Abstract
Objectives
mRNA for serotonin 2C receptor (5-HT2CR) undergoes editing which results in numerous isoforms. More highly edited isoforms exhibit decreased function. We recently found greater 5-HT2CR editing in suicide victims with prior bipolar disorder (BPD) or schizophrenia (SZ) compared with non-suicide patients and normal controls (NC). This study compares suicides and non-suicides with major depressive disorder (MDDSuic and MDDNoSuic) and non-suicide NC.
Methods
mRNA editing was assessed in prefrontal cortex of 24 MDDSuic, 21 MDDNoSuic, and 56 NC using next generation sequencing. mRNA expression of 5-HT2CR and editing enzymes (ADAR1-2) was assessed by real-time PCR.
Results
Editing was lower in MDDNoSuic than in MDDSuic, which did not differ from NC. No differences in the 5-HT2CR or ADAR1 expression were detected. ADAR2 expression was higher in NC than in MDD subjects, but did not differ between MDDNoSuic and MDDSuic.
Conclusions
Our findings suggest the presence of two factors associated with 5-HT2CR editing. One factor, which probably stems from decreased ADAR2 expression, is linked to MDD and is associated with less editing. The other, seen also in our previous study of suicide in BP and SZ, is linked to suicide alone and is associated with more editing and, therefore, less receptor function.
Keywords: RNA editing, ADAR, serotonin receptor, major depressive disorder, suicide
Introduction
The lives of at least 5%, and perhaps 15% or more, of people with major depressive disorder (MDD), bipolar disorder (BPD), schizophrenia (SZ) and borderline personality disorder end in suicide (Hawton and Van Heeringen 2009). Conversely, over 90% of suicide victims meet DSM criteria for at least one psychiatric illness, most commonly MDD (Cavanagh et al. 2003). Diagnosis and treatment of depression are pivotal to preventing suicide. However, the response to antidepressant medications is typically slow and frequently inadequate. Furthermore, although antidepressant treatments decrease risk for suicide, the mechanism is unclear. Neuronal effects, such as enhanced expression of BDNF (Chen et al. 2001) and increased hippocampal neurogenesis (Boldrini et al. 2009), are believed to mediate antidepressant activity, but these effects have been observed largely in treated individuals who committed suicide.
The serotonin 2C receptor (5-HT2CR) warrants consideration as a target for novel antidepressant and anxiolytic treatments that may help to prevent suicidal behavior. It contributes to the control of mood, sleep, appetite, motor activity, endocrine secretion and sexual function (Serretti et al. 2004). Animal and in vitro studies suggest a therapeutic potential of 5-HT2CR ligands for treatment of mood and anxiety disorders (Serretti et al. 2004; Millan 2005). Direct activation of 5-HT2CR increases anxiety-like behaviors in humans and animals (Gatch 2003), and its indirect activation by specific serotonin reuptake inhibitors (SSRIs) may contribute to the transient anxiogenic effects of these compounds (Bagdy et al. 2001). Many antidepressants, including several SSRIs and tricyclic antidepressants, also act directly on the 5-HT2CR (Palvimaki et al. 1996; Chanrion et al. 2008); the clinical implications of this are unclear. Chronic administration of SSRIs can desensitize 5-HT2CR, and both desensitization and blockade of 5-HT2CRs can increase dopaminergic and noradrenergic activity in the brain (Di Giovanni et al. 1999; Bristow et al. 2000). Thus, inhibition of 5-HT2CR may be key to the antidepressant effect of SSRIs (Esposito 2006), or it may limit their suppression of suicide, or both.
Recent studies implicate increased editing of 5-HT2CR mRNA in suicide (Niswender et al. 2001; Gurevich et al. 2002; Iwamoto and Kato 2003; Akbarian 2008; Dracheva et al. 2008). RNA editing is a post-transcriptional process that can be broadly defined as any site-specific alteration in an RNA sequence (Gott and Emeson 2000). The most prevalent type of human RNA editing converts adenosine residues into inosine in double-stranded (ds) RNAs (A-to-I editing) through catalysis by specific editing enzymes, adenosine deaminases acting on RNA (ADAR1 and ADAR2) (Bass 2002; Valente and Nishikura 2005). A-to-I editing most frequently targets repetitive RNA sequences located within introns and 5′ or 3′ untranslated regions (UTRs); however, the biological significance of noncoding RNA editing remains largely undetermined (Nishikura 2010). Only a limited number of protein-coding pre-mRNA transcripts are known to undergo editing, which results in recoding and subsequent alterations of function. Most of these transcripts encode proteins that are involved in neurotransmission (Gott and Emeson 2000). The 5-HT2CR mRNA can be edited at five closely-spaced adenosine residues [A, B, E (also known as C'), C, and D sites] that can alter codons for three amino acids in the putative second intracellular loop of the receptor, a region involved in coupling to G-proteins (Burns et al. 1997; Werry et al. 2008). Combinatorial editing at these five positions can generate up to 32 mRNA variants encoding 24 different receptor isoforms. The unedited Ile156-Asn158-Ile160 (INI) isoform possesses considerable constitutive and agonist-stimulated activity. In contrast, when the 5-HT2CR is edited, its coupling to G-proteins and its affinity for serotonin are drastically reduced. The extent of editing correlates with 5-HT2CR functional activity: more highly edited isoforms exhibit the least function (Fitzgerald et al. 1999; Herrick-Davis et al. 1999; Niswender et al. 1999; Wang et al. 2000b; Quirk et al. 2001; Berg et al. 2001; Marion et al. 2004).
We recently found greater 5-HT2CR editing in the dorsolateral prefrontal cortex (DLPFC) of suicide victims who had suffered from BPD or SZ than in subjects with the same diagnoses who died by other means (Dracheva et al. 2008). This suggested that enhanced levels of 5-HT2CR editing are associated specifically with suicide. Most commonly, however, suicide occurs in the context of depression. We undertook the present study to compare suicides and non-suicides with MDD. Similar to our previous study, we detected more editing of 5-HT2CR in suicide than in non-suicide MDD subjects. Thus, in three major psychiatric diseases that comprise some 75% of suicides, higher levels of editing are consistently associated with suicide. In non-psychiatric comparison subjects, editing levels were similar to those in suicides with MDD and non-suicides with SZ or BPD. Thus, regardless of the contributions of underlying disease, suicide is probably associated with lower function of 5-HT2CR in the DLPFC.
Methods
Specimens
Specimens from the DLPFC (BA9) were from the Stanley Medical Research Institute (SMRI; depression/suicide cohort and Array Collection; total RNA preparations) and the Maryland Brain Collection (MBC; tissue samples). These were derived from subjects with MDD who died by suicide (MDDSuic) or by other means (MDDNoSuic) and from non-suicide, psychiatrically normal comparison subjects (NC) (Table I; Supplementary Table 1 available online). Details of the procedures that were employed in these Brain Banks for tissue collection, neuropathologic examination (to rule out degenerative and neurologic disease), and retrospective clinical diagnoses based on DSM-IV criteria have been described elsewhere (Torrey et al. 2000; Pandey et al. 2007). NC comprised subjects without a history of serious mental illness, except that substance abuse or dependence were not exclusionary. Eight to nine subjects in each diagnostic group had a history of moderate to heavy use, abuse, or dependence on alcohol, illicit drugs, or both (Table I). Antidepressant and antipsychotic medications had been prescribed, respectively, to 17 and seven of 20 MDDNoSuic, 15 and four of 17 MDDSuic, and one and none of 55 NC. The medication histories of the remaining subjects are unknown; among these, toxicology detected antidepressants in one NC, one MDDNoSuic and two MDDSuic cases, but no additional cases with antipsychotics. Dates of medication are unknown, but postmortem toxicological examinations detected prescribed antidepressant drugs in 11 of 14 tested subjects. On the other hand, antipsychotic drugs were detected in only two of five subjects tested to whom they were prescribed, and in one of these, the detected drug was not the same as the one reportedly prescribed.
Table I.
Demographic information of the study cohort. PMI, pH, and age at death are shown as Mean ± SEM.
| Diagnosis/ Manner of death |
Cohort | Subjects number |
Age at death (years) |
PMI (hr) | Brain pH | Sex | Race | Number of subjects with | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Antidepressants* | Alcohol use** | Drug use*** | Smoking | ||||||||
| NC | SMRI (Array Collection) | 34 | 43.8 ± 1.3 | 29.5 ± 2.2 | 6.60 ± 0.05 | 25M; 9F | 34W | 0 | 2 | 1 | 5 |
| SMRI; (Depression /SuicideCohort) | 11 | 47.1 ± 3.8 | 25.1 ± 3.4 | 6.62 ± 0.05 | 7 M;4 F | 10W; 1 H | 1 | 4 | 1 | 4 (4 UK) | |
| MBC | 11 | 50.6 ± 5.0 | 18.2 ± 1.3 | 6.66 ± 0.06 | 7 M;4 F | 10W; 1 B | 1 | 2 | 0 | 1 | |
| Total | 56 | 45.8 ± 1.5 | 26.5 ± 1.6 | 6.62 ± 0.03 | 39M;17F | 54W; 1B; 1H | 2 | 8 | 2 | 10 (4 UK) | |
| MDDNoSuic | SMRI; (Depression /Suicide Cohort) | 11 | 44.8 ± 2.3 | 32.6 ± 5.9 | 6.65 ± 0.03 | 7 M;4 F | 11W | 9 | 3 (1 UK) | 2 (1 UK) | 4 (5 UK) |
| MBC | 10 | 44.3 ± 6.3 | 17.0 ± 2.8 | 6.64 ± 0.18 | 6M;4 F | 7W; 3 B | 10 | 3 | 2 | 2 | |
| Total | 21 | 44.6 ± 3.2 | 25.2 ± 3.7 | 6.65 ± 0.06 | 13M; 8F | 18W; 3B | 19 | 6(1 UK) | 4 (1 UK) | 6 (5 UK) | |
| MDDSuic | SMRI; (Depression /Suicide Cohort) | 11 | 41.9 ± 3.4 | 27.5 ± 4.2 | 6.67 ± 0.05 | 7 M; 4 F | 10W; 1 H | 9 | 3 (1 UK) | 2 (1 UK) | 2 (6 UK) |
| MBC | 13 | 44.2 ± 5.3 | 21.8 ± 1.0 | 6.60 ± 0.06 | 9 M; 4 F | 13W | 9 (4 UK) | 4 | 2 (1 UK) | 3 | |
| Total | 24 | 43.2 ± 3.2 | 24.5 ± 2.1 | 6.64 ± 0.04 | 16M; 8F | 23W; 1H | 18 | 7 (1 UK) | 3 (2 UK) | 5 (6 UK) | |
UK, unknown; M, male; F, female; W, white; H, Hispanic; B, Black.
Prescribed medications or revealed by toxicology.
Heavy use (past or present) for SMRI; abuse/dependence for MBC.
Moderate to heavy use (past or present) for SMRI; abuse/dependence for MBC.
RNA and cDNA
Total RNA was extracted from the MBC tissue samples using Ambion ToTally RNA Kit (Applied Biosystems). Only high quality RNA samples that yielded RNA integrity numbers (RINs) ≥ 6.0 by Agilent Bioanalyzer were used in the study The average RIN for the MBC cohort was 7.7 ± 0.1 (Mean ± SEM). Although we did not have individual RIN values for the SMRI cases, poor RNA quality (based on assessment with Bioanalyzer) is one of the exclusion criteria for the SMRI specimens. cDNA was synthesized using High Capacity cDNA Reverse Transcription (RT) kit (Applied Biosystems) and equal quantities of RNA from each subject (1 μg of RNA per 10 μl of RT reaction).
Analysis of 5-HT2CR editing
cDNA from each of the 101 subjects was used as a template to amplify by PCR the region that contains all five editing sites (region of editing) as described previously (Dracheva et al. 2008), except that Next Generation Sequencing (NGS) was employed. See Supplementary material available online for details.
Analysis of 5-HT2CR and ADARs mRNA expression
mRNA expression of the 5-HT2CR, ADAR1, and ADAR2 splicing variants was measured by quantitative real-time PCR (qPCR) mostly as described (Dracheva et al. 2009). See Supplementary methods, Supplementary Table 3, and Supplementary Figure 1, all available online, for details.
Statistical analysis
All statistical analyses were performed using SAS Version 9.2.
Editing
For each subject, the frequencies of all 32 5-HT2CR mRNA editing variants were calculated as a proportion of the number of NGS reads for this variant to the total number of NGS reads obtained for this subject. The efficiency of editing at each editing site (A, B, E, C, or D) was computed as the sum of frequencies of all variants that were edited at this site. The frequency of editing at the E or C site was computed as the sum of frequencies of all variants that were edited at either site or both.
Frequencies for each of the six most common editing variants (ABCD, ABD, NONE, A, AD, ACD) and frequency of editing at the E or C sites were then compared among the three study groups using a separate Analysis of Covariance (ANCOVA) model for each variant. Five possible covariates were considered: age, sex, PMI, race and cohort (Table I). Mallows' Cp was used to select the best model involving a subset of these covariates (Lance 2005).
5-HT2CR editing sites are closely spaced, and it has been determined that in rodent brain editing at one site infl uenced the observed frequency of editing at other sites (Du et al. 2006; Enstero et al. 2009). Similarly, our analysis in the human DLPFC showed that editing at each of the five sites is interdependent on editing at each of the other sites (see Results). Therefore, when analyzing the editing efficiencies, a single analysis was done using a repeated measures model that incorporated efficiencies over all sites (A, B, E, C, or D). In this analysis, there was one outcome measure — efficiency — with five levels corresponding to the five editing sites. The analysis was performed using a random-intercept model with unstructured covariance and denominator degrees of freedom computed with formulas detailed by Kenward and Roger (1997).
Among all possible covariates, PMI and cohort were selected and retained as covariates in all statistical modeling of editing parameters. To control for multiple comparisons of the three study groups made within each ANCOVA and the repeated measures model, a Tukey–Kramer adjustment was made.
Gene expression
The differences in the relative expression of the 5-HT2CR and ADARs splicing variants among the study groups were analyzed by ANCOVA with five possible covariates (see above) considered for inclusion. Mallow's Cp model selection criteria revealed age and cohort, and age and PMI as significant covariates for 5-HT2CR and ADARs analyses, respectively. Those covariates were included in the corresponding ANCOVA models.
Comparison between the traditional and NGS methods
NGS analysis of the specimens from 34 NC subjects (SMRI Array Collection) was compared with our earlier results using traditional editing analysis (∼ 45 clones per subject) with the same subjects and brain region (Dracheva et al. 2008). For each of the detected mRNA variants, the mean and standard deviation of its frequencies by the two measures were compared by paired t-test and Wilcoxon signed rank test for paired data, respectively.
Results
Comparison between NGS and the traditional method of editing analysis
Although the traditional cloning and sequencing method is assumed to provide unambiguous results (Burns et al. 1997; Sodhi et al. 2005), this method relies on sampling of a limited population of cloned transcripts (usually 20–100), and is thus prone to random effects that may obscure differences between experimental groups. NGS technology has recently been used for 5-HT2CR editing measurements in the rodent brain (Morabito et al. 2010; Abbas et al. 2010). In order to explore the potential of the NGS method for assessing 5-HT2CR editing in human postmortem brain, we compared the NGS analysis of specimens from 34 NC subjects from the SMRI Array Collection with our earlier results using the traditional approach with the same subjects and brain region (Dracheva et al. 2008).
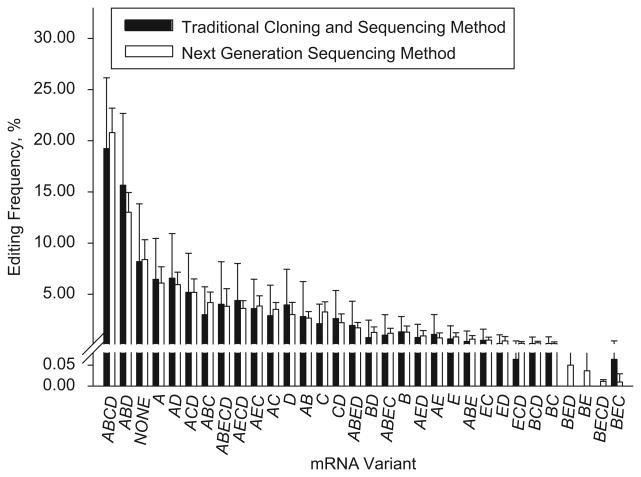
The results of both methods are shown in Figure 1. The NGS approach detected all 32 mRNA variants, compared with only 29 by the traditional method. While no differences between the means of editing frequencies obtained by both methods were detected, the standard deviations were significantly smaller for NGS (P< 0.0001). The average coefficient of variation (CV) of the frequencies in the NGS analysis was approximately one-third that of the traditional method (Mean CVs were 0.56 and 1.60 for NGS and the traditional method, respectively).
Figure 1.
Comparison of the 5-HT2CR mRNA editing analyses performed by the traditional and NGS methods. Shown are Means ± SDs of frequencies in the postmortem DLPFC samples from 34 NC (SMRI Array Collection). 45.7 ± 0.3 clones vs. 733,133.0 ± 42,419.0 reads were analyzed by the traditional and NGS methods, respectively.
Overall analysis of RNA editing
Since the NGS method clearly demonstrated an advantage over the traditional approach, we used NGS to assess the 5-HT2CR editing in the DLPFC. On average, 561,291 ± 55,851 (Mean ± SE) reads were obtained for each subject, thus providing reliable (with 99% probability) detection of even the rarest editing variants, with abundance of < 0.001% (Morabito et al. 2010). The mean values of frequencies of the mRNA variants and efficiencies of editing at each of the five editing sites within each of the study groups (NC, MDDSuic, MDDNoSuic) are shown in Table II and in Supplementary Table 2, respectively In addition, because previous studies suggested that editing at the E or C sites by themselves or in combination with other sites is sufficient to markedly reduce 5-HT2CR function (Niswender et al. 1999; Wang et al. 2000b; Tohda et al. 2010), we computed and analyzed frequencies of editing at the E or C sites. As in our earlier studies of human DLPFC (Dracheva et al. 2008), only six of the variants, which contained editing combinations ABCD, ABD, NONE, A, AD, and ACD, were observed at ∼5% or greater frequency in all three study groups (Table II). Taken together these six variants constituted the majority of the 5-HT2CR transcripts (∼60%).
Table II.
Frequencies for the observed 5-HT2CR mRNA variants. Shown are Means ± SEM. The frequencies of the variants that were significantly different among the groups are highlighted.
| mRNA variant | NC (N = 56) | MDDNoSuic (N = 21) | MDDSuic (N = 24) |
|---|---|---|---|
| ABCD | 21.24 ± 0.31 | 19.85 ± 0.77 | 22.26 ± 0.66 |
| ABD | 13.04 ± 0.22 | 13.08 ± 0.38 | 12.80 ± 0.45 |
| None | 8.18 ± 0.23 | 8.80 ± 0.61 | 7.29 ± 0.25 |
| A | 5.82 ± 0.19 | 6.27 ± 0.27 | 6.40 ± 0.82 |
| AD | 5.67 ± 0.15 | 6.28 ± 0.34 | 5.27 ± 0.23 |
| ACD | 5.15 ± 0.15 | 5.26 ± 0.16 | 5.02 ± 0.21 |
| ABC | 4.30 ± 0.13 | 4.41 ± 0.20 | 4.61 ± 0.20 |
| ABECD | 3.89 ± 0.22 | 3.37 ± 0.40 | 3.48 ± 0.34 |
| AECD | 3.88 ± 0.10 | 3.78 ± 0.18 | 3.99 ± 0.20 |
| AEC | 3.84 ± 0.12 | 3.76 ± 0.15 | 4.05 ± 0.19 |
| AC | 3.48 ± 0.08 | 3.51 ± 0.14 | 3.39 ± 0.16 |
| C | 3.19 ± 0.11 | 3.10 ± 0.16 | 3.24 ± 0.40 |
| D | 3.00 ± 0.13 | 3.61 ± 0.28 | 2.97 ± 0.18 |
| AB | 2.81 ± 0.09 | 2.71 ± 0.13 | 2.80 ± 0.14 |
| CD | 2.20 ± 0.10 | 2.03 ± 0.14 | 2.01 ± 0.10 |
| ABED | 1.73 ± 0.06 | 1.45 ± 0.09 | 1.69 ± 0.10 |
| BD | 1.38 ± 0.06 | 1.48 ± 0.10 | 1.42 ± 0.08 |
| B | 1.27 ± 0.06 | 1.42 ± 0.14 | 1.18 ± 0.07 |
| ABEC | 1.21 ± 0.06 | 1.14 ± 0.10 | 1.13 ± 0.08 |
| AED | 0.87 ± 0.06 | 0.85 ± 0.06 | 0.81 ± 0.05 |
| E | 0.78 ± 0.05 | 0.81 ± 0.07 | 0.78 ± 0.08 |
| AE | 0.74 ± 0.05 | 0.81 ± 0.09 | 0.76 ± 0.05 |
| ABE | 0.60 ± 0.04 | 0.56 ± 0.06 | 0.60 ± 0.04 |
| EC | 0.55 ± 0.04 | 0.56 ± 0.05 | 0.91 ± 0.04 |
| ED | 0.40 ± 0.05 | 0.25 ± 0.03 | 0.33 ± 0.04 |
| BCD | 0.26 ± 0.02 | 0.30 ± 0.04 | 0.24 ± 0.02 |
| ECD | 0.23 ± 0.02 | 0.22 ± 0.03 | 0.27 ± 0.03 |
| BC | 0.20 ± 0.02 | 0.25 ± 0.03 | 0.21 ± 0.03 |
| BE | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.03 ± 0.01 |
| BED | 0.04 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 |
| BEC | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| BECD | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| ★E or C | 58.82 ± 0.52 | 56.35 ± 1.09 | 59.87 ± 0.48 |
Computed as a sum of frequencies of all variants that were edited at either E or C site or both.
Interdependence of editing at different sites
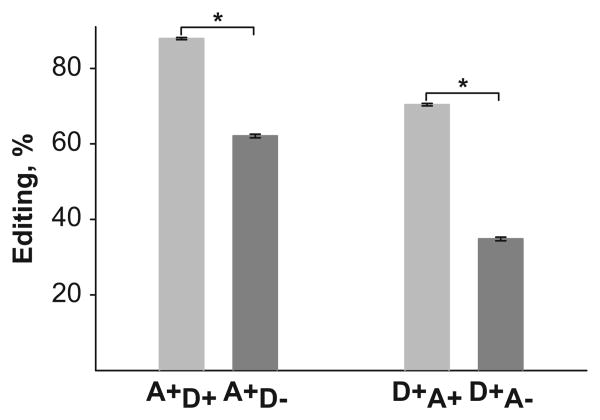
Because of the previous findings in rodents (Du et al. 2006; Enstero et al. 2009), and because all five 5-HT2CR editing sites are situated in close proximity to each other, we asked if editing at one site could influence the frequency of editing at each of the other four sites in the human DLPFC. For each subject, we calculated the percent of editing at each site in the context of +/− editing at each of the other four sites as described (Du et al. 2006). For example, A+ D+ is the percent of the A sites edited in the context of the D site edited, i.e. the ratio of A+ D+ to (A+ D+ + A− D+), where (+) denotes an editing event (from A to I residue) and (–) denotes an absence of an editing event (a genomic A residue). Similarly, A+ D− is the ratio of A+ D− to (A+ D− + A− D−). When the entire cohort (N = 101) was analyzed, a strong interdependence of editing at different pairs of sites was observed (Supplementary Table 4). The data indicated that editing at any site is dependent on editing at any other site (all t values ≥ 10.76, all P values ≤ 2.16 × 10−18, paired Student t-test). For example, editing at the A site is 87.90% when site D is edited, and is significantly lower (62.09%) when site D is not edited. Similarly, editing at the D site is 70.39% when site A is edited and only 34.83% when site A is not edited (Figure 2). Negative correlations between editing at different sites were also observed (negative Δs is Supplementary Table 4). For example, editing at the B site is 38.3% when site E is edited; however, it reaches 54.79% when site E is not edited. Similarly, editing at the E site is 13.78% when site B is edited and is significantly higher (23.79%) when site B is not edited. The most striking finding was a strong interdependence between the A and D sites, because it has been established that these two sites are edited by different enzymes (ADAR1 vs. ADAR2). A similar relationship was also observed in rodent brain (Enstero et al. 2009). Comparable results were obtained when interdependence of editing at different pairs of sites was analyzed separately for subjects from three different study groups (NC, N = 56; MDDNoSuic, N = 21; MDDSuic, N = 24; all t values ≥ 3.62, all p values < 0.0015).
Figure 2.
Interdependence of editing between A and D sites. (+) denotes an editing event and (−) denotes an absence of an editing event. Subscripts refer to the editing status of the context site, i.e. A+ D+ is the ratio of A+ D+ to (A+ D+ + A− D+), where edited D site is the context site. Shown is an analysis for the entire cohort (N = 101). The results demonstrate that editing at the A site is dependent on the status of editing at the D site and vice versa. *Difference between editing frequencies at a site when the status of editing at another site is (+) or (−) (P values < 10−86, by paired Student t-test).
Comparison of editing among study groups
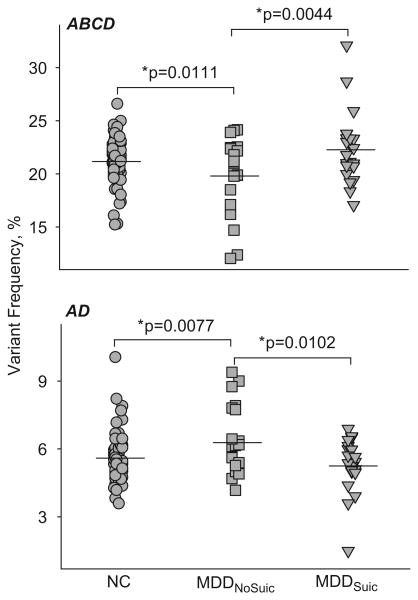
ANCOVA demonstrated that 5-HT2CR editing differed significantly among the groups (Table III). Specifically, the sum of frequencies of all transcripts edited at the E or C sites, and the frequency of the most abundant mRNA variant, ABCD (Figure 3), which collectively represent a population of the low-functioning receptor isoforms (Niswender et al. 1999; Wang et al. 2000b; Tohda et al. 2010), were lower in MDDNoSuic compared with NC and MDDSuic. Similarly, editing efficiency at sites A, B, E, C, and D (analyzed as repeated measures) was lower in MDDNoSuic compared with two other groups. In contrast, the frequencies of the nonedited (NONE) variant and the AD variant that encode high-functioning receptor isoforms (Niswender et al. 1999; Wang et al. 2000b; Tohda et al. 2010) were higher in MDDNoSuic than in NC or MDDSuic (Figure 3). There were no significant differences between NC and MDDSuic. Frequencies of other transcripts did not differ among the groups.
Table III.
Analyses of editing parameters by ANCOVA. PMI and Cohort (SMRI-Array Collection, SMRI-Depression /Suicide Cohort, or MBC) were used as covariates. Significant P values are shown in bold.
| Editing Parameters | Comparison among three groups | MDDSuic vs. MDDNoSuic | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| F | df | P | MDDNoSuic vs. NC | P (Tukey–Kramer adjusted) | MDDSuic vs. NC | |
| *A,B,C,D,E | 4.99 | 2,93 | 0.009 | 0.011 | 0.041 | 0.835 |
| E or C | 7.38 | 2,93 | 0.001 | 0.004 | 0.003 | 0.999 |
| ABCD | 6.44 | 2,93 | 0.002 | 0.011 | 0.004 | 0.970 |
| ABD | 0.18 | 2,93 | 0.8317 | 0.971 | 0.929 | 0.820 |
| NONE | 5.05 | 2,93 | 0.008 | 0.094 | 0.007 | 0.608 |
| A | 1.54 | 2,93 | 0.220 | 0.335 | 0.985 | 0.245 |
| AD | 6.08 | 2,93 | 0.003 | 0.008 | 0.010 | 0.983 |
| ACD | 0.37 | 2,93 | 0.693 | 0.791 | 0.697 | 0.9892 |
Analyzed as repeated measures.
Figure 3.
Distribution of frequencies of the low-functioning ABCD and high-functioning AD 5-HT2CR mRNA variants among the study groups. Upper panel, the frequency of ABCD was lower in MDDNoSuic compared with NC or MDDSuic, but did not differ between NC and MDDSuic. Lower panel, the frequency of AD was higher in MDDNoSuic compared with NC or MDDSuic, but did not differ between NC and MDDSuic.
The variety of antidepressants (including serotoninselective and mixed serotonin and catecholamine re-uptake inhibitors) was too great, and the number of medication-naïve MDD subjects too small, for meaningful subgroup analysis (Supplementary Table 1). However, by history and by toxicology, antidepressant use was not different between the MDDSuic and the MDDNoSuic. Alcohol and illicit drug use was more prominent in both MDD groups compared to NC, but did not differ between the two MDD groups. The history of smoking did not differ among the three study groups.
5-HT2C R mRNA expression
Two TaqMan assays were employed to detect each of the two 5-HT2CR mRNA splice variants that are expressed in the DLPFC — 5-HT2CRsp1 and 5-HT2CRsp2 — that result from splicing at the alternative (sp1) and regular (sp2) splice sites, respectively (see Supplementary methods). The alternative splice variant, 5-HT2CRsp1, does not contain the region of editing (Canton et al. 1996; Flomen et al. 2004). We found no significant differences among the three study groups (all P values ≥ 0.130).
ADAR mRNA expression
ADAR1 and ADAR2 are alternatively spliced, generating multiple mRNA variants (see Supplementary methods and Supplementary Figure 1). Because of the large distances between different ADARs' transcription initiation and splicing sites, it is unachievable to generate qPCR assays for each individual transcript. Thus, we attempted to obtain a complete analysis of the ADARs' transcription by using a number of pre-designed (Applied Biosystems) and custom TaqMan assays (Supplementary Table 3). We found no significant differences between MDDNoSuic and MDDSuic groups for any of the ADAR1 or ADAR2 expression assays (Supplementary Table 5). Therefore, these two groups were combined for comparisons between MDD and NC. The results are summarized in Table IV. Although significant differences between MDD and NC were detected with an assay that measures the combined expression of variants 1, 2, 4, and 5 of ADAR1, we found no differences between groups by other ADAR1 assays (combined 1, 4, and 5, combined 2 and 3, variant 4, combined 1, 2, 3, 4, and 5). MDD and NC differed significantly by ADAR2 assays that measure combined 1 and 4 and combined 2 and 3 variants (Table IV). In both assays, higher ADAR2 expression was detected in NC than in MDD.
Table IV.
Analysis of ADAR1 and ADAR2 mRNA expression by ANCOVA. Comparison between diagnoses (NC vs. MDD) is presented. PMI and age were used as covariates. The expression was assessed using TaqMan assays described in Supplementary Methods, Supplementary Table 3, and Supplementary Figure 1, all available online. The expression of target transcripts was normalized to geometric mean of five endogenous controls (B2M, GUSB, PPIA, RPLPO, HPRT1). Shown are Mean ± SEM of relative expression values. Significant differences (P < 0.05) are in bold face.
| Assay | Relative mRNA Expression NC (N = 55) | Relative mRNA Expression MDD (N = 45) | F | df | P |
|---|---|---|---|---|---|
| ADAR1 V1, 2, 3, 4, 5 | 1.09 ± 0.02 | 1.14 ± 0.03 | 1.73 | 96 | 0.191 |
| ADAR1 V1, 2, 4, 5 | 1.01 ± 0.01 | 0.96 ± 0.01 | 2.76 | 96 | 0.007 |
| ADAR1 V1, 4, 5 | 0.94 ± 0.02 | 0.92 ± 0.02 | 0.63 | 96 | 0.532 |
| ADAR1 V2, 3 | 1.03 ± 0.03 | 0.97 ± 0.04 | 1.19 | 96 | 0.239 |
| ADAR1 V4 | 1.05 ± 0.02 | 1.07 ± 0.02 | 0.55 | 96 | 0.584 |
| ADAR2 V1, 4 | 1.20 ± 0.03 | 0.99 ± 0.03 | 4.66 | 96 | 0.000 |
| ADAR2 V2, 3 | 1.17 ± 0.03 | 1.03 ± 0.03 | 3.88 | 96 | 0.000 |
Discussion
In this study, we compared 5-HT2CR RNA editing in the DLPFC of subjects with MDD who died by suicide, subjects with MDD who died by other means, and non-suicide NC subjects. We used NGS technology on the Illumina platform to provide quantitative estimates of variant transcript frequencies and efficiencies of editing at the five editing sites. The NGS technology has recently been used for 5-HT2CR editing measurements in the rodent brain (Abbas et al. 2010; Morabito et al. 2010), but it has never been tested in human postmortem tissue. We initially compared the NGS approach to the most commonly used low-throughput traditional approach (sequencing of 45 independent clones) in 34 human postmortem specimens from the DLPFC. The NGS approach showed substantially increased precision and sensitivity of the measurements, thus facilitating an accurate comparison of editing data among different study groups. In addition, significantly smaller coefficients of variation in editing frequencies were obtained in the NGS analysis compared with the traditional method, thus indicating that 5-HT2CR editing is regulated more tightly than was previously realized.
We then used NGS to assess 5-HT2CR editing in the entire study cohort (101 specimens). Using these measurements, we first examined whether editing at one site could influence editing at each of the other four sites. Studies with mouse knockout models showed that ADAR1 and ADAR2 have distinct actions on 5-HT2CR pre-mRNA, which are likely due to the recognition of specific secondary structures of the RNA that are formed between the exon sequence around the editing sites and a downstream intronic complementary sequence (Higuchi et al. 2000; Wang et al. 2000a; Hartner et al. 2004). As a result, the A site is predominantly edited by ADAR1, while the site D is mostly edited by ADAR2. The other sites have a potential to be edited by both enzymes. It was also suggested that ADAR1 and ADAR2 exhibit cross talk with each other and, therefore, the relative expression of the different ADAR enzymes may ultimately influence the pattern of editing (reviewed in Werry et al. (2008)). The mechanism underlying this cross talk is unclear, but may involve perturbation of the dsRNA structure in the process of editing. Our study demonstrated a strong interdependence of editing at different pairs of sites. In most cases there was a positive interconnection between the two sites, suggesting that editing at one site facilitates editing at another site. However, negative relationships were also observed (between B and E and between D and E sites), implying that in this case editing at one site impedes editing at another site. Although the A and D sites are edited by different enzymes, a very strong interdependence was observed between editing at these two sites, supporting the evidence of an association between ADAR1 and ADAR2 activities. Interdependence of editing among the sites was previously observed in rodent brain (Du et al. 2006; Enstero et al. 2009); however, the relationships identified in those studies were considerably different than those detected in humans, probably because of differences between humans and rodents in the nucleotide sequence of 5-HT2CR pre-mRNA in the region of editing (Werry et al. 2008).
We then investigated whether variations in 5-HT2CR editing in the DLPFC are associated with a history of depression or suicide by comparing editing among three groups, MDDNoSuic, MDDSuic and NC. We found less editing in MDDNoSuic than in MDDSuic, while the latter did not differ from NC. Compared with MDDSuic and NC, MDDNoSuic showed lower editing efficiency across all sites, lower frequencies of the mRNA variants that encode low-functioning (hypoactive) protein isoforms (ABCD and transcripts edited at the E or C sites) and higher frequencies of the variants that encode high-functioning isoforms (NONE and AD). Because a relatively large proportion of 5-HT2CR is edited to hypoactive forms regardless of diagnosis, the additive effect of these alterations, although they involve only a few percent of all 5-HT2CR, should be significantly higher activities, both agonist-stimulated and constitutive, in the DLPFC of non-suicide depressed subjects than in subjects from two other groups.
Our earlier study of BPD and SZ produced similar results, i.e., more editing in suicides than in non-suicides with the same diagnoses (Dracheva et al. 2008). However, in that earlier study, editing levels were “normal” (i.e., comparable to those in NC) in the non-suicides with BPD or SZ, and greater than “normal” in the suicides. Both studies show that, compared with non-suicides with the same diagnosis, suicides express higher levels of mRNA variants that encode hypoactive 5-HT2CR protein isoforms, and lower levels of variants that encode high-activity isoforms. This leads us to two alternative hypotheses: (1) the baseline state of editing in these disorders is the non-suicidal state; it is below “normal” in MDDNoSuic and “normal” in BPDNoSuic and SZNoSuic. Higher levels of editing occur in certain individuals or at certain times and are associated with increased risk of suicide. (2) The baseline state of editing is the suicidal state; it is “nor mal” in MDDSuic and above “normal” in BPDSuic and SZSuic. Lower levels of editing are present in certain individuals or at certain times, and protect against suicide.
Our findings of differences in ADAR expression between NC and MDD seemingly favor the first hypothesis. Although the analysis of ADAR1 rendered mixed results that do not allow any definite conclusions, our data strongly suggest decreased expression of ADAR2 in MDD (both suicides and non-suicides) compared to NC. No differences in ADAR1 or ADAR2 expression were detected among diagnoses or between suicides and non-suicides in our earlier study of BPD and SZ. Thus, we can speculate that below “normal” editing that we observed in MDDNoSuic results from underexpression of ADAR2 in MDD. Because of a strong association between ADAR1 and ADAR2 discussed above, decreased expression of ADAR2 should influence not only editing at the D site, but at all five editing sites.
The serotonin system displays multiple abnormalities in MDD and is the primary target of the most common treatments (Smith et al. 1997; Mann et al. 2000; Moreno et al. 2002; Mann 2003, 2009; Ernst et al. 2009). Conversely, abnormalities of the serotonin system are less prominent in SZ and BPD, and serotonin receptors, including 5-HT2CR, are secondary targets of common treatments, particularly atypical antipsychotics (Di et al. 2002). Thus, suppression of editing (and by inference, augmentation of function) of 5-HT2CR in MDDNoSuic may represent a compensatory adaptation (via decrease in ADAR2 expression) for impaired serotonergic input to their DLPFC (or a response to treatment that accomplishes the same thing).
The biological mechanism that contributes to higher 5-HT2CR editing (and therefore, hypoactive receptors) in suicide subjects with a history of psychiatric illness compared with non-suicide psychiatric patients is not clear. Increased expression of ADAR1 has been recently demonstrated in MDD subjects who died of suicide compared with NC and non-suicide MDD subjects (Simmons et al. 2010). However, we did not reproduce this finding in our study. In addition to the level of ADARs' mRNA expression, differences may also arise from changes in the levels of ADAR proteins and/or their enzymatic activities, or from other factors involved in regulating the editing process. We have begun to explore this by testing expression of several other putative regulators of editing (specifically the expression of the noncoding small nucleolus RNA HBII-52, and mRNA for helicase A (RHA)) (Yang et al. 2004; Vitali et al. 2005). We detected no differences among the diagnoses or between suicides and non-suicides (Lyddon and Dracheva, unpublished observations). RNA editing is a complex process, which undoubtedly involves many components that have not yet been uncovered.
Although not always consistent, previous pharmacological studies in rodents as well as studies in rodent models of depression indicated that 5-HT2CR editing may constitute a dynamic adaptive mechanism that facilitates maintenance of optimal receptor function (reviewed in Werry et al. (2008)). The editing processes (e.g., activity of editing enzymes) may be properly fine-tuned in some subjects, e.g., NC or non-suicide MDD, with changes in environment and/or circuit activity adequately counteracted by plastic adaptation of 5-HT2CR function via editing, or inadequately regulated in other subjects, in whom failure to adapt 5-HT2CR function increases the risk for suicide. It remains to be determined whether increased 5-HT2CR editing is a stable trait or a temporal state.
Our study is limited by a number of concerns inherent in most postmortem human brain research. One problem is that some of the subjects in each group were abusing drugs or alcohol, although, since our samples were fairly well-balanced in this regard, we would expect the main effect on the data, if any, would be to add variance and thus to make it less likely to find a difference between diagnostic groups. For heuristic purposes, we re-examined the data after excluding cases with historical or toxicological evidence of abuse or dependence. Although the smaller sample size reduced statistical power and significance, the pattern and magnitude of the differences among groups was essentially unchanged (Supplementary Figure 2).
Our study's most significant shortcoming is that essentially all of the MDD subjects were receiving antidepressant medications, so we cannot tell whether medication, rather than an innate response to depression, accounts for the suppression of editing in the non-suicide MDD cases. It will be extremely difficult to address this issue in an autopsy study, because when medication-naïve subjects in whom a diagnosis of MDD can be established come to autopsy, it is almost always because of suicide. Multiple lines of evidence, however, argue against the possibility that editing is suppressed by antidepressant medications: (1) similar to results obtained in earlier studies (reviewed in Werry et al. (2008)), a recent report in mice that employed NGS (and therefore, achieved high precision in assessing 5-HT2CR editing levels) showed that chronic treatments with antidepressant drugs resulted in alterations that were in opposite direction of those observed in non-suicide MDD subjects (Abbas et al. 2010). Specifically, in that study editing at the A and B sites was increased in the striatum and hippocampus following fluoxetine and only in the hippocampus following amitriptyline treatments. Neither drug influenced editing in the neocortex; in addition, no editing alterations were observed in any brain region following olanzapine or clozapine treatment. (2) Antidepressant use was not different between the MDDSuic and the MDDNoSuic subjects in our study. However, a significant difference in editing was detected between the groups. Of course, in a post mortem study (a necessity for studying suicide), we cannot determine whether low levels of editing were present before treatment, or whether they are correlated with an anti-suicidal effect of treatment. (3) In our previous 5-HT2CR editing study of SZ and BPD patients with and without suicide, history of antidepressant treatment among suicide victims had no discernable impact on editing, suggesting that editing is unaffected by antidepressant treatment, or at least by antidepressant treatment that fails to prevent suicide. Also, no differences were observed between non-psychiatric controls and SZ or BPD who did not commit suicide, although more than half of the non-suicide bipolar patients received antidepressant medications and all non-suicide schizophrenia patients received antipsychotic medications (Dracheva et al. 2008). Based on this collective evidence, we believe that the editing differences between MDDSuic and MDDNoSuic subjects that we find are either unrelated to medications or, in some cases, may be related to the efficacy of the medications in preventing suicide.
To summarize, our findings suggest the presence of two factors that are associated with 5-HT2CR editing in the DLPFC. One factor is linked to depression and is negatively associated with 5-HT2CR editing efficiency (and by inference, positively associated with the receptor function). This factor appears to stem from decreased expression of at least one editing enzyme, ADAR2. The other factor, seen also in our study of suicide in BP and SZ (Dracheva et al. 2008), is linked to suicide and is associated with more editing and less receptor function. The biological underpinning of this suicide-associated factor remains to be determined. Because suicide is a rare and unpredictable event, collections of autopsy specimens build slowly. By combining material from three excellent collections, we have achieved the largest postmortem editing study reported to date. However, compared with biological studies of live individuals, the sample size is relatively small, and the findings require confirmation in independent cohorts.
Supplementary Material
Supplementary Table 1. Demographic Information of the study cohort.
Supplementary Table 2. Efficiency of editing at the five editing sites of the 5-HT2CR mRNA.
Supplementary Table 3. Description of the TaqMan assays used in the study.
Supplementary Table 4. Interdependence of editing among different editing sites.
Supplementary Table 5. Analysis of ADARs mRNA expression by ANCOVA.
Supplementary Figure 1. Schematic representation of the five ADAR1 and four ADAR2 mRNA splice variants.
Supplementary Figure 2. Frequencies of the 5-HT2CR editing transcripts in all subjects and excluding subjects with evidence of substance abuse or dependence.
Acknowledgments
We are grateful to Mr Scott Navarrett and Ms Katie Lutz for their superb technical assistance and to Dr D.J. Jamison for his help with bioinformatics. Postmortem brain tissue was donated by The Stanley Medical Research Institute courtesy of Drs Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken. This study was supported by VA Merit award to SD, by grants from the Hope for Depression Foundation and the American Foundation for Suicide Prevention (SD), and by the VISN3 Mental Illness Research and Education Clinical Center (MIRECC) (SD, LJS).
Footnotes
Statement of interest: None to declare.
References
- Abbas AI, Urban DJ, Jensen NH, Farrell MS, Kroeze WK, Mieczkowski P, et al. Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method. Nucleic Acids Res. 2010;38:e118. doi: 10.1093/nar/gkq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S. Approaching the molecular pathology of suicide. Biol Psychiatry. 2008;64:643–644. doi: 10.1016/j.biopsych.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Bagdy G, Graf M, Anheuer ZE, Modos EA, Kantor S. Anxiety-like effects induced by acute fluoxetine, sertraline or m-CPP treatment are reversed by pretreatment with the 5-HT2C receptor antagonist SB-242084 but not the 5-HT1A receptor antagonist WAY-100635. Int J Neuropsychopharmacol. 2001;4:399–408. doi: 10.1017/S1461145701002632. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KA, Cropper JD, Niswender CM, Sanders-Bush E, Emeson RB, Clarke WP. RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br J Pharmacol. 2001;134:386–392. doi: 10.1038/sj.bjp.0704255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M, Underwood MD, Hen R, Rosoklija GB, Dwork AJ, John MJ, et al. Antidepressants increase neural progenitor cells in the human hippocampus. Neuropsychopharmacology. 2009;34:2376–2389. doi: 10.1038/npp.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow LJ, O'Connor D, Watts R, Duxon MS, Hutson PH. Evidence for accelerated desensitisation of 5-HT(2C) receptors following combined treatment with fl uoxetine and the 5-HT(1A) receptor antagonist, WAY 100,635, in the rat. Neuropharmacology. 2000;39:1222–1236. doi: 10.1016/s0028-3908(99)00191-4. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Canton H, Emeson RB, Barker EL, Backstrom JR, Lu JT, Chang MS, et al. Identification, molecular cloning, and distribution of a short variant of the 5-hydroxytryptamine2C receptor produced by alternative splicing. Mol Pharmacol. 1996;50:799–807. [PubMed] [Google Scholar]
- Cavanagh JT, Carson AJ, Sharpe M, Lawrie SM. Psychological autopsy studies of suicide: a systematic review. Psychol Med. 2003;33:395–405. doi: 10.1017/s0033291702006943. [DOI] [PubMed] [Google Scholar]
- Chanrion B, Mannoury la CC, Gavarini S, Seimandi M, Vincent L, Pujol JF, et al. Inverse agonist and neutral antagonist actions of antidepressants at recombinant and native 5-hydroxytryptamine2C receptors: differential modulation of cell surface expression and signal transduction. Mol Pharmacol. 2008;73:748–757. doi: 10.1124/mol.107.041574. [DOI] [PubMed] [Google Scholar]
- Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry. 2001;50:260–265. doi: 10.1016/s0006-3223(01)01083-6. [DOI] [PubMed] [Google Scholar]
- Di Giovanni G, De Deurwaerdere P, Di Mascio M, Di M V, Esposito E, Spampinato U. Selective blockade of serotonin-2C/2B receptors enhances mesolimbic and mesostriatal dopaminergic function: a combined in vivo electrophysiological and microdialysis study. Neuroscience. 1999;91:587–597. doi: 10.1016/s0306-4522(98)00655-1. [DOI] [PubMed] [Google Scholar]
- Di M V, Cacchio M, Di GC, Di GG, Esposito E. Biochemical evidence that the atypical antipsychotic drugs clozapine and risperidone block 5-HT(2C) receptors in vivo. Pharmacol Biochem Behav. 2002;71:607–613. doi: 10.1016/s0091-3057(01)00714-6. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Lyddon R, Barley K, Marcus SM, Hurd YL, Byne WM. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology. 2009;34:2237–2251. doi: 10.1038/npp.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, Patel N, Woo DA, Marcus SM, Siever LJ, Haroutunian V. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. Mol Psychiatry. 2008;13:1001–1010. doi: 10.1038/sj.mp.4002081. [DOI] [PubMed] [Google Scholar]
- Du Y, Davisson MT, Kafadar K, Gardiner K. A-to-I pre-mRNA editing of the serotonin 2C receptor: comparisons among inbred mouse strains. Gene. 2006;382:39–46. doi: 10.1016/j.gene.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Enstero M, Daniel C, Wahlstedt H, Major F, Ohman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, Mechawar N, Turecki G. Suicide neurobiology. Prog Neurobiol. 2009;89:315–333. doi: 10.1016/j.pneurobio.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Esposito E. Serotonin-dopamine interaction as a focus of novel antidepressant drugs. Curr Drug Targets. 2006;7:177–185. doi: 10.2174/138945006775515455. [DOI] [PubMed] [Google Scholar]
- Fitzgerald LW, Iyer G, Conklin DS, Krause CM, Marshall A, Patterson JP, et al. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology. 1999;21:82–90S. doi: 10.1016/S0893-133X(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Flomen R, Knight J, Sham P, Kerwin R, Makoff A. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 2004;32:2113–2122. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatch MB. Discriminative stimulus effects of m-chlorophenylpiperazine as a model of the role of serotonin receptors in anxiety. Life Sci. 2003;73:1347–1367. doi: 10.1016/s0024-3205(03)00422-3. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Hawton K, Van Heeringen K. Suicide. Lancet. 2009;373:1372–1381. doi: 10.1016/S0140-6736(09)60372-X. [DOI] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Niswender CM. Serotonin 5-HT2C receptor RNA editing alters receptor basal activity: implications for serotonergic signal transduction. J Neurochem. 1999;73:1711–1717. doi: 10.1046/j.1471-4159.1999.731711.x. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci Lett. 2003;346:169–172. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- Lance CE. Mallows' Cp Statistics. In: Everitt BS, Howell DC, editors. Encyclopedia of statistics in behavioral science. Chichester: John Wiley & Sons; 2005. pp. 1119–1120. [Google Scholar]
- Mann JJ. Neurobiology of suicidal behaviour. Nat Rev Neurosci. 2003;4:819–828. doi: 10.1038/nrn1220. [DOI] [PubMed] [Google Scholar]
- Mann JJ, Huang YY, Underwood MD, Kassir SA, Oppenheim S, Kelly TM, et al. A serotonin transporter gene promoter polymorphism (5-HTTLPR) and prefrontal cortical binding in major depression and suicide. Arch Gen Psychiatry. 2000;57:729–738. doi: 10.1001/archpsyc.57.8.729. [DOI] [PubMed] [Google Scholar]
- Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, et al. Candidate endophenotypes for genetic studies of suicidal behavior. Biol Psychiatry. 2009;65:556–563. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion S, Weiner DM, Caron MG. RNA editing induces variation in desensitization and trafficking of 5-hydroxytryptamine 2c receptor isoforms. J Biol Chem. 2004;279:2945–2954. doi: 10.1074/jbc.M308742200. [DOI] [PubMed] [Google Scholar]
- Millan MJ. Serotonin 5-HT2C receptors as a target for the treatment of depressive and anxious states: focus on novel therapeutic strategies. Therapie. 2005;60:441–460. doi: 10.2515/therapie:2005065. [DOI] [PubMed] [Google Scholar]
- Morabito MV, Ulbricht RJ, O'Neil RT, Airey DC, Lu P, Zhang B, et al. High-throughput multiplexed transcript analysis yields enhanced resolution of 5-hydroxytryptamine 2C receptor mRNA editing profiles. Mol Pharmacol. 2010;77:895–902. doi: 10.1124/mol.109.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno FA, Rowe DC, Kaiser B, Chase D, Michaels T, Gelernter J, et al. Association between a serotonin transporter promoter region polymorphism and mood response during tryptophan depletion. Mol Psychiatry. 2002;7:213–216. doi: 10.1038/sj.mp.4000962. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, et al. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- Palvimaki EP, Roth BL, Majasuo H, Laakso A, Kuoppamaki M, Syvalahti E, et al. Interactions of selective serotonin reuptake inhibitors with the serotonin 5-HT2c receptor. Psychopharmacology (Berlin) 1996;126:234–240. doi: 10.1007/BF02246453. [DOI] [PubMed] [Google Scholar]
- Pandey GN, Dwivedi Y, Ren X, Rizavi HS, Roberts RC, Conley RR. Cyclic AMP response element-binding protein in post-mortem brain of teenage suicide victims: specific decrease in the prefrontal cortex but not the hippocampus. Int J Neuropsychopharmacol. 2007;10:621–629. doi: 10.1017/S1461145706007231. [DOI] [PubMed] [Google Scholar]
- Quirk K, Lawrence A, Jones J, Misra A, Harvey V, Lamb H, et al. Characterisation of agonist binding on human 5-HT2C receptor isoforms. Eur J Pharmacol. 2001;419:107–112. doi: 10.1016/s0014-2999(01)00943-8. [DOI] [PubMed] [Google Scholar]
- Serretti A, Artioli P, De Ronchi D. The 5-HT2C receptor as a target for mood disorders. Expert Opin Ther Targets. 2004;8:15–23. doi: 10.1517/14728222.8.1.15. [DOI] [PubMed] [Google Scholar]
- Simmons M, Meador-Woodruff JH, Sodhi MS. Increased cortical expression of an RNA editing enzyme occurs in major depressive suicide victims. NeuroReport. 2010;21:993–997. doi: 10.1097/WNR.0b013e32833f11c3. [DOI] [PubMed] [Google Scholar]
- Smith KA, Fairburn CG, Cowen PJ. Relapse of depression after rapid depletion of tryptophan. Lancet. 1997;349:915–919. doi: 10.1016/s0140-6736(96)07044-4. [DOI] [PubMed] [Google Scholar]
- Sodhi MS, Airey DC, Lambert W, Burnet PW, Harrison PJ, Sanders-Bush E. A rapid new assay to detect RNA editing reveals antipsychotic-induced changes in serotonin-2C transcripts. Mol Pharmacol. 2005;68:711–719. doi: 10.1124/mol.105.014134. [DOI] [PubMed] [Google Scholar]
- Tohda M, Hang PT, Kobayashi N, Matsumoto K. Serotonin 2C receptor (5-HT2CR) mRNA editing-induced downregulation of 5-HT2CR function in Xenopus oocytes: the significance of site C editing. J Pharmacol Sci. 2010;113:362–367. doi: 10.1254/jphs.10094fp. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium 1. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000a;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 2000b;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Brain Res Mol Brain Res. 2004;124:70–78. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Demographic Information of the study cohort.
Supplementary Table 2. Efficiency of editing at the five editing sites of the 5-HT2CR mRNA.
Supplementary Table 3. Description of the TaqMan assays used in the study.
Supplementary Table 4. Interdependence of editing among different editing sites.
Supplementary Table 5. Analysis of ADARs mRNA expression by ANCOVA.
Supplementary Figure 1. Schematic representation of the five ADAR1 and four ADAR2 mRNA splice variants.
Supplementary Figure 2. Frequencies of the 5-HT2CR editing transcripts in all subjects and excluding subjects with evidence of substance abuse or dependence.