Abstract
The title solvate, C20H14N4O4S·C3H7NO, comprises a stereogenic centre but the centrosymmetric space group causes the presence of the racemate in the crystal. The spiro-joined fragments are almost orthogonal, with a dihedral angle of 86.8 (2)° between the mean planes of the pyrane ring and the dihydroindolone ring system. The atoms of the indolinone bicycle are coplanar, with an r.m.s. deviation of 0.005 Å. In the crystal, pairs of N—H⋯O hydrogen bonds link the molecules into centrosymmetric dimers which are linked to the dimethylformamide solvent molecules by further N—H⋯O hydrogen bonds. N—H⋯N hydrogen bonds link neighbouring dimers into [010] chains.
Related literature
A three-component condensation of 1-R-4-hydroxy-2-oxo-1,2-dihydroquinolines, isatin and malononitrile gave satisfactory yield of 4,3′-spiro[(6-R-2-amino-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3-carbonitrile)-2′-oxindoles], see: Ukrainets et al. (2009 ▶). For van der Waals radii, see: Zefirov (1997 ▶) and for puckering parameters, see: Zefirov et al. (1990 ▶). For mean bond lengths, see: Bürgi & Dunitz (1994 ▶).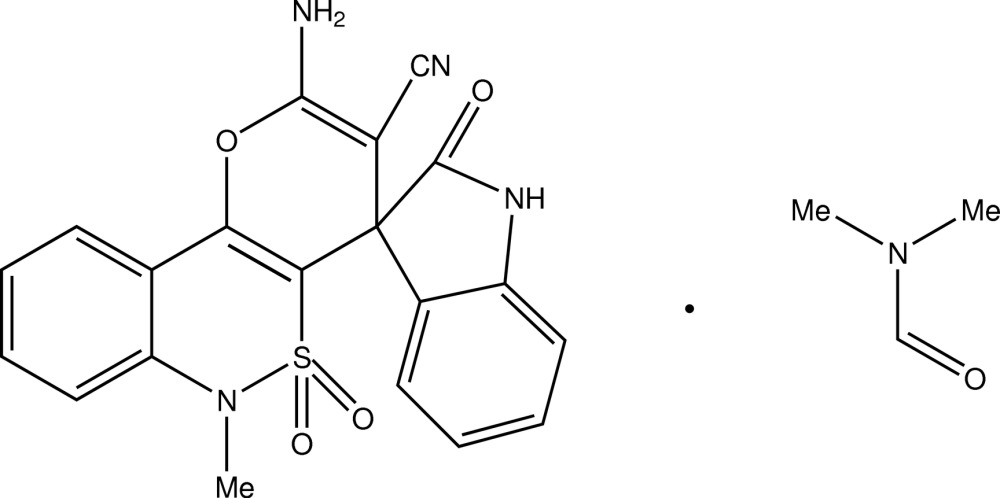
Experimental
Crystal data
C20H14N4O4S·C3H7NO
M r = 479.51
Orthorhombic,

a = 17.2493 (12) Å
b = 9.6046 (5) Å
c = 27.7664 (17) Å
V = 4600.1 (5) Å3
Z = 8
Mo Kα radiation
μ = 0.19 mm−1
T = 293 K
0.30 × 0.02 × 0.02 mm
Data collection
Agilent Xcalibur"3 diffractometer
Absorption correction: multi-scan (CrysAlis RED; Agilent, 2011 ▶) T min = 0.946, T max = 0.996
30060 measured reflections
4049 independent reflections
2147 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.072
wR(F 2) = 0.194
S = 0.99
4049 reflections
323 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.44 e Å−3
Δρmin = −0.27 e Å−3
Data collection: CrysAlis CCD (Agilent, 2011 ▶); cell refinement: CrysAlis RED (Agilent, 2011 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: XP in SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814013634/kp2471sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814013634/kp2471Isup2.hkl
CCDC reference: 1007876
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O5i | 0.82 (5) | 2.01 (5) | 2.797 (7) | 160 (5) |
| N3—H3NA⋯O1ii | 0.86 (5) | 2.22 (5) | 3.051 (6) | 162 (4) |
| N3—H3NB⋯N4iii | 0.83 (4) | 2.37 (4) | 3.159 (6) | 160 (4) |
Symmetry codes: (i)  ; (ii)
; (ii) 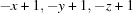 ; (iii)
; (iii)  .
.
supplementary crystallographic information
S1. Comment
A three-component condensation of 1-R-4-hydroxy-2-oxo-1,2-dihydro- quinolines, isatin, and malononitrile gave a satisfactory yields of 4,3'- spiro[(6-R-2-amino-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]quinoline-3- carbonitrile)-2'-oxindoles] (Ukrainets et al., 2009). This is an interesting reaction takes place more easily with sulfo analogues of 1-R-4-hydroxy- 2-oxo-1,2-dihydroquinolines. For example, 1-methyl-4-oxo-3,4-dihydro-1H- 2λ6,1-benzothiazine-2,2-dione was already 15 minute after the start of the reaction forms the corresponding 2'-amino-6'-methyl-2-oxo-1,2-dihydro-6'H– spiro[indole-3,4'-pyrano[3,2-c][2,1]benzothiazine]3'-carbonitrile-5,5'- dioxide (I), which was isolated as a solvate of DMF. The spiro-joined tricyclic and bicyclic fragments of I are turned relatively to each other in such way that the dihedral angle between mean planes of the pyrane ring and dihydroindolone bicycle is 86.8°. At that the C1—C2 bond is elongated as compared with its mean value (Bürgi & Dunitz, 1994) 1.511 Å. The bicyclic fragment is slightly non-planar, the five-membered heterocycle adopts an envelope conformation with deviation of the C1 atom by 0.14 Å. In addition the dihedral angle between planar N1—C1(=O1)—C2 fragment and aromatic ring is 11.5 °. The pyrane ring adopts a boat conformation (the puckering parameters (Zefirov et al., 1990) are: S=0.24, Θ=73.0°, Ψ=9.1°). Deviations of the O2 and C2 atoms from the mean plane of the remaining atoms of this ring are 0.10 Å and 0.20 Å, respectivey. The benzothiazine ring adopts a twist-boat conformation (the puckering parameters are S=0.57, Θ=47.2°, Ψ=25.9°). Deviations of the S1 and C9 atoms from the mean plane of the remaining atoms of this ring are -0.76 Å and -0.23 Å, respectivey. The steric repulsion between methyl group and atoms of the C10···C15 ring (shortened intramolecular contacts are: H20b···C11 2.71 Å, H11···C20 2.50 Å as compared with van der Waals radii sum ((Zefirov, 1997) 2.87 Å and H11···H20b 2.26 Å (2.34 Å)) results in elongation of the N2—C10 bond up to 1.415 (6) Å as compared with its mean value 1.371 Å. In the crystal the molecules form the centrosymmetric dimers (Fig. 2) by N3—H3Na···O1' (1 - x, 1 - y, 1 - z) intermolecular hydrogen bonds (Table 1). Each monomer of such dimer is bonded with DMF solvate molecule by the N1—H···O5' (0.5 + x, y, 1.5 - z) hydrogen bond. N3—H3Nb···N4' (1 - x, -y, 1 - z) hydrogen bond is observed between neighboring dimers (Table 1).
S2. Experimental
A mixture of 1-methyl-4-oxo-3,4-dihydro-1H-2λ6,1-benzothiazine-2,2- dione (2.11 g, 0.01 mol), isatin (1.47 g, 0.01 mol), malononitrile (0.66 g, 0.01 mol), and triethylamine (1.4 ml, 0.01 mol) in methanol (20 ml) was refluxed for 15 min, and cooled. The precipitated were off, washed with methanol, and crystallized from DMF-H2O (1:1). Prepared 3.21 g (67%) solvate crystals of the pyranobenzothiazine with DMF; mp 437-439 K (decomp.).
S3. Refinement
All hydrogen atoms were located from electron density difference maps and were refined in the riding motion approximation with Uiso constrained to be 1.5 times Ueq of the carrier atom for the methyl group and 1.2 times Ueq of the carrier atom for the other atoms. Hydrogen atoms of the aminogroups are refined using isotropic approximation.
Figures
Fig. 1.
The title compound with atomic membering. All atoms are shown with displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
Hydrogen bonded, centrosymmetric dimers with dimethyl foramid solvate connected by hydrogen bonds.The hydrogen bonds are shown by dashed lines.
Crystal data
| C20H14N4O4S·C3H7NO | Dx = 1.385 Mg m−3 |
| Mr = 479.51 | Melting point = 437–439 K |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2361 reflections |
| a = 17.2493 (12) Å | θ = 3.2–21.6° |
| b = 9.6046 (5) Å | µ = 0.19 mm−1 |
| c = 27.7664 (17) Å | T = 293 K |
| V = 4600.1 (5) Å3 | Stick, colourless |
| Z = 8 | 0.30 × 0.02 × 0.02 mm |
| F(000) = 2000 |
Data collection
| Agilent Xcalibur"3 diffractometer | 4049 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2147 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| Detector resolution: 16.1827 pixels mm-1 | θmax = 25.0°, θmin = 2.8° |
| ω scans | h = −20→17 |
| Absorption correction: multi-scan (CrysAlis RED; Agilent, 2011) | k = −11→11 |
| Tmin = 0.946, Tmax = 0.996 | l = −33→33 |
| 30060 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.072 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.194 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0901P)2] where P = (Fo2 + 2Fc2)/3 |
| 4049 reflections | (Δ/σ)max < 0.001 |
| 323 parameters | Δρmax = 0.44 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Experimental. Absorption correction: CrysAlis RED, Agilent Technologies, Version 1.171.36.24 (release 03-12-2012 CrysAlis171 .NET) (compiled Dec 3 2012,18:21:49) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.37888 (7) | 0.59466 (11) | 0.64282 (4) | 0.0534 (4) | |
| O1 | 0.58546 (17) | 0.4943 (3) | 0.59913 (12) | 0.0611 (9) | |
| O2 | 0.41460 (16) | 0.4674 (3) | 0.51036 (9) | 0.0464 (7) | |
| O3 | 0.30029 (19) | 0.5566 (3) | 0.65291 (13) | 0.0712 (10) | |
| O4 | 0.4331 (2) | 0.5859 (3) | 0.68127 (12) | 0.0748 (10) | |
| O5 | 0.1982 (3) | 0.3611 (7) | 0.7857 (2) | 0.157 (2) | |
| N1 | 0.5617 (3) | 0.3404 (4) | 0.66047 (15) | 0.0579 (11) | |
| H1N | 0.605 (3) | 0.330 (5) | 0.6722 (19) | 0.073 (18)* | |
| N2 | 0.3820 (2) | 0.7522 (3) | 0.62123 (14) | 0.0569 (10) | |
| N3 | 0.4424 (2) | 0.2711 (5) | 0.47288 (15) | 0.0554 (11) | |
| H3NB | 0.457 (2) | 0.189 (4) | 0.4712 (15) | 0.048 (14)* | |
| H3NA | 0.434 (2) | 0.322 (4) | 0.4479 (17) | 0.057 (15)* | |
| N4 | 0.5120 (2) | 0.0310 (4) | 0.56205 (14) | 0.0653 (12) | |
| N5 | 0.3135 (3) | 0.3806 (7) | 0.8231 (3) | 0.1135 (19) | |
| C1 | 0.5437 (2) | 0.4109 (4) | 0.62037 (16) | 0.0475 (11) | |
| C2 | 0.4597 (2) | 0.3692 (4) | 0.60525 (15) | 0.0414 (10) | |
| C3 | 0.4343 (3) | 0.2863 (4) | 0.64885 (15) | 0.0448 (11) | |
| C4 | 0.3633 (3) | 0.2320 (4) | 0.66055 (17) | 0.0527 (12) | |
| H4 | 0.3212 | 0.2404 | 0.6398 | 0.063* | |
| C5 | 0.3562 (3) | 0.1631 (4) | 0.70495 (19) | 0.0661 (14) | |
| H5 | 0.3082 | 0.1289 | 0.7146 | 0.079* | |
| C6 | 0.4193 (4) | 0.1461 (5) | 0.73416 (17) | 0.0686 (14) | |
| H6 | 0.4136 | 0.0968 | 0.7628 | 0.082* | |
| C7 | 0.4917 (3) | 0.2001 (5) | 0.72248 (17) | 0.0669 (14) | |
| H7 | 0.5344 | 0.1891 | 0.7426 | 0.080* | |
| C8 | 0.4966 (3) | 0.2713 (4) | 0.67924 (16) | 0.0498 (11) | |
| C9 | 0.4128 (2) | 0.4982 (4) | 0.59449 (15) | 0.0394 (10) | |
| C10 | 0.3533 (2) | 0.7780 (4) | 0.57434 (18) | 0.0504 (11) | |
| C11 | 0.3224 (3) | 0.9077 (4) | 0.5625 (2) | 0.0643 (14) | |
| H11 | 0.3168 | 0.9758 | 0.5861 | 0.077* | |
| C12 | 0.3004 (3) | 0.9344 (5) | 0.5162 (2) | 0.0743 (16) | |
| H12 | 0.2806 | 1.0216 | 0.5084 | 0.089* | |
| C13 | 0.3070 (3) | 0.8350 (5) | 0.4807 (2) | 0.0685 (14) | |
| H13 | 0.2911 | 0.8548 | 0.4495 | 0.082* | |
| C14 | 0.3368 (2) | 0.7075 (4) | 0.49171 (17) | 0.0511 (11) | |
| H14 | 0.3412 | 0.6405 | 0.4677 | 0.061* | |
| C15 | 0.3609 (2) | 0.6756 (4) | 0.53878 (16) | 0.0444 (11) | |
| C16 | 0.3960 (2) | 0.5445 (4) | 0.55022 (15) | 0.0394 (10) | |
| C17 | 0.4408 (2) | 0.3341 (4) | 0.51557 (15) | 0.0415 (10) | |
| C18 | 0.4615 (2) | 0.2827 (4) | 0.55921 (15) | 0.0404 (10) | |
| C19 | 0.4899 (2) | 0.1437 (4) | 0.56126 (15) | 0.0459 (11) | |
| C20 | 0.3838 (3) | 0.8663 (4) | 0.65669 (18) | 0.0827 (18) | |
| H20C | 0.3317 | 0.8908 | 0.6655 | 0.124* | |
| H20B | 0.4091 | 0.9458 | 0.6429 | 0.124* | |
| H20A | 0.4116 | 0.8367 | 0.6848 | 0.124* | |
| C21 | 0.2672 (5) | 0.3931 (5) | 0.7875 (2) | 0.133 (3) | |
| H21 | 0.2881 | 0.4311 | 0.7595 | 0.159* | |
| C22 | 0.3923 (5) | 0.4117 (12) | 0.8231 (5) | 0.251 (8) | |
| H22A | 0.4055 | 0.4587 | 0.7938 | 0.376* | |
| H22B | 0.4216 | 0.3270 | 0.8256 | 0.376* | |
| H22C | 0.4040 | 0.4707 | 0.8501 | 0.376* | |
| C23 | 0.2835 (7) | 0.3142 (14) | 0.8647 (5) | 0.259 (7) | |
| H23A | 0.2449 | 0.2474 | 0.8553 | 0.389* | |
| H23B | 0.2605 | 0.3825 | 0.8855 | 0.389* | |
| H23C | 0.3248 | 0.2676 | 0.8814 | 0.389* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0699 (9) | 0.0464 (7) | 0.0440 (7) | 0.0084 (6) | −0.0028 (6) | −0.0073 (5) |
| O1 | 0.054 (2) | 0.072 (2) | 0.057 (2) | −0.0097 (16) | −0.0099 (17) | 0.0087 (17) |
| O2 | 0.065 (2) | 0.0423 (15) | 0.0317 (16) | 0.0125 (13) | −0.0046 (14) | 0.0015 (13) |
| O3 | 0.065 (2) | 0.071 (2) | 0.077 (3) | 0.0050 (17) | 0.0236 (19) | −0.0025 (17) |
| O4 | 0.102 (3) | 0.066 (2) | 0.056 (2) | 0.0198 (18) | −0.034 (2) | −0.0182 (17) |
| O5 | 0.067 (3) | 0.285 (7) | 0.118 (5) | 0.007 (4) | 0.030 (3) | −0.012 (4) |
| N1 | 0.048 (3) | 0.073 (3) | 0.053 (3) | 0.004 (2) | −0.012 (2) | 0.012 (2) |
| N2 | 0.070 (3) | 0.041 (2) | 0.060 (3) | 0.0093 (18) | −0.009 (2) | −0.0175 (18) |
| N3 | 0.083 (3) | 0.046 (2) | 0.037 (3) | 0.013 (2) | −0.006 (2) | −0.004 (2) |
| N4 | 0.086 (3) | 0.051 (2) | 0.059 (3) | 0.017 (2) | −0.001 (2) | 0.002 (2) |
| N5 | 0.074 (4) | 0.165 (5) | 0.102 (5) | −0.014 (4) | 0.002 (4) | −0.012 (4) |
| C1 | 0.049 (3) | 0.050 (3) | 0.044 (3) | 0.005 (2) | −0.005 (2) | −0.001 (2) |
| C2 | 0.041 (3) | 0.043 (2) | 0.040 (3) | 0.0064 (18) | −0.002 (2) | 0.0000 (18) |
| C3 | 0.053 (3) | 0.040 (2) | 0.041 (3) | 0.005 (2) | −0.002 (2) | −0.0063 (19) |
| C4 | 0.066 (3) | 0.038 (2) | 0.054 (3) | −0.003 (2) | 0.004 (3) | −0.007 (2) |
| C5 | 0.086 (4) | 0.057 (3) | 0.055 (3) | −0.009 (3) | 0.007 (3) | 0.001 (3) |
| C6 | 0.104 (4) | 0.067 (3) | 0.035 (3) | −0.007 (3) | 0.001 (3) | 0.008 (2) |
| C7 | 0.088 (4) | 0.068 (3) | 0.044 (3) | −0.002 (3) | −0.010 (3) | 0.010 (2) |
| C8 | 0.055 (3) | 0.051 (2) | 0.043 (3) | 0.006 (2) | −0.007 (2) | 0.006 (2) |
| C9 | 0.039 (2) | 0.039 (2) | 0.040 (2) | 0.0001 (18) | −0.002 (2) | −0.0038 (19) |
| C10 | 0.045 (3) | 0.043 (2) | 0.063 (3) | 0.0048 (19) | 0.001 (2) | −0.001 (2) |
| C11 | 0.069 (3) | 0.049 (3) | 0.074 (4) | 0.017 (2) | 0.002 (3) | −0.001 (3) |
| C12 | 0.075 (4) | 0.058 (3) | 0.090 (5) | 0.028 (3) | 0.007 (3) | 0.016 (3) |
| C13 | 0.066 (3) | 0.067 (3) | 0.072 (4) | 0.020 (3) | −0.008 (3) | 0.020 (3) |
| C14 | 0.050 (3) | 0.053 (3) | 0.051 (3) | 0.009 (2) | 0.000 (2) | 0.006 (2) |
| C15 | 0.039 (3) | 0.043 (2) | 0.050 (3) | 0.0034 (18) | 0.001 (2) | 0.003 (2) |
| C16 | 0.038 (2) | 0.040 (2) | 0.040 (3) | 0.0008 (17) | −0.002 (2) | −0.0032 (19) |
| C17 | 0.047 (3) | 0.039 (2) | 0.039 (3) | 0.0045 (18) | −0.001 (2) | −0.001 (2) |
| C18 | 0.043 (2) | 0.034 (2) | 0.044 (3) | 0.0069 (17) | 0.000 (2) | −0.0010 (18) |
| C19 | 0.051 (3) | 0.051 (3) | 0.035 (3) | 0.002 (2) | 0.002 (2) | 0.000 (2) |
| C20 | 0.115 (5) | 0.053 (3) | 0.081 (4) | 0.005 (3) | −0.012 (3) | −0.025 (3) |
| C21 | 0.104 (7) | 0.184 (8) | 0.110 (7) | 0.023 (6) | 0.044 (6) | −0.010 (6) |
| C22 | 0.105 (7) | 0.364 (17) | 0.284 (16) | −0.088 (9) | 0.044 (8) | −0.183 (13) |
| C23 | 0.178 (11) | 0.392 (19) | 0.207 (14) | 0.006 (11) | 0.028 (10) | 0.131 (13) |
Geometric parameters (Å, º)
| S1—O4 | 1.422 (3) | C2—C3 | 1.514 (6) |
| S1—O3 | 1.432 (3) | C2—C18 | 1.525 (6) |
| S1—N2 | 1.628 (4) | C3—C4 | 1.370 (6) |
| S1—C9 | 1.732 (4) | C3—C8 | 1.374 (6) |
| O1—C1 | 1.228 (5) | C4—C5 | 1.404 (6) |
| O2—C17 | 1.366 (4) | C5—C6 | 1.367 (7) |
| O2—C16 | 1.370 (5) | C6—C7 | 1.390 (7) |
| O5—C21 | 1.231 (8) | C7—C8 | 1.384 (6) |
| N1—C1 | 1.339 (6) | C9—C16 | 1.339 (5) |
| N1—C8 | 1.404 (6) | C10—C11 | 1.394 (6) |
| N2—C10 | 1.415 (6) | C10—C15 | 1.400 (6) |
| N2—C20 | 1.474 (5) | C11—C12 | 1.365 (7) |
| N3—C17 | 1.331 (5) | C12—C13 | 1.375 (7) |
| N4—C19 | 1.148 (5) | C13—C14 | 1.363 (6) |
| N5—C21 | 1.277 (8) | C14—C15 | 1.406 (6) |
| N5—C22 | 1.391 (9) | C15—C16 | 1.432 (5) |
| N5—C23 | 1.417 (11) | C17—C18 | 1.357 (6) |
| C1—C2 | 1.560 (6) | C18—C19 | 1.423 (6) |
| C2—C9 | 1.510 (5) | ||
| O4—S1—O3 | 117.4 (2) | C8—C7—C6 | 116.2 (5) |
| O4—S1—N2 | 108.0 (2) | C3—C8—C7 | 122.5 (5) |
| O3—S1—N2 | 109.9 (2) | C3—C8—N1 | 110.3 (4) |
| O4—S1—C9 | 109.15 (19) | C7—C8—N1 | 127.2 (5) |
| O3—S1—C9 | 109.5 (2) | C16—C9—C2 | 124.8 (4) |
| N2—S1—C9 | 101.58 (19) | C16—C9—S1 | 117.4 (3) |
| C17—O2—C16 | 119.9 (3) | C2—C9—S1 | 117.8 (3) |
| C1—N1—C8 | 111.3 (4) | C11—C10—C15 | 119.9 (5) |
| C10—N2—C20 | 119.5 (4) | C11—C10—N2 | 120.5 (4) |
| C10—N2—S1 | 119.3 (3) | C15—C10—N2 | 119.5 (4) |
| C20—N2—S1 | 116.5 (3) | C12—C11—C10 | 119.7 (5) |
| C21—N5—C22 | 126.3 (9) | C11—C12—C13 | 121.4 (4) |
| C21—N5—C23 | 116.4 (7) | C14—C13—C12 | 119.7 (5) |
| C22—N5—C23 | 116.9 (10) | C13—C14—C15 | 121.0 (4) |
| O1—C1—N1 | 126.4 (4) | C10—C15—C14 | 118.3 (4) |
| O1—C1—C2 | 125.6 (4) | C10—C15—C16 | 120.1 (4) |
| N1—C1—C2 | 108.0 (4) | C14—C15—C16 | 121.5 (4) |
| C9—C2—C3 | 115.7 (3) | C9—C16—O2 | 120.8 (3) |
| C9—C2—C18 | 107.0 (3) | C9—C16—C15 | 126.0 (4) |
| C3—C2—C18 | 112.9 (3) | O2—C16—C15 | 113.3 (3) |
| C9—C2—C1 | 110.0 (3) | N3—C17—C18 | 128.6 (4) |
| C3—C2—C1 | 100.9 (3) | N3—C17—O2 | 109.8 (4) |
| C18—C2—C1 | 110.3 (3) | C18—C17—O2 | 121.5 (4) |
| C4—C3—C8 | 120.9 (4) | C17—C18—C19 | 117.9 (4) |
| C4—C3—C2 | 130.5 (4) | C17—C18—C2 | 123.0 (3) |
| C8—C3—C2 | 108.6 (4) | C19—C18—C2 | 119.0 (3) |
| C3—C4—C5 | 117.7 (5) | N4—C19—C18 | 178.5 (5) |
| C6—C5—C4 | 120.5 (5) | O5—C21—N5 | 127.8 (6) |
| C5—C6—C7 | 122.1 (5) | ||
| O4—S1—N2—C10 | 160.2 (3) | O3—S1—C9—C2 | −97.2 (3) |
| O3—S1—N2—C10 | −70.5 (4) | N2—S1—C9—C2 | 146.6 (3) |
| C9—S1—N2—C10 | 45.4 (4) | C20—N2—C10—C11 | −3.9 (6) |
| O4—S1—N2—C20 | −44.3 (4) | S1—N2—C10—C11 | 150.8 (3) |
| O3—S1—N2—C20 | 85.0 (4) | C20—N2—C10—C15 | 171.6 (4) |
| C9—S1—N2—C20 | −159.1 (3) | S1—N2—C10—C15 | −33.6 (5) |
| C8—N1—C1—O1 | 170.6 (4) | C15—C10—C11—C12 | −0.5 (7) |
| C8—N1—C1—C2 | −9.1 (5) | N2—C10—C11—C12 | 175.0 (5) |
| O1—C1—C2—C9 | −47.7 (5) | C10—C11—C12—C13 | 0.9 (8) |
| N1—C1—C2—C9 | 132.0 (4) | C11—C12—C13—C14 | −0.7 (8) |
| O1—C1—C2—C3 | −170.4 (4) | C12—C13—C14—C15 | 0.1 (7) |
| N1—C1—C2—C3 | 9.3 (4) | C11—C10—C15—C14 | −0.1 (6) |
| O1—C1—C2—C18 | 70.0 (5) | N2—C10—C15—C14 | −175.6 (4) |
| N1—C1—C2—C18 | −110.3 (4) | C11—C10—C15—C16 | 177.1 (4) |
| C9—C2—C3—C4 | 54.1 (6) | N2—C10—C15—C16 | 1.5 (6) |
| C18—C2—C3—C4 | −69.7 (5) | C13—C14—C15—C10 | 0.3 (7) |
| C1—C2—C3—C4 | 172.7 (4) | C13—C14—C15—C16 | −176.8 (4) |
| C9—C2—C3—C8 | −124.9 (4) | C2—C9—C16—O2 | 6.7 (6) |
| C18—C2—C3—C8 | 111.3 (4) | S1—C9—C16—O2 | −174.0 (3) |
| C1—C2—C3—C8 | −6.4 (4) | C2—C9—C16—C15 | −170.9 (4) |
| C8—C3—C4—C5 | 1.2 (6) | S1—C9—C16—C15 | 8.4 (6) |
| C2—C3—C4—C5 | −177.7 (4) | C17—O2—C16—C9 | 9.0 (5) |
| C3—C4—C5—C6 | −2.9 (7) | C17—O2—C16—C15 | −173.1 (3) |
| C4—C5—C6—C7 | 2.7 (7) | C10—C15—C16—C9 | 11.0 (6) |
| C5—C6—C7—C8 | −0.6 (7) | C14—C15—C16—C9 | −171.9 (4) |
| C4—C3—C8—C7 | 0.8 (6) | C10—C15—C16—O2 | −166.7 (4) |
| C2—C3—C8—C7 | 179.9 (4) | C14—C15—C16—O2 | 10.3 (5) |
| C4—C3—C8—N1 | −177.5 (4) | C16—O2—C17—N3 | 169.2 (4) |
| C2—C3—C8—N1 | 1.6 (5) | C16—O2—C17—C18 | −11.4 (6) |
| C6—C7—C8—C3 | −1.1 (7) | N3—C17—C18—C19 | 0.8 (7) |
| C6—C7—C8—N1 | 176.9 (4) | O2—C17—C18—C19 | −178.6 (3) |
| C1—N1—C8—C3 | 5.0 (5) | N3—C17—C18—C2 | 177.5 (4) |
| C1—N1—C8—C7 | −173.2 (4) | O2—C17—C18—C2 | −1.9 (6) |
| C3—C2—C9—C16 | −144.0 (4) | C9—C2—C18—C17 | 14.6 (5) |
| C18—C2—C9—C16 | −17.2 (5) | C3—C2—C18—C17 | 143.0 (4) |
| C1—C2—C9—C16 | 102.6 (5) | C1—C2—C18—C17 | −105.0 (4) |
| C3—C2—C9—S1 | 36.8 (4) | C9—C2—C18—C19 | −168.8 (3) |
| C18—C2—C9—S1 | 163.6 (3) | C3—C2—C18—C19 | −40.3 (5) |
| C1—C2—C9—S1 | −76.7 (4) | C1—C2—C18—C19 | 71.7 (5) |
| O4—S1—C9—C16 | −146.6 (3) | C17—C18—C19—N4 | −25 (22) |
| O3—S1—C9—C16 | 83.5 (3) | C2—C18—C19—N4 | 158 (22) |
| N2—S1—C9—C16 | −32.7 (4) | C22—N5—C21—O5 | 176.6 (8) |
| O4—S1—C9—C2 | 32.7 (3) | C23—N5—C21—O5 | 4.2 (12) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O5i | 0.82 (5) | 2.01 (5) | 2.797 (7) | 160 (5) |
| N3—H3NA···O1ii | 0.86 (5) | 2.22 (5) | 3.051 (6) | 162 (4) |
| N3—H3NB···N4iii | 0.83 (4) | 2.37 (4) | 3.159 (6) | 160 (4) |
Symmetry codes: (i) x+1/2, y, −z+3/2; (ii) −x+1, −y+1, −z+1; (iii) −x+1, −y, −z+1.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: KP2471).
References
- Agilent (2011). CrysAlis CCD and CrysAlis RED Agilent Technologies Ltd, Yarnton, England.
- Bürgi, H.-B. & Dunitz, J. D. (1994). Structure Correlation, Vol. 2, pp. 767–784. Weinheim: VCH.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Ukrainets, I. V., Red’kin, R. G., Sidorenko, L. V. & Turov, A. V. (2009). Chem. Heterocycl. Compd, 45, 1478–1484.
- Zefirov, Yu. V. (1997). Kristallografiya, 42, 936–958.
- Zefirov, N. S., Palyulin, V. A. & Dashevskaya, E. E. (1990). J. Phys. Org. Chem. 3, 147–154.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536814013634/kp2471sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814013634/kp2471Isup2.hkl
CCDC reference: 1007876
Additional supporting information: crystallographic information; 3D view; checkCIF report




