Abstract
A common task in biological research is to predict function for proteins by comparing sequences between proteins of known and unknown function. This is often done using pair-wise sequence alignment algorithms (e.g. BLAST). A problem with this approach is the assumption of a simple equivalence between a minimum sequence similarity threshold and the function similarity between proteins. This assumption is based on the binary concept of homology in that proteins are or not homologous. The relationship between sequence and function however is more complex as well as pertinent for predicting protein function, e.g. evaluating BLAST alignments or developing training sets for profile models based on functional rather than homologous groupings. Our motivation for this study was to model sequence and function similarity between proteins to gain insights into the “sequence-function similarity relationship between proteins for predicting function. Using our model we found that function similarity generally increases with sequence similarity but with a high degree of variability. This result has implications for pair-wise approaches in that it appears sequence similarity must be very high to ensure high function similarity. Profile models which enable higher sensitivity are a potential solution. However, multiple sequences alignments (a necessary prerequisite) are a problem in that current algorithms have difficulty aligning sequences with very low sequence similarity, which is common in our data set, or are intractable for high numbers of sequences. Given the importance of predicting protein function and the need for multiple sequence alignments, algorithms for accomplishing this task should be further refined and developed.
General Terms: Experimentation, Biostatistics, Bioinformatics, Multiple Sequence Alignment
1. INTRODUCTION
Predicting protein function, or protein annotation, is a crucial step toward a deep understanding of cellular processes that will provide the foundation of modern research in biology and biomedicine [10,11,12]. The most common annotation method is pair-wise protein sequence comparison to “transfer” or predict function based on a minimum level of sequence similarity between proteins of known and unknown function [4,6,17,21]. The most popular pair-wise approach is BLAST [1]. There are also approaches based on comparisons between multiple sequences. The general consensus regarding these “profile” models, such as HMM’s [7,8], is that they are highly sensitive and accurate. However, profile models depend on algorithms which generate multiple sequence alignments. These have been shown to be inaccurate when aligning proteins of very low sequence similarity or otherwise intractable [3,19,22].
Although useful, there is a logical discrepancy with pair-wise methods in that an implicit equivalence relation is assumed between a minimum threshold of sequence similarity and the function similarity between proteins. This assumption is based on the binary concept of homology in that proteins are or are not homologous (related by common descent). The more appropriate answer is whether or not sequence similarity implies a degree of function similarity. In practice, little information about the functions themselves or variability in the underlying relationship between sequence and function similarity between proteins and classes of functions is considered in pair-wise approaches. Given the high estimates of annotation error rates in public databases for predicted functions [5,9,23], and the number of proteins needing annotations this matter deserves further investigation.
Here we explicitly model the relationship between protein sequence similarity and function similarity; what we call the sequence-function similarity relationship. What our model helps assess is the continuous range of likelihoods that a protein has a particular function based on its sequence similarity to a protein of known function. The implication of our model is that pairwise (and profile) methods may be informed by this exercise. Some previous studies have investigated this relationship [6,16] but the literature is sparse for non-enzymes functions and for specific details to inform pair-wise approaches.
To model the sequence-function similarity relationship we first needed to numerically quantify the distance between functions. This can be done by relating measures of function specificity between terms in the Gene Ontology (GO, [2]). The GO describes a diversity of protein functions (GO terms) in a controlled fashion and represents function specificity as a hierarchical directed acyclic graph. The structure of the GO, combined with a numerical measure of function (i.e. term) specificity (e.g. Information Content [14,15,20]), enables the measurement of distance between two functions by finding common “ancestor” terms. Function similarity, unlike function specificity which is a property of a single GO term, is a property of two GO terms. The function similarity between two GO terms is quantified by relating the term specificities of the two terms and the specificity of their common ancestor term (see Methods). Using this function similarity metric, the relationship between sequence similarity and function similarity can be modeled.
2. METHODS
2.1 Information Content
The Information Content (IC) of a GO term is related to the probability of discovering a particular GO term in a data set (e.g. of proteins). The definition of IC is:
| (1) |
Where t is a particular GO term (assigned to a protein) and p is the probability of that term occurring in a data set. The frequency of a GO term is the number of times it or any of its child terms occurs. The approach of using IC in the GO was originally demonstrated by [14].
2.2 Function Similarity
The function similarity between two GO terms is calculated by determining the ratio of function specificity between their most specific common ancestor and the mean specificity of the two terms:
| (2) |
Where Fspecm is the mean function specificity of the two GO terms and Fspeca is the function specificity of their most specific common ancestor. Our Fsim measure is based on IC, i.e. Relative Information Content (RIC), in a manner similar to [13].
2.3 Data Sets
We selected RefSeq proteins from the Entrez database [18] with GO evidence codes indicated experimentally confirmed functions (EXP, IDA, IPI, IMP, IGI, IEP, see http://www.geneontology.org/GO.evidence.shtml). There were 23,101 proteins for which we calculated the IC of their GO terms. From this set we selected single-function proteins determined by choosing only proteins with a single evidence code (any type).
2.4 Sequence Similarity
Protein sequences were compared using BLAST [1] under default parameters with the exception of low-complexity filtering turned off. Sequence similarity between proteins was determined using the reverse reciprocal bit score (RRBS) which is based on the BLAST bit score. The formula for RRBS is:
| (3) |
Where bitscore is the BLAST similarity statistic between proteins [1]. The RRBS metrics was originally developed by [20] to account for the non-reciprocal nature of BLAST bit scores. We take the square root to help with statistical modeling.
2.5 Statistical Model
Statistical modeling was performed using Generalized Linear Models (GLM) on the open-source R statistical environment. The “family=quasi(link=logit, variance=u(1−u)” parameter was used for the glm function in R.
3. RESULTS
3.1 Gold-Standard Protein Data Set
We excluded non-specific GO terms (IC>6.6) as these tended to confound the model. We also selected proteins with a single GO term. Using these filters we retained 1,424 proteins which still covers 27.8% of GO terms with experimental evidence.
3.2 Sequence-Function Relationship
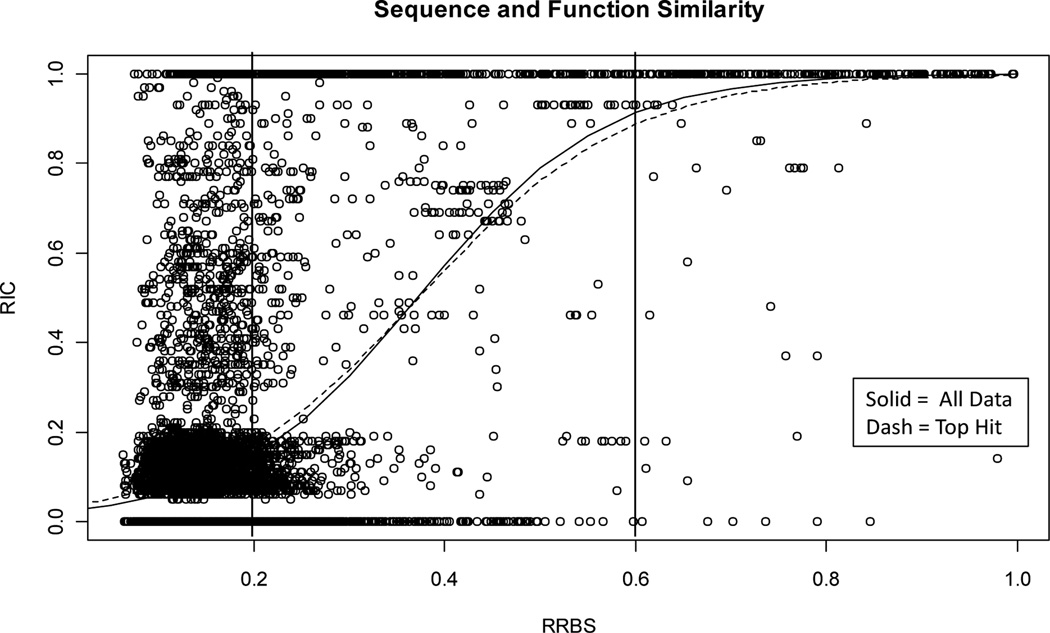
We produced models based on data sets of all BLAST hits for each protein (All pairs) and also when retaining only the best hit for each protein (Top hit). We found that the use of a low-complexity filter had a slight detrimental effect (data not shown). The difference between the All pairs and Top hit model was not very large. Our model therefore refers to the All pairs model. Using the models we found that at high sequence similarity (RRBS>0.6) the function similarity is generally high (mean RIC=0.93, STD=0.22). This indicated good potential “power” for pair-wise methods in predicting function similarity in this range (Figure 1). In contrast, the low sequence similarity range (RRBS<=0.2) generally indicates low function similarity (mean RIC=0.03, STD=0.18), although some protein pairs retain high function similarity however (RIC~1.0, Figure 1). In the “moderate” sequence similarity range (0.2<=RRBS<=0.6) the variability in the data around the model implies that any two proteins can have very similar or distant function similarity (mean RIC=0.33, but STD=0.43), see also Figure 1. This result suggests that accurate prediction of high function similarity between proteins in this moderate range of sequence similarity is difficult, at least with pair-wise methods like BLAST.
Figure 1.
Model fits (All Data and Top Hit). Points represent the RRBS and RIC value for protein pairs. Vertical lines indicate regions of low, moderate, and high sequence similarity.
3.3 Sequence-Function Relationship by GO Category
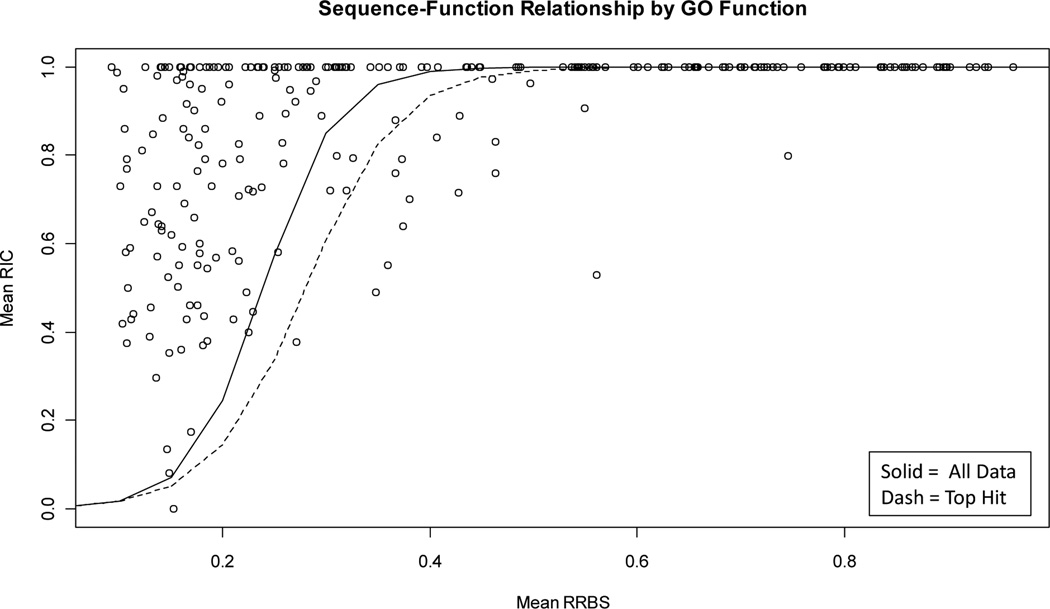
To characterize categories of functions we conducted an analysis on GO terms rather than protein pairs (i.e. GO terms are associated with multiple protein pairs). We first grouped protein pairs by common GO ancestor then calculated the mean RIC and RRBS for the protein pairs grouped by common GO ancestor. We also kept track of the number of pairs associated with each GO ancestor (ancestor frequencies). In all, there were 253 terms identified as GO ancestors. We then fitted weighted GO ancestor model of the mean RIC and mean RRBS with the weights being the ancestor frequencies. We did this for All pairs and Top hit data (Figure 3).
The degree to which function categories (GO ancestors) deviated from the GO ancestor model was determined by calculating residuals normalized by the standard deviation:
| (4) |
Where k is an arbitrary constant and u is the model prediction (i.e. mean) at a particular RRBS value. After calculating residuals, we ranked GO terms by their normalized residual values (hence to need to estimate k). GO terms which lie well above or below the GO ancestor model indicate categories of functions where relatively low sequence similarity is required on average to achieve high function similarity or vice versa. In our analysis we found that many more GO terms like above the GO ancestor model (229) than below it (24) due to more frequent GO terms having greater weight in the model (Figure 3). To investigate significantly diverging terms we selected the bottom 50% percentile (residual>=0.14) of GO terms lying below the GO ancestor model (12 total) and the 90% percentile (residual>=0.47) of GO terms which lie above the GO ancestor model (25 total). The choice of percentage differs because our intent was to select similar size data sets (12 vs 25). A caveat with this approach is that some GO ancestors have few protein pairs and some have many, especially those which lie below the model. This makes it hard to draw general inferences although some broad insights are garnered.
The top 50% of terms below the model appears enriched for enzymes (“catalytic activitiy”, GO term=GO:0003824) in that 75.0% of terms here are enzymes verus 49.4% of terms overall. It therefore appears that enzymes require higher levels of sequence similarity to achieve high function similarity as opposed to functions which lie below the model. Pair-wise methods may perform better here given that higher sequence similarity appears to be needed. The top 90% of terms which lie above the GO ancestor model appear enriched for “binding” proteins (“binding”, GO term=GO:0005488) given that 32.0% of terms in this range are binding proteins versus 25.3% of terms overall. In contrast to functions which lie below the GO ancestor model, these functions have much less sequence similarity associated with high function similarity. It may be more difficult to apply pairwise methods like BLAST to these functions give the lower levels of sequence similarity between these functions on average.
4. DISCUSSION
4.1 Implications for Pair-Wise Methods
Our results illuminate a potential limitation in pair-wise methods like BLAST; the sequence similarity between two proteins must be quite high for them by have high function similarity (based on the average case). However, our results also demonstrate that the sequence-function similarity relationship varies greatly by function category (GO term). Some categories of function contain groups of protein sequences that are much more dissimilar to one another versus the average case. The use of a strict, all-inclusive, sequence similarity threshold when using pair-wise methods would be too conservative when attempting to annotate these categories of proteins. A logical next step to address this is to develop profile models (e.g. HMM’s) for these function categories given the consensus opinion that they are more accurate and sensitive than pair-wise methods [7,8].
4.2 Profile Models for Annotation
Profile models for predicting protein function should be based on groups of proteins whose functions are determined by biological experimentation to avoid “circular” reasoning (i.e. training the profile model on proteins which themselves have predicted function), as on our data set. The problem we face however is that the number of proteins with experimental characterizations of function is relatively small compared to the number of possible functions (there are over 8000 GO terms). For example, in our data set almost half of GO terms are represented by three proteins or less. This makes to difficult to develop profile models as the models estimate the probabilities of positionspecific residues in protein sequences [7]. A potential way to develop larger data sets is to use less-specific GO terms. By the “true-path rule” of the GO hierarchy, the path from a term to the root must always be true; or it is true that child terms “are” their parent terms (although more specific representations of protein function). For example, in applying the true-path rule to our data set the GO term “isomerase activity” (GO term=GO:0016853) can be represented by over 300 proteins, if proteins associated with all of its child terms are included in the set.
This in turn presents another challenge. Profile models depend on highly accurate multiple sequence alignment (MSA) of proteins. Consider that isomerases (GO term=GO:0016853), a category of proteins in our data set related by a common function, appear to have very low sequence similarity among them (mean RRBS=0.04, STD=0.11) and are also of highly variable length (mean=410, STD=259). The sequence similarity between isomerases is also highly variable in that some protein sequences are highly similar to one another while others are very dissimilar. Note that existing benchmarks for evaluating MSA algorithms appear to be clearly limited in regard to the potential diversity and variability in protein sequences [3], as we see here for isomerases. Given the apparent low sequence similarity, identifying the few common or similar residues between the sequences is imperative for developing profile models such as for isomerases. However, according to previous evaluations, existing MSA algorithms may perform poorly or are inaccurate when aligning groups of proteins with very low degrees of sequences similarity and of highly variable lengths [3,19,22], which is likely due to various heuristics employed to improve the tractability of these algorithms. A potential approach could be a more exhaustive alignment search method using parallel computing. Nonetheless, if MSA algorithms are to be successful they must be able to produce accurate multiple alignments under the conditions we describe here.
Figure 2.
Sequence and function similarity relationship by GO term. GO terms above or below the line indicate functions which may require more sequence similarity between proteins to achieve high function similarity than average.
ACKNOWLEDGMENTS
We thank Elizabeth Stewart, Gregory Hather, Natali Kolker, and Gerald Van Belle for their critical and insightful discussions.
Funding: NIH (5R01 GM076680-02 and UO1 DK072473), NSF (DBI-0544757 and NSF-07140) and Seattle Children’s Research Institute internal funds to Eugene Kolker.
Footnotes
Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. To copy otherwise, to republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.
Contributor Information
Roger Higdon, Seattle Children’s Research Institute, 1900 Ninth Avenue, Seattle, WA 98101, roger.higdon@seattlechildrens.org.
Brenton Louie, Seattle Children’s Research Institute, 1900 Ninth Avenue, Seattle, WA 98101, brenton.louie@seattlechildrens.org.
Eugene Kolker, Seattle Children’s Research Institute, 1900 Ninth Avenue, Seattle, WA 98101, eugene.kolker@seattlechildrens.org.
REFERENCES
- 1.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackshields G, Wallace IM, Larkin M, Higgins DG. Analysis and comparison of benchmarks for multiple sequence alignment. In Silico Biology. 2006;6(4):321–339. [PubMed] [Google Scholar]
- 4.Bork P. Powers and pitfalls in sequence analysis: the 70% hurdle. Genome Research. 2000;10(4):398–400. doi: 10.1101/gr.10.4.398. [DOI] [PubMed] [Google Scholar]
- 5.Brenner SE. Errors in genome annotation. Trends in Genetics. 1999;15(4):132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 6.Devos D, Valencia A. Practical limits of function prediction. Proteins: Structure, Function, and Genetics. 2000;41(1):98–107. [PubMed] [Google Scholar]
- 7.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 8.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. Journal of Molecular Biology. 2001;313(4):903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 9.Jones C, Brown A, Baumann U. Estimating the annotation error rate of curated GO database sequence annotations. BMC Bioinformatics. 2007;8(1):170. doi: 10.1186/1471-2105-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolker E, Makarova KS, Shabalina S, et al. Identification and functional analysis of 'hypothetical' genes expressed in Haemophilus influenzae. Nucl. Acids Res. 2004;32(8):2353–2361. doi: 10.1093/nar/gkh555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolker E, Picone AF, Galperin MY, et al. Global profiling of Shewanella oneidensis MR-1: Expression of hypothetical genes and improved functional annotations. PNAS. 2005;102(6):2099–2104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolker E, Purvine S, Galperin MY, et al. Initial Proteome Analysis of Model Microorganism Haemophilus influenzae Strain Rd KW20. J. Bacteriol. 2003;185(15):4593–4602. doi: 10.1128/JB.185.15.4593-4602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin D. An Information-Theoretic Definition of Similarity; Proceeding of the 15th International Conference on Machine Learning; 1998. pp. 296–304. [Google Scholar]
- 14.Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19(10):1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 15.Louie B, Bergen S, Higdon R, Kolker E. Quantifying Protein Function Specificity in the Gene Ontology. Standards in Genomic Sciences. doi: 10.4056/sigs.561626. (in press),. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louie B, Higdon R, Kolker E. A Statistical Model of Protein Sequence Similarity and Function Similarity Reveals Overly-Specific Function Predictions. PLoS ONE. 2009;4(10):e7546. doi: 10.1371/journal.pone.0007546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louie B, Tarczy-Hornoch P, Higdon R, Kolker E. Validating annotations for uncharacterized proteins in Shewanella oneidensis. Omics: A Journal of Integrative Biology. 2008;12(3):211–215. doi: 10.1089/omi.2008.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucl. Acids Res. 2005;33(suppl_1):D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClure MA, Vasi TK, Fitch WM. Comparative analysis of multiple protein-sequence alignment methods. Molecular Biology and Evolution. 1994;11(4):571–592. doi: 10.1093/oxfordjournals.molbev.a040138. [DOI] [PubMed] [Google Scholar]
- 20.Pesquita C, Faria D, Bastos H, Ferreira A, Falcao A, Couto F. Metrics for GO based protein semantic similarity: a systematic evaluation. BMC Bioinformatics. 2008;9(Suppl 5):S4. doi: 10.1186/1471-2105-9-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12(2):85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucl. Acids Res. 1999;27(13):2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valencia A. Automatic annotation of protein function. Current Opinion in Structural Biology. 2005;15(3):267–274. doi: 10.1016/j.sbi.2005.05.010. [DOI] [PubMed] [Google Scholar]