Abstract
Myelodysplastic syndromes (MDS) are a group of hematopoietic malignancies characterized by ineffective hematopoiesis. Recently, we identified MDS-associated microRNAs (miRNAs) that are down-regulated in MDS. This study examines possible explanations for that observed down-regulation of miRNA expression in MDS. Since genomic losses are insufficient to explain the down-regulation of all our MDS-associated miRNAs, we explored other avenues. We demonstrate that these miRNAs are predominantly intragenic, and that, in many cases, they and their host genes are expressed in a similar pattern during myeloid maturation, suggesting their co-regulation. This co-regulation is further supported by the down-regulation of several of the host genes in MDS and increased methylation of the shared promoters of several miRNAs and their respective host genes. These studies identify a role of hypermethylation of miRNA promoters in the down-regulation of MDS-associated miRNAs, unifying research on miRNAs in MDS and epigenetic regulation in MDS into a common pathway.
Keywords: Myelodysplastic syndrome, microRNAs, hypermethylation, host genes, myeloid maturation
Introduction
Myelodysplastic syndromes (MDS) are a group of hematopoietic malignancies characterized by ineffective hematopoiesis, peripheral cytopenias, dysplasia of hematopoietic cells and a propensity to transform to acute myeloid leukemia (AML) [1]. MDS is a disease of the elderly, with a median age of 76 years, with a reported incidence of 75 per 100 000 over the age of 65 [2]. Array comparative genomic hybridization (aCGH) in addition to traditional metaphase cytogenetics studies has identified numerous karyotypic and cryptic gains and losses in MDS [3–5]. In addition, MDS has been associated with a global hypermethylation state and mutations in components of the spliceosome [6–10]. However, the mechanisms leading to MDS pathogenesis and disease progression remain to be fully elucidated.
MicroRNAs (miRNAs) are small, endogenous, non-coding RNAs, which act as translational repressors and thereby serve as master regulators of cell identity and differentiation [11]. miRNAs have been shown to govern normal human hematopoiesis, and aberrant expression of miRNAs has been reported in a number of hematological malignancies [12,13]. We have previously presented miRNA expression profiles of bone marrow samples from patients with MDS, identifying eight miRNAs that are down-regulated in MDS samples compared to normal controls [14]. We herein discuss several possible mechanisms of the down-regulation of these MDS-associated miRNAs, with a particular focus on the study of methylation of miRNA promoters in MDS.
Materials and methods
Cell lines and reagents
The acute promyelocytic leukemia (APL) cell lines, HL60 and NB4, were purchased from the American Type Culture Collection (ATCC, Manassas, VA). HL60 and NB4 cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) and RPMI medium, respectively, both supplemented with 10% fetal calf serum, 1 mg/mL l-glutamine and 1% penicillin/streptomycin (Invitrogen/Gibco, Carlsbad, CA). The cells were treated either with 1 μM all-trans retinoic acid (ATRA, purchased from Sigma, St. Louis, MO) in dimethylsulfoxide (DMSO, purchased from Sigma) or DMSO alone as a vehicle control for 7 days. ATRA was prepared in DMSO, stored as a 1 mM solution at −20 °C.
Flow cytometry
Flow cytometry studies were performed in the Research Flow Cytometry Laboratory at the Nashville Veterans Association Hospital. A phycoerythrin (PE)-conjugated antibody against CD11b (clone D12; BD Biosciences, Franklin Lakes, NJ) was used in this study in addition to 7-aminoactinomycin D (7-AAD) for viability (BD Biosciences). Cells were incubated with the antibody on ice for 30 min. Cells were analyzed on a three-laser, eight-color FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data analysis was performed using FACSDiva software (Becton Dickinson).
RNA isolation
RNA was isolated using a MirVana microRNA isolation kit (Invitrogen/Ambion, Austin, TX) on days 0, 2, 4 and 7 of ATRA treatment according to the vendor instructions.
Quantitative real time polymerase chain reaction amplification of miRNAs and their host genes
Analysis of host gene expression was performed by reverse transcription on 100 ng of each RNA sample using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), and the resultant cDNAs were assayed by quantitative real-time polymerase chain reaction (qRT-PCR) using TaqMan Gene Expression Assays (Applied Biosystems). For analysis of miRNA levels, 10 ng of total RNA was reverse transcribed using a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems), and cDNAs were assessed by qRT-PCR using TaqMan miRNA Assays (Applied Biosystems) on a Bio-Rad CFX instrument (Hercules, CA). All results were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression (host genes) or to U6 (miRNAs). Each time point was further normalized to the corresponding vehicle control samples.
Patient samples
Left-over flow cytometry specimens and paraffin-embedded particle preparations were obtained under appropriate Institutional Review Board approval. Left-over flow cytometry specimens were obtained as freshly frozen (FF) cell pellets after red cell lysis at 37 °C using PharmLyse Lysis Buffer (BD Biosciences). The pellets were frozen in minimal phosphate-buffered saline. A total of 14 FF samples with a diagnosis of MDS or probable MDS (four definitive MDS, 10 where the pathology report and review of the slides were suggestive, but not diagnostic, of MDS) and 12 normal controls (four negative controls, eight where the pathology report favored a reactive cause for the patient’s cytopenias) were studied as the training set. Additionally, 26 MDS and 20 normal control formalin-fixed paraffin-embedded (FFPE) particle preparations were selected as a validation set. Clinical history, including any treatment at the time of sample procurement, was obtained from a review of the electronic medical record (see Supplementary Table 1 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.790542). The MDS samples included six patients currently receiving regular cycles of DNA methyltransferase inhibitors (DNMTIs; either azacytidine or decitabine) as well as 20 patients who were not undergoing active treatment with DNMTIs. The average age of the patients with MDS not currently on DNMTIs was 66.4 years (range 47–87), with 13 males and seven females. Although there was a range of MDS subtypes encompassed in this group, 60% were categorized as refractory cytopenia with multilineage dysplasia. The average age of the patients currently undergoing DNMTI therapy was 65.7 years (range 56–78) and the patients were all male, including a range of MDS subtypes and responses to therapy. Patients with 5q– syndrome were excluded from this study. Normal controls were chosen from bone marrow aspirations performed for staging of lymphoma which were negative for involvement by any hematolymphoid malignancy. The average age of the normal controls was 52.2 years (range 12–80), with 10 males and 12 females.
DNA isolation
Genomic DNA (gDNA) was isolated from FF bone marrow cell pellets (obtained as remnant flow cytometry samples) using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA) according to the vendor’s instructions. Genomic DNA from the FFPE particle preparations was extracted using a RecoverAll™ Total Nucleic Acid Isolation FFPE kit (Applied Biosystems) per the vendor’s instructions.
DNA bisulfite treatment and pyrosequencing analysis
Bisulfite treatment of DNA samples was performed using a methylSEQr Bisulfite Conversion Kit (Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. PCR amplification of CpG islands present in the promoter regions of the miRNA host genes was performed on 30 ng of the modified DNA sample. All PCR primers and sequencing primers were designed using PSQ assay design software (Qiagen, Valencia, CA), except the primers for WWP2, SRSF2, ZNF207 and SFRP2 genes, which were purchased pre-designed from Qiagen (see Supplementary Table 2 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.790542). Each PCR reaction was designed to produce products shorter than 250 bp to eliminate obstacles that may occur as a result of DNA degradation in FFPE tissues. PCR products were run on 2% agarose gel to assess the quality of the products. The PCR products were then subjected to quantitative pyrosequencing analysis using a Biotage PyroMark MD system (Qiagen) following the protocol provided by the manufacturer.
Statistical analysis
Statistical analysis was performed using a χ2 distribution test to quantify the probability of all MDS-associated miRNAs showing an intragenic location, a two-tailed Pearson correlation coefficient for the comparison of methylation profiles in FF and FFPE samples, a Pearson correlation coefficient for trends on miRNA and host gene expression during myeloid maturation, and a Student’s t-test for comparison of both MDS versus normal control samples and treated MDS versus normal control samples. Samples for which both the miRNA relative expression levels (after normalizing to a control sample to allow comparison across plates as well as a sample of blood only to control for blood contamination of the particle preparations) [14] and the methylation status were studied numbered 20 samples. These were rank ordered by expression levels (quantized into three bins) and their methylation percentage graphed. For each bin, the p-value for hypermethylation was calculated by a one-sample t-test compared to a hypothetical mean of 5% methylation, based upon the well-known convention of assigning hypermethylation to percentages over 10%.
Results
Chromosomal losses and MDS
One possible mechanism for down-regulation of miRNAs in MDS is genomic losses. Since the karyotypes of the patients studied in this original work were quite varied, including many with a normal karyotype, the down-regulation of miRNAs could not be overtly correlated to metaphase cytogenetic aberrancies. Unfortunately, due to the limited availability of the fresh frozen tissue used in the training set and the archival nature of the samples used in the validation sets, we were unable to perform aCGH on the exact samples examined in the original study to look for more subtle losses of genomic material. We therefore compared the locations of our miRNAs (miR-103-1, miR-140, miR-150, miR-342, miR-378, miR-483, miR-632 and miR-636) to recurrent sites of chromosomal losses in published studies of patients with MDS by aCGH [3–5,15]. Importantly, none of the MDS-associated miRNAs are located on chromosomes 7, 8 or 20, sites of recurrent aberrations in MDS (Table I, column 2). The genomic losses at 5q32, 14q32.2 and 19q13.33, which have been recurrently found in MDS, might contribute to the down-regulation we observe in miR-378, miR-342 and miR-150, respectively (Table I, column 4), but cannot explain the full range of underexpressed miRNAs. In particular, the patients in our initial study were carefully selected to not have overt karyotypic deletions of the long arm of chromosome 5 (only four of a total of 42 patients from the initial study had any cytogenetic evidence of aberrations of chromosome 5) [14]. Additionally, the aCGH aberrations found at chromosomes 14 and 19 were found in rare patients with MDS, and cannot account for a pattern of down-regulation seen across the MDS cohort.
Table I.
Location of miRNAs down-regulated in MDS (columns 2 and 3), correlation with loci reported to demonstrate losses by aCGH (column 4) [3,4], association with promoter methylation status demonstrated in this study by pyrosequencing (column 5) and correlation with expression of host genes determined by transcriptional profi ling using RMA analysis of MILE study data (column 6) [14,21].
| miRNA | Location | Host gene | aCGH | Promoter methylation status |
Transcriptional profiling |
|---|---|---|---|---|---|
| hsa-mir-103-1 | 5q34 | PANK3 (intron 5) | No sig. difference | No sig. difference | |
| hsa-mir-140 | 16q22.1 | WWP2 (intron 15–16) | Hypermethylated | Down (p < 0.05) | |
| hsa-mir-150 | 19q13.33 | (RPS11) | Loss | No sig. difference | No sig. difference |
| hsa-mir-342 | 14q32.2 | EVL (intron 3) | Loss | No sig. difference | Down (p < 0.05) |
| hsa-mir-378 | 5q32 | PPARGC1B (intron 1) | Loss | Hypermethylated | No sig. difference |
| hsa-mir-483 | 11p15.5 | IGF2 (intron 2–4) | No sig. difference | No sig. difference | |
| hsa-mir-632 | 17q11.2 | ZNF207 (exon 1) | Hypermethylated * | Down (p < 0.05) | |
| hsa-mir-636 | 17q25.1 | SRSF2 (intron 1, exon 2–3) | No sig. difference | No sig. difference |
Representative data are shown, see Figure 2.
MDS, myelodysplastic syndromes; aCGH, array comparative genomic hybridization; RMA, robust multichip analysis; MILE, Microarrays in Leukemia; sig., significant.
Coordinated expression patterns of microRNAs and their host genes during ATRA-mediated myeloid maturation of APL cell lines
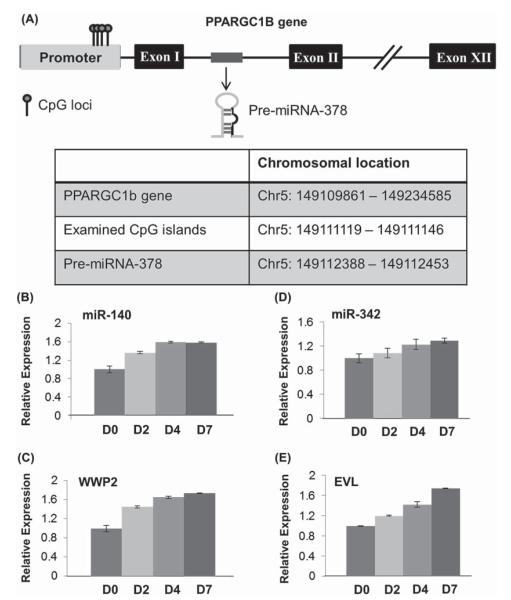
In our previous work, we discovered eight miRNAs that are underexpressed in MDS [14]. Surprisingly, all eight of these MDS-associated miRNAs were located within a protein-coding gene in the genome, or host gene (Table I, column 3). A schematic of a host gene, PPARGC1B, including the location of the intragenic miRNA-378, is shown in Figure 1(A). miRNAs are typically located in intergenic regions, often grouped together in polycistrons which are co-transcribed [16]. Less commonly, intragenic miRNAs are embedded within a host gene, believed to be regulated by the host gene promoter [17,18]. Since only approximately 40% of known miRNAs are located within host genes [16], the finding that all of the MDS-associated miRNAs are intragenic was highly significant, with a χ2 value of < 0.0001.
Figure 1.
(A) Schematic representation of the host gene PPARGC1B showing location of individual CpG loci studied and the coding region for miRNA-378 within the first exon. Chromosomal locations of PPARGC1B gene, CpG islands and pre-miRNA-378 hairpin are shown in the table. Average methylation of locus 4 was 13% in patients with MDS but 3% in controls (p-value = 0.02). (B–E) Expression of miRNAs and mRNAs in HL60 cells during ATRA-induced myeloid maturation on experimental days 0, 2, 4 and 7 (D0, D2, D4, D7) by qRT-PCR. (B) Expression of miR-140. (C) Expression of WWP2, host gene of miR-140 (Pearson correlation coefficient between WWP2 and miR-140 expression, R = 0.99). (D) Expression of miR-342. (E) Expression of EVL, host gene of miR-342 (Pearson correlation coefficient between EVL and miR-342 expression, R = 0.97).
To determine the expression patterns of the miRNAs and their host genes during myeloid maturation, we used a readily available, standard model of myeloid maturation [19,20]. Supraphysiologic levels of ATRA are known to induce maturation of promyelocytic leukemias, which provides a well-known model of myeloid maturation [19]. Accordingly, the APL cell lines, HL60 and NB4, were treated with ATRA over the course of 7 days. Maturation was confirmed both morphologically by microscopy of Wright stained cytospin preparations of the cells and immunophenotypically by an increase in CD11b expression by flow cytometry (representative HL60 data shown in Supplementary Figure 1 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.790542). Expression changes in the miRNAs and mRNA levels of host genes during ATRA-mediated maturation were assessed by qRT-PCR. Concomitant up-regulation of miRNAs and their host genes during the course of myeloid maturation was observed for the following pairs: PANK3/miR-103, WWP2/miR-140, EVL/miR-342, ZNF207/miR-632, SFRS2/miR-636 [selected examples shown, Figures 1(B)–1(E); full data are available in the Supplementary Table 3 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.790542]. These findings demonstrate the similar expression patterns of the MDS-associated miRNAs and their host genes. miR-181b, which demonstrated minimally decreased expression during the 7-day experiment, was examined as a negative control, since it is known to show relatively stable expression changes during myeloid maturation from previous cell line-based studies [20].
Host genes are underexpressed in patients with MDS
With the similarity of the expression patterns of host genes and their embedded miRNAs in cell line models of maturation, the host genes themselves should be underexpressed in MDS if co-regulated with the MDS-associated miRNAs. Large multicenter microarray studies have been published which include patients with MDS and normal controls. We conducted a robust multichip analysis (RMA) of the publicly available data set GSE13159 (from the Microarrays in Leukemia [MILE] group), comparing the 206 MDS patient samples with 74 normal controls [14,21]. Although for most of the host genes there was no significant difference in expression between patients with MDS and controls, WWP2 (host gene for miR-140), EVL (host gene for miR-342) and ZNF207 (host gene for miR-632) demonstrated significantly decreased expression in the RMA analysis (p-value < 0.05) (Table I, column 6). The magnitude of these decreases in expression seen in MDS compared to controls was less than two-fold, of the same order of magnitude as the subtle changes seen in miRNA expression, supporting the co-regulation of the host genes and their associated miRNAs.
Host gene promoter methylation in patients with MDS
To study promoter methylation of the host gene/miRNA pairs, we conducted pyrosequencing of bisulfite-converted genomic DNA (gDNA) from bone marrow aspirate specimens. Our initial sample set was composed of prospectively collected FF samples. This initial set represented 14 cases of MDS or probable MDS and 12 cases of non-MDS diagnoses. In this initial set, pyrosequencing identified individual loci in the IGF2 and WWP2 promoter regions that were statistically hypermethylated in MDS compared to controls (data not shown). These tantalizing data required further cases of definitive MDS to fully examine the methylation status of these promoter regions. We therefore evaluated the feasibility of the assay using FFPE particle preparations, using primers carefully designed to amplify only short regions of gDNA to address the shearing problem encountered with fixed samples. There was a significant correlation in methylation status between the set of paired samples for each bone marrow aspirate (R2 = 0.98) (Table II). Therefore, FFPE samples proved an adequate source of gDNA for pyrosequencing for the regions interrogated.
Table II.
Comparison of methylation levels in paired fresh frozen or paraffin-embedded samples (R2 = 0.98).
| IGF2 DMR0 RUN1 |
Fresh frozen |
FFPE |
||
|---|---|---|---|---|
| Pos. 8 | Pos. 18 | Pos. 8 | Pos. 18 | |
| CM10–74 | 45 | 100 | 31 | 100 |
| CM10–172 | 49 | 100 | 47 | 100 |
| CM10–430 | 30 | 45 | 21 | 41 |
| CM10–650 | 50 | 100 | 48 | 100 |
| CM10–765 | 65 | 100 | 57 | 100 |
| CM10–1597 | 48 | 100 | 43 | 87 |
| CM10–1807 | 50 | 100 | 44 | 100 |
IGF2, insulin-like growth factor 2; Pos., position; FFPE, formalin-fixed paraffin embedded.
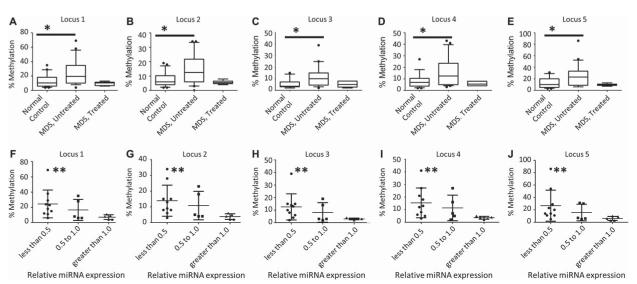
We therefore examined an additional 22 normal controls and 26 patients with MDS by using gDNA extracted from archival FFPE tissues. For each host gene promoter, 4–7 CpG sites were evaluated (Table III). The SFRP2 gene promoter was included as a positive control, since it was previously reported to have higher methylation levels in MDS [6]. Statistical analysis of the pyrosequencing data confirmed the increased methylation in the SFRP2 gene promoter in patients with MDS (Table III). Host genes for miR-103, miR-150, miR-342, miR-558 and miR-639 did not demonstrate any significant methylation in either the MDS or the control samples. However, significantly increased promoter methylation (p-value < 0.05 by Student’s t-test) was identified in patients with MDS not currently undergoing treatment with DNMTIs for the WWP2/miR-140, PPARGC1B/miR-378 and ZNF207/miR-632 host gene/miRNA pairs, while IGF2/miR-483 and SFRS2/miR-636 promoters demonstrated increased methylation in MDS which neared significance [Table III, Figures 2(A)–2(E) for whisker plot of results for the five methylation regions in the ZNF207 promoter; full data available in the Supplementary Table 4 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.790542]. Interestingly, the six samples from patients currently undergoing regular cycles of treatment with either azacytidine or decitabine showed normalization of their methylation status for WWP2/miR-140, PPARGC1B/miR-378 and ZNF207/miR-632, confirming the presence and activity of the therapeutics. Therefore, promoter hypermethylation was seen for several host gene/miRNA pairs. In addition, treatment with DNMTIs can lead to normalization of the promoter methylation.
Table III.
Compiled methylation results for all methylation regions examined by pyrosequencing*.
| CpG regions methylated in MDS versus controls (p-values†) |
|||||||
|---|---|---|---|---|---|---|---|
| miRNA/host gene | Region 1 | Region 2 | Region 3 | Region 4 | Region 5 | Region 6 | Region 7 |
| PANK3/miR-103 | NA | NA | NA | NA | NA | NA | NA |
| WWP2/miR-140 | 0.023 | 0.007 | 0.011 | 0.005 | 0.023 | 0.043 | 0.032 |
| RPS11/miR-150 | NA | NA | NA | NA | NA | NA | NA |
| EVL/miR-342 | NA | NA | NA | NA | NA | NA | NA |
| PPARGC1B/miR-378 | 0.036 | 0.048 | 0.047 | 0.021 | |||
| IGF2/miR-483 | 0.087 | 0.415 | 0.052 | 0.123 | 0.288 | 0.448 | 0.111 |
| ZNF207/miR-632 | 0.017 | 0.013 | 0.009 | 0.029 | 0.020 | ||
| SFRS2/miR-636 | 0.091 | 0.054 | 0.147 | 0.167 | 0.319 | ||
| SFRP2 (positive control) | 0.016 | 0.006 | 0.003 | 0.008 | 0.015 | 0.013 | 0.185 |
Regions examined by pyrosequencing were as follows: PANK3 (Chr5: 167982853–167982916), WWP2 (Chr16: 69795297–69795233), RPS11 (Chr19: 49998667–49998742), EVL (Chr14: 100484676–100484715), PPARGC1B (Chr5: 149111119–149111146), IGF2 (Chr11: 2151538–2151580 and 2143160–2143212), ZNF207 (Chr17: 30677150–30677195), SFRS2 (Chr17: 74734100–74734123) and SFRP2 (Chr4: 154712680–154712715).
p-Values obtained from two-tailed t-test analysis of MDS samples (no recent DNMTI therapy) compared to normal controls.
MDS, myelodysplastic syndromes; DNMTI, DNA methyltransferase inhibitor; NA, essentially no significant methylation in either MDS or control samples.
Figure 2.
(A–E) Methylation of five CpG sites on the ZNF207/miR-632 promoter (Chr17: 30677150–30677195) showing hypermethylation of MDS samples (n = 20) in this region compared to normal controls (n = 22) and compared to samples from patients with MDS actively receiving DNMTIs (n = 6). p-Values (*) from Student’s t-test are 0.017, 0.013, 0.009, 0.029 and 0.020 for loci 1 through 5, respectively (see also Table III). Plots demonstrate the median (line) with 1st and 3rd quartiles within the box and 10–90 percentile whiskers. (F–J) Comparison of miR-632 relative expression and methylation status for 20 MDS and control samples demonstrating significantly increased methylation for those samples demonstrating less than 0.5 relative expression by one-sample t-test (**p-values are 0.0081, 0.019, 0.046, 0.023 and 0.021 for loci 1 through 5, respectively). Plots demonstrate mean with standard deviation.
Of the archival tissues used for this study, 20 samples (both MDS and normal controls) had sufficient RNA to have been studied for their miRNA expression as well. For those samples that had less than 0.5 relative expression, there was a statistically significant increase in methylation over an unmethylated state (by convention less than 10% methylation) [Figures 2(F)–2(J) for the sample loci in ZNF207 shown in Figures 2(A)–1(E)].
Discussion
We previously identified several miRNAs that are underexpressed in MDS bone marrow samples compared to normal control marrow samples [14]. In this study, we have sought explanations for that observed down-regulation of miRNA expression in MDS. We have identified that some miRNAs may be underexpressed due to genomic losses. However, this explanation, from aCGH data, seems insufficient to explain the differential expression of all the MDS-associated miRNAs, although smaller deletions cannot be entirely excluded.
We have therefore also demonstrated that several intragenic miRNAs and their host genes demonstrate similar expression patterns during myeloid maturation, suggesting their co-regulation. If miRNAs and their host genes are co-regulated and the miRNAs are underexpressed in MDS, then the host genes, too, should be down-regulated. We analyzed the transcriptional profiles of patients with MDS and controls to determine the corresponding expression of the host genes in MDS [14,21]. Statistically significant under-expression of several host genes – WWP2, EVL and ZNF207 – was identified. These data support the co-regulation of miRNAs and their host genes.
Although there are a myriad of potential regulatory processes which might result in decreased transcription of the miRNAs and their host genes in MDS, one potential cause is epigenetic regulation, which is known to play a prominent role in MDS. Specifically, global hypermethylation has been identified in MDS, and the only Food and Drug Administration (FDA)-approved drugs for the treatment of non-5q– subtypes of MDS are DNMTIs, although the exact mechanism of their therapeutic effect is unclear [6,22–25]. We examined the methylation status of selected CpG loci in the promoters of miRNAs/host genes and found increased promoter methylation of miR-140/WWP2, miR-378/PPARGC1B and miR-632/ZNF207. The larger validation methylation studies were largely performed on paraffin-embedded tissues, demonstrating the ability of pyrosequencing to acquire reasonable methylation data from this sample type. Interestingly, six of the MDS patient samples used in the pyrosequencing studies were from patients currently undergoing therapy with DNMTIs. These samples demonstrated low levels of methylation compared to the other patients with MDS, similar to that of the normal controls. This normalization of methylation status may serve as a pharmacodynamic marker of therapy. However, the number of patients on DNMTIs was too small to discriminate responders from non-responders in this study.
Several of the host genes themselves are either known to be implicated or are related to families of genes known to play roles in MDS or hematopoiesis. Recently, mutations at P95 of SRSF2, host gene of miR-636 and a component of the splicing machinery, have been identified in patients with MDS [10]. These mutations, as well as mutations in other spliceosome components, have been implicated in the pathogenesis of MDS [7–9,26].
Although a definitive host gene for miR-150 has not been identified, RPS11 lies in proximity to the miRNA, although oriented in the opposite direction. It is tantalizing to hypothesize that these two genetic loci may be related, since other ribosomal proteins have been implicated in several acquired and constitutional disorders of bone marrow failure, including 5q– syndrome, Diamond–Blackfan syndrome, dyskeratosis congenital, Shwachman–Diamond syndrome and Treacher–Collins syndrome [27–30].
Insulin-like growth factor 2 (IGF2), host gene of miR-483, is a known imprinted gene whose product is a critical factor in supporting hematopoiesis [31]. Aberrant imprinting of IGF2 and its reciprocally imprinted gene at an adjacent locus, H19, has been implicated in congenital growth disorders, Beckwith–Wiedemann syndrome and Silver–Russell syndrome, as well as neoplastic diseases such as Wilm’s tumor, rhabdomyosarcoma, hepatoblastoma, colorectal carcinoma and breast carcinoma [32] Biallelic expression of IGF2 is typical of the immature fraction in bone marrow, but the role of IGF2 in MDS is yet unclear [33–36]. The characteristically higher methylation levels of its promoter found in this study may be useful in the development of pharmacodynamic markers in response to therapy with DNMTIs.
The host genes for miR-103, miR-140, miR-342, miR-378 and miR-632 also may themselves have biological significance in MDS, although there is currently no described relationship. PANK3, host gene for miR-103-1, is a pantothenate kinase, important in the biosynthesis of coenzyme A, a key cofactor in all metabolically active cells. WWP2, host gene for miR-140, is an E3 ubiquitin ligase in the NEDD4-like protein family that has been shown to regulate PTEN (phosphatase and tensin homolog deleted on chromosome 10) ubiquitination [37]. EVL, a member of the Ena/VASP-like protein family which function in actin cytoskeletal remodeling, is the host gene for miR-342, and may play a role in RAD51-mediated repair of double-strand DNA breaks through homologous recombination [38]. PPARGC1B, host gene for miR-378, is a transcription factor activator implicated in the pathogenesis of diabetes and obesity [39]. Lastly, ZNF207, host gene for miR-632, is a zinc finger transcription regulator with poorly understood biology.
In conclusion, we have identified that there is hypermethylation of several MDS-associated miRNA promoters that might contribute to the down-regulation of MDS-associated miRNAs and their associated host genes. Treatment with DNMTIs appears to result in normalization of the methylation at these sites. This study, therefore, unites the significant findings of miRNA dysregulation in MDS with research on hypermethylation in MDS into a common pathway of pathogenesis. Overall, it is unclear whether the down-regulated host gene or the miRNA is the major contributor to MDS phenotype. As we have outlined above, some of the host genes may conceivably play a role in MDS pathogenesis. However, since miRNAs are each master regulators of various cellular processes, including hematopoietic differentiation, through their repression of multiple mRNAs, they may have wider effects in MDS pathobiology. The specific mechanisms or pathways through which miRNA dysregulation causes the MDS phenotype remain one of the most important unanswered questions in this field [40]. This work provides a bridge between the epigenetic phenomenon of hypermethylation in MDS and individually identified markers, both protein-encoding genes and miRNAs, of the MDS phenotype.
Supplementary Material
Acknowledgements
We thank Dr. Christine Eischen for her thoughtful reading of the manuscript and Drs. Adam Seegmiller and Mary Ann Thomspon Arildsen for additional graphical and statistical suggestions. We also gratefully acknowledge the early assistance in the mining of published methylation data which was performed by Dr. Maria Figueroa, although that work is not specifically cited in this research.
Th is study was supported by the Doris Duke Charitable Foundation, Clinical Scientist Development Award (A.S.K.), CTSA grant UL1 RR024975 from NCRR/NIH (A.S.K.), American Cancer Society Institutional Research Grant #IRG-58-009-51 (A.S.K.) and the CCSG 5P30 CA068485 (histology).
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at www.informahealthcare.com/lal.
Supplementary material available online Tables and figure showing further results
References
- [1].Swerdlow SH, Campo E, Harris NL, et al., editors. World Health Organization classification of tumours of haematopoietic and lymphoid tissues. 4th ed IARC Press; Lyon, France: 2008. [Google Scholar]
- [2].Cogle CR, Craig BM, Rollison DE, et al. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117:7121–7125. doi: 10.1182/blood-2011-02-337964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Starczynowski DT, Vercauteren S, Sung S, et al. Copy number alterations at polymorphic loci may be acquired somatically in patients with myelodysplastic syndromes. Leuk Res. 2011;35:444–447. doi: 10.1016/j.leukres.2010.08.003. [DOI] [PubMed] [Google Scholar]
- [4].Starczynowski DT, Vercauteren S, Telenius A, et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112:3412–3424. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]
- [5].Vercauteren SM, Sung S, Starczynowski DT, et al. Array comparative genomic hybridization of peripheral blood granulocytes of patients with myelodysplastic syndrome detects karyotypic abnormalities. Am J Clin Pathol. 2010;134:119–126. doi: 10.1309/AJCPH27ZIZEJLORF. [DOI] [PubMed] [Google Scholar]
- [6].Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Patnaik MM, Lasho TL, Hodnefield JM, et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119:569–572. doi: 10.1182/blood-2011-09-377994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- [11].Obernosterer G, Tafer H, Martinez J. Target site effects in the RNA interference and microRNA pathways. Biochem Soc Trans. 2008;36:1216–1219. doi: 10.1042/BST0361216. [DOI] [PubMed] [Google Scholar]
- [12].Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- [13].Pelosi E, Labbaye C, Testa U. MicroRNAs in normal and malignant myelopoiesis. Leuk Res. 2009;33:1584–1593. doi: 10.1016/j.leukres.2009.04.039. [DOI] [PubMed] [Google Scholar]
- [14].Erdogan B, Facey C, Qualtieri J, et al. Diagnostic microRNAs in myelodysplastic syndrome. Exp Hematol. 2011;39:915–926. e2. doi: 10.1016/j.exphem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- [15].Tiu RV, Gondek LP, O’Keefe CL, et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood. 2011;117:4552–4560. doi: 10.1182/blood-2010-07-295857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ronchetti D, Lionetti M, Mosca L, et al. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci USA. 1980;77:2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fatica A, Rosa A, Ballarino M, et al. Role of microRNAs in myeloid differentiation. Biochem Soc Trans. 2008;36:1201–1205. doi: 10.1042/BST0361201. [DOI] [PubMed] [Google Scholar]
- [21].Mills KI, Kohlmann A, Williams PM, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114:1063–1072. doi: 10.1182/blood-2008-10-187203. [DOI] [PubMed] [Google Scholar]
- [22].Kaminskas E, Farrell A, Abraham S, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- [23].Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- [24].Kantarjian HM, O’Brien S, Huang X, et al. Survival advantage with decitabine versus intensive chemotherapy in patients with higher risk myelodysplastic syndrome: comparison with historical experience. Cancer. 2007;109:1133–1137. doi: 10.1002/cncr.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Silverman LR, McKenzie DR, Peterson BL, et al. Further analysis of trials with azacitidine in patients with myelodysplastic syndrome: studies 8421, 8921, and 9221 by the Cancer and Leukemia Group B. J Clin Oncol. 2006;24:3895–3903. doi: 10.1200/JCO.2005.05.4346. [DOI] [PubMed] [Google Scholar]
- [26].Damm F, Thol F, Kosmider O, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26:1137–1140. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- [27].Boultwood J. The role of haploinsufficiency of RPS14 and p53 activation in the molecular pathogenesis of the 5q- syndrome. Pediatr Rep. 2011;3(Suppl. 2):e10. doi: 10.4081/pr.2011.s2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Czibere A, Bruns I, Junge B, et al. Low RPS14 expression is common in myelodysplastic syndromes without 5q- aberration and defines a subgroup of patients with prolonged survival. Haematologica. 2009;94:1453–1455. doi: 10.3324/haematol.2009.008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pellagatti A, Hellstrom-Lindberg E, Giagounidis A, et al. Haploinsufficiency of RPS14 in 5q- syndrome is associated with deregulation of ribosomal- and translation-related genes. Br J Haematol. 2008;142:57–64. doi: 10.1111/j.1365-2141.2008.07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang CC, Lodish HF. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- [32].Murrell A, Ito Y, Verde G, et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PloS One. 2008;3:e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bock O, Schlue J, Kreipe H. Reduced expression of H19 in bone marrow cells from chronic myeloproliferative disorders. Leukemia. 2003;17:815–816. doi: 10.1038/sj.leu.2402830. [DOI] [PubMed] [Google Scholar]
- [34].Hofmann WK, Takeuchi S, Frantzen MA, et al. Loss of genomic imprinting of insulin-like growth factor 2 is strongly associated with cellular proliferation in normal hematopoietic cells. Exp Hematol. 2002;30:318–323. doi: 10.1016/s0301-472x(01)00797-4. [DOI] [PubMed] [Google Scholar]
- [35].Morison IM, Eccles MR, Reeve AE. Imprinting of insulin-like growth factor 2 is modulated during hematopoiesis. Blood. 2000;96:3023–3028. [PubMed] [Google Scholar]
- [36].Tessema M, Langer F, Bock O, et al. Down-regulation of the IGF-2/H19 locus during normal and malignant hematopoiesis is independent of the imprinting pattern. Int J Oncol. 2005;26:499–507. [PubMed] [Google Scholar]
- [37].Maddika S, Kavela S, Rani N, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13:728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Takaku M, Ueno H, Kurumizaka H. Biochemical analysis of the human ENA/VASP-family proteins, MENA, VASP and EVL, in homologous recombination. J Biochem. 2011;149:721–729. doi: 10.1093/jb/mvr029. [DOI] [PubMed] [Google Scholar]
- [39].Zorzano A, Hernandez-Alvarez MI, Palacin M, et al. Alterations in the mitochondrial regulatory pathways constituted by the nuclear cofactors PGC-1alpha or PGC-1beta and mitofusin 2 in skeletal muscle in type 2 diabetes. Biochim Biophys Acta. 2010;1797:1028–1033. doi: 10.1016/j.bbabio.2010.02.017. [DOI] [PubMed] [Google Scholar]
- [40].Chen C. microRNAs in myelodysplastic syndromes: how much do we know and not know? Leuk Res. 2013;37:241–242. doi: 10.1016/j.leukres.2012.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.