Abstract
Adolescent idiopathic scoliosis (AIS) is a common disorder with strong evidence for genetic predisposition. Quantitative trait loci (QTLs) for AIS susceptibility have been identified on chromosomes. We performed a genome-wide genetic linkage scan in 7 multiplex families using 400 marker loci with a mean spacing of 8.6 centiMorgans (cM). We used Genehunter Plus to generate linkage statistics, expressed as homogeneity (HLOD) scores, under dominant and recessive genetic models. We found a significant linkage signal on chromosome 12p, whose support interval extends from near 12pter, spanning approximately 10 million bases or 31 cM. Fine mapping within the region using 20 additional markers reveals maximum HLOD = 3.7 at 5 cM under a dominant inheritance model, and a split peak maximum HLOD = 3.2 at 8 and 18 cM under a recessive inheritance model. The linkage support interval contains 95 known genes. We found evidence suggestive of linkage on chromosomes 1, 6, 7, 8, and 14. This study is the first to find evidence of an AIS susceptibility locus on chromosome 12. Detection of AIS susceptibility QTLs on multiple chromosomes in this and other studies demonstrate that the condition is genetically heterogeneous.
Keywords: AIS, axial skeletal growth, chromosome 12p, genetic heterogeneity, susceptibility locus
INTRODUCTION
The Scoliosis Research Society defines adolescent idiopathic scoliosis (AIS) as a lateral spinal curvature ≥10° on plane radiographs, developing after age 11 and without any identifiable underlying secondary cause. AIS is a common condition, estimated to affect 2%–3% of the population.1 There is a 2:1 female preponderance of scoliotic curves exceeding 10°, but the incidence of smaller curves is equal in males and females.
Manifestations of AIS and its treatment are manifold and life-threatening when curves exceed 70°.2,3 Management varies according to curve magnitude.4 Treatment for patients with small curves (<25°) is observation until maturity. Currently, the standard of care for curves between 25° and 45° in a growing patient requires use of an underarm thoraco-lumbar-sacral brace worn 22 hours per day.5 Curves exceeding 45° in a growing child require surgical intervention, consisting of posterior spinal fusion and/or anterior fusion and instrumentation.
Various hypotheses regarding AIS pathogenesis have been proposed,6–10 with multifactorial causation the current majority opinion. In spite of limited insight into mechanism, there is strong evidence that genetic predisposition contributes to AIS. Several research groups have identified quantitative trait loci for scoliosis to various autosomes and to the X chromosome.11–18
We undertook linkage mapping of seven previously unreported families with multiple AIS-affected individuals. We found evidence of a novel AIS susceptibility locus on chromosome 12p. The chromosome 12 susceptibility locus is detected under both dominant and recessive models, and is localized to a 9 million base region harboring approximately 100 known genes.
MATERIALS AND METHODS
Members of 7 multigeneration, multiplex AIS families of European ancestry were recruited at the Hospital for Special Surgery, New York, New York. Clinical phenotype was validated by clinical evaluation and radiographic analysis. AIS was diagnosed if onset of scoliosis was age 11 years or greater, there was no identifiable secondary cause of scoliosis, and there was documentation of ≥10° spinal curvature by the Cobb method. All subjects enrolled in the study had a complete physical examination by two investigators (CLR [pediatric orthopedist] and PFG [clinical geneticist]). Radiographs were performed when the angle of trunk rotation measured by scoliometer was 5° or greater, as is the current standard of clinical practice. None of the patients had skeletal evidence of Marfan syndrome. All subjects provided informed consent/assent, and the study was Institutional Review Board-approved.
DNA was prepared by standard methods from whole blood. The whole genome linkage scan was performed using the Marshfield screening set 16, which includes 400 microsatellite markers with average spacing of 8.6 cM (details available at http://research.marshfieldclinic.org/genetics/sets/combo.html). Genotyping was performed as described previously.19 Purpose-developed software packages called SCORER and IMAGER were used for allele calling, genotype checking, and data management.20 Preliminary data integrity checks were performed to verify the Hardy-Weinberg equilibrium of each marker, identify and exclude contiguous regions of homozygosity, incompatible genotypes, gender errors, marker dropout, and individual dropout.
We performed our primary parametric linkage analysis with Genehunter Plus21,22 under both dominant and recessive inheritance models. Other model assumptions included a population AIS frequency between 1% and 3% and penetrance between 0.60 and 1.00. We used PEDCHECK23 and RELPAIR24 to confirm consistency with Mendelian inheritance and to trace and correct errors. Single-point and multi-point analyses were carried out for the 7 families together and homogeneity LOD (HLOD) score was determined according to combined recombination information and heterogeneity within the families. A heterogeneity parameter was calculated in GENEHUNTER to indicate the probability of the same phenotype resulting from different genes or alleles. Loci with HLOD score higher than 2.3 and 3.4 were considered as suggestive and significant for linkage.25 Fine structure mapping with 20 additional microsatellite markers was chosen on the basis of information and even spacing within the candidate region.
The first secondary analysis omitted families 1 and 5, which were the outliers in the heterogeneity analysis. The second included all 7 families and was performed using Superlink v. 1.6.26,27
RESULTS
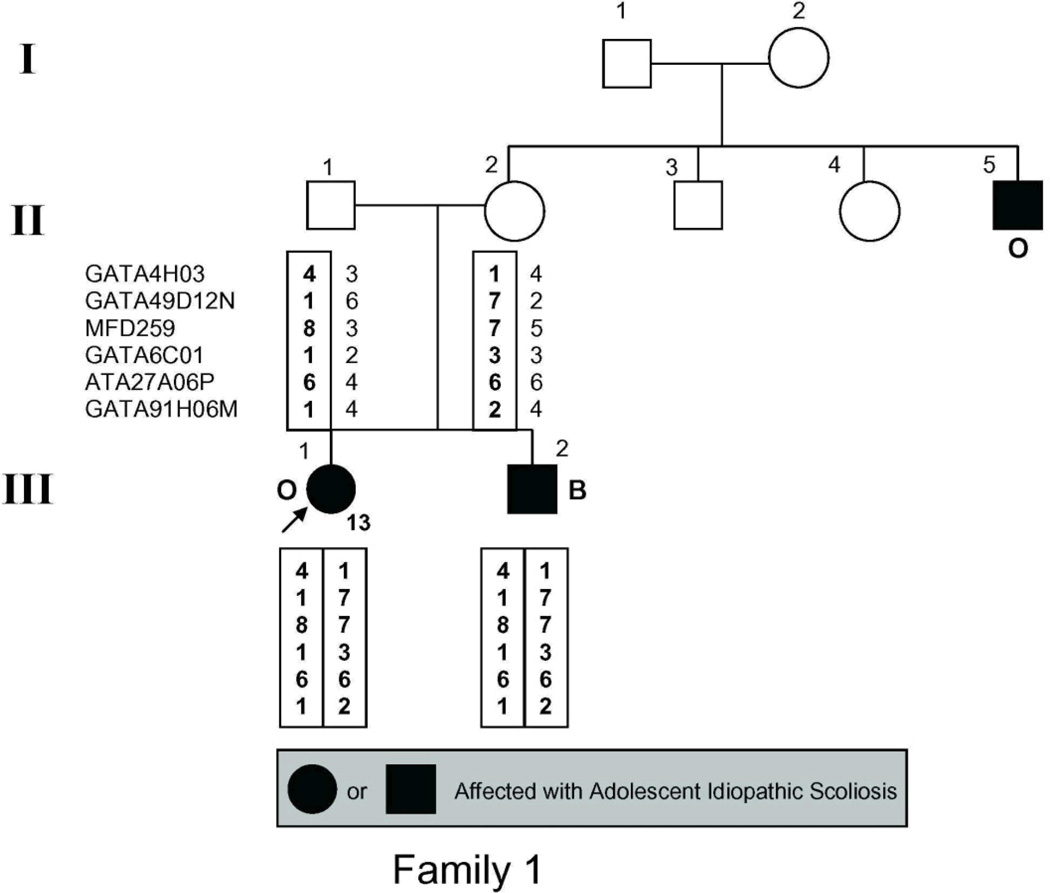
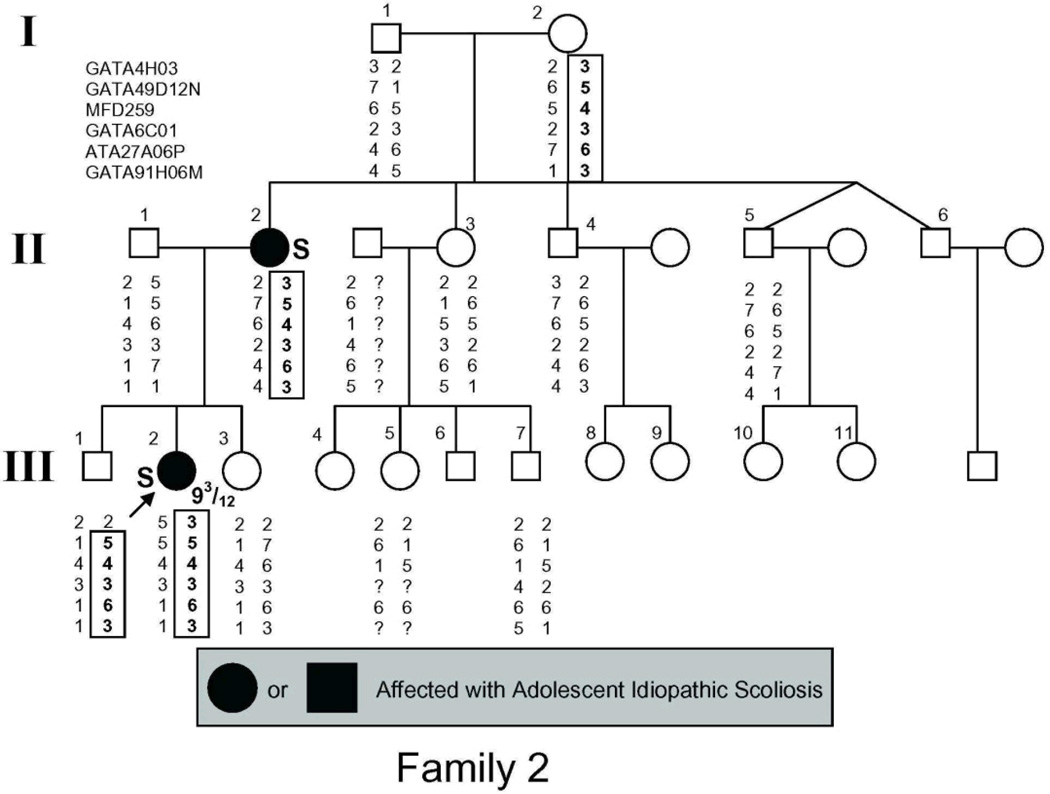
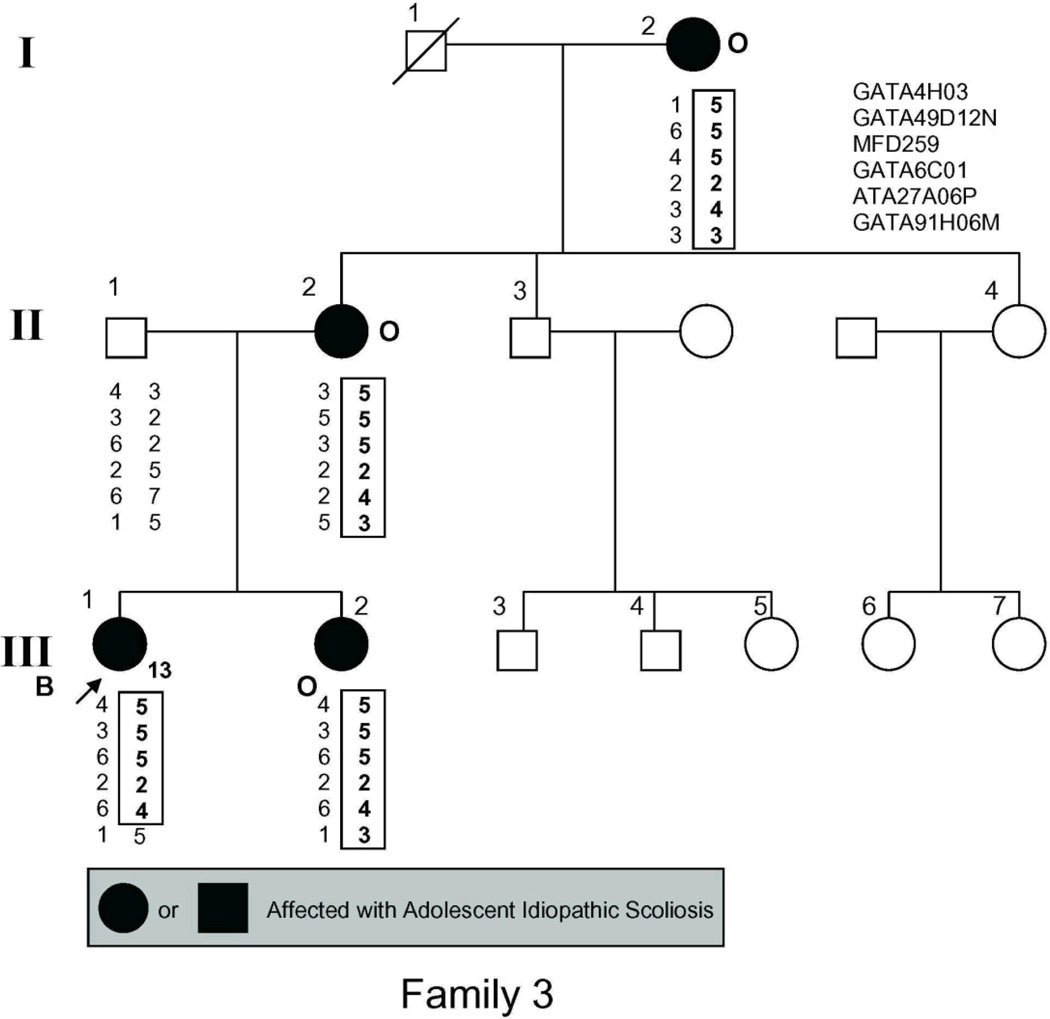
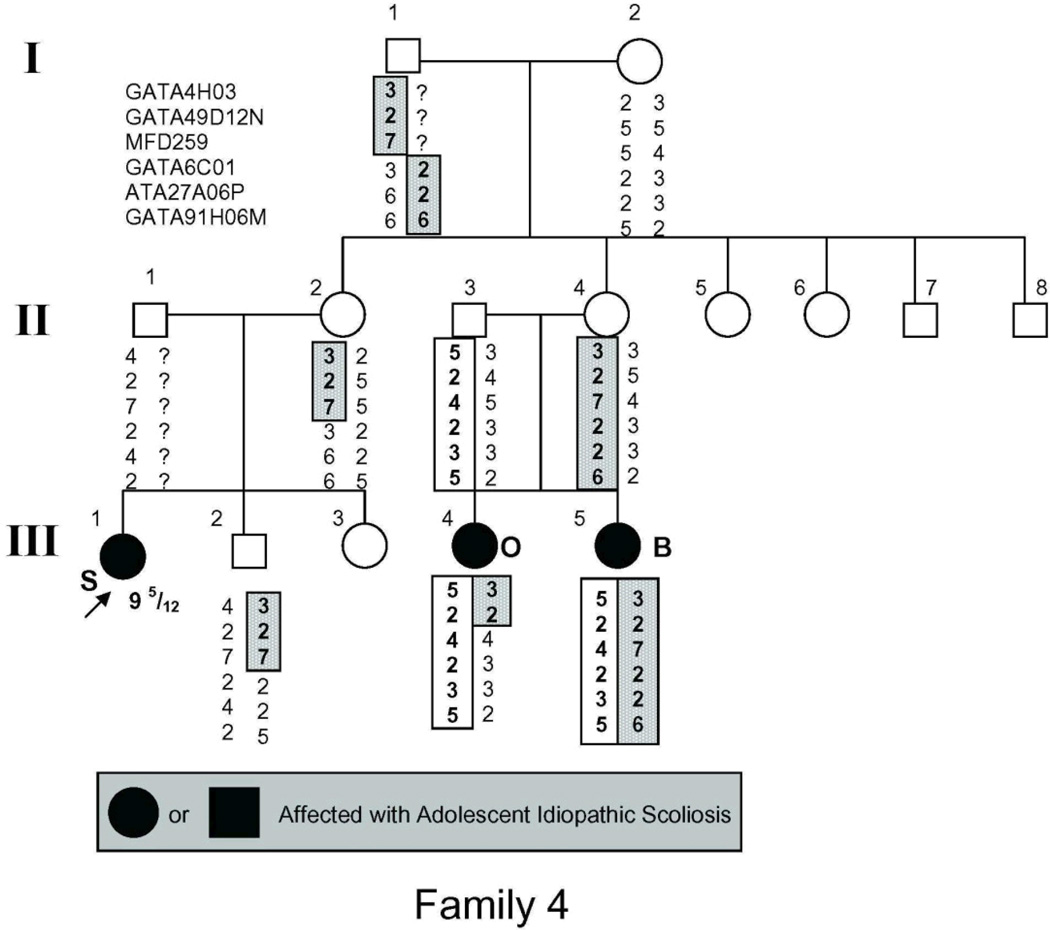
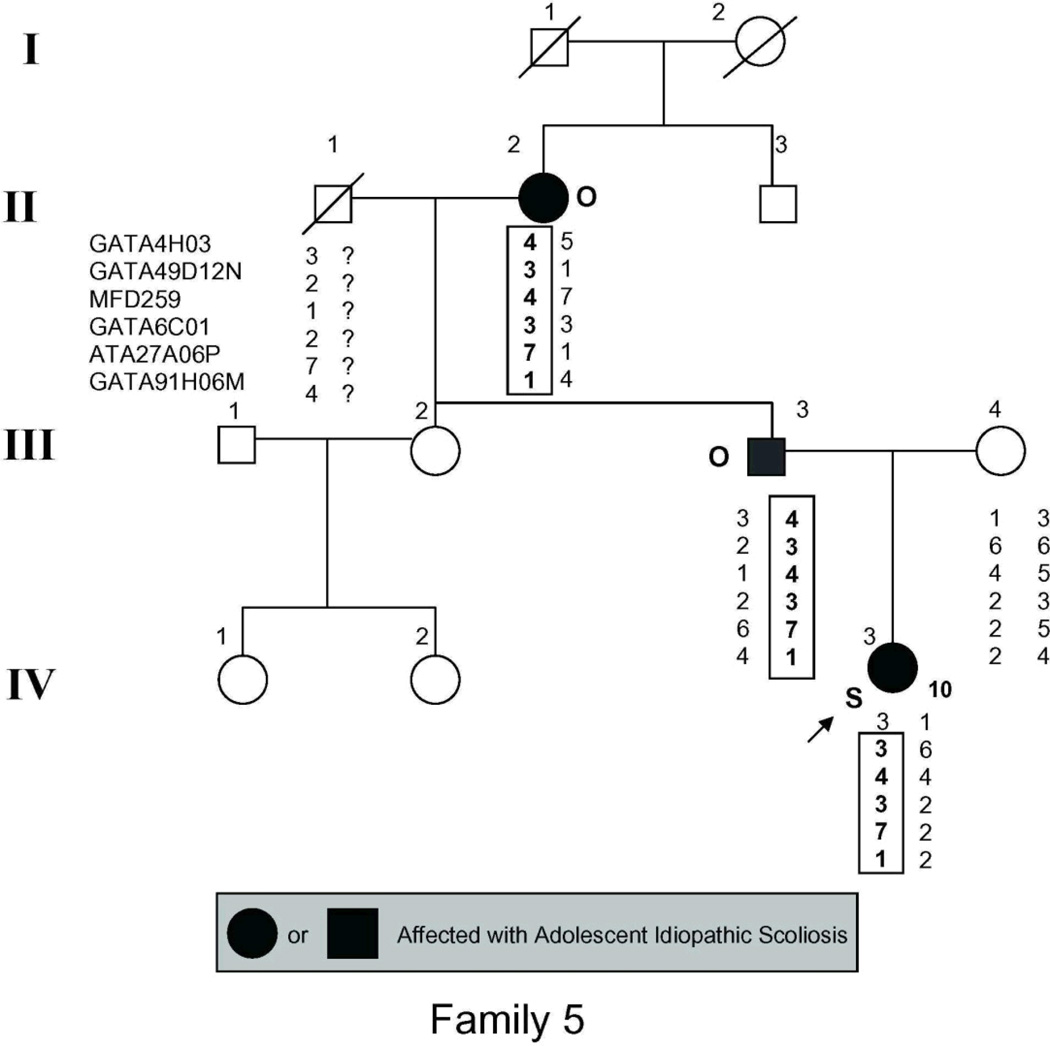
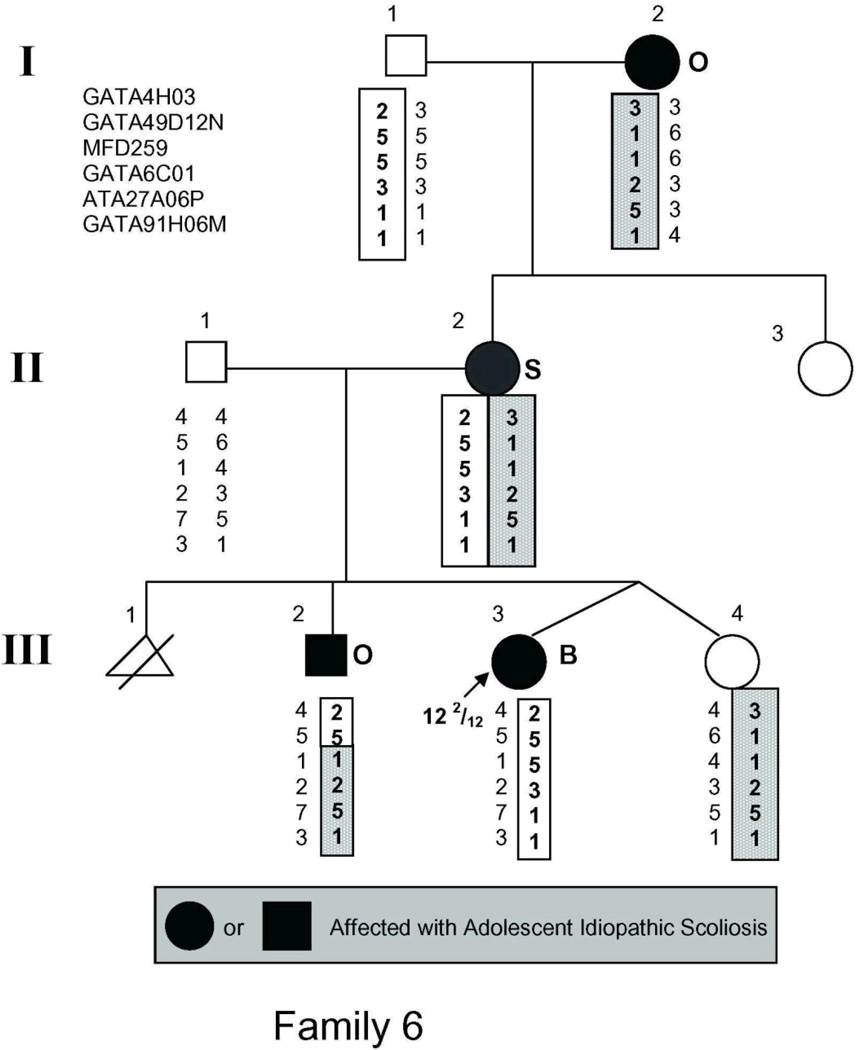
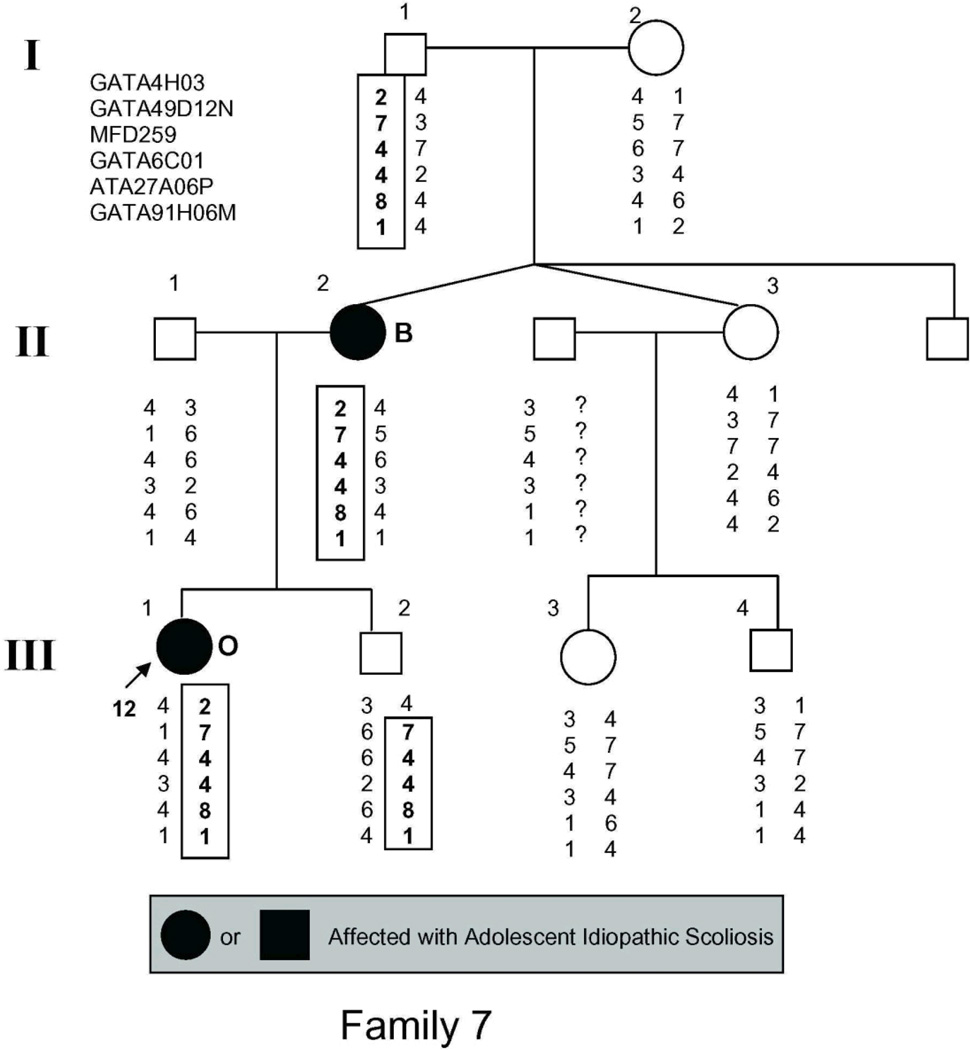
The study population (Fig. 1) included 54 individuals, 21 of whom had AIS by the above criteria and 2 more of whom had spinal curvature that approached but fell short of the 10° threshold for affected status and were there fore classified as unaffected in the linkage analysis.
Figure 1.
Pedigrees of families studied. Management was determined by curve magnitude. Curves ≤25° were observed (O), curves between 25° and 40° were treated with bracing (B), and curves >40° were surgically corrected (S). Circles = females. Squares = males. Shaded circles or squares indicate affected. Index cases are indicated by an arrow.
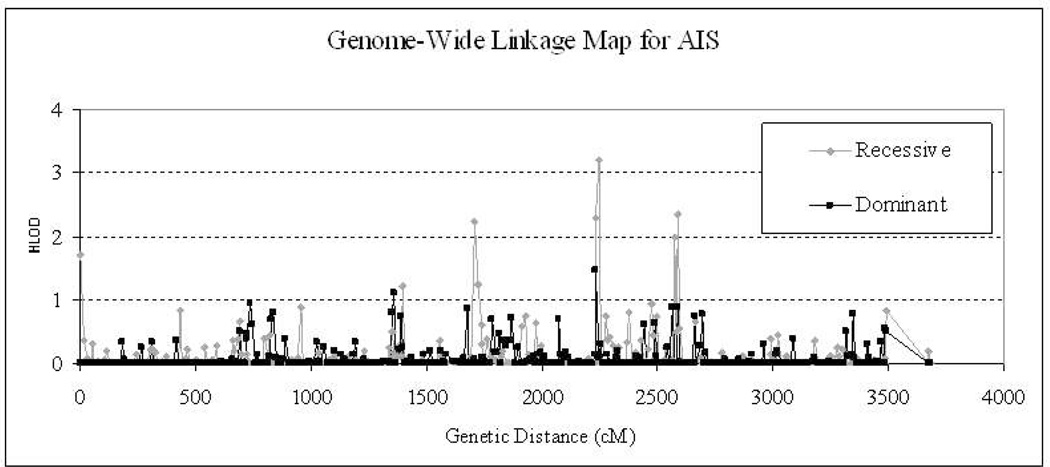
The results of the best model’s genome-wide linkage analysis are displayed in Fig. 2. A significant linkage signal, with HLOD = 3.5 with homogeneity (α = 1.0), disease frequency of 0.02, and penetrance of 0.99 was observed at GATA49D12 under a recessive model. We observed several other linkage signals with HLOD >1.0, including chromosome 1 (marker GTTTT002P: HLOD [recessive] = 1.7), chromosome 6 (marker ATA6C09P: HLOD [dominant] = 1.1), chromosome 7 (marker GATA24F3ZP: HLOD [recessive] = 1.2), chromosome 8 (markerUTZ129L: HLOD [recessive] = 1.2), chromosome 12 (marker D12S372: HLOD [recessive] = 2.0; HLOD [dominant] = 1.5), chromosome 14 (marker GGAA30H04ZP: HLOD [recessive] = 2.0). The HLOD maximum in the dominant genome-wide scan was 1.5 at GATA4H03.
Figure 2.
Genome-wide linkage profiles with dominant and recessive models. The length of whole genome consists of all 24 chromosomes sequentially and is represented in centiMorgans (cM). The profile in red stands for the result of recessive model, while the black one represents the dominant model. HLOD is the homozygous LOD score which combines multi-family information based on homogeneity of all families at every locus.
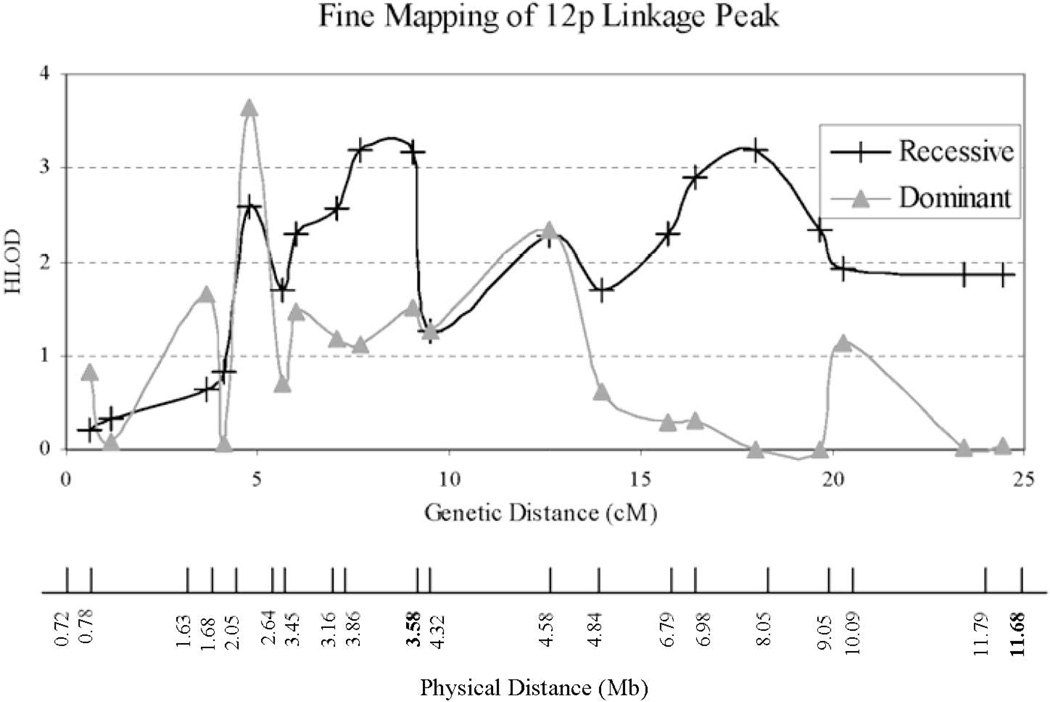
To further investigate the significant linkage signal at GATA49D12, we performed fine structure linkage mapping of chromosome 12p with an additional 20 microsatellite markers listed in Table 1. The results are shown in Fig. 3. The dominant model yields a linkage peak with HLOD = 3.7 (α = 1.0) at 5 cM, while the recessive model reveals a pair of peaks with HLOD = 3.2 (α = 1.0) at 8 and 18 cM. The “pair” of recessive linkage peaks more likely represents a single locus at 13 cM.
Table 1.
Markers Used for Fine Mapping
| Marker | Position (bases) | HLOD recessive | HLOD dominant | Source |
|---|---|---|---|---|
| D12S341 | 719,580 | 0.2 | 0.8 | deCode |
| D12S94 | 780,851 | 0.3 | 0.1 | deCode |
| D12S1608 | 1,629,617 | 0.6 | 1.7 | deCode |
| D12S1656 | 1,677,650 | 0.8 | 0.1 | deCode |
| D12S100 | 2,046,914 | 2.6 | 3.7 | deCode |
| D12S1615 | 2,640,746 | 1.7 | 0.7 | deCode |
| D12S1626 | 3,165,986 | 2.6 | 1.2 | deCode |
| D12S372 | 3,457,710 | 2.3 | 1.5 | Mfld screen set 16 |
| D12S1652 | 3,583,169 | 3.2 | 1.5 | deCode |
| D12S1685 | 3,862,066 | 3.2 | 1.1 | deCode |
| D12S1725 | 4,315,907 | 1.3 | 1.3 | deCode |
| D12S1624 | 4,581,357 | 2.3 | 2.3 | deCode |
| D12S314 | 4,838,740 | 1.7 | 0.6 | deCode |
| D12S1623 | 6,792,183 | 2.3 | 0.3 | deCode |
| D12S1625 | 6,975,843 | 2.9 | 0.3 | deCode |
| GATA49D12N | 8,048,975 | 3.2 | 0.0 | Mfld screen set 16 |
| D12S1695 | 9,046,784 | 2.4 | 0.0 | deCode |
| D12S1674 | 10,087,431 | 1.9 | 1.1 | deCode |
| D12S1697 | 11,685,667 | 1.9 | 0.0 | deCode |
| D12S89 | 11,793,428 | 1.9 | 0.0 | deCode |
Figure 3.
Fine mapping results of 12p3 region. Both physical and genetic distances from 12pter are given. Single-point analysis yields both LOD and HLOD of 3.2 at 12p (GATA49D12N). The heterogeneity parameter alpha is 1.0. NPL score is 1.38 with p-value of 0.053. The dominant model yields a linkage peak with HLOD = 3.7 (α = 1.0) at 5 cM, while the recessive model reveals a pair of peaks with HLOD = 3.2 (α = 1.0) at 8 and 18 cM. Marker positions are those provided by deCode. The exact chromosome position can be found in Table 2. The genetic and physical distances are not linearly related due to varying recombination rates along the length of the chromosomes. Moreover, there are discrepancies in marker order between the genetic and physical maps shown as boldfaced physical map position labels.
We performed a heterogeneity analysis to estimate how many families contributed to the observed linkage signals on chromosome 12p. For the recessive model, we found that α = 1, indicating that all the families contribute to the linkage signal. For the dominant model, α ranges between 0.3 and 0.8, with families 2, 3, 4, 6, and 7 contributing to the linkage signal. Secondary analyses, restricting the linkage analysis omitting families 1 and 5, and using Superlink gave virtually identical results (data not shown).
The candidate interval extending from D12S1608 at 1,677,650 base pairs from pter to D12S1674 at 10,087,431 base pairs, corresponding to the 12p13.3 cytogenetic band. The region is gene rich and includes 95 known genes and a comparable number of uncharacterized known and potential transcripts.
DISCUSSION
We report significant evidence of linkage of AIS to chromosome 12p based on data from 7 newly reported multiplex kindreds. By limiting the study to families in which there is more than one affected member, and by choosing families with at least one member with a curve exceeding 25°, we have designed our study to maximize the chance of finding an oligogenic inheritance pattern.28 Moreover, our sample displays a high degree of homogeneity, with all the families contributing to the observed recessive linkage and 5 of the 7 families contributing to the observed dominant linkage. While none of the pedigrees appears to fit an autosomal recessive mode of inheritance, a pseudodominant mode of inheritance is possible, which can result in an affected offspring through the union of a homozygous and a heterozygous individual. We hypothesize an additive model of inheritance with contributions from both dominant and recessive models. The data do not allow us to determine whether the same locus is being found under both recessive and dominant models, or to establish whether the linkage peak represents a single locus or several tightly linked loci. Although we do not believe that any of our families are consanguineous, all are of European descent and have resided in the New York metropolitan area, so it is possible that some share common ancestors. Investigation of 7 unrelated southern Chinese families in Hong Kong revealed a significant major AIS susceptibility gene at 19p13.3.12 As in our study, although the families were apparently unrelated, common ancestry could not be excluded. Occult consanguinity was reported in a genealogical study of AIS families recruited in Salt Lake City, in which 127 of 131 apparently unrelated probands were found to share common ancestry.29 Moreover, the Salt Lake City kindreds shared significantly more common ancestry than a control cohort matched by birth year.
Several other research groups have studied AIS genetics (Table 2). The most progress has been made on chromosome 8, where CHD718 and SNTG130 have been proposed as candidate genes. Previous work has also suggested the possibility that genes contributing to AIS may, when mutated, be responsible for known genetic syndromes. CHD7 mutations are responsible for the CHARGE (coloboma of the eye, heart defects, atresia of the choanae, retardation of growth and/or development, genital and/or urinary abnormalities, and ear abnormalities) syndrome.31 The chromosome 17 locus includes a region duplicated in Charcot-Marie-Tooth type 1A syndrome.13 Scoliosis occurs in approximately one third of patients with Charcot-Marie-Tooth, likely secondary to a neuromuscular etiology.32
Table 2.
Summary of Prior AIS Linkage Studies
| Reference | N (families/subjects)* | Location | Methods | Comments |
|---|---|---|---|---|
| 18 | 52/123 (affected) | 1p, 8q, 10q | “Affecteds only” nonparametric linkage, TDT | CHD7 identified as candidate gene on chromosome 8 |
| 16 | 202/1198 | 1,6,8,9,16 and17 | Genomic screen and model independent analysis | As threshold for scoliosis curvature increased, chromosomes 9 and 16 became more significant |
| 17 | 202/1198 | 5p13, 13q13.3, 13q32 | Nonparametric sib pair, linkage disequilibrium | Kyphoscoliotic subgroup analysis |
| 11 | 1/14 | 6q distal 10q 18q | Two point parametric and nonparametric multipoint linkage | Genome wide search in one family (7 affected members), followed by testing of “hot spots” in a second large family |
| 13 | 1/17 | 17p11 | Parametric linkage | Overlaps duplication in CMT type 1A |
| 12 | 7/52 | 19p13.3 | Parametric and nonparametric linkage | Recruited patients in whom scoliosis developed in adolescence |
| 15 | 202/1198 | 19p13 | Nonparametric sib pair, parametric linkage | Subgroup analysis limited to kindreds in which at least 1 member had curve ≥30° |
| 14 | 202/1198 | Xq23, Xq26.1 | Nonparametric sib pair, parametric linkage | Estimate that 15% of AIS families display X-linkage |
| 41 | 25/116(affected) | 9q31.2-q34.2; 17q25.3-qtel | Two point parametric and nonparametric multipoint | Confirmation of ref. 15 |
NR = not reported.
Taken as a whole, the existing literature points to genetic heterogeneity, with multiple genes involved in AIS pathogenesis, and to oligogenic causation, with a major gene segregating in most affected families. Both these generalizations are conditioned in part by methodology, reflecting recruitment of distinct populations and more severely affected subjects.
The candidate region for the chromosome 12 susceptibility locus is gene rich, including 95 known genes and a similar number of uncharacterized known or potential transcripts. Some of the genes in the interval are attractive candidates for AIS susceptibility. These include genes encoding ion channels (KCNA1,33 KCNA5,33 KCNA6,33 SCNN1A34), bone derived growth factors (GDF3),35 growth factors regulating muscle (FGF6)36 and neurons (NTF3),37 enzymes involved in protein processing (USP5,38 SPSB239), and components of the extracellular matrix (MFAP5).40 Their participation in neuromuscular and connective tissue development makes these plausible positional candidate genes.
The strength of linkage mapping as an investigative strategy is that it allows us to identify candidate genes by genomic location, without prior knowledge of function. However, our study also has important limitations. Our findings are based on a small number of families. Given ample evidence of the genetic heterogeneity of AIS, it is uncertain how frequently our findings can be generalized to other affected kindreds. The large number of genes included within the candidate interval is another important limitation of our study. Further fine mapping studies in additional families will ultimately allow the interval to be further refined and limited, reducing the number of potential candidate genes.
In spite of recent progress in AIS genetics, we know little regarding the developmental processes that operate during adolescent axial skeletal growth. Limited mechanistic understanding prevents us from predicting which patients will have progressive disease as they mature. Discovering the mechanisms that mediate vertebral growth and development and understanding how their disruption leads to spinal curvature remains the ultimate goal of this and similar investigations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Heart, Lung, and Blood Institute, Mammalian Genotyping Service- NO1-HV-48141, Beatrice and Samuel A. Seaver Foundation, and Marshfield Clinic Research Foundation.
The authors thank Marshfield Clinic Research Foundation for its support through the assistance of Alice Stargardt in the preparation of this manuscript. This paper is Madison GRECC manuscript 2007-04.
Footnotes
All authors state that they have no conflicts of interest.
REFERENCES
- 1.Weinstein SL. The thoracolumbar spine. In: Turek SL, Weinstein SL, Buckwalter JA, editors. Turek's Orthopaedics: Principles and Their Application. 5th ed. Lippincott, Philadelphia, USA: 1994. pp. 447–484. [Google Scholar]
- 2.Nachemson A. A long term follow-up study of non-treated scoliosis. Acta Orthop Scand. 1968;39:466–476. doi: 10.3109/17453676808989664. [DOI] [PubMed] [Google Scholar]
- 3.Enneking WF, Harrington P. Pathological changes in scoliosis. J Bone Joint Surg Am. 1969;51:165–184. [PubMed] [Google Scholar]
- 4.Weinstein SL, Ponseti IV. Curve progression in idiopathic scoliosis. J Bone Joint Surg Am. 1983;65:447–455. [PubMed] [Google Scholar]
- 5.Nachemson AL, Peterson LE. Effectiveness of treatment with a brace in girls who have adolescent idiopathic scoliosis. A prospective, controlled study based on data from the Brace Study of the Scoliosis Research Society. J Bone Joint Surg Am. 1995;77:815–822. doi: 10.2106/00004623-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Sahlstrand T, Petruson B. A study of labyrinthine function in patients with adolescent idiopathic scoliosis. I. An electro-nystagmographic study. Acta Orthop Scand. 1979;50:759–769. doi: 10.3109/17453677908991307. [DOI] [PubMed] [Google Scholar]
- 7.Moreau A, Wang DS, Forget S, et al. Melatonin signaling dysfunction in adolescent idiopathic scoliosis. Spine. 2004;29:1772–1781. doi: 10.1097/01.brs.0000134567.52303.1a. [DOI] [PubMed] [Google Scholar]
- 8.Morcuende JA, Minhas R, Dolan L, et al. Allelic variants of human melatonin 1A receptor in patients with familial adolescent idiopathic scoliosis. Spine. 2003;28:2025–2028. doi: 10.1097/01.BRS.0000083235.74593.49. [DOI] [PubMed] [Google Scholar]
- 9.Lowe T, Lawellin D, Smith D, et al. Platelet calmodulin levels in adolescent idiopathic scoliosis: do the levels correlate with curve progression and severity? Spine. 2002;27:768–775. doi: 10.1097/00007632-200204010-00016. [DOI] [PubMed] [Google Scholar]
- 10.Wynne-Davies R. Familial (idiopathic) scoliosis. A family survey. J Bone Joint Surg Br. 1968;50:24–30. [PubMed] [Google Scholar]
- 11.Wise CA, Barnes R, Gillum J, et al. Localization of susceptibility to familial idiopathic scoliosis. Spine. 2000;25:2372–2380. doi: 10.1097/00007632-200009150-00017. [DOI] [PubMed] [Google Scholar]
- 12.Chan V, Fong GC, Luk KD, et al. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. 2002;71:401–406. doi: 10.1086/341607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salehi LB, Mangino M, De Serio S, et al. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum Genet. 2002;111:401–404. doi: 10.1007/s00439-002-0785-4. [DOI] [PubMed] [Google Scholar]
- 14.Justice CM, Miller NH, Marosy B, et al. Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine. 2003;28:589–594. doi: 10.1097/01.BRS.0000049940.39801.E6. [DOI] [PubMed] [Google Scholar]
- 15.Alden KJ, Marosy B, Nzegwu N, et al. Idiopathic scoliosis: identification of candidate regions on chromosome 19p13. Spine. 2006;31:1815–1819. doi: 10.1097/01.brs.0000227264.23603.dc. [DOI] [PubMed] [Google Scholar]
- 16.Miller NH, Justice CM, Marosy B, et al. Identification of candidate regions for familial idiopathic scoliosis. Spine. 2005;30:1181–1187. doi: 10.1097/01.brs.0000162282.46160.0a. [DOI] [PubMed] [Google Scholar]
- 17.Miller NH, Marosy B, Justice CM, et al. Linkage analysis of genetic loci for kyphoscoliosis on chromosomes 5p13, 13q13.3, and 13q32. Am J Med Genet A. 2006;140:1059–1068. doi: 10.1002/ajmg.a.31211. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Gordon D, Zhang D, et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007;80:957–965. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber JL, Broman KW. Genotyping for human whole-genome scans: past, present, and future. Adv Genet. 2001;42:77–96. doi: 10.1016/s0065-2660(01)42016-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Heil JD, Dickinson WK, et al. Computer system for large scale STRP genotyping. Am J Hum Genet A. 1997;61:A302. [abstract]. [Google Scholar]
- 21.Kruglyak L, Daly MJ, Reeve-Daly MP, et al. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 22.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epstein MP, Duren WL, Boehnke M. Improved inference of relationships for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 26.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(suppl 1):S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 27.Fishelson M, Geiger D. Optimizing exact genetic linkage computations. J Comput Biol. 2004;11:263–275. doi: 10.1089/1066527041410409. [DOI] [PubMed] [Google Scholar]
- 28.Axenovich TI, Zaidman AM, Zorkoltseva IV, et al. Segregation analysis of idiopathic scoliosis: demonstration of a major gene effect. Am J Med Genet. 1999;86:389–394. [PubMed] [Google Scholar]
- 29.Ogilvie JW, Braun J, Argyle V, et al. The search for idiopathic scoliosis genes. Spine. 2006;31:679–681. doi: 10.1097/01.brs.0000202527.25356.90. [DOI] [PubMed] [Google Scholar]
- 30.Bashiardes S, Veile R, Allen M, et al. SNTG1, the gene encoding gamma1-syntrophin: a candidate gene for idiopathic scoliosis. Hum Genet. 2004;115:81–89. doi: 10.1007/s00439-004-1121-y. [DOI] [PubMed] [Google Scholar]
- 31.Hall BD. Choanal atresia and associated multiple anomalies. J Pediatr. 1979;95:395–398. doi: 10.1016/s0022-3476(79)80513-2. [DOI] [PubMed] [Google Scholar]
- 32.Karol LA, Elerson E. Scoliosis in patients with Charcot-Marie-Tooth disease. J Bone Joint Surg Am. 2007;89:1504–1510. doi: 10.2106/JBJS.F.01161. [DOI] [PubMed] [Google Scholar]
- 33.Brandt T, Strupp M. Episodic ataxia type 1 and 2 (familial periodic ataxia/vertigo) Audiol Neurootol. 1997;2:373–383. doi: 10.1159/000259262. [DOI] [PubMed] [Google Scholar]
- 34.Chang SS, Grunder S, Hanukoglu A, et al. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- 35.Levine AJ, Brivanlou AH. GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development. 2006;133:209–216. doi: 10.1242/dev.02192. [DOI] [PubMed] [Google Scholar]
- 36.Fiore F, Sebille A, Birnbaum D. Skeletal muscle regeneration is not impaired in Fgf6 −/− mutant mice. Biochem Biophys Res Commun. 2000;272:138–143. doi: 10.1006/bbrc.2000.2703. [DOI] [PubMed] [Google Scholar]
- 37.Ernfors P, Lee KF, Kucera J, et al. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson KD, Tashayev VL, O'Connor LB, et al. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 39.Kile BT, Schulman BA, Alexander WS, et al. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 40.Gibson MA, Hatzinikolas G, Kumaratilake JS, et al. Further characterization of proteins associated with elastic fiber microfibrils including the molecular cloning of MAGP-2 (MP25) J Biol Chem. 1996;271:1096–1103. doi: 10.1074/jbc.271.2.1096. [DOI] [PubMed] [Google Scholar]
- 41.Ocaka L, Zhao C, Reed JA, et al. Assignment of two loci for autosomal dominant adolescent idiopathic scoliosis to chromosomes 9q31.2-q34.2 and 17q25.3-qtel. J Med Genet. 2008;45:87–92. doi: 10.1136/jmg.2007.051896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.