Abstract
The human Y chromosome is intriguing not only because it harbours the master-switch gene determining gender but also because of its unusual evolutionary trajectory. Previously an autosome, Y chromosome evolution has been characterized by massive gene decay. Recent whole-genome and transcriptome analyses of Y chromosomes in humans and other primates, in Drosophila species as well as in plants have shed light on the current gene content of the Y, its origins and its long-term fate. Comparative analysis of young and old Y chromosomes have given further insights into the evolutionary and molecular forces triggering Y degeneration and its evolutionary destiny.
Sex chromosomes are derived from autosomes and have evolved independently many times in different lineages. For example, the human X and Y chromosomes originated about 200-300 million years ago in eutherian mammals1,2 after the split of monotremes, and sex chromosomes evolved independently in birds, snakes, and multiple times in other reptiles, amphibians and fish; they also formed repeatedly in many invertebrate taxa and plants3-5. Sex chromosomes carry the master-sex determining genes, are subject to unique evolutionary forces6,7, and play a prominent role in many evolutionary processes, such as speciation8, adaptation9,10 and genomic conflict11. Some Y chromosomes such as those of human and Drosophila, and some species of plants on which this article will focus are known to undergo a process termed Y chromosome degeneration in which the Y chromosome looses most of its original genes over evolutionary time3-7.
Two features set Y chromosomes apart from the rest of the genome; these are a lack of recombination on the Y over some or most of its length6, and male-limited transmission of the non-recombining segment10. Investigating Y chromosomes is challenging; their lack of recombination prevents classical linkage mapping studies, and their high content of repetitive and ampliconic sequences has excluded them from most genome sequencing projects12. Classical genetics studies have shown that Y chromosomes often encode nearly no genes13, and indeed, some species have completely lost their Y14,15.
Recent genomic advances, however, have provided newly detailed views of Y evolution and revealed many novel and often surprising insights12. Analyses of Y chromosome sequences have shed light on the DNA sequence composition and gene content of both recently formed and ancient Y chromosomes in both animals and plants. These studies have revealed the evolutionary forces operating on Y chromosomes, provided novel insights into molecular and evolutionary mechanisms that initiate Y differentiation, or determine its long-time survival (in particular, the human Y), and these recent insights will be the focus of this article.
I first present a brief summary of whole-genome analyses of the genomic architecture of old Y chromosomes in primates and Drosophila, followed by a description of the evolutionary processes that drive Y chromosome differentiation. I then present recent findings from genome-wide investigations of the molecular processes initiating Y degeneration in recently formed Y chromosomes of plants and Drosophila, and conclude with a discussion of the long-term destiny of Y chromosomes, and promising future directions of Y chromosome research.
Genomic composition of old Y chromosomes
Old Y chromosomes, such as those of D. melanogaster and human, are often highly heterochromatic and have a large amount of repetitive and ampliconic DNA15, making them notoriously difficult to sequence12. The genes carried by the sex chromosomes of humans and other mammals overlap, suggesting that they all evolved from a common ancestral chromosome >200 MY ago, and all members of Drosophila share a homologous ancestral sex chromosome pair that formed >60 MY ago; thus, the Y chromosomes of these model species are evolutionarily old and the X and Y are highly diverged from each other. To date only three primate species – human, chimpanzee and rhesus macaque - have had most of their Y chromosome euchromatic DNA sequenced16-21 (Box 1). In addition, detailed molecular and bioinformatics analyses have allowed identification of most of the protein-coding genes located on the completely heterochromatic Y chromosome of Drosophila melanogaster22-30, but the vast majority of non-exonic DNA on the Drosophila Y has not been sequenced (Box 2). Comparative analysis of primate Y chromosomes and Drosophila melanogaster Y chromosomes have revealed common features that are shared between their Y chromosomes and evolved independently in these clades; the most striking characteristics being chromosome-wide decay in functional genes on the Y.
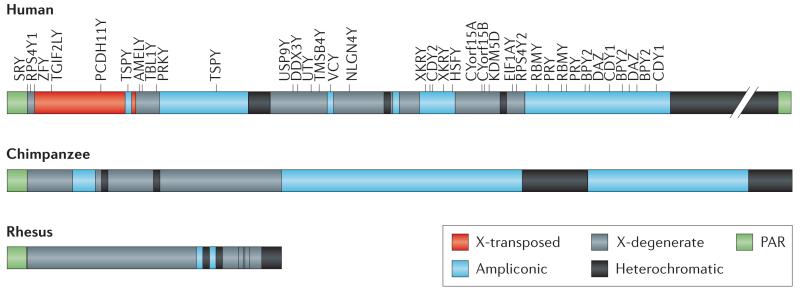
Box 1. Primate Y chromosome as revealed from genomic studies.
Genomic and genetic analyses have revealed that the primate sex chromosomes are composed of a heterogeneous mix of sequences16,20-22 with different evolutionary trajectories:
Pseudoautosomal regions (PARs)
These sequences flank the male-specific region of the Y (MSY), and have homologs on the X chromosome. The PARs of the sex chromosomes recombine during meiosis102. The human Y has two PARs. The rhesus and chimpanzee Y chromosomes only have one PAR that corresponds to the short-arm PAR in human, and the precise boundary between that PAR and the MSY is identical in the three species.
Heterochromatin
The human Y contains a large heterochromatic block (roughly 40-Mb long), which has not been sequenced. Both the chimpanzee and rhesus Y have much less heterochromatin.
X-transposed region
This is a 3.4 Mb region that is unique to humans and transposed to the Y chromosome about 3-4 MY ago, and contains only 2 genes.
X-degenerate region
The X-degenerate region is a deteriorated version of the ancestral autosome that formed the Y. It contains 16 single-copy genes in humans with homologs on the X that mostly have housekeeping functions16,18. Genes located in the X-degenerate regions are well conserved among primates. Specifically, all of the 16 X-degenerate genes are shared between human and rhesus, while chimpanzees have lost four of them through inactivating mutations.
Ampliconic regions
The ampliconic region is a highly repetitive sequence. In humans, ampliconic regions contain about 60 genes that belong to 9 different gene families all with male-specific function, often located in palindromes17,103 (see Box 3). While humans and chimpanzee contain a substantial amount of ampliconic DNA, this sequence feature is almost absent from rhesus and the euchromatic segment of the MSY is notably smaller in rhesus compared to that of the human and chimpanzee; only 0.5 Mb of the rhesus MSY euchromatin is ampliconic, compared to 10.2 Mb and 14.7 Mb in human and chimpanzee, respectively19-21. Also, fewer of the ampliconic regions are in palindromes in rhesus; the human and chimpanzee MSYs have 8 and 19 palindromes that span 5.5 Mb and 7.5 Mb, respectively, while the rhesus MSY has only three palindromes which collectively span 437 kb. Unlike in humans, most palindromes in chimpanzee exist in multiple copies, so that each palindrome arm has multiple potential partners for both intra- and interpalindrome gene conversion. Despite the elaborate structure of the chimpanzee Y, there are no chimpanzee-specific ampliconic genes, yet three out of nine multi-copy, testis-expressed gene families present in human are pseudogenized or are simply absent in chimpanzee. Thus, the gene repertoire of the chimpanzee MSY is considerably smaller and simpler than that of the human MSY. The chimpanzee MSY contains only two-thirds as many distinct genes or gene families as the human MSY, and only half as many protein-coding transcription units.
Box 2. The Drosophila melanogaster Y chromosome.
The ~40 Mb Y chromosome of Drosophila is completely heterochromatic, and only parts of the coding sequence of Y-linked protein-coding genes have been sequenced104. Male D. melanogaster lacking a Y chromosome are viable but sterile, indicating that Y-linked genes have a role in spermatogenesis22. Investigations of Y chromosome rearrangements identified six fertility factors on the Drosophila Y22,23, and several of these fertility factors span several mega-bases of DNA, and are over 100 times larger than the average Drosophila gene23. This dramatic increase in gene size is due to gigantic introns that consist largely of repetitive DNA and results in the formation of giant lampbrush-like loops during their transcription in spermatogenesis105. Bioinformatics analysis of the D. melanogaster genome identified 13 protein-coding genes (one of which is in multiple copies) and two RNA-coding genes, most of which have no homologs on the X; instead, their closest paralogs are autosomal suggesting that these genes transposed onto the Y from an autosomal ancestral copy 24-26. Comparison of the Y chromosome gene content across the 12 sequenced Drosophila species revealed that only three of the D. melanogaster Y-linked genes are Y-linked in all species, and only 5 of the genes were inferred to be present in the most recent common ancestor of the 12 Drosophila species 63 MY ago29 and the other genes were acquired more recently27. Thus, the gene content of the Drosophila Y chromosome shows low level of conservation, and gene gains had a prominent role in the evolution of the Drosophila Y chromosome.
The only homologous region between the X and the Y chromosome in D. melanogaster is the rDNA locus, a tandemly repeated array consisting of hundreds of units encoding ribosomal RNA genes, which physically accounts for about 10% of the entire Y chromosome28. The lack of homology between the X and the Y has led to the suggestion that the Y in Drosophila is in fact derived from a supernumerary B chromosome after loss of the ancestral Y30, but can also simply mean that all original genes were lost from the Y over time. Interestingly, although the Drosophila Y chromosome is degenerated, heterochromatic, and contains very few genes, increasing evidence suggests that it plays an important role in regulating the expression of numerous, possibly hundreds to thousand of autosomal and X-linked genes, which is presumably related to epigenetic modification of chromatin state by the Y chromosome31-33. That is, the Y chromosome of D. melanogaster – which accounts for 20 % of the haploid genome of a male fly, is entirely heterochromatic, and might function as a sink for proteins necessary for the establishment of heterochromatin. Indeed, the Y chromosome exerts an epigenetic effect in PEV (position effect variegation; the silencing of genes due to their proximity to heterochromatin); an extra Y in the karyotype (XX-Y females and XY-Y males) suppresses the variegation while a missing Y (X0 males) enhances PEV106. The Y might interfere with gene expression either by recruiting proteins involved in chromatin remodeling (PEV) or by disturbing some key transcription factors107, 108.
In humans, the euchromatic part of the Y chromosome is about 23-Mb in size and contains 78 protein-coding genes, compared to 150-Mb of euchromatin and about 800 protein-coding genes on the X. Similarly, the D. melanogaster X contains about 22 Mb of euchromatic sequence, and harbors about 2200 genes, while the ~40 Mb Y chromosome is completely heterochromatic and contains only 13 known protein-coding genes. Y chromosomes instead often contain a large fraction of repetitive and heterochromatic DNA. Genome sequencing and transcriptome analysis has also confirmed that many genes of male related function are located on the Y chromosome in both primates18 and Drosophila22-25. Furthermore, sequencing of the human Y chromosome has revealed large ampliconic regions that contain genes belonging to different gene families16. These gene families commonly reside in palindromes and gene conversion between members of a gene family may have aided their long term survival on the Y17 (Box 3). Sequencing also revealed the presence of evolutionary strata (see below). Also interesting is that gene gain has played an important role in Drosophila Y chromosome evolution27, and the Drosophila Y chromosome influences expression of possibly hundreds of autosomal and X-linked genes31-33. Whole-genome and transcriptome analysis has also revealed some other surprising twists and interesting characteristics of these chromosomes (for details see Box 1 and Box 2), but largely conform to the classical view of Y chromosomes representing highly degenerate versions of the X. Both the primate and the Drosophila Y chromosome have lost most of their ancestral genes that were initially present on the autosome that formed the Y millions of years ago, and most of the remaining genes, have a male-specific function.
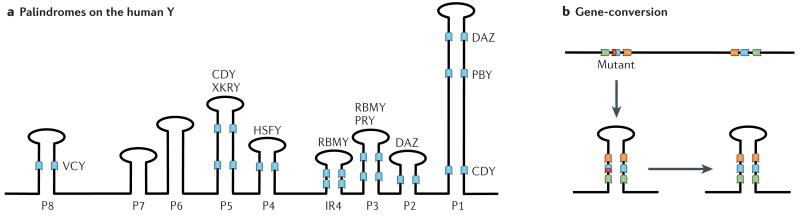
Box 3. Palindromes and gene conversion on the primate Y A.
Genome sequence analysis of the human Y revealed that a substantial fraction of the chromosome is occupied by large repeat units (amplicons). Amplicons can be organized as tandem arrays, or as inverted repeats (palindromes). In humans, there are eight palindromes that make up 54% (5.5 Mb) of the ampliconic sequence (P1-P8 (see below)), and most contain several protein-coding genes belonging to different gene families (such as the DAZ gene cluster in P1 and P2 or the CDY gene cluster in P1 and P5).
B. Palindromes allow for intrachromosomal gene conversion between members of a gene family, through pairing of homologous genes that reside within palindromes on the Y. This may allow the restoration of a mutation-free gene copy at a locus with a deleterious mutation (depicted in red) through a gene conversion with another copy of the same gene family, and has been shown to oppose Y degeneration by analytical and computer simulation methods109,110. A consequence of gene conversion between the arms of a palindrome is a high amount of sequence identity between repeat units. In humans, 60% of ampliconic sequences (including all 8 palindromes) show 99.9% or greater intrachromosomal sequence identity. A side effect of the repetitive structure of amplicons is that they are evolutionary rather unstable and have undergone dramatic restructuring across primates, and have high mutation rates. Indeed, several cases of human male sterility are associated with deletions of Y-linked chromosomal segments that are the result of recombination events between repeat regions111.
Evolutionary forces on the Y chromosome
Sex chromosomes originate from autosomes34,35. They can arise in species with separate sexes where sex is determined by environmental cues (such as in turtles where temperature determines the sex of developing embryos) and can also arise in hermaphrodites (i.e. individuals with both male and female sex organs). Figure 1 illustrates a potential path leading to heteromorphic sex chromosomes, but others are possible34. The first step in the evolution of Y chromosomes is likely to be the acquisition of a male-determining gene on one member of a pair of autosomes that ultimately will become the sex chromosomes (for example, a male-determining gene forming a proto-Y chromosome). Genetic sex determination with otherwise homomorphic sex chromosomes is observed in many taxa in amphibians, fish, reptiles and many invertebrates15,36. For heteromorphic sex chromosomes to originate after the acquisition of a male-determining gene, recombination needs to become suppressed between the homomorphic proto-sex chromosomes5,34. This allows the Y chromosome to evolve independently of its X homologue.
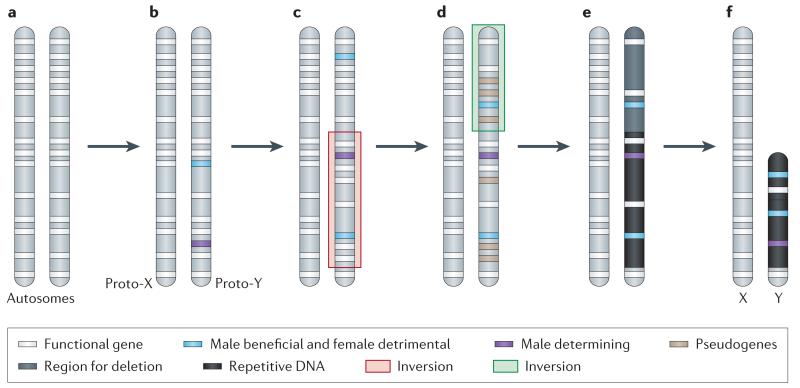
Figure 1. A Model for the differentiation of sex chromosomes.
a. Sex chromosomes formed from ordinary autosomes, containing identical sets of genes b. A potential first step in the evolution of heteromorphic sex chromosomes is the acquisition of a sex-determining locus on a proto-sex chromosome, such as a male-determining gene. The emergence of separate sexes and sex chromosomes from a hermaphrodite ancestor requires a male- and a female-sterility mutation to occur on the proto-sex chromosome6. c. Accumulation of sexually antagonistic mutations close to the sex-determining region select for suppression of recombination on the proto-sex chromosomes which can be achieved by chromosomal inversion. d. The non-recombining region can increase, if other mutations with sex-specific fitness effects accumulate on proto-sex chromosomes. Furthermore, a lack of recombination results in the accumulation of loss-of function mutations at Y-linked genes (pseudogenization). e. Lack of recombination also results in an accumulation of repetitive DNA, which can lead to an increase of the size of the evolving Y. f. Large segments of non-functional DNA can be deleted in old Y chromosomes, and reduce their physical size. The evolutionary outcome of this process are heteromorphic sex chromosomes, where the X chromosome largely resembles the autosome from which it derived, and the Y chromosomes has lost most of its ancestral genes, and may instead have accumulated repetitive DNA.
The potential role of sexually antagonistic mutations
Sexually antagonistic mutations37 — which are beneficial to one sex, but detrimental to the other5,34 — are thought to provide the selective force to suppress recombination between nascent sex chromosomes. Sexually antagonistic mutations are more likely to become established in a population if they are more often transmitted through the sex that they benefit10, and restriction of recombination between the nascent proto-X and proto-Y chromosomes may be a consequence of selection favoring genetic linkage between sexually antagonistic mutations and the sex-determining region. Evidence from mammals suggests that the elimination of recombination might be achieved through chromosomal inversions on the proto-sex chromosomes1,2,38,39. Inversions are known to locally suppress recombination in heterozygotes38. Thus, an inversion on one of the proto-sex chromosomes can repress recombination between the proto-X and proto-Y in males, and allow them to accumulate mutations independently and differentiate. Suppression of recombination can evolve in multiple steps along the proto-sex chromosomes, as has happened in mammals1, birds40 or plants41,42. In particular, comparison between genes in humans located in the X-degenerate region on the Y (see Box 1 for details) and their homologs along the X has revealed that groups of X-Y gene pairs show heterogeneous levels of sequence divergence1,2,43. This implies that the Y did not stop recombining with the X chromosome at once over its entire length, but instead in multiple successive steps. The regions along the sex chromosomes that stopped recombining at distinct time points are called evolutionary strata, with the oldest stratum in humans (stratum 1) dating back over 240 MY and the youngest stratum (stratum 5) originating only 30 MY ago. SRY, the master male-determining gene in mammals, is located in the oldest stratum1, consistent with predictions that the emergence of a sex-determining gene can trigger the differentiation of homomorphic sex chromosomes.
The effects of a lack of recombination on the Y
A variety of evolutionary models have been proposed to explain the degeneration of the Y44-49, and a common feature of these models is that the net efficacy of natural selection is strongly reduced on a non-recombining chromosome (Box 4). Natural populations are subject to recurrent mutations, some of which increase survival or reproductive success (beneficial mutations), but most mutations have negative fitness consequences (deleterious mutations). Natural selection results in beneficial mutations becoming incorporated into the genome of a species, while deleterious ones are selected against (allowing and maintaining adaptation50). On a recombining chromosome, selection can act independently on each mutation. In the absence of recombination, however, selection operates on entire chromosomes. An entire chromosome will be fixed in the population if a beneficial mutation arises on it, or an entire chromosome will be purged if it carries a deleterious mutation51.
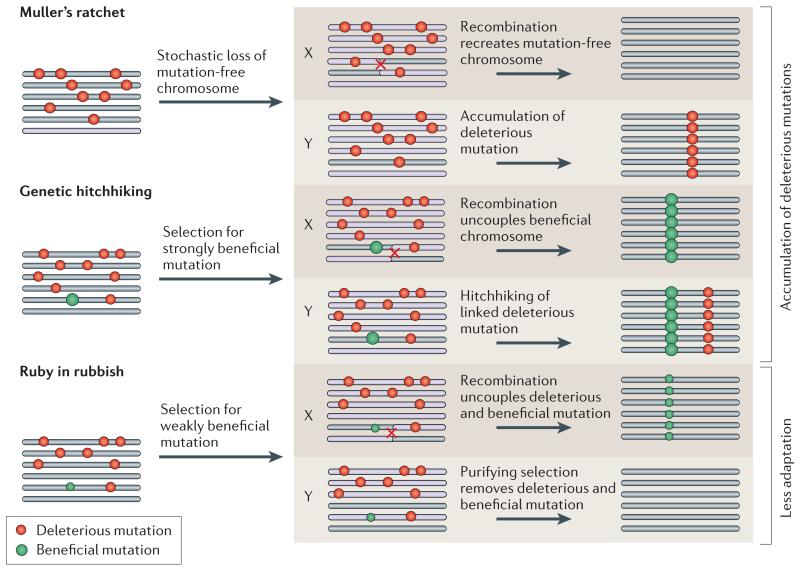
Box 4. Population processes of Y chromosome degeneration.
Gene decay on a non-recombining Y chromosome is a consequence of either an accumulation of deleterious mutations at ancestral genes (Muller’s ratchet46 and genetic hitchhiking44), or results from a lower rate of adaptation on the Y relative to homologous X-linked genes (ruby in the rubbish45). The cartoon shows a population of six recombining proto-X chromosomes, or six non-recombining proto-Y chromosomes that are subject to beneficial and deleterious mutations.
Muller’s ratchet
This mechanism refers to the irreversible accumulation of deleterious mutations in a finite non-recombining population. In finite populations chromosomes may accumulate mutations due to stochastic effects. Recombination on the X allows the recreation of mutation-free chromosomes, while this loss is irreversible on a non-recombining Y chromosome, and will result in the fixation of a particular deleterious mutation on the Y112.
Genetic hitchhiking
Newly arising beneficial mutations can occur on a chromosome that also contains deleterious mutations. Due to recombination, beneficial alleles can fix on the X chromosome without dragging along linked deleterious mutations. On a non-recombining Y chromosome, however, the fixation of the beneficial mutation will simultaneously fix the linked deleterious mutation. Genetic hitchhiking requires that the selective advantage of the beneficial mutation outweighs the effect of the linked deleterious allele, so that the Y chromosome containing the beneficial mutation has a net selective advantage47. If this is not the case, the ruby in the rubbish process occurs (see below).
Ruby in the rubbish
Y chromosomes may undergo less adaptive evolution relative to the X chromosome, due to the linkage of beneficial alleles with deleterious mutations. Beneficial mutations of weak effect can become uncoupled from linked, strongly deleterious mutations by recombination on the X chromosome, and fix in the population. In contrast, on non-recombining Y chromosomes they will be eliminated by purifying selection. X-linked genes will continue to adapt and incorporate beneficial mutations, while the Y chromosome will lag behind Eventually, it can become advantageous for the male to no longer express its maladapted Y-linked genes and silence or inactivate them.
The consequence of chromosome-wide linkage is that mutations at different loci that segregate in the population can influence the fixation or loss of linked mutations (for more details see Box 4). Thus, Y chromosomes are expected to accumulate deleterious mutations, and incorporate fewer beneficial mutations. Consistent with these theoretical expectations, non-recombining Y chromosomes generally show lower levels of adaptation, i.e. both increased rates of accumulation of deleterious mutations and lower rates of adaptive evolution relative to recombining regions of the genome in various species52-56. However, while the general properties of these models are well understood theoretically, their relative contribution to the observed degeneration of Y chromosomes in natural populations is less clear6. In particular, the evolutionary dynamics of these models and the speed at which they cause gene decay on an evolving Y strongly depends on both selection coefficients and mutation rates for beneficial and deleterious alleles34,47,57, parameters which are often poorly characterized.
The effect of male-limited transmission
Another unusual feature of Y chromosomes is that they are always transmitted from father to son and, unlike any other part of the genome, are never passed through females. Male-limited transmission implies that the Y is an ideal part of the genome to carry genes that increase male fitness5,34, since male-beneficial mutations on the Y are always transmitted through the sex in which they are advantageous and are sheltered from counter-selection in females if they are sexually antagonistic10 (i.e. good for males but harmful to females). Thus, predictions for the gene content of Y chromosomes are clear; gene decay dominates, and surviving Y-linked genes are enriched for male-beneficial functions. Whole-genome sequence analyses in primates and Drosophila have confirmed that the gene content of ancient Y chromosomes reflects these evolutionary forces (see Box 1 & 2). That is the vast majority of the ancestral genes have been lost from the Y, and many remaining genes, or genes that have been recruited secondarily to the Y, harbor male-specific functions18,23.
Initial stages of Y chromosome evolution
Little can be learned about the evolutionary forces and molecular mechanisms driving gene decay from gene-poor ancient Y chromosomes. Recent genomic studies in young plant Y and Drosophila neo-Y chromosomes, where degeneration is in progress, have revealed new insights into the origin of non-recombining Y chromosomes, and how and why they degenerate.
Y chromosome evolution in Drosophila neo-sex chromosomes
Neo-sex chromosomes of Drosophila offer a powerful opportunity to study Y chromosome degeneration in action in a comparative framework58 (Figure 2). Drosophila neo-sex chromosomes are formed by fusions of autosomes with the ancestral sex chromosomes that are shared among all members of the Drosophila genus (see Figure 2). Male Drosophila completely lack recombination; Y-fused chromosomes – called neo-Y’s – are therefore male-limited and sheltered from recombination, and thus subject to the same evolutionary forces as a true Y chromosome. The age of the fusion roughly correlates with the level of degeneration on the neo-Y with the caveat that the exact rate of degeneration depends on several species-specific factors, such as effective population size, number of genes present on the neo-Y, or generation time. Several Drosophila species have recently formed “neo-sex chromosomes” which, depending on how long they have been segregating as sex chromosomes, are at a different stage in their transition from an ordinary autosome to a heteromorphic pair of sex chromosomes. Drosophila neo-Y chromosomes thus allow the study of processes of gene decay on an evolving Y chromosome in a comparative chronological order. In particular, three Drosophila species with neo-sex chromosomes of different ages and different levels of degeneration have been characterized at the genomic level (D. pseudoobscura, D. miranda and D. albomicans, Figure 2). Interestingly, the eutherian Y chromosome derives largely from a single autosomal region added to the ancestral sex chromosomes after the split from marsuplials 80-130 MY ago43, i.e. most of the human Y is also a ‘neo-Y chromosome’.
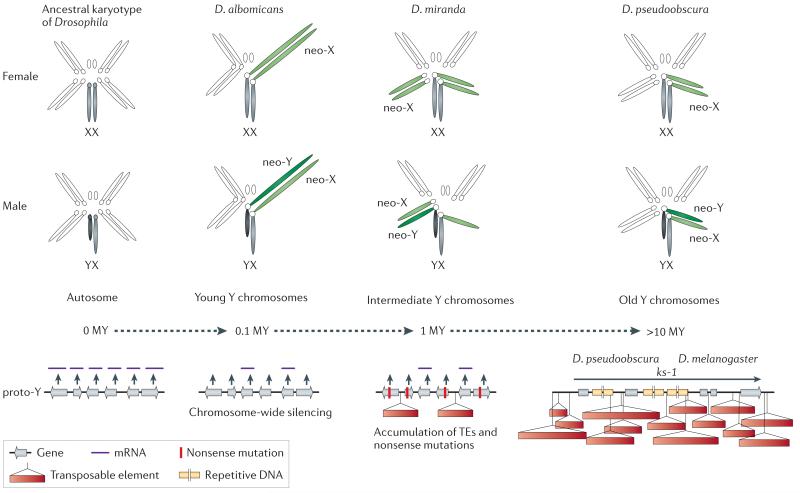
Figure 2. Neo-sex chromosomes in Drosophila.
Neo-sex chromosomes (in green) are formed by fusions of autosomes with the ancestral sex chromosomes (in grey), and the neo-X and neo-Y carry identical gene sets at the time of their origination (0 MY). To date, three Drosophila species with neo-Y’s of different age have been studied in detail, and provide a dynamic picture of the molecular changes associated with Y differentiation, and their karyotype is shown. Chromosome-wide down-regulation of protein-coding genes before accumulation of nonsense mutations at coding sequences is observed on the neo-Y in D. albomicans, which originated about 0.1 MY ago. Thus, gene decay on the neo-Y appears to be initiated by chromosome-wide silencing, possibly as a result of epigenetic modifications causing a change in chromatin structure. A general alteration of the genome architecture of the neo-Y has occurred within only 1 MY in D. miranda. In particular, the neo-Y of this species is characterized by massive decay in gene function, including the fixation of stop codons and frame shift mutations, large deletions and a general accumulation of repetitive transposable element DNA. The loss of gene function is accompanied by the beginning of heterochromatinization of the neo-Y. After 15 MY, almost no sequence similarity between the neo-Y and its former homolog can be detected, and the neo-Y has become entirely heterochromatic. This evolutionary stage has been reached in D. pseudoobscura, and resembles the general architecture of the ancestral Y in Drosophila. Genes on the ancestral Y of Drosophila contain massive introns, filled with repetitive DNA. Y-linked genes in Drosophila, such as ks-1, are characterized by huge introns, filled with repetitive satellite DNA.
The neo-Y chromosome of D. pseudoobscura
D. pseudoobscura has a neo-sex chromosome pair that was formed about 15 MY ago. The neo-Y of D. pseudoobscura resembles the architecture of the ancestral Y chromosome of Drosophila and is entirely heterochromatic. It has not been fully assembled yet, but it appears to harbor only a handful of active genes59. Its former homolog, a neo X, has maintained most of its roughly 3000 ancestral protein-coding genes, and evolved all the features typical of an X chromosome, i.e. it is fully dosage compensated, and has evolved the gene content typical for an X (such as an excess of ovary genes, and a deficiency of testis genes, as observed for the ancestral X chromosome in Drosophila60-62). Thus, an autosome can evolve into a fully differentiated sex chromosome pair within only 15 MY, and a non-recombining Y can loose thousands of genes within that time period.
The Neo-Y chromosome of D. miranda
The D. miranda neo-sex chromosome originated only about 1MY ago52, and its neo-Y shows features characteristic of both its autosomal origin and those of a degenerate Y. Cytological analysis shows that the neo-Y is partly heterochromatic63. Recent analysis of the genome and transcriptome of D. miranda has provided a comprehensive view of the evolutionary forces operating on this young neo-Y64-66. Specifically, it is still largely homologous to the neo-X; over 70% of neo-X sequences have homologs on the neo-Y and over 90% of the protein-coding genes are present on the neo-Y66. However, the neo-Y shows massive accumulation of repetitive DNA, with almost half of the neo-Y sequence consisting of transposable-element (TE) derived DNA53,65,67. Further, of the roughly 3000 genes initially present on the ancestral neo-Y, over 40% have already become non-functional, and have acquired frame-shift mutations, stop codons, or are entirely deleted66,68. Thus, the whole protein-coding gene complement of the neo-Y has undergone dramatic, genome-wide degeneration, in accordance with theoretical expectations of a reduced effectiveness of natural selection on a non-recombining chromosome51. In addition, whole-transcriptome analysis shows that gene decay at the DNA level is accompanied by chromosome-wide gene silencing. Specifically, a large fraction of neo-Y genes - about 50% - is expressed at a lower level from the neo-Y relative to the neo-X64-66. Thus, Y degeneration can proceed very rapidly on these neo-Y chromosomes in Drosophila, with over a thousand genes having lost their function within only 1 MY. However, since a dosage compensation machinery exists in Drosophila60, Y degeneration may proceed at a faster rate than it would if a new sex chromosome was formed de novo in the genome of a species without dosage compensation already in place.
Initiation of neo-Y degeneration in D. albomicans
Many neo-Y genes are pseudogenized and down-regulated in D. miranda, and the neo-Y is accumulating transposable element DNA and has partly become heterochromatic. However, the sequence of events is unclear. Does an accumulation of nonsense mutations on the neo-Y result in the production of many mal-functional proteins from a degenerating neo-Y, which in turn selects for down-regulation of these genes? Gene silencing could potentially be achieved through recruitment of TEs on the neo-Y, resulting in transcriptionally inactive heterochromatin, or through the accumulation of mutations in regulatory regions. Alternatively, does gene silencing and/or heterochromatin formation occur first, and down-regulated neo-Y genes then accumulate nonsense mutations neutrally? Genome and transcriptome analysis of the younger neo-Y of D. albomicans (less than 0.1MY old) partially answered these questions. In this species, the neo-Y (which contains almost 5000 genes) shows almost no evidence of degeneration at the DNA sequence level, with less than 2% of protein-coding genes being pseudogenes69. Intriguingly, however, a large fraction of these neo-Y genes - about 30% - are expressed at a lower level than their neo-X homologs, despite little decay of those genes at the protein level70. This suggests that transcriptional down-regulation precedes degeneration of protein-coding genes. The formation of heterochromatin is a plausible explanation for this chromosome-wide down-regulation but it is also possible that the accumulation of mutations in promoter and intronic regulatory sequences can lead to lower expression. It will be of interest to identify the molecular changes resulting in down-regulation on the D. albomicans neo-Y.
The role of gene function in Y chromosome degeneration
The study of neo-Y chromosomes has also allowed insight into the sequence of events leading to masculinization of Y genes. Intriguingly, the shrinking gene repertoire on the neo-Y of D. miranda shows the first signs of specialization of gene content for male-specific roles66. In particular, genes are not lost randomly from the neo-Y with regard to gene function71. Instead, genes with male-beneficial fitness effects are significantly more likely to be retained on the neo-Y when compared with random neo-Y genes71, while genes with female-beneficial effects are preferentially lost66,71. Further, genes with inferred male-specific function (those with male-beneficial, or male-beneficial female-detrimental effects) undergo increased rates of adaptive protein evolution on the neo-Y66. Also, neo-Y genes with reproductive function have evolved male-tissue specific expression or increased their expression on the neo-Y in male tissue66. Thus, in this model system at least, masculinization accompanies the early stages of Y chromosome evolution. This supports the hitchhiking model of Y chromosome degeneration (see Box 4), i.e. the fixation of male-beneficial mutations can drag along segregating deleterious mutations. This model was criticized initially on the basis that it might be difficult for beneficial mutations to overcome the deleterious effects of linked detrimental mutations34. However, the observed masculinization of the D. miranda neo-Y demonstrates that beneficial mutations can fix on a gene-rich young Y chromosome66, and may drag along linked deleterious mutations to fixation. Indeed, patterns of neutral variation in D. miranda neo-Y chromosomes in natural populations support adaptive evolution occurring on the neo-Y72.
The initial stages of sex chromosome evolution in plants
Unlike animals, most land plants are co-sexual, and male and female reproductive functions are found within a single individual. Only a small number of plant species (about 6%) have evolved separate sexes, which is sometimes associated with the emergence of heteromorphic sex chromosomes73,74. Most plant sex chromosomes are evolutionary young73, and provide an interesting contrast to animal sex chromosomes.
Recent transcriptome analysis in Silene latifolia, the white campion, has provided some initial insights into the evolution of plant sex chromosomes75,76. The S. latifolia X and Y chromosome were formed about 10 MY ago, may contain roughly 4,000 genes, and are morphologically distinguishable. The Y is significantly larger than the X, which may be caused by an accumulation of repetitive DNA77. Also, YY plants are non-viable, suggesting that severely deleterious mutations have already accumulated on the S. latifolia Y chromosome77. While the Y chromosome of this species has not been sequenced yet, recent transcriptome analysis has identified about 400 genes pairs from the X and Y75,76. Rates of protein evolution are elevated for Y-linked genes in Silene compared to their X-linked homologs75,76. In addition, levels of gene expression are significantly lower at Y-linked genes relative to their X homologs75,76. This indicates that the efficacy of purifying selection is reduced on the Silene Y chromosome, and deleterious mutations have started to accumulate, similar to young animal Y or neo-Y chromosomes. However, the rate of gene loss may be lower in plants than that of Drosophila or humans, and over 80% of Y-linked genes may still be functional75,76. This could indicate that Y chromosomes generally degenerate much more slowly in plants than animals75,76. A potential explanation for this differences lies in the haploid expression from Y chromosome-bearing gamets in plants but not animals. That is, in plants pollen express many genes, whereas in animals, sperm show very limited gene expression78. Expression in the haploid stage may result in stronger selection pressure to maintain Y-linked genes in plants, while Y-linked genes are generally sheltered by their X homologs in animals.
Molecular characterization of young sex chromosomes in several plant species is ongoing79, and recently, the first Y-linked genomic sequence was published from papaya42. Sex determination in papaya is controlled by sex chromosomes, with two different Y chromosomes controlling male and hermaphrodite development. Comparison between the hermaphrodite-specific Y sequence with the corresponding X-linked regions revealed two large-scale inversions forming two evolutionary strata, which likely caused recombination suppression between the X and Y at 7 MY and 2 MY ago. A dramatic increase in size of the Y was detected mostly due to retrotransposon insertions on the Y42. Most of the genes present in the ancestral chromosome region that formed the sex chromosomes are still present on the current papaya Y, but gene decay proceeded more rapidly on the Y relative to the X-linked region indicating potential degradation of the Y42. In some plant species, such as Rumex sp., Y degeneration has progressed even further, resembling the Y of many animals. Thus, while gene loss on plant Y chromosomes might be retarded relative to animals, degeneration of non-recombining chromosomes is a wider phenomena shared between kingdoms.
Long-term survival of the Y
The process of Y chromosome degeneration has prompted the suggestion that continuing gene loss will lead to the eventual disappearance of the human Y80-83. These predictions are based on a naive model of a constant rate of gene loss from the Y chromosome. However, recent theoretical and experimental studies have clearly demonstrated that Y degeneration does not proceed in this simple linear fashion21,84, and refute these sensational claims of human Y extinction (Figure 3).
Figure 3. Evolutionary dynamics of Y degeneration.
a. Population genetic processes to explain gene loss on a non-recombining chromosome do not predict continuous gene decay and eventual extinction of the Y84,85. A simple model of constant gene decay is shown by the red dashed line. Instead, analytical approximations (green line) and computer simulations (asterisk) under the Muller’s ratchet and genetic hitchhiking model suggest that gene-rich non-recombining chromosomes degenerate rapidly initially, but gene decay slows down over time and ultimately comes to a complete halt once a threshold number of Y-linked genes has been reached. In addition, empirical evidence from Drosophila neo-Y chromosome sequencing and primate Y chromosome gene content are consistent with this non-linear degeneration.
Predicting the future of the Y using theoretical models
The evolutionary forces that drive gene loss on a non-recombining chromosome are well understood (see above). Recent theoretical studies modeling these evolutionary forces and population simulation studies have investigated the temporal dynamics of these models on an evolving Y, and have demonstrated that the rate of gene decay is expected to decrease over evolutionary time and should come very close to a complete halt on an old, gene-poor Y chromosome84,85. The reason for the predicted rapid decline in the rate of gene loss lies in the fact that old Y chromosomes have many fewer active genes. Fewer functional loci on the Y implies that the total number of beneficial and deleterious mutations – which are proportional to the number of functional genes on the Y – that can simultaneously segregate in the population is dramatically decreased. Interference among segregating mutations drives Y degeneration (see Box 4), and fewer segregating mutations means less interference among them. Thus, eventually, the very same processes that caused gene loss on the Y chromosome are expected to become exceedingly slow84, 85, and evolutionary theory does not predict that a Y chromosome would drive itself to extinction.
Neo-sex chromosomes and the future of Y chromosomes
Empirical observations of degenerating Y chromosomes of different ages are also inconsistent with a linear model of the rate of gene loss from the Y chromosome. If Y chromosomes degenerate at a constant rate, a Y chromosome twice as old should only carry half as many genes as the younger one. Probably the best empirical estimates of gene loss from Y chromosomes can be obtained in comparative data from neo-Y chromosomes in Drosophila species (see Figure 2). In D. miranda the neo-Y formed about 1 MY ago. The neo-Y of this species has lost about 40% of its ancestral genes66 – starting with ~3000 and now reaching ~1700 genes – and a constant rate of gene decay would imply that this neo-Y should go extinct after roughly 2-3 MY. However, if one assumes that the same forces act upon all neo-Y chromosomes resulting in similar rates of decay, empirical data from the D. pseudoobscura neo-Y show the contrary. The neo Y chromosome of this species was formed over 15 MY ago and still has over a dozen genes left out of the original ~3000, and this number of genes is similar to that of the ancestral Y of D. melanogaster, which is over 60 MY old27. Thus, if one extrapolates from these three species, Y degeneration in Drosophila clearly does not follow a linear path; instead, gene decay is very rapid initially, but eventually equilibrates at a small number of stable genes. If anything, the ancestral Y chromosome of Drosophila is actually gaining genes, instead of losing them27, and over half of the genes currently present on the D. melanogaster Y were acquired less than 60 MY ago, after the radiation of the Drosophila genus27.
Primate Y chromosomes and the future of Y chromosome
Comparative genomic studies in primates provide further empirical evidence against human Y extinction21. Recent genomic data has shown that the rhesus macaque, which split from humans 30 MY ago, has an almost identical Y gene set as humans21. This implies that the gene content of the human Y has been stable in the past 30MY and that the last common ancestor had already reached an equilibrium Y chromosome gene number (Figure 3). A linear rate of decay would have instead predicted that human and rhesus would show little gene overlap on their Y’s since they should have lost genes independently since their split. This empirical data thus refutes the hypothesis of the imminent loss of the human Y.
Interestingly, in some organisms, such as most nematodes, the Y chromosome has entirely disappeared (i.e. they have XX/X0 sex determination86), which is the evolutionary end point of all erosion of genetic activity from the Y. However, for the complete elimination of the Y to occur, an alternative way of sex determination must have evolved (such as by determining sex from the ratio of X chromosomes to autosomes), and any genes required for male function must have been translocated to other chromosomes. Thus, while the Y chromosome can ultimately be lost, it will do so only if alternative sex determination mechanisms and male fertility functions on other chromosomes have evolved first, and the Y can disappear without any negative fitness consequences. Otherwise, Y chromosomes can be a stable and important component of the genome in many species.
Conclusions and perspectives
Like many areas in biology, research on sex chromosome differentiation will greatly benefit from the genomic revolution. Many new models of sex chromosome evolution, at all stages of differentiation (that is nascent Y chromosomes as well as highly differentiated Y’s) can now be tackled at the DNA sequence and chromatin level, and sex-specific transcription profiles can be obtained. Though repetitive DNA and the ampliconic structure of many old Y chromosomes continues to pose a serious challenge into deciphering their genome sequence, and currently require very laborious and expensive approaches, new sequencing technologies that will provide much longer reads should allow us to make progress in sequencing such challenging sequences, including Y chromosomes.
Furthermore, studying a diverse set of Y chromosomes will allow us to test if certain features identified in model organisms are a general characteristics of Y chromosomes. For example, are amplicons unique to primates, or do they enable survival of Y-linked genes in other taxonomic groups as well? Do other taxa have similar mega-base sized introns like Drosophila? Are evolutionary strata, which have already been found in both animals and plants, a general feature of sex chromosome evolution? In addition, such data will allow us to investigate further whether the processes that are currently hypothesized to drive Y chromosome evolution do, in fact, do so. That is for example whether sexually antagonistic mutations do indeed accumulate along the recombining portion of the sex chromosomes, and drive the suppression of recombination along the proto-sex chromosomes.
It will also be of great interest to study more Y chromosomes from plants, particularly young ones, to establish whether retardation of Y chromosome degeneration is a general property in plants. Haploid Y-bearing cells in animals (sperm) show little gene expression but are expressed in plants (pollen), and haploid purifying selection may slow down Y degeneration in plants.
Another challenge in our understanding is why not all homomorphic sex chromosomes stop recombining with each other, and become heteromorphic over long evolutionary times. For example, birds have homologous sex chromosomes which formed about 120 MY ago87, and are similar in age to the mammalian sex chromosomes88. Yet, while all mammals and most bird lineages have highly differentiated sex chromosomes, in some groups of birds, such as ratites, the sex chromosomes remain homomorphic89. Similar differences in the progression from homomorphic to heteromorphic sex chromosomes are seen among snake lineages. Comparisons of the genomic sequences and transcriptomes between taxa with homomorphic and heteromorphic sex chromosomes should provide clues to this puzzle. Both birds and snakes have female-heterogametic sex chromosomes, that is, females have a non-recombining W chromosome. Contrasting patterns of evolution on Y versus W chromosomes should reveal whether W chromosomes show similar patterns of degeneration, but specialization of their gene content to reflect the female-limited transmission of a W chromosome.
There is also a high occurrence of nascent sex chromosomes in fish, reptiles and amphibians90-97, and tackling their patterns of sequence evolution at the molecular level will greatly increase our understanding of the molecular basis and evolutionary processes of the beginnings of Y degeneration. Also, there are several closely related groups in fish, reptiles and amphibians that differ in their male- or female heterogamety98, and examination of these species groups will allow us to contrast evolutionary forces occurring on Y vs. W chromosomes.
Finally, invertebrates, which comprise the vast majority of animal species, have a multitude of sex determining mechanisms, and Y and W chromosomes have evolved independently many times in many different taxa15. Recent efforts have started to characterize transitions of sex chromosomes in insects99-101, but many other systems with interesting biology and karyotypes provide a treasure trove to study sex chromosome evolution15. To conclude, a comparative analysis of Y chromosomes at different stages of differentiation across the tree of life will give further insights into the characteristics and the evolutionary forces that act upon sex chromosomes.
Acknowledgements
Funded by NIH grants (R01GM076007 and R01GM093182) and a Packard Fellowship to D.B
Glossary
- Sex Chromosomes
A pair of chromosome that determines the sex of an individual.
- Y chromosome degeneration
The process of gene loss from the Y chromosome.
- Genomic Conflict
Conflict of different genes within an organism, which arises when genes inside a genome are not transmitted by the same rules (such as mitochondria and nuclear genes), or when a particular gene’s transmission is increased to the detriment of other parts of the rest of the genome.
- Homomorphic Sex Chromosomes
A pair of chromosomes that is morphologically indistinguishable.
- Heteromorphic Sex Chromosomes
A pair of chromosomes that is morphologically distinct.
- Proto-Sex Chromosomes
A new pair of chromosomes that (recently) acquired a sex-determining function, but otherwise contains identical genes.
- Dosage compensation
A process that balances expression of sex-linked and autosomal genes in the heterogametic sex.
- Male heterogamety
A species where females have two X chromosomes, and males have an X and a Y chromosome.
- Female heterogamety
A species where males have two Z chromosomes, and females have a Z and a W chromosome.
- Recombination
The breaking and rejoining of DNA strands to form a new combination of genetic information.
- Sexually antagonistic mutation
A mutation that is beneficial to one sex, but detrimental to the other.
- Genetic linkage
The tendency of genes that are proximal to each other on a chromosome to be inherited together.
- Chromosomal Inversion
A chromosome rearrangement where a segment of a chromosome is reversed end to end.
- B chromosome
A supernumerary chromosome that is not essential for the life of a species, and present only in some of the individuals of a species.
- Beneficial Mutation
A mutation that increases the survivorship or fecundity (fitness) of its carrier.
- Deleterious Mutation
A mutation that decreases the survivorship or fecundity (fitness) of its carrier.
- Muller’s ratchet
The irreversible accumulation of deleterious mutations in an asexual population.
- Genetic Hitchhiking
The fixation of a deleterious mutation that is linked to a beneficial allele.
- Ruby in the Rubbish Model
The selective elimination of a beneficial mutation, that is linked to a deleterious allele.
- Heterochromatin
A tightly packed form of DNA, that is typically genetically inactive, and contains repetitive sequences and few genes.
- Palindrome
A DNA sequence composed of two inverted repeats (arms) separated by a short spacer.
- Position effect variegation
Position effect variegation (PEV). The variable, heritable suppression of genes by their juxtaposition to heterochromatin or telomeres, or by movement of a gene into a different nuclear domain or chromosomal context.
about the author
Doris Bachtrog is an Associate professor in the Department of Integrative Biology and the Center of Evolutionary Theoretical Biology at the University of California Berkeley. Bachtrog received her Ph.D. in 2002 and held a faculty appointment at University of California San Diego, USA, between 2005 and 2008. Her research encompasses the evolution of sex chromosomes in Drosophila, with a particular focus on processes and molecular mechanisms of Y degeneration and dosage compensation of the X.
Footnotes
Competing interests statement
The author declares no competing financial interests.
FURTHER INFORMATION
Link 1: [http://ib.berkeley.edu/labs/bachtrog/]
Online summary
-
○Recently developed genomic technologies have shed light on the genomic composition of the ancient Y chromosomes of some primates and Drosophila melanogaster, and shown that Y chromosomes in these species largely conform to the previous held view of being degenerate
-
○The presence of evolutionary strata confirmed by genome sequencing of the sex chromosomes support that Y chromosome degeneration occurred through successive arrest of recombination over time. In addition, the enrichment of Y chromosomes for genes of male beneficial functions suggest that sexually antagonistic mutations may play a role in Y chromosome evolution.
-
○Genome sequencing of young Y chromosomes in plants and neo-Y chromosomes in Drosophila have provided insight into the molecular processes that trigger initiation of Y degeneration. Empirical evidence suggest that gene silencing occurs before pseudogenization
-
○Empirical observations in Drosophila neo-sex chromosomes, primate Y chromosomes and theoretical models and computer simulations show that degeneration is not a linear process and so Y chromosomes in these species will likely not completely degenerate in the future
References
- 1.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 2.Ross MT, et al. The DNA sequence of the human X chromosome. Nature (London) 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth B. The evolution of sex chromosomes. Science. 1990;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 4.Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin Cummings; Menlo Park, CA: 1983. [Google Scholar]
- 5.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. [Google Scholar]
- 6.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–72. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachtrog D, et al. Are all sex chromosomes created equal? Trends in Genetics. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–43. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- 10.Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 11.Werren JH, Beukeboom LW. Sex determination, sex ratios, and genetic conflict. Annual Review of Ecology and Systematics. 1998;29:233–261. [Google Scholar]
- 12.Page DC, et al. Reconstructing sex chromosome evolution. Genome Biology. 2010;11 [Google Scholar]
- 13.Burgoyne PS. The mammalian Y chromosome: a new perspective. Bioessays. 1998;20:363–6. doi: 10.1002/(SICI)1521-1878(199805)20:5<363::AID-BIES2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Castillo ER, Marti DA, Bidau CJ. Sex- and neo-sex chromosomes in Orthoptera: a review. Journal of Orthoptera Research. 2010;19:213–231. [Google Scholar]
- 15.White MJD. Animal Cytology and Evolution. Cambridge University Press; 1973. [Google Scholar]
- 16.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. The paper showed that the genome sequence of the non-recombining region of the human Y is a mosaic of different sequences, including X-transposed, X-degenerate and ampliconic regions.
- 17.Rozen S, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–6. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 18.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–80. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 19.Hughes J, et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 20.Hughes JF, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. This paper shows that the chimpanzee Y contains twice as many palindromes as humans, yet has lost large fractions of Y-linked genes since divergence from the last common ancestor with humans.
- 21.Hughes JF, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–U124. doi: 10.1038/nature10843. The paper shows that the gene content of the rhesus Y chromosome is highly similar to that of humans, which refutes the claim that continous gene loss is leading to the extinction of the human Y.
- 22.Brosseau GE. Genetic analysis of the male fertility factors on the Y-chromosome of Drosophila melanogaster. Genetics. 1960;45:257–274. doi: 10.1093/genetics/45.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatti M, Pimpinelli S. Cytological and genetic analysis of the Y-chromosome of Drosophila melanogaster. 1 Organization of the fertility factors. Chromosoma. 1983;88:349–373. [Google Scholar]
- 24.Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 2000;97:13239–44. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho AB, Dobo BA, Vibranovski MD, Clark AG. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:13225–30. doi: 10.1073/pnas.231484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vibranovski MD, Koerich LB, Carvalho AB. Two new Y-linked genes in Drosophila melanogaster. Genetics. 2008;179:2325–7. doi: 10.1534/genetics.108.086819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. This study shows that the gene content of the Drosophila Y is highly dynamic, and that the ancestral Y is gaining genes at a higher rate than loosing it.
- 28.McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-z. [DOI] [PubMed] [Google Scholar]
- 29.Williams SM, Robbins LG, Cluster PD, Allard RW, Strobeck C. Superstructure of the Drosophila ribosomal gene family. Proc Natl Acad Sci U S A. 1990;87:3156–60. doi: 10.1073/pnas.87.8.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends in Genetics. 2009;25:270–277. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. This is the first of a series of papers showing that variation on the Drosophila Y chromosome influences the transcription of 100s-1000s of genes in the Drosophila genome.
- 32.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, et al. Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc Natl Acad Sci U S A. 2012;109:9941–6. doi: 10.1073/pnas.1207367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. This paper provides a classic review on the evolution of sex chromosomes and dosage compensation.
- 35.Lahn BT, Pearson NM, Jegalian K. The human Y chromosome, in the light of evolution. Nat Rev Genet. 2001;2:207–16. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- 36.Quinn AE, Sarre SD, Ezaz T, Graves JAM, Georges A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biology Letters. 7:443–448. doi: 10.1098/rsbl.2010.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innocenti P, Morrow EH. The Sexually Antagonistic Genes of Drosophila melanogaster. Plos Biology. 2010;8 doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkpatrick M. How and Why Chromosome Inversions Evolve. Plos Biology. 2010;8 doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaitre C, et al. Footprints of inversions at present and past pseudoautosomal boundaries in human. Genome Biol Evol. 2009;1:56–66. doi: 10.1093/gbe/evp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handley L, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 2004;167:367–76. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas M, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A. 2012;109:13710–5. doi: 10.1073/pnas.1207833109. The first sequence of a plant Y chromosome reveals evolutionary strata, an accumulation of repetitive DNA and gene loss on a plant Y.
- 43.Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JA. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80-130 million years ago. Cytogenet Cell Genet. 2001;92:74–9. doi: 10.1159/000056872. [DOI] [PubMed] [Google Scholar]
- 44.Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orr HA, Kim Y. An adaptive hypothesis for the evolution of the Y chromosome. Genetics. 1998;150:1693–8. doi: 10.1093/genetics/150.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc. Natl. Acad. Sci. U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bachtrog D, Gordo I. Adaptive evolution of asexual populations under Muller’s ratchet. Evolution Int J Org Evolution. 2004;58:1403–13. doi: 10.1111/j.0014-3820.2004.tb01722.x. [DOI] [PubMed] [Google Scholar]
- 48.Peck JR. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics. 1994;137:597–606. doi: 10.1093/genetics/137.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice WR, Chippindale AK. Sexual recombination and the power of natural selection. Science. 2001;294:555–559. doi: 10.1126/science.1061380. [DOI] [PubMed] [Google Scholar]
- 50.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; Oxford: 1930. [Google Scholar]
- 51.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–94. [PubMed] [Google Scholar]
- 52.Bachtrog D, Charlesworth B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 53.Bachtrog D. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nature Genetics. 2003;34:215–219. doi: 10.1038/ng1164. [DOI] [PubMed] [Google Scholar]
- 54.Betancourt AJ, Presgraves DC. Linkage limits the power of natural selection in Drosophila. Proc Natl Acad Sci U S A. 2002;99:13616–20. doi: 10.1073/pnas.212277199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filatov D. Substitution Rates in a New Silene latifolia Sex-Linked Gene, SlssX/Y. Mol Biol Evol. 2004 doi: 10.1093/molbev/msi003. [DOI] [PubMed] [Google Scholar]
- 56.Bartolomé C, Charlesworth B. Evolution of amino-acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics. 2006;174:2033–44. doi: 10.1534/genetics.106.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordo I, Charlesworth B. The degeneration of asexual haploid populations and the speed of Muller’s ratchet. Genetics. 2000;154:1379–1387. doi: 10.1093/genetics/154.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lucchesi JC. Gene Dosage Compensation and the Evolution of Sex Chromosomes. Science. 1978;202:711–716. doi: 10.1126/science.715437. [DOI] [PubMed] [Google Scholar]
- 59.Carvalho A, Clark A. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108–10. doi: 10.1126/science.1101675. [DOI] [PubMed] [Google Scholar]
- 60.Marin I, Franke A, Bashaw GJ, Baker BS. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature. 1996;383:160–163. doi: 10.1038/383160a0. [DOI] [PubMed] [Google Scholar]
- 61.Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Assis R, Zhou Q, Bachtrog D. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 2012 doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinemann M, Steinemann S. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica. 1998;102-103:409–20. [PubMed] [Google Scholar]
- 64.Bachtrog D. Expression profile of a degenerating neo-Y chromosome in Drosophila. Curr Biol. 2006;16:1694–9. doi: 10.1016/j.cub.2006.07.053. [DOI] [PubMed] [Google Scholar]
- 65.Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–5. doi: 10.1126/science.1225385. This is the first whole-genome and transcriptome analysis of a young, evolving sex chromosome.
- 67.Steinemann M, Steinemann S. Degenerating Y chromosome of Drosophila miranda: a trap for retrotransposons. Proc Natl Acad Sci U S A. 1992;89:7591–5. doi: 10.1073/pnas.89.16.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bachtrog D. Sex chromosome evolution: Molecular aspects of Y chromosome degeneration in Drosophila. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Q, et al. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics. 2012;13:109. doi: 10.1186/1471-2164-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Q, Bachtrog D. Chromosome-Wide Gene Silencing Initiates Y Degeneration in Drosophila. Current Biology. 2012;22:522–525. doi: 10.1016/j.cub.2012.01.057. [DOI] [PubMed] [Google Scholar]
- 71.Kaiser VB, Zhou Q, Bachtrog D. Nonrandom Gene Loss from the Drosophila miranda Neo-Y Chromosome. Genome Biology and Evolution. 2011;3:1329–1337. doi: 10.1093/gbe/evr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bachtrog D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat Genet. 2004;36:518–22. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- 73.Ming R, Bendahmane A, Renner SS. Annual Review of Plant Biology. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- 74.Westergaard M. The mechanism of sex determination in flowering plants. Adv. Genet. 1958;9:217–281. doi: 10.1016/s0065-2660(08)60163-7. [DOI] [PubMed] [Google Scholar]
- 75.Chibalina MV, Filatov DA. Plant Y Chromosome Degeneration Is Retarded by Haploid Purifying Selection. Current Biology. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- 76.Bergero R, Charlesworth D. Preservation of the Y Transcriptome in a 10-Million-Year-Old Plant Sex Chromosome System. Current Biology. 2011;21:1470–1474. doi: 10.1016/j.cub.2011.07.032. These two recent papers (75, 76) suggest that degeneration of a Y chromosome in plants might be retarded relative to animals, due to haploid selection in Y-carrying gametophytes.
- 77.Bernasconi G, et al. Silene as a model system in ecology and evolution. Heredity. 2009;103:5–14. doi: 10.1038/hdy.2009.34. [DOI] [PubMed] [Google Scholar]
- 78.Schafer M, Nayernia K, Engel W, Schafer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–52. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- 79.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–U237. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 80.Graves J. The degenerate Y chromosome - can conversion save it? Reprod Fertil Dev. 2004;16:527–34. doi: 10.10371/RD03096. [DOI] [PubMed] [Google Scholar]
- 81.Aitken R, Marshall Graves J. The future of sex. Nature. 2002;415:963. doi: 10.1038/415963a. [DOI] [PubMed] [Google Scholar]
- 82.Graves J. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–14. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 83.Sykes B. Adam’s Curse: A future without men. Norton, W. W. & Company; 2004. [Google Scholar]
- 84.Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–25. doi: 10.1534/genetics.107.084012. This is atheoretical study examining the temporal dynamics of processes of Y degeneration that shows that rates of gene loss are highly non-linear on a degenerating Y chromosomes, predicting a stable gene content of old Y chromosomes.
- 85.Engelstaedter J. Muller’s Ratchet and the Degeneration of Y Chromosomes: A Simulation Study. Genetics. 2008;180:957–967. doi: 10.1534/genetics.108.092379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hodgkin J. Primary sex determination in the nematode C. elegans. Development. 1987;101:5–16. doi: 10.1242/dev.101.Supplement.5. [DOI] [PubMed] [Google Scholar]
- 87.van Tuinen M, Hedges SB. Calibration of avian molecular clocks. Mol Biol Evol. 2001;18:206–13. doi: 10.1093/oxfordjournals.molbev.a003794. [DOI] [PubMed] [Google Scholar]
- 88.Veyrunes F, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–73. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ellegren H. Evolution of the avian sex chromosomes and their role in sex determination. Trends in Ecology and Evolution. 2000;15:188–192. doi: 10.1016/s0169-5347(00)01821-8. [DOI] [PubMed] [Google Scholar]
- 90.Peichel C, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–24. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 91.Ezaz T, Sarre SD, O’Meally D, Graves JAM, Georges A. Sex Chromosome Evolution in Lizards: Independent Origins and Rapid Transitions. Cytogenetic and Genome Research. 2009;127:249–260. doi: 10.1159/000300507. [DOI] [PubMed] [Google Scholar]
- 92.Mank JE, Avise JC. Evolutionary Diversity and Turn-Over of Sex Determination in Teleost Fishes. Sexual Development. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- 93.Sarre SD, Ezaz T, Georges A. Annual Review of Genomics and Human Genetics. 2011;12:391–406. doi: 10.1146/annurev-genom-082410-101518. [DOI] [PubMed] [Google Scholar]
- 94.Quinn AE, Sarre SD, Ezaz T, Graves JAM, Georges A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biology Letters. 2011;7:443–448. doi: 10.1098/rsbl.2010.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura M. Sex determination in amphibians. Semin Cell Dev Biol. 2009;20:271–82. doi: 10.1016/j.semcdb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 96.Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- 97.Graves JAM, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Doorn GS, Kirkpatrick M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 2010;186:629–45. doi: 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pease JB, Hahn MW. Sex Chromosomes Evolved from Independent Ancestral Linkage Groups in Winged Insects. Molecular Biology and Evolution. 2012;29:1645–1653. doi: 10.1093/molbev/mss010. [DOI] [PubMed] [Google Scholar]
- 100.Verhulst EC, van de Zande L, Beukeboom LW. Insect sex determination: it all evolves around transformer. Curr Opin Genet Dev. 2010;20:376–83. doi: 10.1016/j.gde.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 101.Sanchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52:837–56. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 102.Otto SP, et al. About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends in Genetics. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 103.Iwase M, Satta Y, Hirai H, Hirai Y, Takahata N. Frequent gene conversion events between the X and Y homologous chromosomal regions in primates. BMC Evolutionary Biology. 2010;10 doi: 10.1186/1471-2148-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carvalho AB, et al. Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: how far can we go? Genetica. 2003;117:227–37. doi: 10.1023/a:1022900313650. [DOI] [PubMed] [Google Scholar]
- 105.Bonaccorsi S, Pisano C, Puoti F, Gatti M. Y-chromosome loops in Drosophila melanogaster. Genetics. 1988;120:1015–1034. doi: 10.1093/genetics/120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics. 1989;122:793–800. doi: 10.1093/genetics/122.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang P-P, Hartl DL, Lemos B. Y Not a Dead End: Epistatic Interactions Between Y-Linked Regulatory Polymorphisms and Genetic Background Affect Global Gene Expression in Drosophila melanogaster. Genetics. 2010;186:109–U221. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marais GAB, Campos PRA, Gordo I. Can Intra-Y Gene Conversion Oppose the Degeneration of the Human Y Chromosome? A Simulation Study. Genome Biology and Evolution. 2010;2:347–357. doi: 10.1093/gbe/evq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Connallon T, Clark AG. Gene Duplication, Gene Conversion and the Evolution of the Y Chromosome. Genetics. 2010;186:277–U451. doi: 10.1534/genetics.110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hughes JF, Rozen S. Genomics and genetics of human and primate y chromosomes. Annu Rev Genomics Hum Genet. 13:83–108. doi: 10.1146/annurev-genom-090711-163855. [DOI] [PubMed] [Google Scholar]
- 112.Charlesworth B, Charlesworth D. Rapid fixation of deleterious alleles can be caused by Muller’s ratchet. Genet. Res. 1997;70:63–73. doi: 10.1017/s0016672397002899. [DOI] [PubMed] [Google Scholar]