermatogenesis (Ge et al., 2008; Johnson et al., 2008; Cool & Capel, 2009). To determine the roles of MEIG1 in germ cells and testis somatic cells, Meig1flox/flox mice were crossed with the Hsp2a-Cre mice to disrupt the Meig1 gene only in male germ cells. The Hsp2a-Cre directs expression of Cre recombinase under the control of the mouse Hsp2a (heat shock protein 2 [Hsp70-2]) promoter (Dix et al., 1996). Hsp2a-Cre sequences of the transgene are flanked by Acrv1 (acrosomal vesicle protein 1 [SP-10]) insulator elements that are believed to tether the Acrv1 gene to the nuclear matrix in somatic cells to prevent transcription (Reddi et al., 2003; Abhyankar et al., 2007). Our initial immunochemistry experiments also confirmed that the Cre recombinase was expressed in the spermatocytes. When these transgenic mice are bred with mice containing a loxP-flanked sequence of interest, Cre-mediated recombination is expected to result in deletion of the floxed sequences in leptotene/zygotene spermatocytes of the male offspring. Thus, the Hsp2a-Cre mice are useful for generating conditional mutations for studying mammalian spermatocyte development, gametogenesis and meiosis (Inselman et al., 2010).
Similar to the Cmv-Cre; Meig1flox/flox mice, all Hsp2a-Cre; Meig1flox/flox males tested were infertile, and severe spermiogenesis defects occurred in these mice. Sperm number was significantly reduced, and the epididymides from these mice only contained debris and degenerating spermatozoa. The manchette structure was disrupted or absent, the ‘9 + 2’ axonemes were not present in the remaining flagella, and the redundant nuclear envelope was enlarged. It has been reported that expression of Hsp2a-Cre may be ‘leaky’ and recombination of loxP-flanked sequences can occur in some cells in somatic tissues (Inselman et al., 2010). Our PCR using DNA isolated from tail slips and multiple somatic tissues indicates that the loxP-flanked sequence remained intact in these tissues, distinguishing them from the Cmv-Cre; Meig1flox/flox mice, in which the loxP-flanked sequence was deleted globally (Schwenk et al., 1995). PCR using DNA isolated from testicular somatic cells and germ cells demonstrated that the floxed region was deleted in germ cells. A trace amount of intact floxed region was also amplified in the germ cells of the Hsp2a-Cre; Meig1flox/flox mice. This may be due to early-stage germ cells when the Cre recombinase is not active. These early-stage germ cells may not transcribe sufficient Meig1 mRNA and translate enough MEIG1 protein, which explains why Meig1 mRNA and protein were not detected in the Hsp2a-Cre; Meig1flox/flox mice.
The Prm-Cre mice have a transgene comprising the mouse protamine 1 promoter and Cre recombinase coding sequence, which mediates the efficient recombination of a cre target transgene in the male germ line, but not in other tissues (O'Gorman et al., 1997). Compared with the Hsp2a-Cre mice, the Cre recombinase is expressed at a later stage in the Prm-Cre mice, which was confirmed in initial immunochemistry experiment using specific antibody against Cre. As the Meig1-deficient mice showed a phenotype post-meiotically, we wished to know if disruption of Meig1 after meiosis also gives rise to spermatogenesis defects by crossing the Meig1flox/flox mice with the Prm-Cre mice. Even though expression of both Meig1 mRNA and protein was significantly reduced, the Prm-Cre; Meig1flox/flox males were fertile, and testis histology, sperm number, morphology and motility were also normal. This suggests that MEIG1 protein translated during the meiosis stage is sufficient for the spermatids to support normal spermatogenesis.
A major challenge in conditional gene targeting is that Cre expression can sometimes be ‘leaky’, even though its expression is driven by tissue/cell-specific promoters. The reduction in Meig1 mRNA and protein that we observed in the testis of Meig1flox/flox; Prm1-Cre males might be explained by the fact that some Cre protein was expressed at an earlier stage than expected from the known pattern of Prm1 promoter activity. This unscheduled Cre expression could have partially disrupted the Meig1 gene. If that is the case, the residual Meig1 that was expressed was clearly sufficient to support normal spermiogenesis. An alternative explanation is that the reduction in Meig1 mRNA and protein is the result of turnover of these molecules, and the failure to replace them in the later stages of spermiogenesis due to disruption of Meig1 expression in the Meig1flox/flox; Prm1-Cre testis.
Besides the germs cells, somatic Sertoli cells are another major population in the seminiferous tubules (Griswold, 1998). These cells have been proposed to maintain the integrity and compartmentalization of the seminiferous epithelium, secrete fluid to form a tubular lumen, participate in spermiation, and phagocytosis (Fisher, 2002; Kawasaki et al., 2002; Vernet et al., 2008; Lie et al., 2009), and these functions are essential for normal spermatogenesis. Even though Meig1 mRNA and protein were underdetectable in the testis of Hsp2a-Cre; Meig1flox/flox by Northern blot and Western blot analyses, PCR revealed that intact Meig1 is present in the somatic cells of Hsp2a-Cre; Meig1flox/flox (Supplemental Figure 2). As Meig1 is present in the Sertoli cells, we wished to know if MEIG1 also regulates spermatogenesis through a role on this cell type. Amh-Cre mice were crossed with the Meig1flox/flox mice so that Meig1 was disrupted only in the Sertoli cells. The Amh-Cre transgenic mice express Cre recombinase directed by the Sertoli cell-specific promoter elements of the anti-Mullerian hormone (Amh) gene (Holdcraft & Braun, 2004). When Amh-Cre transgenic males are bred with female mice containing a loxP-flanked sequence of interest, Cre-mediated recombination will result in deletion of the flanked sequence specifically in Sertoli cells. These Amh-Cre transgenic mice have been useful in generating conditional knockouts in Sertoli cells for studying several genes that play roles in male embryonic sexual differentiation and the regulation of spermatogenesis (Elliott et al., 2010; Meng et al., 2011). PCR results indicated that the floxed region of Meig1 gene was deleted in somatic cells in the testis, but not in the germ cells. Given that the testicular somatic cell preparation also contains small numbers of Leydig cells and peritubular cells, and the Meig1 gene in these cells remains intact in the Amh-Cre; Meig1flox/flox mice, a trace amount of Meig1 expression can be expected (Supplemental Figure 6). Amh-Cre; Meig1flox/flox males showed normal fertility, produced normal number of spermatozoa and had normal testicular histology. These findings suggest that MEIG1 does not play a fundamental role in spermatogenesis in the Sertoli cells.
In conclusion, our results demonstrate that MEIG1 regulates spermiogenesis through an action in germ cells, not in Sertoli cells, and that the Meig1 gene must be active during a discrete period in spermatogenesis after which it is dispensable.
Supplementary Material
Figure S1. Breeding strategy to generate Hsp2a-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Hsp2a-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Hsp2a-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S2. Identification of the Hsp2a-Cre; Meig1flox/flox mice. (A) Mice genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel) and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the deletion allele. The deletion bands were only present in the total testis and germ cells. Cmv-Cre; Meig1flox/flox mouse is a control.
Figure S3. Breeding strategy to generate Prm-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Prm-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Prm-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S4. Identification of the Prm-Cre; Meig1flox/flox mice. (A) Mice genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel), and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the deletion allele. Notice that the deletion band is only present in germ cells. Cmv-Cre; Meig1flox/flox mouse is a control.
Figure S5. Breeding strategy to generate Amh-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Amh-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Amh-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S6. Identification of the Amh-Cre; Meig1flox/flox mice. (A) Genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel), and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the allele with the deletion. Notice that the deletion band is only present in the total testis and somatic cells. Cmv-Cre; Meig1flox/flox mouse is a control.
Acknowledgments
We thank Drs. Mitch Eddy and Greg Buchold (National Institute of Environmental Health Sciences) for their comments and suggestions.
This work was supported by VCU PRIP award (to Zhibing Zhang), National Institutes of Health Grants HD37416 (to J.F.S.), HD 045783 (to J.C.H.) and 5F31HD062314 (to DRN-G). Tissue processing and staining were performed in Histology Core Facility of Virginia Commonwealth University. Transmission electron microscopy was performed in the Imaging Core of Virginia Commonwealth University (5P30NS047463) and the advanced microscopy facility of the University of Virginia.
Footnotes
Supporting Information: Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- Bouma GJ, Affourtit JP, Bult CJ, Eicher EM. Transcriptional profile of mouse pre-granulosa and Sertoli cells isolated from early-differentiated fetal gonads. Gene Expr Patterns. 2007;7:113–123. doi: 10.1016/j.modgep.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Chang YF, Lee-Chang JS, Panneerdoss S, MacLean JA, 2nd, Rao MK. Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. Biotechniques. 2011;51:341–342. 344. doi: 10.2144/000113764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Moses A, Malkov M, Shalom S, Ever L, Don J. A switch in the phosphorylation state of the dimeric form of the Meg1 protein correlates with progression through meiosis in the mouse. Cell Growth Differ. 1997;8:711–719. [PubMed] [Google Scholar]
- Cool J, Capel B. Mixed signals: development of the testis. Semin Reprod Med. 2009;27:5–13. doi: 10.1055/s-0028-1108005. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Rosario-Herrle M, Gotoh H, Mori C, Goulding EH, Barrett CV, Eddy EM. Developmentally regulated expression of Hsp70-2 and a Hsp70-2/lacZ transgene during spermatogenesis. Dev Biol. 1996;174:310–321. doi: 10.1006/dbio.1996.0076. [DOI] [PubMed] [Google Scholar]
- Don J, Wolgemuth DJ. Identification and characterization of the regulated pattern of expression of a novel mouse gene, meg1, during the meiotic cell cycle. Cell Growth Differ. 1992;3:495–505. [PubMed] [Google Scholar]
- Don J, Winer MA, Wolgemuth DJ. Developmentally regulated expression during gametogenesis of the murine gene meg1 suggests a role in meiosis. Mol Reprod Dev. 1994;38:16–23. doi: 10.1002/mrd.1080380104. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, Kinchen JM, Zhang J, Lysiak JJ, Ravichandran KS. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2010;467:333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ever L, Steiner R, Shalom S, Don J. Two alternatively spliced Meig1 messenger RNA species are differentially expressed in the somatic and in the germ-cell compartments of the testis. Cell Growth Differ. 1999;10:19–26. [PubMed] [Google Scholar]
- Fisher D. New light shed on fluid formation in the seminiferous tubules of the rat. J Physiol. 2002;542:445–452. doi: 10.1113/jphysiol.2002.018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Chen G, Hardy MP. The role of the Leydig cell in spermatogenic function. Adv Exp Med Biol. 2008;636:255–269. doi: 10.1007/978-0-387-09597-4_14. [DOI] [PubMed] [Google Scholar]
- Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- Inselman AL, Nakamura N, Brown PR, Willis WD, Goulding EH, Eddy EM. Heat shock protein 2 promoter drives Cre expression in spermatocytes of transgenic mice. Genesis. 2010;48:114–120. doi: 10.1002/dvg.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Thompson DL, Jr, Varner DD. Role of Sertoli cell number and function on regulation of spermatogenesis. Anim Reprod Sci. 2008;105:23–51. doi: 10.1016/j.anireprosci.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Nakagawa A, Nagaosa K, Shiratsuchi A, Nakanishi Y. Phosphatidylserine binding of class B scavenger receptor type I, a phagocytosis receptor of testicular sertoli cells. J Biol Chem. 2002;277:27559–27566. doi: 10.1074/jbc.M202879200. [DOI] [PubMed] [Google Scholar]
- Lie PP, Mruk DD, Lee WM, Cheng CY. Epidermal growth factor receptor pathway substrate 8 (Eps8) is a novel regulator of cell adhesion and the blood-testis barrier integrity in the seminiferous epithelium. FASEB J. 2009;23:2555–2567. doi: 10.1096/fj.06-070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- Meng J, Greenlee AR, Taub CJ, Braun RE. Sertoli cell-specific deletion of the androgen receptor compromises testicular immune privilege in mice. Biol Reprod. 2011;85:254–260. doi: 10.1095/biolreprod.110.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi PP, Shore AN, Shapiro JA, Anderson A, Stoler MH, Acharya KK. Spermatid-specific promoter of the SP-10 gene functions as an insulator in somatic cells. Dev Biol. 2003;262:173–182. doi: 10.1016/s0012-1606(03)00349-x. [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Eldar T, Karminsky OD, Itach SB, Pietrokovski S, Don J. Meig1 deficiency causes a severe defect in mouse spermatogenesis. Dev Biol. 2010;338:158–167. doi: 10.1016/j.ydbio.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner R, Ever L, Don J. MEIG1 localizes to the nucleus and binds to meiotic chromosomes of spermatocytes as they initiate meiosis. Dev Biol. 1999;216:635–645. doi: 10.1006/dbio.1999.9520. [DOI] [PubMed] [Google Scholar]
- Tabuchi Y, Takasaki I, Kondo T. Identification of genetic networks involved in the cell injury accompanying endoplasmic reticulum stress induced by bisphenol A in testicular Sertoli cells. Biochem Biophys Res Commun. 2006;345:1044–1050. doi: 10.1016/j.bbrc.2006.04.177. [DOI] [PubMed] [Google Scholar]
- Vernet N, Dennefeld C, Klopfenstein M, Ruiz A, Bok D, Ghyselinck NB, Mark M. Retinoid X receptor beta (RXRB) expression in Sertoli cells controls cholesterol homeostasis and spermiation. Reproduction. 2008;136:619–626. doi: 10.1530/REP-08-0235. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sapiro R, Kapfhamer D, Bucan M, Bray J, Chennathukuzhi V, et al. A sperm-associated WD repeat protein orthologous to Chlamydomonas PF20 associates with Spag6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Biol. 2002;22:7993–8004. doi: 10.1128/MCB.22.22.7993-8004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Moss SB, Jones BH, Ho C, Wang H, Kishida T, Gerton GL, Radice GL, Strauss JF., 3rd Haploinsufficiency for the murine orthologue of Chlamydomonas PF20 disrupts spermatogenesis. Proc Natl Acad Sci USA. 2004;101:12946–12951. doi: 10.1073/pnas.0404280101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kostetskii I, Tang W, Haig-Ladewig L, Sapiro R, Wei Z, et al. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod. 2006;74:751–759. doi: 10.1095/biolreprod.105.049254. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shen X, Gude DR, Wilkinson BM, Justice MJ, Flickinger CJ, Herr JC, Eddy EM, Strauss JF., 3rd MEIG1 is essential for spermiogenesis in mice. Proc Natl Acad Sci USA. 2009;106:17055–17060. doi: 10.1073/pnas.0906414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Breeding strategy to generate Hsp2a-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Hsp2a-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Hsp2a-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S2. Identification of the Hsp2a-Cre; Meig1flox/flox mice. (A) Mice genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel) and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the deletion allele. The deletion bands were only present in the total testis and germ cells. Cmv-Cre; Meig1flox/flox mouse is a control.
Figure S3. Breeding strategy to generate Prm-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Prm-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Prm-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S4. Identification of the Prm-Cre; Meig1flox/flox mice. (A) Mice genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel), and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the deletion allele. Notice that the deletion band is only present in germ cells. Cmv-Cre; Meig1flox/flox mouse is a control.
Figure S5. Breeding strategy to generate Amh-Cre; Meig1flox/flox mice. Meig1flox/flox males were crossed with Amh-Cre females, the resulting females carrying Cre allele were backcrossed with the Meig1flox/flox males to generate Amh-Cre; Meig1flox/flox mice. Meig1flox/flox males were used as controls.
Figure S6. Identification of the Amh-Cre; Meig1flox/flox mice. (A) Genotyping by PCR. DNA isolated from tail slips was used for PCR with a primer set flanking the left loxP site (X) (upper panel), and a primer set to amplify the Cre recombinase gene (lower panel). The lower band in the upper panel represents the wild-type allele; the upper band is the floxed allele. (B) PCR using the primer set flanking the floxed region. The upper band represents the intact allele; the lower band represents the allele with the deletion. Notice that the deletion band is only present in the total testis and somatic cells. Cmv-Cre; Meig1flox/flox mouse is a control.


