While transitions of sex determination mechanisms are frequent in species with homomorphic sex chromosomes1–3, heteromorphic sex chromosomes are thought to represent a terminal evolutionary stage, due to chromosome-specific adaptations such as dosage compensation or an accumulation of sex-specific mutations1,4. Here, we show that an autosome of Drosophila, the dot chromosome, was ancestrally a differentiated X chromosome. We analyze the whole-genome of true fruit flies, flesh flies and soldier flies to show that genes located on the dot chromosome of Drosophila are X-linked in outgroup species while Drosophila X-linked genes are autosomal. We further date this chromosomal transition to early Drosophilid evolution by genome sequencing of other Drosophilidae. Our results unravel several puzzling aspects of the biology of the Drosophila dot chromosome as remnants of its former life as a sex chromosome, such as its minor feminizing role in sex determination5 or its targeting by a chromosome-specific regulatory mechanism6, and we show that patterns of biased gene expression of the dot during early embryogenesis, oogenesis and spermatogenesis resemble that of the current X. Thus, while sex chromosomes are not necessarily evolutionary traps and can revert back to an autosomal inheritance, the highly specialized genome architecture of this former X chromosome suggests severe fitness costs must be overcome for such a turnover to occur.
Sex is an important and conserved feature, yet sex determination mechanisms are labile in many taxa with non-differentiated, homomorphic sex chromosomes, and often vary among closely related species, or among individuals within a species1. Highly differentiated, heteromorphic sex chromosomes (that is, a degenerate, gene-poor Y chromosome, and an often dosage compensated X), however, appear to represent an evolutionary end point and become a permanent fixture of the genome in many species groups1,3,4. Special adaptations on highly evolved sex chromosomes, such as dosage compensation or inactivation during male meiosis, an accumulation of sex-determining and sex-specific genes, or inviability of YY individuals, prevent the reversal of heteromorphic sex chromosomes back to autosomes1,4, and sex-chromosome differentiation is viewed as an evolutionary one-way street. Indeed, the heteromorphic sex chromosomes of both mammals and birds originated independently from an ancestor with homomorphic sex chromosomes over 100 MY ago, and the stable inheritance of the sex chromosomes in these two clades reflects their highly specialized genome architecture7. In contrast, homomorphic sex chromosomes in fish, amphibians and reptiles often show high rates of turnover between species2,3. While observations in vertebrates support the notion that heteromorphic sex chromosomes present an ‘evolutionary trap’ inert to turnover7, little is known about such transitions in other taxa. Here, we uncover a sex chromosome reversion within Diptera (flies), in the genetic model organism Drosophila.
In many higher Diptera (suborder Brachycera) including Drosophila, the basic karyotype (2n=12) consists of 5 pairs of large euchromatic rods (named Muller elements A–E in Drosophila; each containing well over 2000 genes), and a smaller heterochromatic dot chromosome8 (element F in Drosophila; only containing about 100 genes; see Fig. 1). The gene content of Muller elements is highly conserved across Diptera9. One of the large rods (element A) segregates as the X chromosome in Drosophila, and sex is determined by the dose of the X-linked gene Sex-Lethal (Sxl), with diploid XX embryos developing into females, and haploid XY into males10. The identity of the sex chromosomes and the master sex determination gene is not conserved across other Diptera families11; instead, a small and often heterochromatic chromosome pair segregates as the sex chromosome in several other Brachycera12. No genes have yet been isolated from the sex chromosomes in outgroup species, and the relationship with Drosophila chromosomes, if any, is unclear.
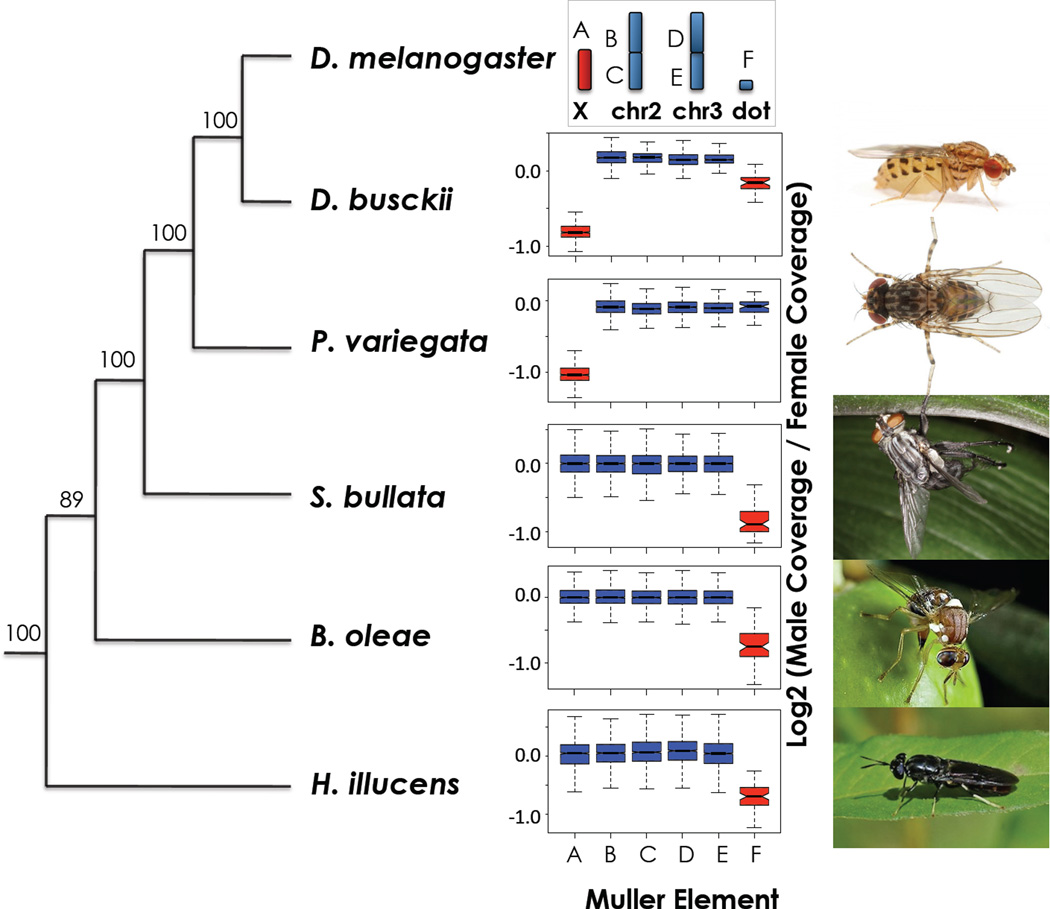
Figure 1. Sex chromosomes in higher Diptera revealed by genome analysis.
Evolutionary relationship inferred from 185 conserved protein-coding genes (93134 amino-acids) using PhyML, and male/female coverage ratio across chromosome elements (Muller elements A–F) in the Diptera species studied. X chromosomes (in red) have only half the read coverage in males vs. females. Boxes extend from the first to the third quartile and whiskers to the most extreme data point within 1.5 times the interquantile range.
We used whole-genome sequencing to identify sex-linked genes in species from several higher Diptera families, including the black soldier fly Hermetia illucens (Stratiomyidae, a basal Brachycera), the olive fruit fly Bactrocera oleae (Tephritidae), the grey fleshfly Sarcophaga bullata (Sarcophagidae), the Stegana Phorida variegata (Drosophilidae, a sister clade to Drosophila), as well as the basal Drosophila D. busckii (see Fig. 1, Fig. S1). Paired-end Illumina genomic reads were obtained from single males and females of each species, assembled, and the resulting scaffolds were mapped to the different Muller elements according to their homology to D. melanogaster genes (Table S1). Male and female reads were mapped back to the scaffolds, and male to female (M/F) coverage was used to detect sex-linked sequences, with X-linked scaffolds only showing half the coverage in males compared to females13. Consistent with Muller-A being the ancestral sex chromosome in all Drosophilidae, M/F coverage is only half that relative to autosomes in D. busckii and P. variegata (Fig. 1, Table S2, Fig. S2–S4). In the more distantly related outgroups B. oleae, S. bullata and H. illucens, M/F coverage shows that element A is an autosome; instead, element F, the “dot chromosome” of Drosophila, has significantly reduced M/F coverage in outgroup species (Fig. 1, Table S2, Fig. S5–S7). This implies that element F is a heteromorphic sex chromosome pair in several Brachycera, with a completely degenerate Y. The phylogenetic pattern in fact suggests that the dot was a sex chromosome in an ancestor of Drosophila, and only reverted to an autosome in the lineage leading to Drosophilidae.
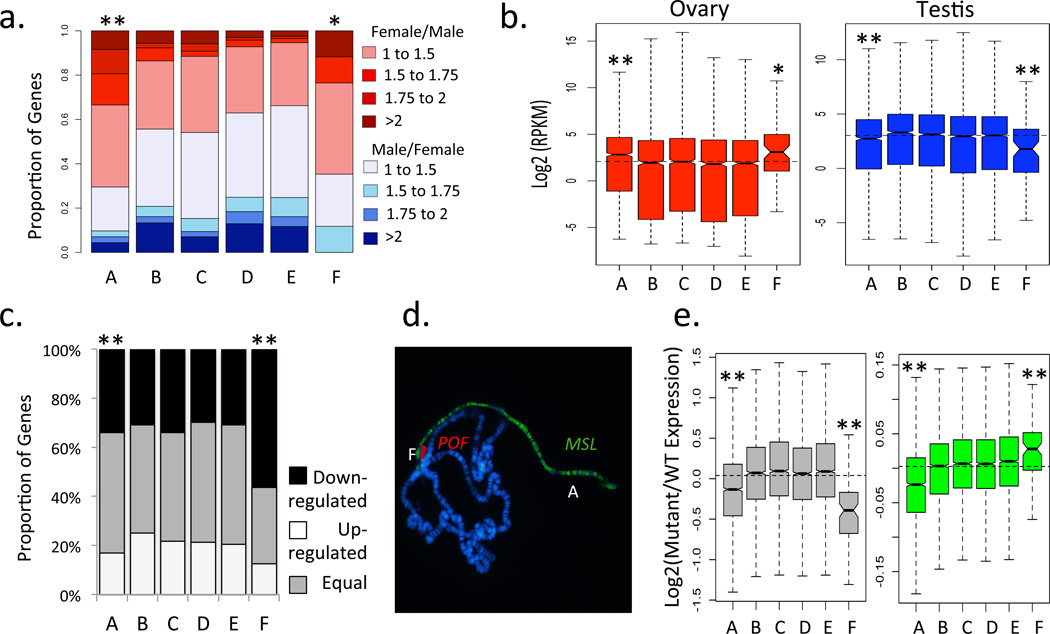
Certain features of their genome architecture distinguish X chromosomes from autosomes: they contain sex-determining factors (Sxl in Drosophila)10, show a non-random gene content (a deficiency of male and excess of female genes in Drosophila)14, are transcriptionally inactivated during spermatogenesis15, chromosome haploidy is not lethal, and chromosome-wide regulatory mechanisms ensure balanced levels of gene expression in the heterogametic sex (the MSL-complex in Drosophila)16. Interestingly, the dot chromosome of D. melanogaster harbors several peculiar characteristics that have puzzled drosophilists for decades, but can now be understood as possible remnants of its history as an X chromosome. Specifically, the dot has a minor role in sex determination and contains feminizing factors5, with increased dosage of the dot chromosome shifting 2X:3A (autosome) intersex individuals towards female development5. Consistent with its feminizing effect, genes located on the dot chromosome generally show higher expression in female compared to male embryos during early development, similar to X-linked genes (Fig. 2a). There is also a deficit of testis and an excess of ovary expression on the dot, resembling sex-biased expression profiles of the current Drosophila X (Fig. 2b). In addition, genes on the dot chromosome are down-regulated during male meiosis (Fig 2c), mimicking the phenomenon of male germline X inactivation. Furthermore, flies with only one copy of the dot chromosome are viable and fertile, and the dot is targeted by the chromosome-specific protein POF6 (Painting of fourth), which is involved in transcriptional regulation of genes located on the dot chromosome6. This resembles the MSL-complex, the only other known protein complex that specifically targets a chromosome, and POF may be part of a putative ancestral mechanisms of dosage compensation17,18 (Fig. 2d). In some Drosophila species, POF shows male-specific binding to the X chromosome in addition to binding to the dot, further supporting its involvement in dosage compensation17. Some components of the MSL complex are necessary for normal expression of genes located on the dot19, consistent with interactions of the regulatory network for the current and former sex chromosome of Drosophila (Fig. 2e). Thus, many features of the dot chromosome in Drosophila resemble unique characteristics of the current X chromosome that distinguish it from autosomes, and can be interpreted as signatures of its former life as a differentiated X chromosome. Female-biased expression during early embryogenesis, an excess of ovary genes and a deficiency of testis genes on Muller F (but not Muller A) are observed in Diptera species were the dot segregates as the X (Fig. 3). This confirms that these peculiarities of the dot were present in the X-linked ancestor, and also shows that Muller A only acquired them once it became sex-linked in the lineage leading to Drosophila.
Figure 2. Properties of the dot chromosome in Drosophila melanogaster that resemble that of an X.
a. Zygotic transcription, before the onset of MSL-dosage compensation, is female-biased for genes located on the dot (data from ref. 28). b. Genes located on the dot chromosome show increased transcription in ovary, and decreased transcription in testis (data from ref. 29). c. An excess of genes located on the dot is down-regulated during male meiosis (data from ref. 30). d. Chromosome-specific transcriptional regulation of genes located on the dot chromosome by the protein POF. e. Mis-regulation of genes located on the dot chromosome in mutants for some components of the MSL dosage compensation complex (data from ref. 19). Levels of significance (based on resampling for panels b and e and using Chi-square tests for panels a and c) are represented by * (p<0.05) and ** (p<0.01). Boxes (panels b and e) as in Fig. 1.
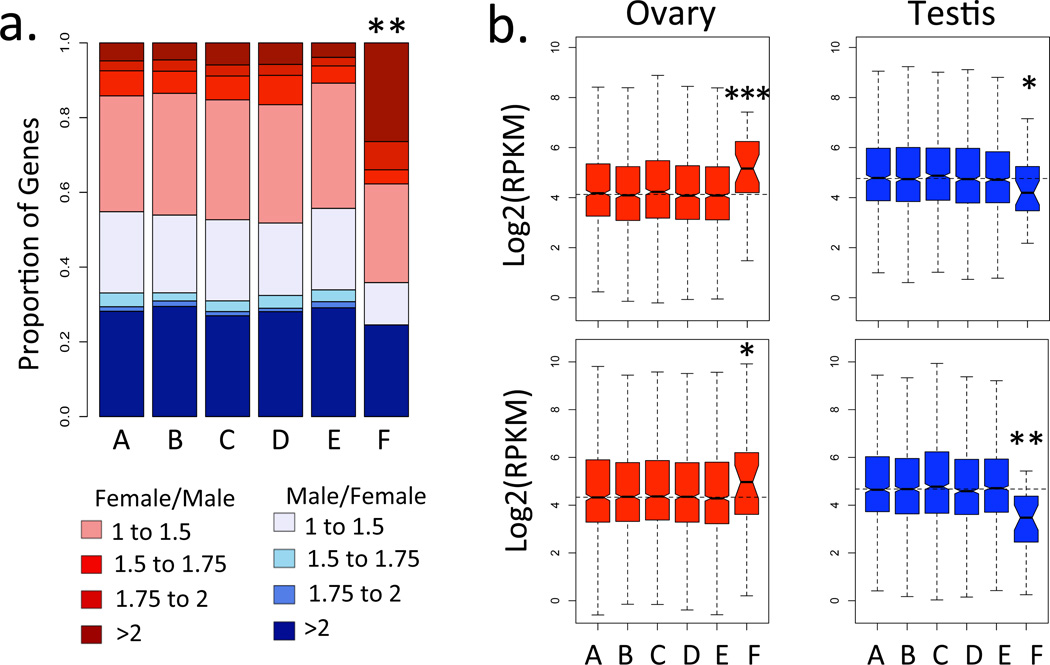
Figure 3. Gene expression in early embryos and adult gonads in outgroup Diptera species.
a. Zygotic transcription is female-biased for genes located on the X-chromosome (element F) of B. oleae, but not the autosomal element A. FPKM values were estimated by RNA-sequencing of male and female stage 5 embryos. b. Over-expression in ovary and under-expression in testis on the X-chromosome (element F) but not the autosomal element A in B. oleae (top) and S. bullata (bottom). FPKM values were estimated through RNA-sequencing of dissected testis and ovaries. Levels of significance (based on Chi-square tests for panel a and using one-tailed Wilcoxon-tests for panels b) are represented by * (p<0.05), ** (p<0.01) and *** (p<0.001). Boxes (panel b) as in Fig. 1.
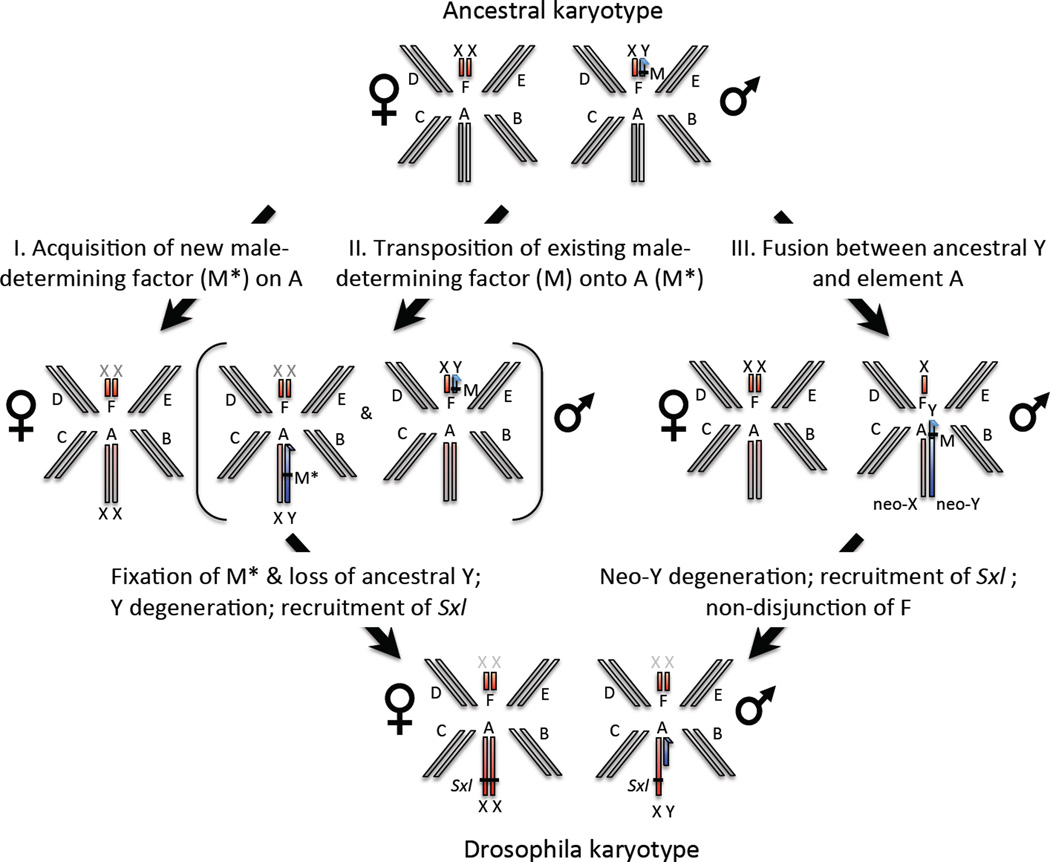
How could this transition have happened? Sex in most outgroup species of Drosophila is determined by the presence of a factor on the Y chromosome (M-factor) causing maleness20. Such a dominant-Y system with element F as the sex chromosome could evolve into the Drosophila system (dose-dependent sex determination with element A as the sex chromosome), through various intermediate steps1, and we outline three possible paths that involve mutational events that have been observed in Diptera. This sex chromosome transition could be initiated by a single epistatic mutation on element A (M*) that makes individuals male regardless of their sex chromosome karyotype, or through a translocation of the existing male-determining factor (M) onto element A (Fig. 4). Novel sex determining genes occurring on different chromosomes or translocations of M-factors onto autosomes have been observed in houseflies21 or humpbacked flies22. The fixation of the new male-determining gene on element A would lead to reversal of the ancestral X (Muller F) to an autosome, and the ancestral Y would be completely lost. Loss of the Y is only possible if the ancestral Y chromosome carried no essential male-fertility genes, as seems to be the case in several Diptera species21, or if those male-fertility genes move to another chromosome23. The emergence of a male-determining gene on element A would cause male-limited transmission of this chromosome and - since higher Diptera males generally lack recombination24 - set in motion genome-wide degeneration of the non-recombining proto-Y. Eventually, Sxl was recruited as a dose-dependent sex determination gene in Drosophila, and MSL-mediated dosage compensation evolved on element A16. The current karyotype of Drosophila could also have evolved through a chromosomal fusion between the ancestral Y chromosome and element A (Fig. 4). This would create a male-limited neo-Y (the fused element A) that initially is identical to the neo-X (the unfused element A). The non-recombining neo-Y would undergo chromosome-wide degeneration, and Y-autosome fusions coupled with neo-Y degeneration have happened repeatedly in several Drosophila species and other Diptera12,25. This fused neo-Y chromosome would form the current Y of Drosophila, and male-fertility genes, if present on the ancestral Y, could be preserved on the chimeric Y. Both element F and element A would simultaneously segregate as X chromosomes, and eventually Sxl would take over the sex determining function on element A, and a non-disjunction event could restore diploidy for element F in both sexes. Thus, under both the translocation of an existing or the emergence of a new M-factor scenario, the current Y chromosome of Drosophila shares no homology to the Y of its ancestor, while under the chromosomal fusion model, the current Drosophila Y is a chimera of the ancestral Y and the degenerated element A and may thus harbor some genes that are also Y-linked in outgroup species. We detect no Y-linked protein-coding genes that are shared between Drosophila and Diptera where Muller F is the sex chromosome (Table S3–S5, Fig S8–S10), providing some support against a fusion between the new and ancestral Y. However, it should be noted that the gene content of the Y is generally poorly conserved even within Drosophila26, and the high turnover of Y-linked genes may obscure any homology between the Y-chromosome of Drosophilids and that of their outgroups.
Figure 4. Turnover of sex chromosomes in Drosophila.
Hypothetical transition from an ancestral karyotype with element F segregating as a sex chromosome, to the karyotype observed in Drosophila where element A is the sex chromosome. In outgroup species, maleness is determined by the presence of a factor on the Y chromosome (M-factor). Element A could either have acquired a new, epistatic M*-factor (scenario I), or the existing M-factor could have transposed to element A (scenario II). This transition could also have been initiated by a fusion of the ancestral Y to element A (scenario III). Degeneration of the male-limited, non-recombining element A, followed by recruitment of Sxl for sex determination would create the ancestral karyotype of Drosophila.
Cytological and genetic studies have shown that the dot chromosome of D. busckii is fused to both the X and Y chromosome27. Interestingly, this basal species in the Drosophila genus also seems to lack MSL-mediated dosage compensation; instead, POF binds to the fused elements A and F in males only17, and it had been suggested that the A-F fusion represents the ancestral state of Drosophila18. Our data, however, clearly show that D. busckii harbors a recent, secondary fusion: P. variegata, an outgroup species, only has element A as its sex chromosome, and the reduction in M/F coverage for element F in D. busckii is much less pronounced than for element A. This suggests that reads from the Y-fused homolog of element F are still similar enough to map to the element F portion of the X. To conclude, our whole-genome analysis reveals that heteromorphic sex chromosomes are not necessarily a terminal stage in sex chromosome evolution. Instead, the dot chromosome of Drosophila provides an example of a highly differentiated sex chromosome reverting back to an autosome, and several odd properties of the dot, such as its feminizing effect, its non-random gene content, and the presence of chromosome-specific regulation of gene expression, are possible remnants of its former life as a sex chromosome. Such chromosome-specific adaptations of an X chromosome, and male-beneficial genes that may have accumulated on the ancestral Y, impose strong fitness constraints that will limit turnover of highly evolved sex chromosomes in nature.
Methods
S1.1 Sample collection
Drosophila busckii individuals from an inbred isofemale line originally caught in Tallinn (Estonia) in 2000 were provided by Jan Larsson. Wild-caught male and female Phortica variegata were collected in Bari, Italy by Siju Kunhi Purayil and Ilona Kadowm, shipped in ethanol, and kept at −20°C until DNA extraction. Pupae of Sarcophaga bullata were obtained from Carolina Biological, and incubated at room temperature until adults emerged. These were immediately sexed and used for DNA extraction. Bactrocera oleae males and females were provided by Pinelopi Mavragani from a stock maintained at the Aristotle University of Thessaloniki, shipped in ethanol and kept at −20°C until DNA extraction. Hermetia illucens were provided by Urs Schmidt-Ott, shipped in ethanol, and kept at −20°C until DNA extraction.
S1.2 Library preparation and sequencing
DNA was extracted from a single male and a single female individual from each species using the Puregene Core Kit A (Qiagen). Library preparation and paired-end sequencing of Bactrocera oleae, Phortica variegata, Sarcophaga bullata and Hermetia illucens were performed at the Beijing Genomics Institute (en.genomics.cn/). The D. busckii libraries were prepared in the Bachtrog Lab following the standard Illumina protocol, and sequenced at the Berkeley sequencing facility. For each species, libraries and sequencing were performed for a single male and a single female separately. The raw sequenced data consists of 90bps paired-end reads, with an insert size of 500bps for all libraries. All the reads generated for this analysis have been deposited at the NCBI Short Reads Archive under bioproject SRP021047.
S1.3 Genome Assembly
We obtained 100bps paired-end reads for one male and one female of each species, of which the first 10bps were trimmed. For each species, the male and female genomic reads were assembled using SOAPdenovo (soap.genomics.org.cn) with default parameters and a Kmer value of 31. Gaps in the assembly were further reduced using GapCloser (soap.genomics.org.cn). Only scaffolds longer than 1000bps were kept for further analysis. Statistics for the assemblies are presented in Table S1. In the case of D. busckii, pooling male and female reads for the assembly led to reduced coverage of element F in both males and females (see below and Fig. S2). This is likely due to the hybrid assembly of the neo-X and neo-Y (element F fused to the X, and element F fused to the Y, respectively), as the neo-Y is not yet fully degenerated and some neo-Y-derived reads are included in the assembly. Because this hybrid assembly differs from the true sequence of the neo-X, female reads will not fully map to it, leading to decreased female coverage. This bias disappeared when only female reads were used for the assembly (Fig. S3). This female-only assembly was therefore used for all further analyses. It should be noted that this does not affect our conclusions, as the male to female (M/F) coverage ratio is reduced for elements A and F independently of the assembly used in the analysis.
S1.4 Mapping of the Genomic scaffolds to Muller Elements
We downloaded all Drosophila melanogaster CDS sequences from Flybase (www.flybase.org) and, for each gene, kept only the longest CDS. The resulting D. melanogaster coding sequences were mapped to the genome assembly of each outgroup using blat31 with both a translated query and a translated database, and only the hit with the highest match score was kept for each gene. Hits with a match score below 50 were excluded. A perl script was used to count the number of Drosophila genes per Muller element that mapped to each scaffold. The scaffold was assigned to the Muller element with the largest number of matching genes. When the same number of genes from different Muller elements mapped to a scaffold, a score was calculated for each Muller element by summing the match scores of all the hits for that element, and the scaffold was assigned to the element with the largest score. The rate of concordance of genes on scaffolds (the number of genes that map to the element their scaffold was assigned to vs. total number of genes, calculated using scaffolds carrying at least 3 genes) is high: D. busckii: 96%; P. variegata: 92%; S. bullata:80%; B. oleae: 88%; H. illucens: 71%.
S1.5 Estimation of Male and Female Genomic Coverage
Male and female forward reads were mapped separately to the genomic scaffolds using BWA32 set to the default parameters for paired-end reads. The resulting SAM alignments were used to estimate the male and female coverage depth for each scaffold using SoapCov (soap.genomics.org.cn). Figures S2 to S7 show the male and female coverage distributions for each Muller element for all the species investigated. For each species, we used one-tailed Wilcoxon tests between each element and the rest of the sample to identify systematic reductions in M/F coverage. The results of the tests are presented in Table S2.
S1.6 Building the Phylogenetic Tree
From the blat results (D. melanogaster genes versus outgroup genomic sequence, see section S1.4), we selected the 185 D. melanogaster genes that had an alignment score above 1000 for all of the outgroups species. The genomic regions corresponding to this conserved set of genes were extracted from the genomic scaffolds using a perl script. Genewise was used to infer the protein sequence of the genes in the different outgroups from the corresponding genomic region. To avoid potential biases that could arise by using the D. melanogaster protein sequences as input for Genewise (as Genewise may be more likely to insert errors in the more distant outgroups than in the closer ones), we used Anopheles gambiae protein sequences instead, as A. gambiae is an outgroup to all the species analyzed. For each gene, the inferred protein sequences and the D. melanogaster and A. gambiae protein sequences were aligned using Muscle. We concatenated the Muscle output files, and ran the resulting concatenated alignment through Gblocks to remove gaps and regions of low alignment quality. The final alignment, consisting of 93134 amino acids, was used as input for PhyML to obtain the phylogenetic tree and associated bootstrap values (Fig. S1).
S1.7 Resampling procedure
Differences between elements A and F versus autosomes (elements B–E) were tested by re-sampling n genes 1000 times from the autosomal sample (where n is the number of genes for element A or F present in the sample). Levels of expression of elements A or F were considered to be significantly different from the autosomes if their observed median fell within the 5% one-sided tail of the re-sampled distribution.
S1.8 Expression analysis of adult tissues
B. oleae individuals were obtained from olives collected on the UC Berkeley campus, and S. bullata adults were obtained from pupae purchased from Carolina Biological. Male and female adults were placed in separate vials immediately after emergence, and aged on standard Drosophila food for 4 days. Paired-end RNA-seq reads were obtained from testis, ovary and female and male carcass and whole body. RNA was extracted from each tissue using a Qiagen RNeasy kit following the manufacturer’s protocol. RNA-seq libraries and sequencing were performed at the Beijing Genomics Institute. All the reads generated for this analysis have been deposited at the NCBI Short Reads Archive under bioprojects SRP021043 and SRP021044. For each species, the resulting paired-end 100bps reads were trimmed, pooled and assembled using SOAPdenovo (soap.genomics.org.cn/soapdenovo.html) with a Kmer value of 31 and an insert size of 200bps, and this assembly was further improved using GapCloser (http://soap.genomics.org.cn/soapdenovo.html). Only scaffolds longer than 300bps were kept for further analysis. Scaffolds were mapped to D. melanogaster CDS sequences downloaded from Flybase (www.flybase.org) using blat; only the location with the best mapping score was kept for each scaffold. When more than one scaffold overlapped on the same D. melanogaster gene by more than 20bps, only the scaffold with the highest mapping score was kept for further analysis. When several scaffolds mapped to different parts of the same gene, or had an overlap shorter than 20bps, their sequences were concatenated. The resulting B. oleae and S. bullata gene sequences were assigned to Muller elements according to the genomic location of their D. melanogaster homologs. For each adult tissue, reads were mapped to the B. oleae or S. bullata gene sequences using bowtie234 with default parameters. FPKM values were then estimated from the resulting SAM alignments using Cufflinks with default parameters. Genes with FPKM<10 in both testis and ovary were excluded.
S1.9 Expression analysis of early embryos
In order to assess early embryonic expression, adult individuals of B. oleae were placed in cages with fresh olives. Eggs were periodically collected from the olives and their chorions removed by immersing them in a 50% bleach solution for 1 minute. The dechorionated embryos were staged by observation under a light microscope after immersion in halocarbon oil. Stage 5 embryos were selected, as in D. melanogaster they represent the earliest stage containing primarily zygote-derived RNAs (earlier stages contain mostly maternal RNAs28). RNA and DNA were extracted following the protocol of ref. (28), and the DNA was used to sex the embryos using published sexing primers33. Libraries were made at the Bachtrog lab following the protocol of ref. (28) and 100bp single-end sequencing was performed at the Vincent J. Coates Genomics Sequencing Laboratory, Berkeley. The reads generated for this analysis have been deposited at the NCBI Short Reads Archive under bioproject SRP021044. The first 20 bps of the resulting reads were trimmed before further analysis. Reads were mapped to the B. oleae or S. bullata gene sequences (obtained under S1.8) using bowtie234 with default parameters. FPKM values were then estimated from the resulting SAM alignments using Cufflinks with default parameters. Genes with FPKM<1 in both male and female embryos were excluded.
S1.10 Searching for Drosophila Y-linked genes in Drosophila outgroups
The male DNA-seq reads available for P. variegata, S. bullata, B. oleae and H. illucens were assembled using SOAPdenovo (with a Kmer value of 31), and all scaffolds longer than 200bps were kept for further analysis. One lane of forward female and male DNA-seq reads were mapped back to these male scaffolds separately using BWA (with default parameters) and male and female coverage was estimated using SoapCoverage. We used single-end mapping of the forward read rather than paired-ended mapping because the insert size of the paired-end fragments (500bps) is longer than the minimum size of the fragments. Similarly, RNA-seq reads obtained from male tissues were assembled using SOAPdenovo-trans for S. bullata (whole male, male carcass, testis) and B. oleae (whole male, male carcass, testis), and all transcripts longer than 200bps were kept for further analysis. One lane of forward female and male DNA-seq reads were mapped back separately to the resulting male transcripts using BWA (with default parameters) and male and female coverage was estimated using SoapCoverage. The protein sequences of known ancestral Y-linked genes of Drosophila26 (kl-2, kl-3, ORY, PRY, Ppr-Y, but also CCY and ARY, which may be ancestral) were obtained from NCBI (GI numbers 190608814, 219131049, 16519041, 217416310, 158529626, 190608812 and 281309229, respectively). Genomic scaffolds and transcripts containing homologues to these proteins were identified using tblastn (with an E-value cut-off of 0.01). Sequences identified in our blast search that had no coverage in females, but had coverage higher than 0 in males, were classified as candidate homologues to the Drosophila Y-linked genes (Tables S3 and S4). No such sequences were found in the male transcriptome or genome of S. bullata, B. oleae and H. illucens, the outgroups carrying the ancestral sex chromosome (Muller element F). P. variegata, on the other hand, presents strong evidence of sharing both CCY and kl-2 with other Drosophilids. Primers designed to amplify fragments of the candidate Y-linked P. variegata scaffolds 13109 and 15584, which share homology with kl-2 and CCY respectively, amplified bands of the expected size in male but not female P. variegata, confirming their Y-linkage (Fig. S8).
In Diptera, there is a general tendency for the rDNA to reside on the sex chromosomes, but its location often differs between closely related species35. For example, both the X and the Y of D. melanogaster carry rDNA genes35, and the Y chromosome of D. pseudoobscura, which is non-homologous to the Y of D. melanogaster, has acquired rDNA loci after its formation36. On the other hand, rDNA loci in the Drosophila ananassae subgroup are located on autosomes, and D. simulans, the sister species of D. melanogaster, has lost its Y-linked rDNA cluster35. Thus, the absence or presence of rDNA on sex chromosomes contains limited information on whether the Y chromosomes of Diptera are homologous across families, and we therefore focused our analysis on protein-coding genes. Cytological studies have found that some (but not all) true fruit flies have rDNA loci on their Y (including B. oleae37), while house flies or flesh flies have rDNA clusters on their autosomes38, 39.
S1.11 Candidate Y-linked transcripts of S. bullata and B. oleae and their D. melanogaster homologs
The trimmed whole-body male and female RNA-seq reads of S. bullata and B. oleae, as well as the male and female DNA-seq reads, were mapped back to the assembled male transcriptome of these species (see S1.10) using bowtie2, keeping only reads that mapped fully in the alignment. After filtering out reads with mismatches, the DNA-seq and RNA-seq alignments were used as input for SoapCoverage and Cufflinks, respectively, to estimate the male and female DNA coverage and expression level (in FPKM). Since Y-derived sequences are characterized by their lack of female coverage or expression, scaffolds that had at least 6 male reads mapping, had (female reads / male reads)<0.1 and had (female expression/male expression)<0.1 were classified as candidate Y-linked transcripts (39 transcripts in B. oleae and 87 in S. bullata). PCRs with primers designed for 10 candidate transcripts of S. bullata yielded male-specific bands for 2 transcripts (standard PCR with an annealing temperature of 58°C, see Fig. S9), confirming that the sample contains Y-derived sequences.
In order to check for Y-linkage of these candidate sequences in D. melanogaster, DNA was extracted (using a Qiagen DNeasy kit) from one male and one female D. melanogaster and DNA-seq libraries were made and sequenced at BGI (paired-end with 500bps insert size, 2GB of data per individual). We assembled the resulting trimmed genomic reads with SOAPdenovo (with a Kmer value of 31). The male and female coverage of each genomic scaffold was estimated by mapping the male and female DNA-seq reads against the assembled genome with bowtie2 (keeping only fully mapped reads, and filtering out reads with mismatches) and processing the SAM alignment with SoapCoverage. The candidate Y-linked transcripts of S. bullata and B. oleae were then mapped against the de novo D. melanogaster genome assembly using tblastx (with an E-value cut-off of 0.01, see Table S5). As a control, 12 known Y-linked genes of D. melanogaster were also mapped to this assembly. All 12 D. melanogaster Y-linked genes mapped to scaffolds with male coverage > 0 but no female coverage, confirming that most Y-derived coding sequence is represented in the de novo D. melanogaster assembly, and can be identified by its patterns of male and female coverage. Contrary to this, only 1/39 of the B. oleae and none of the 87 S. bullata candidate Y-derived sequences mapped to D. melanogaster scaffolds with male-specific coverage, supporting a general lack of homology between the Y-chromosome of Drosophilids and that of their outgroups (Table S5). Finally, the single B. oleae candidate that maps to a male-specific D. melanogaster scaffold was shown to yield bands of similar sizes in both sexes when a PCR was performed with primers designed for the scaffold in both B. oleae and D. melanogaster, showing that this fragment is not Y-linked in either species (Fig. S10).
Supplementary Material
Acknowledgments
Funded by NIH grants (R01GM076007 and R01GM093182) and a Packard Fellowship to D.B. All the DNA/RNA-seq reads generated in this study are deposited at NCBI Short Reads Archive (http://www.ncbi.nlm.nih.gov/sra) under bioprojects SRP021043, SRP021044 and SRP021047. We thank Siju Kunhi Purayil, Ilona Kadow, Pinelopi Mavragani, Jan Larsson and Urs Schmidt-Ott for samples, Cor Zonneveld, Luca Mazzon, Darren Obbard, Vitor Aguiar, Nicolas Gompel and Jan Larsson for images, and Qi Zhou and Zaak Walton for technical assistance.
References
- 1.Bull JJ. Evolution of sex determining mechanisms. The Benjamin: Cummings Publishing Company, Inc.; 1983. [Google Scholar]
- 2.Sarre SD, Ezaz T, Georges A. Transitions between sex-determining systems in reptiles and amphibians. Annu Rev Genomics Hum Genet. 2011;12:391–406. doi: 10.1146/annurev-genom-082410-101518. [DOI] [PubMed] [Google Scholar]
- 3.Pokorna M, Kratochvil L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zoological Journal of the Linnean Society. 2009;156:168–183. [Google Scholar]
- 4.Ohno S. Sex Chromosomes and sex-linked genes. Springer Verlag; 1967. [Google Scholar]
- 5.Bridges CB. Sex in relation to chromosomes and genes. Am Nat. 1925;59:127–137. [Google Scholar]
- 6.Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V. Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci U S A. 2001;98:6273–6278. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellott DW, Page DC. Reconstructing the evolution of vertebrate sex chromosomes. Cold Spring Harb Symp Quant Biol. 2009;74:345–353. doi: 10.1101/sqb.2009.74.048. [DOI] [PubMed] [Google Scholar]
- 8.Muller HJ. Bearing of the Drosophila work on systematics. The new systematics. 1940:185–268. [Google Scholar]
- 9.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 10.Cline TW. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 11.Zacharopoulou A. Cytogenetic analysis of mitotic and salivary gland chromosomes in the Medfly Ceratitis capitata. Genome. 1987;29:67–71. [Google Scholar]
- 12.White MJ. Cytological evidence on the phylogeny and classification of the Diptera. Evolution. 1949;3:252–261. doi: 10.1111/j.1558-5646.1949.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 13.Vicoso B, Bachtrog D. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 2011;3:230–235. doi: 10.1093/gbe/evr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis. Proc. Natl. Acad. Sci. USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson J, Svensson MJ, Stenberg P, Mäkitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci USA. 2004;101:9728–9733. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddle NC, Shaffer CD, Elgin SC. A lot about a little dot - lessons learned from Drosophila melanogaster chromosome 4. Biochem. Cell Biol. 2009;87:229–241. doi: 10.1139/o08-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X, Koya SK, Kong Y, Meller VH. Coordinated Regulation of Heterochromatic Genes in Drosophila melanogaster males. Genetics. 2009;182:481–491. doi: 10.1534/genetics.109.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52:837–856. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 21.Dübendorfer A, Hediger M, Burghardt G, Bopp D. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int J Dev Biol. 2002;46:75–79. [PubMed] [Google Scholar]
- 22.Traut W. New Y chromosomes and early stages of sex chromosome differentiation: sex determination in Megaselia. J Genet. 2010;89:307–313. doi: 10.1007/s12041-010-0042-x. [DOI] [PubMed] [Google Scholar]
- 23.Larracuente AM, Noor MA, Clark AG. Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura. Mol Biol Evol. 2010;27:1612–1620. doi: 10.1093/molbev/msq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gethmann RC. Crossing Over in Males of Higer Diptera (Brachycera) J Hered. 1988;79:344–350. doi: 10.1093/oxfordjournals.jhered.a110526. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krivshenko J. New Evidence for the homology of the short euchromatic Elements of the X and Y chromosomes of Drosophila busckii with the microchromosome of Drosophila melanogaster. Genetics. 1959;44:1027–1040. doi: 10.1093/genetics/44.6.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9:e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assis R, Zhou Q, Bachtrog D. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 2012;4:1189–1200. doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-Specific Expression Profiling of Drosophila Spermatogenesis Suggests that Meiotic Sex Chromosome Inactivation Drives Genomic Relocation of Testis-Expressed Genes. PLoS Genetics. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent WJ. BLAT - The BLAST-Like Alignment Tool. Genome Research. 2002;4:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate long-read alignment with Burrows- Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrieli P, Gomulski LM, Bonomi A, Siciliano P, Scolari F, Franz G, Gasperi G. Interchromosomal Duplications on the Bactrocera oleae Y Chromosome Imply a Distinct Evolutionary Origin of the Sex Chromosomes Compared to Drosophila. PloS one. 2011;6:e17747. doi: 10.1371/journal.pone.0017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy V, Monti-Dedieu L, Chaminade N, Siljak-Yakovlev S, Aulard S, Lemeunier F, Montchamp-Moreau C. Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: ananassae and melanogaster. Heredity. 2005;94:388–395. doi: 10.1038/sj.hdy.6800612. [DOI] [PubMed] [Google Scholar]
- 36.Larracuente AM, Noor MA, Clark AG. Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura. Mol Biol Evol. 2010;27:1612–1620. doi: 10.1093/molbev/msq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drosopoulou E, Nakou I, Síchová J, Kubíčková S, Marec F, Mavragani-Tsipidou P. Sex chromosomes and associated rDNA form a heterochromatic network in the polytene nuclei of Bactrocera oleae (Diptera: Tephritidae) Genetica. 2012;140:169–180. doi: 10.1007/s10709-012-9668-3. [DOI] [PubMed] [Google Scholar]
- 38.Parise-Maltempi PP, Avancini RMP. Cytogenetics of the neotropical flesh fly Pattonela intermutans (Diptera, Sarcophagidae) Genetics and Molecular Biology. 2000;23:563–567. [Google Scholar]
- 39.Parise-Maltempi PP, Avancini RMP. Comparative cytogenetic study in Muscidae flies. Braz J Biology. 2007;67:945–950. doi: 10.1590/s1519-69842007000500020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.