Abstract
The title compound, [RuCl2(η6-C6H6)(C12H22ClP)]·CHCl3, was prepared by reaction of [RuCl2(η6-C6H6)]2 with chlorodicyclohexylphosphane in CHCl3 at 323 K under argon. The RuII atom is surrounded by one arene ligand, two Cl atoms and a phosphane ligand in a piano-stool geometry. The phosphane ligand is linked by the P atom, with an Ru—P bond length of 2.3247 (4) Å. Both cyclohexyl rings at the P atom adopt a chair conformation. In the crystal, the RuII complex molecule and the chloroform solvent molecule are linked by a bifurcated C—H⋯(Cl,Cl) hydrogen bond. Intramolecular C—H⋯Cl hydrogen bonds are also observed.
Related literature
For the molecular structure of Ru complexes with the related chlorodiphenylphosphane ligand, see: Jantscher et al. (2009 ▶); Torres-Lubián et al. (1999 ▶).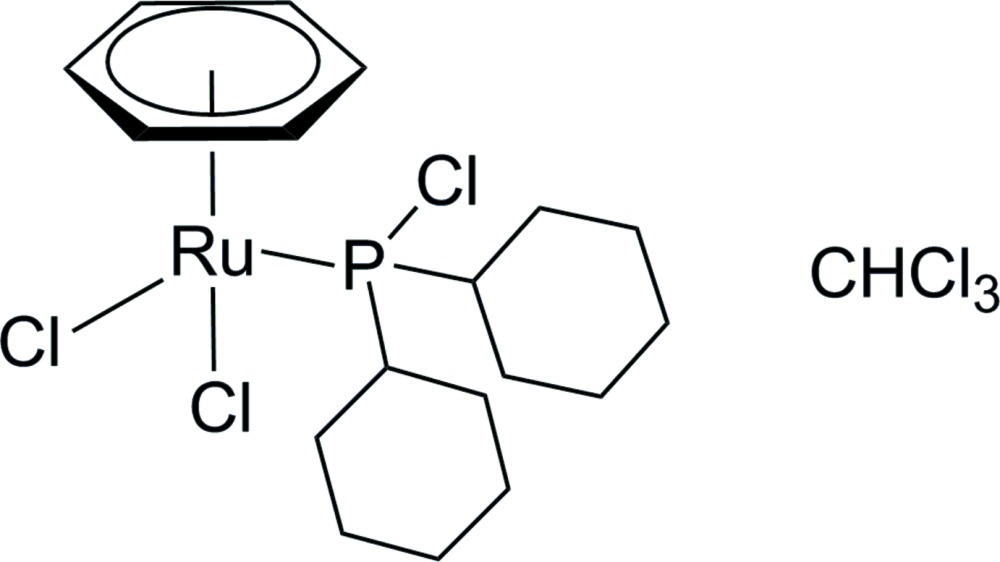
Experimental
Crystal data
[RuCl2(C6H6)(C12H22ClP)]·CHCl3
M r = 602.16
Monoclinic,

a = 7.9717 (1) Å
b = 16.3020 (2) Å
c = 18.0602 (3) Å
β = 91.244 (1)°
V = 2346.45 (6) Å3
Z = 4
Mo Kα radiation
μ = 1.42 mm−1
T = 150 K
0.36 × 0.22 × 0.11 mm
Data collection
Bruker Kappa APEXII DUO diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.630, T max = 0.859
37052 measured reflections
5619 independent reflections
4963 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.019
wR(F 2) = 0.048
S = 1.05
5619 reflections
244 parameters
H-atom parameters constrained
Δρmax = 0.41 e Å−3
Δρmin = −0.37 e Å−3
Data collection: APEX2 (Bruker, 2011 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814012975/is5363sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814012975/is5363Isup2.hkl
CCDC reference: 1006720
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C14—H14B⋯Cl1 | 0.99 | 2.56 | 3.4080 (17) | 144 |
| C18—H18A⋯Cl1 | 0.99 | 2.74 | 3.5366 (17) | 138 |
| C19—H19⋯Cl1i | 1.00 | 2.69 | 3.5539 (17) | 144 |
| C19—H19⋯Cl2i | 1.00 | 2.77 | 3.6119 (18) | 142 |
Symmetry code: (i)  .
.
supplementary crystallographic information
S1. Comment
The half-sandwich (η6–C6H6)-dichlorido(chlorodicyclohexylphosphane)ruthenium(II) complex was formed by reaction of one equivalent of [RuCl2(η6-C6H6)]2 with two equivalents of (Cy2P(1-naphthoyl)) ligand under hydrogenation conditions (CHCl3, 60 bar of H2, 353 K, 3 hrs) as a side product. The cleavage of the 1-naphthoyl group from [RuCl2(η6-C6H6)(Cy2P(1-naphthoyl)] complex forms firstly [RuCl2(η6-C6H6)(Cy2PH)] and subsequent chlorination of the dicyclohexylphosphane unit due to CHCl3 yields the title compound in poor yield. Additionally, we could not observe any trace amount of title compound by using non-chlorinated solvents such as MeOH. The substitution of hydrogen next to phosphane by chlorine coming from solvent molecules is also described for the formation of a Ru-complex with the related chlorodiphenylphosphane ligand by Torres-Lubián et al. (1999). More specifically, the title complex was formed by reaction of [RuCl2(η6-C6H6)]2 with chlorodicyclohexylphosphane in CHCl3 at 323 K under argon in 41% yield. Crystals suitable for X-ray crystal structure analysis could be obtained by crystallization from a chloroform/heptane mixture. In the 31P NMR spectrum of the complex the signal for the phosphorus was observed at 156.3 p.p.m., whereas free ligand signal appears at 128.8 p.p.m.. The title compound shows the three legged piano-stool geometry at the ruthenium centre with the arene, chlorodicyclohexylphosphane and two chlorine ligands in the coordination sphere (Fig. 1). The phosphane ligand is linked by the phosphorus with a Ru—P bond length of 2.3247 (4) Å. Both cyclohexyl rings at the phosphorus atom adopt a chair conformation. The Ru complex is co-crystallized with CHCl3.
S2. Experimental
A 50 ml round bottom flask with inert gas valve was charged with 0.05 mmol (25 mg) [RuCl2(η6-C6H6)]2 and 4 ml CHCl3 under argon atmosphere. To this suspension 0.105 mmol (21 µL) chlorodicyclohexylphosphane was added and the reaction mixture was allowed to react 3 h at 323 K. A clear red brown solution has been formed and the volume was reduced carefully in high vacuum to ca 1 ml. Next 20 ml heptane was added to the reaction mixture and cooled for 1 h with ice bath. The precipitate was washed with heptane (3 × 5 ml) to yield the title compound as an orange brown solid (20 mg, 41%). Red single crystals were grown in CHCl3/heptane mixture at 245 K for 1 day. 1H NMR (300 MHz), CDCl3): δ 5.71 (s, 6H, benzene), 2.79 (m, 2H, Cy), 2.06–1.54 (m, 14H, Cy), 1.27 (br s, 6H, Cy). 13C {1H} NMR (75 MHz, CDCl3): δ 89.8 (RuPh), 40.4 (d, JPC = 9.5 Hz, PCH), 27.2, 26.9, 26.7, 26.4, 26.3, 26.0, 26.0, 25.6 (CH2). 31P {1H} NMR (121 MHz, CDCl3): δ 156.3.
S3. Refinement
H atoms were placed in idealized positions with C—H = 0.95–1.00 Å (CH), 0.99 Å (CH2) and refined using a riding model with Uiso(H) fixed at 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound with 30% displacement ellipsoids. Hydrogen atoms are omitted for clarity.
Crystal data
| [RuCl2(C6H6)(C12H22ClP)]·CHCl3 | F(000) = 1216 |
| Mr = 602.16 | Dx = 1.705 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.9717 (1) Å | Cell parameters from 9877 reflections |
| b = 16.3020 (2) Å | θ = 2.3–27.9° |
| c = 18.0602 (3) Å | µ = 1.42 mm−1 |
| β = 91.244 (1)° | T = 150 K |
| V = 2346.45 (6) Å3 | Prism, orange |
| Z = 4 | 0.36 × 0.22 × 0.11 mm |
Data collection
| Bruker Kappa APEXII DUO diffractometer | 5619 independent reflections |
| Radiation source: fine-focus sealed tube | 4963 reflections with I > 2σ(I) |
| Curved graphite monochromator | Rint = 0.031 |
| Detector resolution: 8.3333 pixels mm-1 | θmax = 27.9°, θmin = 1.7° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | k = −21→21 |
| Tmin = 0.630, Tmax = 0.859 | l = −23→23 |
| 37052 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.019 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.048 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0187P)2 + 1.4791P] where P = (Fo2 + 2Fc2)/3 |
| 5619 reflections | (Δ/σ)max = 0.001 |
| 244 parameters | Δρmax = 0.41 e Å−3 |
| 0 restraints | Δρmin = −0.37 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | −0.2498 (2) | 0.37279 (11) | 0.04879 (10) | 0.0223 (4) | |
| H1 | −0.3279 | 0.3746 | 0.0083 | 0.027* | |
| C2 | −0.2406 (2) | 0.30292 (11) | 0.09373 (10) | 0.0227 (4) | |
| H2 | −0.3181 | 0.2592 | 0.0863 | 0.027* | |
| C3 | −0.1149 (2) | 0.29763 (11) | 0.15045 (10) | 0.0230 (4) | |
| H3 | −0.1069 | 0.2501 | 0.1808 | 0.028* | |
| C4 | −0.0024 (2) | 0.36315 (11) | 0.16134 (10) | 0.0231 (4) | |
| H4 | 0.0864 | 0.3585 | 0.1970 | 0.028* | |
| C5 | −0.0212 (2) | 0.43653 (11) | 0.11906 (10) | 0.0235 (4) | |
| H5 | 0.0498 | 0.4822 | 0.1291 | 0.028* | |
| C6 | −0.1430 (2) | 0.44123 (11) | 0.06332 (10) | 0.0227 (4) | |
| H6 | −0.1552 | 0.4900 | 0.0349 | 0.027* | |
| C7 | 0.2490 (2) | 0.14568 (10) | 0.07869 (9) | 0.0160 (3) | |
| H7 | 0.3484 | 0.1741 | 0.0577 | 0.019* | |
| C8 | 0.2460 (2) | 0.16860 (11) | 0.16112 (9) | 0.0211 (3) | |
| H8A | 0.2381 | 0.2290 | 0.1661 | 0.025* | |
| H8B | 0.1456 | 0.1442 | 0.1837 | 0.025* | |
| C9 | 0.4035 (2) | 0.13820 (11) | 0.20233 (10) | 0.0238 (4) | |
| H9A | 0.5032 | 0.1664 | 0.1828 | 0.029* | |
| H9B | 0.3958 | 0.1518 | 0.2556 | 0.029* | |
| C10 | 0.4238 (2) | 0.04591 (11) | 0.19335 (11) | 0.0276 (4) | |
| H10A | 0.5280 | 0.0278 | 0.2193 | 0.033* | |
| H10B | 0.3280 | 0.0174 | 0.2160 | 0.033* | |
| C11 | 0.4316 (2) | 0.02332 (11) | 0.11175 (11) | 0.0260 (4) | |
| H11A | 0.4399 | −0.0370 | 0.1070 | 0.031* | |
| H11B | 0.5337 | 0.0476 | 0.0905 | 0.031* | |
| C12 | 0.2773 (2) | 0.05349 (10) | 0.06779 (10) | 0.0205 (3) | |
| H12A | 0.1771 | 0.0231 | 0.0840 | 0.025* | |
| H12B | 0.2924 | 0.0421 | 0.0145 | 0.025* | |
| C13 | 0.0709 (2) | 0.13975 (10) | −0.06377 (9) | 0.0161 (3) | |
| H13 | 0.0799 | 0.0791 | −0.0565 | 0.019* | |
| C14 | −0.0879 (2) | 0.15535 (10) | −0.11130 (10) | 0.0200 (3) | |
| H14A | −0.1863 | 0.1318 | −0.0865 | 0.024* | |
| H14B | −0.1060 | 0.2152 | −0.1166 | 0.024* | |
| C15 | −0.0711 (2) | 0.11651 (12) | −0.18795 (10) | 0.0270 (4) | |
| H15A | −0.0682 | 0.0561 | −0.1828 | 0.032* | |
| H15B | −0.1705 | 0.1310 | −0.2190 | 0.032* | |
| C16 | 0.0864 (3) | 0.14489 (13) | −0.22631 (10) | 0.0305 (4) | |
| H16A | 0.0799 | 0.2047 | −0.2356 | 0.037* | |
| H16B | 0.0953 | 0.1167 | −0.2746 | 0.037* | |
| C17 | 0.2405 (2) | 0.12592 (12) | −0.17829 (10) | 0.0246 (4) | |
| H17A | 0.3422 | 0.1453 | −0.2035 | 0.030* | |
| H17B | 0.2503 | 0.0658 | −0.1715 | 0.030* | |
| C18 | 0.2299 (2) | 0.16729 (10) | −0.10279 (9) | 0.0193 (3) | |
| H18A | 0.2282 | 0.2276 | −0.1092 | 0.023* | |
| H18B | 0.3298 | 0.1528 | −0.0720 | 0.023* | |
| C19 | 0.4349 (2) | 0.38768 (11) | 0.85956 (10) | 0.0222 (4) | |
| H19 | 0.3475 | 0.3760 | 0.8970 | 0.027* | |
| Cl1 | 0.01613 (5) | 0.35589 (2) | −0.08323 (2) | 0.01901 (8) | |
| Cl2 | 0.30706 (5) | 0.34441 (2) | 0.04619 (2) | 0.02103 (9) | |
| Cl3 | −0.12314 (5) | 0.11221 (2) | 0.07235 (2) | 0.02052 (8) | |
| Cl4 | 0.54297 (6) | 0.47752 (3) | 0.88609 (3) | 0.03089 (10) | |
| Cl5 | 0.57325 (6) | 0.30361 (3) | 0.85639 (3) | 0.03164 (11) | |
| Cl6 | 0.33498 (7) | 0.40171 (4) | 0.77265 (3) | 0.03799 (12) | |
| P1 | 0.06247 (5) | 0.18613 (2) | 0.02855 (2) | 0.01377 (8) | |
| Ru1 | 0.007356 (16) | 0.324634 (7) | 0.046687 (7) | 0.01356 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0154 (8) | 0.0266 (9) | 0.0251 (9) | 0.0050 (7) | 0.0032 (7) | −0.0038 (7) |
| C2 | 0.0166 (8) | 0.0240 (8) | 0.0279 (9) | −0.0015 (7) | 0.0095 (7) | −0.0046 (7) |
| C3 | 0.0270 (9) | 0.0234 (8) | 0.0190 (8) | 0.0043 (7) | 0.0099 (7) | 0.0008 (7) |
| C4 | 0.0251 (9) | 0.0275 (9) | 0.0170 (8) | 0.0056 (7) | 0.0010 (7) | −0.0062 (7) |
| C5 | 0.0250 (9) | 0.0204 (8) | 0.0253 (9) | 0.0017 (7) | 0.0054 (7) | −0.0085 (7) |
| C6 | 0.0232 (9) | 0.0191 (8) | 0.0260 (9) | 0.0067 (7) | 0.0067 (7) | −0.0004 (7) |
| C7 | 0.0141 (7) | 0.0165 (7) | 0.0173 (8) | 0.0005 (6) | −0.0004 (6) | 0.0005 (6) |
| C8 | 0.0222 (8) | 0.0236 (8) | 0.0176 (8) | 0.0027 (7) | −0.0016 (7) | −0.0017 (7) |
| C9 | 0.0224 (9) | 0.0273 (9) | 0.0213 (9) | −0.0005 (7) | −0.0066 (7) | 0.0002 (7) |
| C10 | 0.0218 (9) | 0.0260 (9) | 0.0344 (10) | −0.0004 (7) | −0.0097 (8) | 0.0074 (8) |
| C11 | 0.0202 (9) | 0.0197 (8) | 0.0378 (10) | 0.0045 (7) | −0.0052 (8) | −0.0008 (8) |
| C12 | 0.0206 (8) | 0.0159 (8) | 0.0248 (9) | 0.0015 (6) | −0.0029 (7) | −0.0004 (7) |
| C13 | 0.0174 (8) | 0.0145 (7) | 0.0165 (8) | −0.0002 (6) | 0.0008 (6) | −0.0024 (6) |
| C14 | 0.0196 (8) | 0.0189 (8) | 0.0213 (8) | −0.0024 (6) | −0.0035 (7) | −0.0011 (6) |
| C15 | 0.0309 (10) | 0.0306 (10) | 0.0192 (9) | −0.0067 (8) | −0.0079 (7) | 0.0000 (7) |
| C16 | 0.0403 (11) | 0.0340 (10) | 0.0171 (9) | −0.0065 (9) | −0.0011 (8) | 0.0025 (8) |
| C17 | 0.0279 (9) | 0.0283 (9) | 0.0178 (8) | 0.0010 (8) | 0.0057 (7) | −0.0024 (7) |
| C18 | 0.0196 (8) | 0.0216 (8) | 0.0168 (8) | 0.0000 (6) | 0.0015 (6) | −0.0015 (6) |
| C19 | 0.0174 (8) | 0.0282 (9) | 0.0210 (8) | 0.0003 (7) | 0.0000 (7) | 0.0001 (7) |
| Cl1 | 0.02313 (19) | 0.01688 (18) | 0.01704 (19) | −0.00012 (15) | 0.00118 (15) | 0.00162 (14) |
| Cl2 | 0.01470 (18) | 0.02085 (19) | 0.0275 (2) | −0.00415 (15) | 0.00011 (16) | −0.00070 (16) |
| Cl3 | 0.01850 (18) | 0.01891 (19) | 0.0243 (2) | −0.00493 (15) | 0.00428 (15) | 0.00082 (15) |
| Cl4 | 0.0287 (2) | 0.0250 (2) | 0.0388 (3) | −0.00293 (18) | −0.0043 (2) | −0.00117 (19) |
| Cl5 | 0.0251 (2) | 0.0273 (2) | 0.0425 (3) | 0.00382 (18) | 0.0000 (2) | −0.0002 (2) |
| Cl6 | 0.0334 (3) | 0.0535 (3) | 0.0266 (2) | 0.0079 (2) | −0.0096 (2) | −0.0028 (2) |
| P1 | 0.01298 (18) | 0.01378 (18) | 0.01459 (19) | −0.00112 (15) | 0.00104 (15) | −0.00075 (15) |
| Ru1 | 0.01285 (6) | 0.01339 (6) | 0.01447 (6) | −0.00015 (5) | 0.00123 (5) | −0.00095 (5) |
Geometric parameters (Å, º)
| C1—C2 | 1.400 (3) | C11—C12 | 1.530 (2) |
| C1—C6 | 1.425 (2) | C11—H11A | 0.9900 |
| C1—Ru1 | 2.1966 (17) | C11—H11B | 0.9900 |
| C1—H1 | 0.9500 | C12—H12A | 0.9900 |
| C2—C3 | 1.420 (3) | C12—H12B | 0.9900 |
| C2—Ru1 | 2.1970 (17) | C13—C18 | 1.530 (2) |
| C2—H2 | 0.9500 | C13—C14 | 1.535 (2) |
| C3—C4 | 1.406 (3) | C13—P1 | 1.8334 (16) |
| C3—Ru1 | 2.1759 (17) | C13—H13 | 1.0000 |
| C3—H3 | 0.9500 | C14—C15 | 1.531 (2) |
| C4—C5 | 1.425 (3) | C14—H14A | 0.9900 |
| C4—Ru1 | 2.1668 (17) | C14—H14B | 0.9900 |
| C4—H4 | 0.9500 | C15—C16 | 1.519 (3) |
| C5—C6 | 1.386 (3) | C15—H15A | 0.9900 |
| C5—Ru1 | 2.2585 (17) | C15—H15B | 0.9900 |
| C5—H5 | 0.9500 | C16—C17 | 1.520 (3) |
| C6—Ru1 | 2.2705 (17) | C16—H16A | 0.9900 |
| C6—H6 | 0.9500 | C16—H16B | 0.9900 |
| C7—C12 | 1.533 (2) | C17—C18 | 1.525 (2) |
| C7—C8 | 1.536 (2) | C17—H17A | 0.9900 |
| C7—P1 | 1.8457 (16) | C17—H17B | 0.9900 |
| C7—H7 | 1.0000 | C18—H18A | 0.9900 |
| C8—C9 | 1.528 (2) | C18—H18B | 0.9900 |
| C8—H8A | 0.9900 | C19—Cl6 | 1.7596 (18) |
| C8—H8B | 0.9900 | C19—Cl4 | 1.7602 (18) |
| C9—C10 | 1.522 (3) | C19—Cl5 | 1.7609 (18) |
| C9—H9A | 0.9900 | C19—H19 | 1.0000 |
| C9—H9B | 0.9900 | Cl1—Ru1 | 2.4036 (4) |
| C10—C11 | 1.522 (3) | Cl2—Ru1 | 2.4110 (4) |
| C10—H10A | 0.9900 | Cl3—P1 | 2.0779 (6) |
| C10—H10B | 0.9900 | P1—Ru1 | 2.3247 (4) |
| C2—C1—C6 | 120.45 (17) | C15—C14—C13 | 110.48 (14) |
| C2—C1—Ru1 | 71.44 (10) | C15—C14—H14A | 109.6 |
| C6—C1—Ru1 | 74.25 (10) | C13—C14—H14A | 109.6 |
| C2—C1—H1 | 119.8 | C15—C14—H14B | 109.6 |
| C6—C1—H1 | 119.8 | C13—C14—H14B | 109.6 |
| Ru1—C1—H1 | 126.4 | H14A—C14—H14B | 108.1 |
| C1—C2—C3 | 119.69 (16) | C16—C15—C14 | 112.07 (15) |
| C1—C2—Ru1 | 71.41 (10) | C16—C15—H15A | 109.2 |
| C3—C2—Ru1 | 70.25 (10) | C14—C15—H15A | 109.2 |
| C1—C2—H2 | 120.2 | C16—C15—H15B | 109.2 |
| C3—C2—H2 | 120.2 | C14—C15—H15B | 109.2 |
| Ru1—C2—H2 | 130.8 | H15A—C15—H15B | 107.9 |
| C4—C3—C2 | 119.52 (17) | C15—C16—C17 | 110.09 (15) |
| C4—C3—Ru1 | 70.76 (10) | C15—C16—H16A | 109.6 |
| C2—C3—Ru1 | 71.86 (10) | C17—C16—H16A | 109.6 |
| C4—C3—H3 | 120.2 | C15—C16—H16B | 109.6 |
| C2—C3—H3 | 120.2 | C17—C16—H16B | 109.6 |
| Ru1—C3—H3 | 129.5 | H16A—C16—H16B | 108.2 |
| C3—C4—C5 | 120.24 (17) | C16—C17—C18 | 111.10 (15) |
| C3—C4—Ru1 | 71.47 (10) | C16—C17—H17A | 109.4 |
| C5—C4—Ru1 | 74.74 (10) | C18—C17—H17A | 109.4 |
| C3—C4—H4 | 119.9 | C16—C17—H17B | 109.4 |
| C5—C4—H4 | 119.9 | C18—C17—H17B | 109.4 |
| Ru1—C4—H4 | 125.7 | H17A—C17—H17B | 108.0 |
| C6—C5—C4 | 119.95 (17) | C17—C18—C13 | 110.17 (14) |
| C6—C5—Ru1 | 72.66 (10) | C17—C18—H18A | 109.6 |
| C4—C5—Ru1 | 67.75 (9) | C13—C18—H18A | 109.6 |
| C6—C5—H5 | 120.0 | C17—C18—H18B | 109.6 |
| C4—C5—H5 | 120.0 | C13—C18—H18B | 109.6 |
| Ru1—C5—H5 | 132.5 | H18A—C18—H18B | 108.1 |
| C5—C6—C1 | 119.83 (17) | Cl6—C19—Cl4 | 110.13 (10) |
| C5—C6—Ru1 | 71.71 (10) | Cl6—C19—Cl5 | 110.11 (10) |
| C1—C6—Ru1 | 68.61 (9) | Cl4—C19—Cl5 | 110.69 (9) |
| C5—C6—H6 | 120.1 | Cl6—C19—H19 | 108.6 |
| C1—C6—H6 | 120.1 | Cl4—C19—H19 | 108.6 |
| Ru1—C6—H6 | 132.6 | Cl5—C19—H19 | 108.6 |
| C12—C7—C8 | 111.62 (14) | C13—P1—C7 | 104.70 (7) |
| C12—C7—P1 | 114.00 (11) | C13—P1—Cl3 | 98.49 (5) |
| C8—C7—P1 | 111.07 (11) | C7—P1—Cl3 | 100.29 (6) |
| C12—C7—H7 | 106.5 | C13—P1—Ru1 | 122.65 (5) |
| C8—C7—H7 | 106.5 | C7—P1—Ru1 | 115.50 (5) |
| P1—C7—H7 | 106.5 | Cl3—P1—Ru1 | 111.79 (2) |
| C9—C8—C7 | 111.31 (14) | C4—Ru1—C3 | 37.77 (7) |
| C9—C8—H8A | 109.4 | C4—Ru1—C1 | 80.04 (7) |
| C7—C8—H8A | 109.4 | C3—Ru1—C1 | 67.77 (7) |
| C9—C8—H8B | 109.4 | C4—Ru1—C2 | 68.03 (7) |
| C7—C8—H8B | 109.4 | C3—Ru1—C2 | 37.89 (7) |
| H8A—C8—H8B | 108.0 | C1—Ru1—C2 | 37.15 (7) |
| C10—C9—C8 | 110.91 (15) | C4—Ru1—C5 | 37.51 (7) |
| C10—C9—H9A | 109.5 | C3—Ru1—C5 | 67.19 (7) |
| C8—C9—H9A | 109.5 | C1—Ru1—C5 | 66.15 (7) |
| C10—C9—H9B | 109.5 | C2—Ru1—C5 | 78.67 (7) |
| C8—C9—H9B | 109.5 | C4—Ru1—C6 | 66.48 (7) |
| H9A—C9—H9B | 108.0 | C3—Ru1—C6 | 78.92 (7) |
| C11—C10—C9 | 110.42 (15) | C1—Ru1—C6 | 37.15 (6) |
| C11—C10—H10A | 109.6 | C2—Ru1—C6 | 66.53 (7) |
| C9—C10—H10A | 109.6 | C5—Ru1—C6 | 35.63 (7) |
| C11—C10—H10B | 109.6 | C4—Ru1—P1 | 115.27 (5) |
| C9—C10—H10B | 109.6 | C3—Ru1—P1 | 90.82 (5) |
| H10A—C10—H10B | 108.1 | C1—Ru1—P1 | 121.93 (5) |
| C10—C11—C12 | 112.06 (15) | C2—Ru1—P1 | 94.15 (5) |
| C10—C11—H11A | 109.2 | C5—Ru1—P1 | 152.59 (5) |
| C12—C11—H11A | 109.2 | C6—Ru1—P1 | 159.01 (5) |
| C10—C11—H11B | 109.2 | C4—Ru1—Cl1 | 150.91 (5) |
| C12—C11—H11B | 109.2 | C3—Ru1—Cl1 | 155.05 (5) |
| H11A—C11—H11B | 107.9 | C1—Ru1—Cl1 | 89.32 (5) |
| C11—C12—C7 | 111.57 (14) | C2—Ru1—Cl1 | 117.20 (5) |
| C11—C12—H12A | 109.3 | C5—Ru1—Cl1 | 113.53 (5) |
| C7—C12—H12A | 109.3 | C6—Ru1—Cl1 | 88.76 (5) |
| C11—C12—H12B | 109.3 | P1—Ru1—Cl1 | 93.366 (15) |
| C7—C12—H12B | 109.3 | C4—Ru1—Cl2 | 91.20 (5) |
| H12A—C12—H12B | 108.0 | C3—Ru1—Cl2 | 119.52 (5) |
| C18—C13—C14 | 112.01 (14) | C1—Ru1—Cl2 | 151.36 (5) |
| C18—C13—P1 | 110.18 (11) | C2—Ru1—Cl2 | 157.35 (5) |
| C14—C13—P1 | 113.20 (11) | C5—Ru1—Cl2 | 90.37 (5) |
| C18—C13—H13 | 107.0 | C6—Ru1—Cl2 | 114.49 (5) |
| C14—C13—H13 | 107.0 | P1—Ru1—Cl2 | 86.505 (14) |
| P1—C13—H13 | 107.0 | Cl1—Ru1—Cl2 | 85.306 (15) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14B···Cl1 | 0.99 | 2.56 | 3.4080 (17) | 144 |
| C18—H18A···Cl1 | 0.99 | 2.74 | 3.5366 (17) | 138 |
| C19—H19···Cl1i | 1.00 | 2.69 | 3.5539 (17) | 144 |
| C19—H19···Cl2i | 1.00 | 2.77 | 3.6119 (18) | 142 |
Symmetry code: (i) x, y, z+1.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: IS5363).
References
- Bruker (2008). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2009). SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2011). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Jantscher, F., Kirchner, K. & Mereiter, K. (2009). Acta Cryst. E65, m941. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Torres-Lubián, R., Rosales-Hoz, M. J., Arif, A. M., Ernst, R. D. & Paz-Sandoval, M. A. (1999). J. Organomet. Chem. 585, 68–82.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S1600536814012975/is5363sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814012975/is5363Isup2.hkl
CCDC reference: 1006720
Additional supporting information: crystallographic information; 3D view; checkCIF report



