Abstract
The zebrafish is one of the leading models for the analysis of the vertebrate visual system. A wide assortment of molecular, genetic, and cell biological approaches is available to study zebrafish visual system development and function. As new techniques become available, genetic analysis and imaging continue to be the strengths of the zebrafish model. In particular, recent developments in the use of transposons and zinc finger nucleases to produce new generations of mutant strains enhance both forward and reverse genetic analysis. Similarly, the imaging of developmental and physiological processes benefits from a wide assortment of fluorescent proteins and the ways to express them in the embryo. The zebrafish is also highly attractive for high-throughput screening of small molecules, a promising strategy to search for compounds with therapeutic potential. Here we discuss experimental approaches used in the zebrafish model to study morpho−genetic transformations, cell fate decisions, and the differentiation of fine morphological features that ultimately lead to the formation of the functional vertebrate visual system.
I. Introduction
The vertebrate central nervous system (CNS) is enormously complex. The human cerebral cortex alone is estimated to contain in excess of 109 neurons (Jacobson, 1991), each characterized by the morphology of its soma and processes, synaptic connections with other cells, receptors expressed on its surface, the neurotransmitters it releases, and numerous other molecular and cellular features. Together these characteristics define cell identity. To understand the development of the CNS, multiple steps involved in the formation of numerous cell identities must be determined. One way to approach this enormously complicated task is to study a relatively simple and accessible region of the CNS. The retina is such a region.
Several characteristics make the retina more approachable than most other areas of the CNS. Most importantly, the retina contains a relatively small number of neuronal cell classes, and these are characterized by stereotypical positions and distinctive morphologies. Even in very crude histological preparations, the identity of individual cells can be frequently and correctly determined based on their location. Cajal noted that the separation of different cells into distinct layers, the small size of dendritic fields, and the presence of layers consisting almost exclusively of neuronal projections are fortuitous characteristics of the retina (Cajal, 1893). In addition, the eye becomes isolated from other parts of the CNS early in embryogenesis, and consequently cell migrations into the retina are limited to the optic nerve and the optic chiasm only (Burrill and Easter, 1994; Watanabe and Raff, 1988). Such anatomical isolation simplifies the interpretation of developmental events within the retina. Taken together, all these qualities make the retina an excellent model system for the studies of vertebrate neuronal development and function.
Teleost retinae have been studied for over a century (Cajal, 1893; Dowling, 1987; Malicki, 2000; Muller, 1857; Rodieck, 1973). The eyes of teleosts in general and zebrafish in particular are large and their neuroanatomy is well characterized. An important advantage of the zebrafish retina for genetic and developmental research is that it is formed and becomes functional very early in development. Neurogenesis in the central retina of the zebrafish eye is essentially complete by 60 hours post fertilization (hpf) (Nawrocki, 1985) and, as judged by behavioral responses to visual stimuli, the zebrafish eye detects light surprisingly early, starting between 2.5 and 3.5 days post fertilization (dpf) (Clark, 1981; Easter and Nicola, 1996). Studies of the zebrafish retina benefit from many general qualities of the system: high fecundity, transparency, embryogenesis that occurs outside the maternal organism, the ease of maintenance in large numbers, the short length of the life cycle, the ability to study haploid development, and most recently the progress in zebrafish genomics, including the genome sequencing project.
The neuronal architecture of the vertebrate retina has been remarkably conserved in evolution. Early investigators noted that even retinae of divergent vertebrate phyla, including teleosts and mammals, display similar organization (Cajal, 1893; Muller, 1857). Gross morphological and histological features of mammalian and teleost retinae display few differences. Accordingly, human and zebrafish retinae contain the same major cell classes organized in the same layered pattern, where light-sensing photoreceptors occupy the outermost layer, while the retinal projection neurons, the ganglion cells, reside in the innermost neuronal layer, proximal to the lens. The retinal interneurons, the amacrine, bipolar, and horizontal cells, localize in between the photoreceptor and ganglion cell layers (Fig. 2). Similarities extend beyond histology and morphology. Pax-2/noi and Chx10/Vsx-2 expression patterns, for example, are very similar in mouse and zebrafish eyes (Liu et al., 1994; Macdonald and Wilson, 1997; Nornes et al., 1990; Passini et al., 1997), and a number of genetic loci display closely related phenotypes in humans and zebrafish alike. These observations stimulated efforts to use the zebrafish as a model of human eye disorders (reviewed in Gross and Perkins, 2008). Consequently, zebrafish eye mutants have been proposed as models of pyruvate dehydrogenase deficiency, choroidemia, achromatopsia, as well as June, Joubert, and Hermansky–Pudlak syndromes (Bahadori et al., 2006; Brockerhoff et al., 2003; Duldulao et al., 2009; Hudak et al., 2010; Krock et al., 2007; Taylor et al., 2004). This is a fortuitous circumstance, considering that throughout the world diseases of the retina affect millions (Cedrone et al., 1997; Dryja and Li, 1995; Hartong et al., 2006; Seddon, 1994). Thus, in addition to being an excellent model for the studies of vertebrate neurogenesis, the zebrafish retina is likely to provide medically relevant insights. In this chapter, following an introduction to zebrafish eye development, we focus on tools currently used to study various aspects of the zebrafish visual system. Since many techniques described in this chapter are also applied to the analysis of other organs, the reader is encouraged to search for more information in other sections of this volume.
Fig 2.
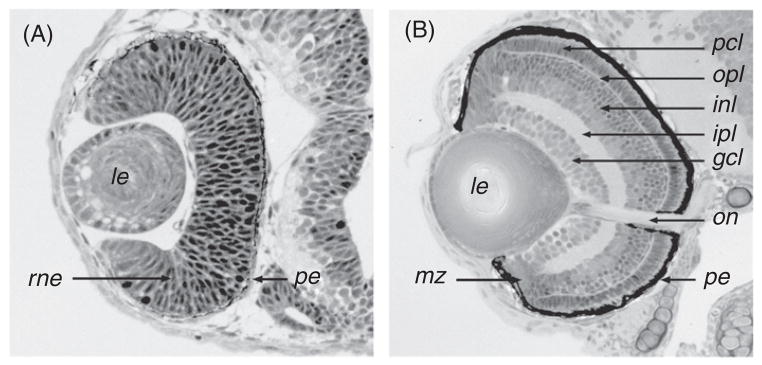
Histology of the zebrafish retina. (A) A section through the zebrafish eye during early stages of neurogenesis at approximately 36 hpf. At this stage, the retina mostly consists of two epithelial layers: the pigmented epithelium and the retinal neuroepithelium. Although some retinal cells are already postmitotic at this stage, they are not numerous enough to form a distinct layer. (B) A section through the zebrafish eye at 72 hpf. With the exception of the marginal zone, where cell proliferation will continue throughout the lifetime of the animal, retinal neurogenesis is mostly completed. The major nuclear and plexiform layers, as well as the optic nerve and the pigmented epithelium, are well differentiated. gcl: ganglion cell layer; inl: inner nuclear layer; ipl: inner plexiform layer; le: lens; mz: marginal zone; on: optic nerve; opl: outer plexiform layer; pcl: photoreceptor cell layer; pe: pigmented epithelium; rne: retinal neuroepithelium.
II. Development of the Zebrafish Retina
A. Early Morphogenetic Events
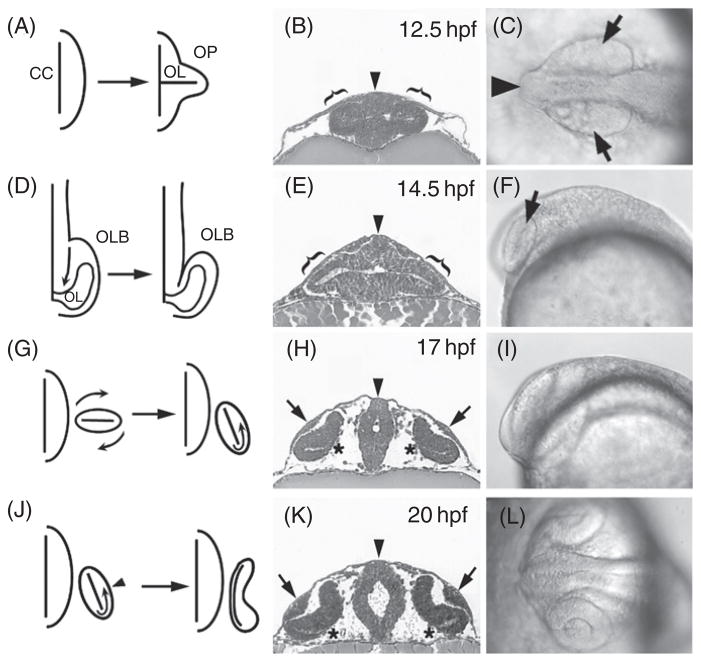
Fate-mapping studies indicate that during early gastrulation the retina originates from a single field of cells positioned roughly between the telencephalic and the diencephalic precursor fields (Woo and Fraser, 1995). During late gastrulation, the anterior and lateral migrations of diencephalic precursors are thought to subdivide the retinal field into two separate primordia (Rembold et al., 2006; Varga et al., 1999). Neurulation in teleosts proceeds somewhat differently than in higher vertebrates. First, the primordium of the CNS does not take the form of a tube (the neural tube), and instead is shaped in the form of a solid rod called the neural keel (Fig. 1B and C) (Kimmel et al., 1995; Lowery and Sive, 2004; Schmitz et al., 1993). Consistent with that, optic vesicles are not present, and the equivalent structures are called optic lobes. These first become evident as bilateral thickenings of the anterior neural keel at about 11.5 hpf, and gradually become more and more prominent (Fig. 1A–C) (Schmitt and Dowling, 1994). They are initially flattened and protrude laterally on both sides of the brain (brackets and arrows, respectively in Fig. 1B and C). At approximately 13 hpf, the posterior portion of the optic lobe starts to separate from the brain, while its anterior part remains attached (Fig. 1D). This attachment will persist throughout eye development, at later stages forming the optic stalk. As its morphogenesis advances, the optic lobe turns around its anteroposterior axis so that its ventral surface becomes directed toward the brain while the dorsal surface starts to face the outside environment (Fig. 1G). Cells forming the outside surface will differentiate into the neural retina. Fate-mapping studies suggest that starting at ca. 15 hpf, cells migrate from the medial to lateral epithelial layer of the optic lobe (Li et al., 2000b). The medial layer becomes thinner and subsequently differentiates as the retinal pigmented epithelium (RPE) (asterisks in Fig. 1H and K). At about the same time, an invagination forms on the lateral (upper, before turning) surface of the optic lobe (Schmitt and Dowling, 1994). This is accompanied by the appearance of a thickening in the epithelium overlying the optic lobe: the lens rudiment (arrows in Fig. 1H). Subsequently, over a period of several hours, both the invagination and the lens placode become increasingly more prominent, transforming the optic lobe into the optic cup (Fig. 1J–L). The choroid fissure forms in the rim of the optic cup next to the optic stalk. The lens placode continues to grow and by 24 hpf it is detached from the epidermis. At the beginning of day 2, the optic cup consists of two closely connected sheets of cells: the pseudos−tratified columnar neuroepithelium (rne) and the cuboidal pigmented epithelium (pe) (Fig. 2A). Starting at about 24 hpf, melanin granules appear in the cells of the pigmented epithelium. In the first half of day 2, concomitant to the expansion of the ventral diencephalon, the eye rotates so that the choroid fissure, which at 24 hpf was pointing above the yolk sac, is now directed toward the heart (Kimmel et al., 1995; Schmitt and Dowling, 1994). Throughout this period, the optic stalk gradually becomes less prominent. In the first half of day 2 as ganglion cells begin to differentiate, the optic stalk provides support for their axons. Later in development, it is no longer present as a distinct structure and its cells may contribute the optic nerve (Macdonald et al., 1997). Lastly, the optic cup rotates around its mediolateral axis (Schmitt and Dowling, 1994). This is the final major morphological transformation in zebrafish eye development.
Fig 1.
Early morphogenetic events leading to the formation of the optic cup. (A) A diagram of a transverse section through anterior neural keel illustrating morphogenetic transformation that leads to the formation of optic lobes. Solid horizontal line represents the ventricular lumen (OL) of the optic lobe. (B) A transverse plastic section through the anterior portion of the neural keel and optic lobes (brackets). (C) Dorsal view of anterior neural keel and optic lobes (arrows) at 12.5 hpf. (D) A schematic representation of anterior neural keel (dorsal view, anterior down). Wing-shaped optic primordia gradually detach from the neural keel starting posteriorly (arrow). (E) A transverse plastic section through anterior neural keel and optic lobes (brackets) at 14.5 hpf. (F) Lateral view of anterior neural keel and optic lobe (arrow) at the same stage. (G) A diagram of dorsoventral reorientation of the optic lobe. (H) A transverse plastic section through neural keel and optic lobes during the reorientation at ca. 17 hpf. At about the same time, lens rudiments start to form (arrows) and the medial layer of the optic lobe becomes thinner as it begins to differentiate into the pigmented epithelium (asterisks). The lateral surface of the optic lobe starts to invaginate. (I) A lateral view of anterior neural keel during optic cup formation. (J) A schematic representation of morphogenetic movements that accompany optic cup formation. Cells migrate (arrow) from the medial to the lateral cell layer around the ventral edge of the lobe. Simultaneously, the initially flat lobe invaginates (arrowhead) to become the concave eye cup. (K) A transverse plastic section through the anterior neural tube during optic cup formation at 20 hpf. Lens rudiments are quite prominent by this stage (arrows). Most of the medial cell layer already displays a flattened morphology, except for the ventralmost regions, which still retain columnar appearance (asterisks). (L) A dorsal view of anterior neural tube and optic lobes at 20 hpf. Vertical arrowheads in B, E, H, and K indicate the midline. CC, central canal; OL, optic lumen; OP, optic primordium; OLB, optic lobe; hpf, hours post fertilization. Except D, C and L, in all panels dorsal is up. Panels A, D, G, and J are based on Easter and Malicki (2002). The remaining panels reprinted from Pujic and Malicki (2001) with permission from Elsevier.
B. Neurogenesis
At the beginning of the second day of development, the zebrafish neural retina still consists of a single sheet of pseudostratified neuroepithelium. Similar to other epithelia, the retinal neuroepithelium is a highly polarized tissue, characterized by apico−basal nuclear movements, which correlate with cell cycle phase (Baye and Link, 2007; Das et al., 2003; Hinds and Hinds, 1974). Nuclei of cells that are about to divide migrate to the apical surface of the neuroepithelium, where both nuclear division and cytokinesis take place. After the division, the newly formed nuclei move back to more basal locations. Although it has been assumed for a long time that dividing cells lose their contact with the basal surface of the neuroepithelium (Hinds and Hinds, 1974), more recent two-photon imaging studies in zebrafish show that this view is most likely incorrect, as a tenuous cytoplasmic process extends toward the basal surface during nuclear division of the neuroepithelial cell (Das et al., 2003). Interestingly, in the brain neuroepithelium, and possibly in the retina, this process splits into two or more prior to the cytokinesis, and the daughter processes are inherited either symmetrically or asymmetrically by the daughter cells (Kosodo et al., 2008).
In between mitotic divisions, the movement of cell nuclei is stochastic most of the time, so that persistent nuclear movements, directed either basally or apically, occur during less than 10% of the cell cycle (Norden et al., 2009). The maximum depth of basally directed translocation is very heterogeneous, ranging from 10% to 90% of neuroepithelial thickness. Interestingly, deeper nuclear migration correlates with divisions that generate post-mitotic cells (Baye and Link, 2007). Mitotic divisions are observed nearly exclusively at the apical surface of the neuroepithelium until about 1.5 dpf. Following that, between 40 and 50 hpf, ca. 50% of mitoses occur in the inner nuclear layer (INL) (Godinho et al., 2005). Very few mitotic divisions are observed in the central retina at later stages.
Despite its uniform morphology, the retinal neuroepithelium is the site of many transformations, apparent in the changes of cell cycle length and in the dynamic characteristics of gene expression patterns. After a period of very slow cell cycle progression during early stages of optic cup morphogenesis, the cell cycle shortens to ca. 10 h by 24 hpf, and later its duration appears even shorter (Baye and Link, 2007; Hu and Easter, 1999; Li et al., 2000a; Nawrocki, 1985). Imaging of individual neuroepithelial cells between 24 and 40 hpf revealed that their cell cycle varies greatly in length from about 4 to 11 h, averaging ca. 6.5 h (Baye and Link, 2007). The significance of changes in the length of the cell cycle, or the genetic mechanisms that regulate them, are not understood.
In parallel to fluctuations of cell cycle length, the expression patterns of numerous genes display dramatic changes in the retinal neuroepithelium during this time. While the transcription of some early expressed genes, such as rx3 or six3, is downregulated, other genes become active. The zebrafish atonal 5 homolog, lakritz, is one interesting example of an important genetic regulator characterized by a dynamic expression pattern. The lakritz gene becomes transcriptionally active in a small group of cells in the ventral retina by 25 hpf, and from there its expression spreads into the nasal, dorsal, and finally temporal eye (Masai et al., 2000). This gradual advance of expression around the retinal surface is noteworthy because it characterizes many other developmental regulators and neuronal differentiation markers (reviewed in Pujic and Malicki, 2004).
Another noteworthy feature of neuroepithelial cells is the orientation of their mitotic spindles. The mitotic spindle position and its role in cell fate determination has been interesting, albeit contentious issue. It has been proposed that in some species the vertical (apico-basal) reorientation of the mitotic spindle characterizes asymmetric cell divisions, which produce cells of different identities: a progenitor cell and a postmitotic neuron for example (Cayouette and Raff, 2003; Cayouette et al., 2001). As such divisions first appear in the neuroepithelium at the onset of neurogenesis, so should vertically oriented mitotic spindles. The analysis of zebrafish neuroepithelial cells found, however, little support for the presence of vertically oriented mitotic spindles: the majority, if not all, of zebrafish neuroepithelial cells divide horizontally (Das et al., 2003).
As the morphogenetic movements that shape and orient the optic cup come to completion, the first retinal cells become postmitotic and differentiate. Gross morphological characteristics of the major retinal cell classes are very well conserved in all vertebrates. Six major classes of neurons arise during neurogenesis: ganglion, amacrine, bipolar, horizontal, interplexiform, and photoreceptor cells. The Müller glia are also generated in the same period. Ganglion cell precursors are the first to become postmitotic in a small patch of ventrally located cells between 27 and 28 hpf (Hu and Easter, 1999; Nawrocki, 1985). The early onset of ganglion cell differentiation is again conserved in many vertebrate phyla (Altshuler et al., 1991). Similar to expression patterns that characterize the genetic regulators of retinal neurogenesis, differentiated ganglion cells first appear in the ventral retina, nasal to the optic nerve (Burrill and Easter, 1995; Schmitt and Dowling, 1996). The rudiments of the ganglion cell layer are recognizable in histological sections by 36 hpf. Approximately 10 h after the first ganglion neuron progenitors exit the cell cycle, cells that contribute to the INL also become postmitotic. Again, this first happens in a small ventral group of cells (Hu and Easter, 1999). By 34–36 hpf, and possibly even earlier, terminal divisions of retinal progenitor cells give rise to pairs of ganglion and photoreceptor cells, indicating that these two cell classes are generated in overlapping windows of time (Poggi et al., 2005).
By 60 hpf, over 90% of neurons in the central retina are postmitotic, and the major neuronal layers are distinguishable by morphological criteria. Cells of different layers become postmitotic in largely non-overlapping windows of time. This is particularly obvious for ganglion cell precursors, most of which, if not all, are postmitotic before the first INL cells exit the cell cycle (Hu and Easter, 1999). This is different from Xenopus, where the times of cell cycle exit for different cell classes overlap extensively (Holt et al., 1988). In contrast to mammals, neurogenesis in teleosts and larval amphibians continues at the retinal margin throughout the lifetime of the organism (Marcus et al., 1999). In adult zebrafish, as well as in other teleosts, neurons are also added in the outer nuclear layer. In contrast to the marginal zone, where many cell types are generated, only rods are added in the outer nuclear layer of the adult (Mack and Fernald, 1995; Marcus et al., 1999).
Photoreceptor morphogenesis starts shortly after the exit of photoreceptor precursor cells from the cell cycle (reviewed in Tsujikawa and Malicki, 2004a). The photore−ceptor cell layer can be distinguished in histological sections by 48 hpf. The expression of visual pigments, opsins, is necessary for photoreceptor outer segment differentiation. Rods are the first to express opsin around 50 hpf, shortly followed by blue and red cones, and somewhat later by short single cones (Raymond et al., 1995; Robinson et al., 1995; Takechi et al., 2003). Photoreceptor outer segments first appear in the ventral patch by 60 hpf, and ribbon synapses of photoreceptor synaptic termini are detectable by 62 hpf (Branchek and Bremiller, 1984; Schmitt and Dowling, 1999). The photoreceptor cell layer of the zebrafish retina contains five types of photoreceptor cells: rods, short single cones, long single cones, and short and long members of double cone pairs. The differentiation of morphologically distinct photoreceptor types becomes apparent by 4 dpf, and by 12 dpf all zebrafish photoreceptor classes can be distinguished on the basis of their morphology (Branchek and Bremiller, 1984).
The photoreceptor cells of the zebrafish retina are organized in a regular pattern, referred to as the “photoreceptor mosaic.” In the adult, cones form regular rows. The spaces between these rows are occupied by rods, which do not display any obvious pattern. Within a single row of cones, double cones are separated from each other by alternating long and short single cones. Adjacent rows of cones are staggered relative to each other so that short single cones of one row are flanked on either side by long single cones of the two neighboring rows (Fadool, 2003; Larison and Bremiller, 1990). In addition to morphology, individual types of photoreceptors are uniquely characterized by spectral sensitivities and visual pigment expressions. Long single cones express blue light-sensitive opsin; short single cones, ultraviolet (UV)-sensitive opsin; double cones, red-sensitive and green-sensitive opsins; whereas rods express rod opsin (Hisatomi et al., 1996; Raymond et al., 1993). The number of opsin genes exceeds the number of photoreceptor types, as two and four independent loci encode red and green opsins, respectively (Chinen et al., 2003). Each green and red opsin gene is expressed in a different subpopulation of double cones. Of the two red opsin genes, LWS-2 is expressed in the central retina, while LWS-1 in the retinal periphery (Takechi and Kawamura, 2005). Similarly, the expression domains of green opsin genes RH2-1 and RH2-2 occupy largely overlapping areas in the central retina, while RH2-3 and RH3-4 are expressed at the retinal circumference in what appears to be non-overlapping regions (Takechi and Kawamura, 2005).
C. Development of Retinotectal Projections
As this aspect of retinal development is discussed at length in an accompanying chapter (Chapter 1), here we comment on some of the most basic observations only. The neuronal network of the retina is largely self-contained. The only retinal neurons that send their projections outside are the ganglion cells. Their axons navigate through the midline of the ventral diencephalon into the dorsal part of the midbrain, the optic tectum. The ganglion cells extend axons shortly after their final mitosis, already while they are migrating toward the vitreal surface (Bodick and Levinthal, 1980). The projections proceed toward the inner surface of the retina and subsequently along the inner limiting membrane toward the optic nerve head. In zebrafish, the first ganglion cell axons exit the eye between 34 and 36 hpf and navigate along the optic stalk and through the ventral region of the brain toward the midline (Burrill and Easter, 1995; Macdonald and Wilson, 1997). At about 2 dpf, the zebrafish optic nerve contains ca. 1800 axons at the exit point from the retina (Bodick and Levinthal, 1980). Cross sections near the nerve head reveal a crescent-shaped optic nerve. Axons of centrally located ganglion cells occupy the outside (dorsal) surface of the crescent whereas the axons of more peripheral (younger) cells localize to the inside (ventral) surface of the optic nerve. With the exception of the axonal trajectories of cells separated by the choroid fissure, axons of neighboring ganglion cells travel together in the optic nerve (Bodick and Levinthal, 1980). In addition to ganglion cell axons, the optic nerve contains retinopetal projections, which appear considerably later, after 5 dpf, and originate in the nucleus olfactoretinalis of the rostral telencephalon (Burrill and Easter, 1994). After crossing the midline, the axonal projections of the ganglion cells split into the dorsal and ventral branches of the optic tract. The ventral branch contains mostly axons of the dorsal retinal ganglion cells, and the dorsal branch mostly of the ventral cells (Baier et al., 1996). The growth cones of the retinal ganglion cells first enter the optic tectum between 46 and 48 hpf. In addition to the optic tectum, the retinal axons innervate nine other, much smaller targets in the zebrafish brain (Burrill and Easter, 1994).
Spatial relationships between individual ganglion cells in the retina are precisely reproduced by their projections in the tectum. The exactitude of this pattern has long fascinated biologists and has been a subject of intensive research in many vertebrate species (Drescher et al., 1997; Fraser, 1992; Sanes, 1993). The spatial coordinates of the retina and the tectum are reversed. The ventral-nasal ganglion cells of the zebrafish retina project to the dorsal-posterior optic tectum whereas the dorsal-temporal cells innervate the ventral-anterior tectum (Karlstrom et al., 1996; Stuermer, 1988; Trowe et al., 1996). By 72 hpf, axons from all quadrants of the retina are in contact with their target territories in the optic tectum.
In summary, development of the zebrafish retina proceeds at a rapid pace. By the end of day 3, all major retinal cell classes have been generated and are organized in distinct layers (Fig. 2B), the photoreceptor cells have developed outer segments, and the ganglion cell axons have innervated their target, the optic tectum. It is also about this time that the zebrafish visual system becomes functional (Clark, 1981; Easter and Nicola, 1996). The brevity of eye morphogenesis and retinal neurogenesis is a major advantage of the zebrafish eye as a model system.
D. Non-Neuronal Tissues
In many vertebrates, the retina is intimately associated with some form of the vascular system (Wise et al., 1971). The mature zebrafish retina features two vessel systems: the choroidal and retinal vasculatures. The first of these tightly surrounds the retinal pigment epithelium, while the second differentiates on the inner surface of the retina (Alvarez et al., 2007; Kitambi et al., 2009). The development of the eye vasculature can be efficiently visualized using transgenic lines. Carriers of the fli-GFP and flk-GFP transgenes are suitable for this purpose (Choi et al., 2007; Lawson and Weinstein, 2002). In these strains, GFP-positive cells first appear in the retinal choroid fissure and the retina toward the end of the first 24 h of embryogenesis (Kitambi et al., 2009). By 48 hpf, a vascular bed forms on the medial surface of the lens (Alvarez et al., 2007; Kitambi et al., 2009). Initially, retinal blood vessels appear to adhere tightly to the lens. As the organism matures, however, vasculature appears to progressively lose contact with the lens and starts to adhere to the vitreal surface of the eye (Alvarez et al., 2007). In contrast to many mammals, including primates, blood vessels do not penetrate the neural retina in zebrafish (Alvarez et al., 2007). In addition to the vasculature, several other non-neuronal ocular tissues, such as the cornea, the iris, the ciliary body, and the lens, have been characterized in the zebrafish in detail (Dahm et al., 2007; Gray et al., 2009; Soules and Link, 2005; Zhang et al., 2009; Zhao et al., 2006).
III. Analysis of Wild-Type and Mutant Visual System
A major goal of eye research in zebrafish is to characterize phenotypes obtained in the course of new generations of forward and reverse genetic studies as well as small-molecule screens. Diverse research approaches are available to study the zebrafish retina. This chapter provides an overview of the available methods. While some techniques are described in detail, the majority are discussed only briefly because of space constraints, and references to sources of more comprehensive protocols are provided. Where applicable, other chapters of this volume are referenced as the source of more complete information. Table I lists some of the most important techniques currently available for the analysis of the zebrafish retina.
Table I.
Techniques Available to Study the Zebrafish Retina and Their Sources/Examples of Use
| Protocol | Goal | Sources/Examples of Use |
|---|---|---|
| Histological analysis | ||
| Electron microscopy | Evaluation of phenotype on a subcellular level | Allwardt et al. (2001), Doerre and Malicki (2002), Kimmel et al. (1981) |
| Light microscopy | Evaluation of phenotype on a cellular level | Malicki et al. (1996), Schmitt and Dowling (1994) |
| Molecular marker analysis | ||
| Antibody staining (whole mount) | Determination of expression pattern on protein level | Schmitt and Dowling (1996) |
| Antibody staining (sections) | Determination of expression pattern on protein level | Pujic and Malicki (2001), Wei and Malicki (2002) |
| In situ hybridization—double labeling | Parallel determination of two expression patterns on transcript level | Hauptmann and Gerster (1994), Jowett (2001), Jowett and Lettice (1994), Strahle et al. (1994) |
| In situ hybridization—frozen sections | Determination of expression pattern on transcript level | Barthel and Raymond (1993), Hisatomi et al. (1996) |
| In situ hybridization—whole mount | Determination of expression pattern on transcript level | Oxtoby and Jowett (1993), Thisse et al. (2004) |
| Gene function analysis | ||
| Implantation | Test of function for a factor (most often diffusible) via the implantation of a bead saturated with this substance | Hyatt et al. (1996), Martinez-Morales et al. (2005) |
| Morpholino knockdown | Test of gene function based on antisense inhibition of its activity | Eisen and Smith (2008), Nasevicius and Ekker (2000), Tsujikawa and Malicki (2004a) |
| Overexpression (DNA injections) | Test of gene function based on enhancement of its activity through DNA injections | Koster and Fraser (2001), Mumm et al. (2006) |
| Overexpression (light-mediated RNA/DNA uncaging) | Identification of gene function through enhancement of its activity in selected tissues at specific developmental stages | Ando et al. (2001), Ando and Okamoto (2003) |
| Overexpression (RNA injections) | Test of gene function based on enhancement of its activity through RNA injections | Macdonald et al. (1995), reviewed in Malicki et al. (2002) |
| Overexpression (UAS–GAL4 system) | Test of gene function in selected tissues using stable transgenic lines | Del Bene et al. (2008), Scheer and Campos-Ortega (1999) |
| TILLING (targeting induced local lesions in genomes) | Identification of chemically induced mutant alleles in a specific genetic locus | Colbert et al. (2001), Wienholds et al. (2002) |
| Zinc finger nucleases | Identification of mutant alleles in a specific locus | Doyon et al. (2008), Meng et al. (2008) |
| Embryological techniques | ||
| Cell labeling (caged fluorophore) | Fate determination for a specific group of cells | Take-uchi et al. (2003) |
| Cell labeling (iontophoretic) | Determination of morphogenetic movements or cell lineage relationships | Li et al. (2000b), Varga et al. (1999), Woo and Fraser (1995) |
| Cell labeling (lipophilic tracers) | Analysis of ganglion cell development (for example, retino tectal projection) | Baier et al. (1996), Malicki and Driever (1999) Mangrum et al. (2002) |
| Cell labeling (fluorescent protein transgenes) | Determination of cell fate and fine differentiation features in living animals | Hatta et al. (2006), Mumm et al. (2006), Neumann and Nuesslein-Volhard (2000) |
| Mitotic activity detection (BrdU) | Identification of mitotically active cell populations; birth dating | Hu and Easter (1999), Larison and Bremiller (1990), Del Bene et al. (2008) |
| Mitotic activity detection (tritiated thymidine) | Identification of mitotically active cell populations; birth dating | Nawrocki (1985) |
| Tissue ablation | Functional test for a field of cells via their removal by surgical means | Masai et al. (2000) |
| Transplantation (whole eye) | Test whether a defect (in axonal navigation, for example) originates within or outside the eye | Fricke et al. (2001) |
| Transplantation (fragment of tissue) | Functional test for a field of cells via their transplantation to an ectopic position by surgical means | Masai et al. (2000) |
| Transplantation (blastomere) | Test of cell autonomy of a mutant phenotype by generating a genetically mosaic embryo | Ho and Kane (1990), Jensen et al. (2001), Malicki and Driever (1999), Jing and Malicki (2009) |
| Behavioral tests | ||
| Optokinetic response | Test of vision based on eye movements; allows for evaluation of visual acuity | Brockerhoff et al. (1995), Clark (1981), Neuhauss et al. (1999) |
| Optomotor response | Test of vision based on swimming behavior | Clark (1981), Neuhauss et al. (1999) |
| Startle response | Simple test of vision based on swimming behavior | Easter and Nicola (1996) |
| Electrophysiological tests | ||
| ERG | Test of retinal function based on the detection of electrical activity of retinal neurons and glia | Avanesov et al. (2005), Brockerhoff et al. (1995) |
| Biochemical approaches | ||
| Co-immunoprecipitation from embryo extracts | Identification of direct and indirect protein binding partners | Insinna et al. (2008), Krock and Perkins (2008) |
| Tandem affinity purification from embryo extracts | Identification of direct and indirect protein binding partners | Omori et al. (2008) |
| Chemical screens | ||
| Screens of small-molecule libraries | Identification of chemicals that affect a developmental process | Kitambi et al. (2008) |
| Genetic screens | ||
| Behavioral | Detection of mutant phenotypes by behavioral tests | Muto et al. (2005), Neuhauss et al. (1999) |
| Histological | Detection of mutant phenotypes via histological analysis of sections | Mohideen et al. (2003) |
| Marker/tracer labeling | Detection of mutant phenotypes via staining with antibodies, RNA probes, or lipophilic tracers | Baier et al. (1996), Guo et al. (1999) |
| Morphological | Detection of mutant phenotypes by morphological criteria | Malicki et al. (1996) |
| Transgene guided | Detection of mutant phenotypes in transgenic lines expressing fluorescent proteins in specific cell populations | Xiao et al. (2005) |
In this table, we primarily cite experiments performed on the retina. Only where references to work on the eye are not available, we refer to studies of other organs. Most forward genetic approaches such as mutagenesis, mapping, and positional cloning methods do not contain visual system-specific features and thus are not listed in this table. These approaches are discussed in depth in other sections of this volume. Entries are listed alphabetically within each section of the table.
After 30 hpf, the observations of retinal development in the zebrafish embryo are hampered by the differentiation of pigment granules in the RPE. In immunohistochemical experiments, for example, the staining pattern is not accessible to visual inspection in whole embryos unless they are sectioned or their pigmentation is inhibited. To inhibit pigmentation, embryos are raised in media containing 1-phenyl-2-thiourea (PTU). Concentrations ranging from 75 to 200 μM are recommended (Karlsson et al., 2001; Westerfield, 2000). Starting between 2 and 3 dpf, embryogenesis is somewhat delayed in PTU-treated embryos, hatching is inhibited, and pectoral fins are abnormal (Karlsson et al., 2001). Appropriate controls have to be included to account for these deviations from normal embryogenesis. An additional disadvantage of using PTU is that it does not inhibit the differentiation of iridophores, which are present on the surface of the eye by 42 hpf, and by 4 dpf are dense enough to impair visualization of retinal cells with fluorescent probes. An alternative to using PTU is to conduct experiments on pigmentation-deficient animals. albino; roy double mutant line is the most useful for this purpose as it lacks both RPE pigmentation and iridophores (Ren et al., 2002). As crossing a mutation of interest into a pigmentation-deficient background takes two generations (or about 6 months), this approach is, however, time consuming.
A. Histological Analysis
Following morphological description, the first and the simplest step in the analysis of a phenotype is histological examination. It allows one to evaluate the major cell classes in the retina at the resolution that whole-mount preparations do not offer. Given the exquisitely precise organization of retinal neurons, histological analysis on tissue sections is frequently very informative. Plastic sections in particular offer very good tissue preservation and thus reveal fine detail. Prior to sectioning, tissue samples are usually embedded in either epoxy (epon, araldite) or in methacrylate (JB4) resins (Polysciences Inc.). Epoxy resins can be used for both light and electron microscopy. Several fixation methods suitable for plastic sections are routinely used (Li et al., 2000b; Malicki et al., 1996). For light microscopy, plastic sections are frequently prepared at 1–8 μm thickness and stained with an aqueous solution of 1% methylene blue and 1% azure II (Humphrey and Pittman, 1974; Malicki et al., 1996; Schmitt and Dowling, 1999).
Following transmitted light microscopy, histological analysis of mutant phenotypes can be performed at a higher resolution using electron microscopy. This allows one to inspect morphological details of subcellular structures, such as the photoreceptor outer segments, cell junctions, cilia, synaptic ribbons, mitochondria, and many other organelles (Allwardt et al., 2001; Doerre and Malicki, 2002; Schmitt and Dowling, 1999; Tsujikawa and Malicki, 2004b). These subcellular features frequently offer insight into the nature of the process being studied (Avanesov et al., 2005; Emran et al., 2010). Electron microscopy can be used in combination with diaminobenzidine (DAB) labeling of specific cell populations. Oxidation of DAB results in the formation of polymers which are chelated with osmium tetroxide and subsequently observed in the electron microscope (Hanker, 1979). Prior to microscopic analysis, cells can be selectively DAB-labeled using several approaches: photoconversion (Burrill and Easter, 1995), antibody staining combined with peroxidase detection (Metcalfe et al., 1990), or retrograde labeling with horseradish peroxidase (HRP) (Metcalfe, 1985).
B. The Use of Molecular Markers
A variety of molecular markers are used to study the zebrafish retina before, during, and after neurogenesis. Endogenous transcripts and proteins are among the most frequently used markers, although smaller molecules, such as neurotransmitters, and neuropeptides can also be used (Avanesov et al., 2005; Cameron and Carney, 2000). During early embryogenesis, the analysis of marker distribution allows one to determine whether the eye field is specified correctly. Several RNA probes are available to visualize the optic lobe during embryogenesis (Table II). Some of them label all cells of the optic lobe uniformly, while others can be used to monitor the optic stalk area (Table II). After the completion of neurogenesis, cell class-specific markers are used to determine whether particular cell populations are specified and occupy correct positions. Some of these markers are listed in Table II. Many transcript and protein detection methods have been described. Detailed protocols for most of these are available and we reference many of them in Table II. Below we discuss in detail the main types of molecular probes used to study the zebrafish visual system.
Table II.
Selected Molecular Markers Available to Study the Zebrafish Retina
| Name | Type | Expression pattern | Referencesa/Sources |
|---|---|---|---|
| Optic lobe, optic stalk markers | |||
| pax2a (pax 2) | RNA probe & Ab (poly) | Nasal retina, optic stalk (≤24 hpf); ON (2 dpf) | Kikuchi et al. (1997), Macdonald et al. (1997), Covance PRB-276P |
| rx1 (zrx1) | RNA probe | Anterior neural keel, optic primordia (≤11 hpf) | Chuang et al. (1999), Pujic and Malicki (2001) |
| rx2 (zrx2) | RNA probe | Anterior neural keel, optic primordia (≤11 hpf) | Chuang et al. (1999), Pujic and Malicki (2001) |
| rx3 (zrx3) | RNA probe | Anterior neural plate (≤9 hpf); optic primordia (≤12 hpf) | Chuang et al. (1999), Pujic and Malicki (2001) |
| six3a (six3) | RNA probe | Neural keel, optic primordia (≤11 hpf) | Pujic and Malicki (2001), Seo et al. (1998) |
| six3b (six6) | RNA probe | Anterior neural keel, optic primordia (≤11 hpf) | Pujic and Malicki (2001), Seo et al. (1998) |
| vax2 | RNA probe | Optic stalk (≤15 hpf); optic stalk, ventral retina (≤18 hpf) | Takeuchi et al. (2003) |
| Ganglion cell markers | |||
| alcamb (neurolin) | RNA probe, Ab (mono & poly) | Ganglion cells (28 hpf, RNA; ≤32 hpf protein) | Fashena and Westerfield (1999), Laessing et al. (1994), Laessing and Stuermer (1996), Zn-5/Zn-8 DSHB and ZIRC |
| cxcr4b | RNA probe | Ganglion cells (30 hpf) | Pujic et al. (2006) |
| gc34 | RNA probe | Ganglion cells (≤36 hpf) | Pujic et al. (2006) |
| L3 | RNA probe | Ganglion cells (30 hpf) | Brennan et al. (1997) |
| Tg (ath5:GFP) | Transgene | Ganglion cells (25 hpf) | Masai et al. (2003, 2005) |
| Tg (brn3c: gap43-GFP) | Transgene | Ganglion cells (42 hpf) | Xiao et al. (2005) |
| Amacrine cell markers | |||
| Ap2α | RNA probe | Amacrine cells (1.5–2 dpf) | Pujic et al. (2006) |
| Ap2β | RNA prove | Amacrine cells (≤36 hpf) | Pujic et al. (2006) |
| Choline acetyltransferase | Ab (poly) | Subset in INL and GCL, IPL (≤5 dpf) | Avanesov et al. (2005), Millipore, cat# AB144P |
| GABA | Ab (poly) | Subset in INL and GCL, IPL (2.5 dpf); ON (2 dpf) | Sandell et al. (1994), Millipore, cat# AB131; Sigma, cat# A2052 |
| GAD67 | Ab (poly) | Subset in INL and few in GCL, IPL (≤7 dpf) | Connaughton et al. (1999), Kay et al. (2001), Millipore, cat# AB9706 |
| Hu C/D | Ab (mono) | INL and GCL (≤3 dpf) | Kay et al. (2001), Link et al. (2000), Invitrogen, cat# A21271 |
| Neuropeptide Y | Ab (poly) | Subset in INL, IPL (≤4 dpf) | Avanesov et al. (2005), ImmunoStar, cat# 22940 |
| Parvalbumin | Ab (mono) | Subset in INL and GCL, IPL (≤3 dpf) | Malicki et al. (2003), Millipore, cat# MAB1572 |
| pax6a (pax6.1) | RNA probe Ab (poly) |
Neuroepithelium (12–34 hpf); INL and GCL (2 dpf); INL (5 dpf) | Hitchcock et al. (1996), Macdonald and Wilson (1997) |
| Serotonin | Ab (poly) | Subset in INL (≤5 dpf) | Avanesov et al. (2005), Sigma, cat# S5545 |
| Somatostatin | Ab (poly) | Subset in INL (≤5 dpf) | Malicki Lab, unpublished data; ImmunoStar, cat# 20067 |
| Substance P | Ab (mono) | Subset in INL (≤5 dpf) | Malicki Lab, unpublished data; AbCam, cat# AB6338 |
| Tyrosine hydroxylase | Ab (mono) | Subset in INL (3–3.5 dpf) | Biehlmaier et al. (2003), Pujic and Malicki (2001), ImmunoStar, cat# 22941; Millipore, cat# MAB318 |
| Bipolar cell markers | |||
| vsx1 | RNA probe | Neuroepithelium (31 hpf); outer INL (50 hpf) | Passini et al. (1997) |
| vsx2 | RNA probe | Neuroepithelium (24 hpf); primarily or exclusively in the bipolar cells (50 hpf) | Passini et al. (1997) |
| Protein kinase C-β1 | Ab (poly) | IPL, OPL (2.5 dpf); bipolar cell somata (4 dpf) | Biehlmaier et al. (2003), Kay et al. (2001), Santa Cruz, cat# sc-209 |
| Tg(nyx::Gal4VP16; UAS::MYFP) | Transgene | ON bipolar cells (2.5 dpf) | Schroeter et al. (2006) |
| Horizontal cell markers | |||
| Cx 52.6 | Ab (poly) | Horizontal cells (≤7 dpf?) | Shields et al. (2007) |
| Cx 55.5 | Ab (poly) | Horizontal cells (≤7 dpf?) | Shields et al. (2007) |
| Horizin | RNA probe | Horizontal cells, weak staining in GCL and inner INL (≤60 hpf) | Pujic et al. (2006) |
| Photoreceptor markers | |||
| Blue opsin | Ab (poly) | Blue cones (≤3 dpf) | Doerre and Malicki (2001), Vihtelic et al. (1999) |
| Blue opsinc | RNA probe | Blue cones (52 hpf) | Chinen et al. (2003), Raymond et al. (1995), Vihtelic et al. (1999) |
| Green opsin | Ab (poly) | Green cones (≤3 dpf) | Doerre and Malicki (2001), Vihtelic et al. (1999) |
| Green opsins (four genes) | RNA probes | Green cones (40–45 hpf) | Chinen et al. (2003), Takechi and Kawamura (2005), Vihtelic et al. (1999) |
| NDRG1 | RNA probe | Photoreceptors (36–48 hpf) | Pujic et al. (2006) |
| Red opsin | Ab (poly) | Red cones (≤3 dpf) | Doerre and Malicki (2001), Vihtelic et al. (1999) |
| Red opsins (two genes) | RNA probes | Red cones (40–45 hpf) | Chinen et al. (2003), Raymond et al. (1995), Takechi and Kawamura (2005) |
| Rod opsin | Ab (poly) | Rods (≤3 dpf) | Doerre and Malicki (2001), Vihtelic et al. (1999) |
| Rod opsinc | RNA probe | Rods (50 hpf) | Chinen et al. (2003), Raymond et al. (1995) |
| Tg (opn1sw1: EGFP) | Transgene | UV cones (≤56 hpf) | Takechi et al. (2003) |
| Tg (xops: GFP) | Transgene | Rods | Fadool (2003) |
| UV opsin | RNA probe | UV cones (56 hpf) | Hisatomi et al. (1996), Takechi et al. (2003) |
| UV opsin | Ab (poly) | UV cones (≤3 dpf) | Doerre and Malicki (2001), Vihtelic et al. (1999) |
| Zpr1 (FRet 43) | Ab (mono) | Double cones in larvae (48 hpf); double cones &bipolar cell subpopulation in the adult | Larison and Bremiller (1990), ZIRC |
| Zpr3 (FRet 11) | Ab (mono) | Rods (50 hpf) | Schmitt and Dowling (1996), ZIRC |
| Zs-4 | Ab (mono) | Rod inner segments (adult), onset unknown | Vihtelic and Hyde (2000), ZIRC |
| Müller glia markers | |||
| cahz (carbonic anhydrase) | RNA probe Ab (poly) | Müller glia (≤4 dpf) | Peterson et al. (1997, 2001) |
| GFAP | Ab (poly) | Müller glia (5 dpf) | Malicki Lab, unpublished data; DAKO, cat# Z0334 |
| Glutamine synthetase | Ab (poly) | Müller glia (60 hpf) | Peterson et al. (2001) |
| Tg (gfap: GFP) | Transgene | Müller glia (48 hpf) | Bernardos and Raymond (2006) |
| Plexiform layer markers | |||
| Phalloidin | Fungal toxin | IPL, OPL, ON (≤60 hpf) | Malicki et al. (2003), Invitrogen, cat# A-12379 |
| Snap-25 | Ab (poly) | OPL, IPL (≤2.5 dpf) | Biehlmaier et al. (2003), StressGen, cat# VAP-SV002 |
| SV2 | Ab (mono) | IPL, OPL (≤2.5 dpf) | Biehlmaier et al. (2003), DSHB |
| Syntaxin-3 | Ab (poly) | OPL (2.5 dpf); faint IPL (5 dpf) | Biehlmaier et al. (2003), Alamone Labs, cat# ANR-005 |
Approximate time of the expression onset is indicated in parenthesis. Sources of commercially available reagents are listed, including catalog numbers where appropriate. Names of markers are listed alphabetically within each section.
DSHB = Developmental Studies Hybridoma Bank (http://dshb.biology.uiowa.edu); ZIRC = Zebrafish International Resource Center (http://zfin.org/zirc/home/guide.php). dpf = days post fertilization; hpf = hours post fertilization; GCL = ganglion cell layer; INL = inner nuclear layer; IPL = inner plexiform layer; OPL = outer plexiform layer; ON = optic nerve.
When references to work performed on zebrafish are not available, experiments on related fish species are cited.
Zn-5 and Zn-8 antibodies both recognize neurolin (Kawahara et al., 2002).
Transcript expression onset was estimated by using goldfish probes (Raymond et al., 1995).
1. Antibodies
Antibody staining experiments can be performed in several ways. Staining of whole embryos is the least laborious. One has to keep in mind, however, that many antibodies produce high background in whole-mount experiments, and the eye pigmentation needs to be eliminated after 30 hpf as described above. At later stages of development, tissue penetration may become an additional problem. This can be circumvented by permeabilizing larvae via increasing detergent concentration above the standard level of 0.5% (2.5% Triton in both blocking and staining solution works well for anti Pax-2 antibody; see Riley et al., 1999) or by enzymatic digestion of embryos (for example collagenase treatment; see Doerre and Malicki, 2002). When background or tissue penetration is a problem, useful alternatives to using whole embryos is staining of either frozen or paraffin sections. Confocal microscopy of retinal sections reduces the background even further, while also enhancing the details of cell architecture.
For cryosectioning, embryos should be fixed as appropriate for a particular antigen and infiltrated in 30% sucrose/phosphate buffered saline (PBS) solution for cryopro−tection. While for many antigens simple overnight fixation in 4% paraformaldehyde (PFA) at 4°C is sufficient, some others require special treatments. For example, anti-gamma aminobutyric acid (GABA) staining of amacrine cells requires fixation in both glutaraldehyde and paraformaldehyde (2% each; see Avanesov et al., 2005; Sandell et al., 1994) (Fig. 3F). Glyoxal-based fixatives (such as Prefer fix supplied by Anatech) may also be useful when testing new antibodies (Dapson, 2007; Pathak et al., 2007). Fixed specimen can be oriented as desired using molds prepared from Eppendorf tubes that are cut transversely into ca. 3–4 mm wide rings. These are then placed flat on a glass slide and filled with embedding medium (Richard-Allan Scientific Inc.). Embryos are placed in the medium, oriented with a needle, and transferred into a cryostat chamber that is cooled to −20°C. Once the medium solidifies, plastic rings are removed with a razor blade.
Fig 3.
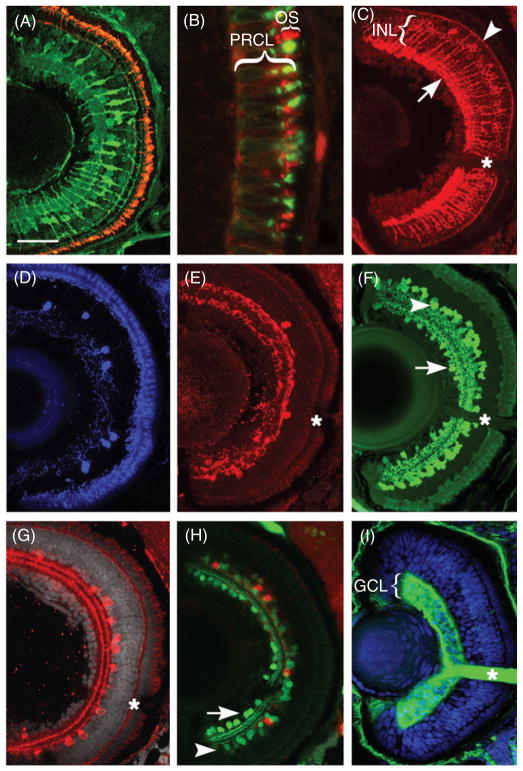
Transverse sections through the center of the zebrafish eye reveal several major retinal cell classes and their subpopulations. (A) Anti-rod opsin antibody detects rod photoreceptor outer segments (red), which are fairly uniformly distributed throughout the outer perimeter of the retina by 5dpf. On the same section, an antibody to carbonic anhydrase labels cell bodies of Müller glia in the INL as well as their radially oriented processes. (B) A higher magnification of the photoreceptor cell layer shows the distribution of rod opsin (red signal) and UV opsin (green signal) in the outer segments (OSs) of rods and short single cones, respectively. (C) A subpopulation of bipolar cells is detected using antibody directed to protein kinase C-β (PKC). While cell bodies of PKC-positive bipolar neurons are situated in the central region of the INL, their processes travel radially into the inner (arrow) and outer (arrowhead) plexiform layers, where they make synaptic connections. (D) Tyrosine hydroxylase-positive interplexiform cells are relatively sparse in the larval retina. (E) Similarly, the distribution of neuropeptide Y is limited to only a few cells per section. (F) The distribution of GABA, a major inhibitory neurotransmitter. GABA is largely found in amacrine neurons in the INL (arrowhead), although some GABA-positive cells are also found in the GCL (arrow). (G) Choline acetyltransferase, an enzyme of acetylcholine biosynthetic pathway, is restricted to a relatively small amacrine cell subpopulation. (H) Antibodies directed to a calcium-binding protein, parvalbumin, recognize another fairly large subpopulation of amacrine cells in the INL (green, arrowhead). Some parvalbumin−positive cells localize also to the GCL and most likely represent displaced amacrine neurons (arrow). By contrast, serotonin-positive neurons (red) are exclusively found in the INL. (I) Ganglion cells stain with the Zn-8 antibody directed to neurolin, a cell surface antigen (Fashena and Westerfield, 1999). In addition to neuronal somata, strong Zn-8 staining exists in the optic nerve (asterisk). In all panels lens is left, dorsal is up. A–H show the retina at 5dpf, while I shows a 3dpf retina. Asterisks indicate the optic nerve. Scale bar equals 50 μm in A and C–I and 10 μm in B. dpf, days post fertilization; GCL, ganglion cell layer; INL, inner nuclear layer; OS, outer segments; PRCL, photoreceptor cell layer. Panels D, G, and H are reprinted from Pujic and Malicki (2004) with permission from Elsevier. (See Plate no. 8 in the Color Plate Section.)
Antibody staining can be efficiently performed on 15–30 μm frozen sections, and analyzed by confocal microscopy. For conventional epifluorescence microscopy, thinner sections may be desired. Upon the application of modified infiltration and embedding protocols, 3 μm sections of the zebrafish embryos can be prepared and analyzed using a conventional microscope equipped with UV illumination (Barthel and Ray−mond, 1990). Some antigens require the application of additional steps during staining protocols, such as antigen retrieval. Sections are immersed in near-boiling solution of 10 mM sodium citrate for 10 min prior to the application of blocking solution. This method significantly improves the labeling of amacrine cell populations by anti-serotonin or anti-choline acetyltransferase antibodies (Fig. 3G and H) (Avanesov et al., 2005). Immersion in cold acetone is another treatment that improves staining with some immunoreagents, such as certain anti-gamma-tubulin antibodies (Pujic and Malicki, 2001).
Alternatively, antibody staining can be performed on plastic sections. Anti-GABA antibodies, for example, work very well with this method. Both epoxy (Epon-812, Electron Microscopy Sciences Inc.) and methacrylate (JB-4, Polysciences Inc.) resins can be used as the embedding medium. This improves the quality of staining, as plastic sections preserve tissue morphology better, compared to frozen ones. In the GABA staining protocol, primary antibody can be detected using avidin–HRP conjugate (Vector Laboratories Inc.) or a fluorophore-conjugated secondary antibody (Fig. 2F and Malicki and Driever, 1999; Sandell et al., 1994). An extensive collection of antibodies that can be used to visualize features of the retina in the adult zebrafish has been also characterized (Yazulla and Studholme, 2001).
2. mRNA Probes
In situ hybridization with most RNA probes works very well on whole embryos (Oxtoby and Jowett, 1993). Following hybridization, embryos are gradually dehydrated in a series of ethanol solutions of increasing concentration, and embedded in plastic as described above (Pujic and Malicki, 2001). An additional fixation step prior to dehydration reduces the leaching of in situ signal (Westerfield, 2000). Expression patterns are then analyzed on 1–5 μm thick sections. Several in situ protocols are available to monitor the expression of two genes simultaneously (Jowett, 2001, and references in Table II; Jowett and Lettice, 1994). In the experiment shown in Fig. 4B, expression patterns of two opsins are detected simultaneously using two different chromogenic substrates of alkaline phosphatase (AP) (Hauptmann and Gerster, 1994). In situ hybridization can also be combined with antibody staining (Novak and Ribera, 2003; Prince et al., 1998). In embryos older than 5 dpf, in situ reagents sometimes do not penetrate to the center of the retina. In such cases, hybridization procedures can be performed more successfully on sections (Hisatomi et al., 1996). Given the small size of zebrafish embryos, in situ hybridization experiments can be performed in a high-throughput fashion using hundreds or even thousands of probes to screen for genes expressed in specific organs, tissues, or even specific cell types (Thisse et al., 2004). In recent years, in situ hybridization could also be performed using robotic devices that carry out most of the tedious steps, including hybridizations and washes (Intavis Bioanalytical Instruments AG). This approach was also applied to the retina and led to the identification of numerous transcripts expressed in subpopula−tions of retinal cells (Pujic et al., 2006). Some of these transcripts can be used as markers of specific retinal cell classes (Table II).
Fig 4.
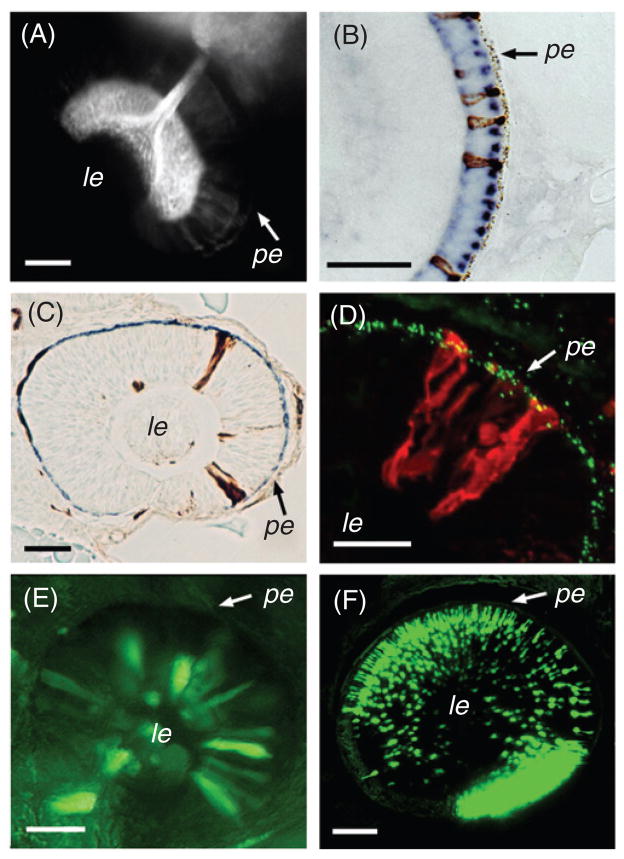
Selected techniques available to study neurogenesis in the zebrafish retina. (A) DiI incorporation into the optic tectum retrogradely labels the optic nerve and ganglion cell somata. (B) A transverse plastic section through the zebrafish retina at 3 dpf. In situ mRNA hybridization using two probes, each targeted to a different opsin transcript and detected using a different enzymatic reaction, visualizes two types of photoreceptor cells. (C) A plastic section through a genetically mosaic retina at ca. 30 hpf. Biotinylated dextran-labeled donor-derived cells incorporate into retinal neuroepithelial sheet of a host embryo and can be detected using HRP staining (brown precipitate). (D) A transverse cryosection through a genetically mosaic zebrafish eye at 36 hpf. In this case, donor-derived clones of neuroepithelial cells are detected with fluorophore-conjugated avidin (red). The apical surface of the neuroepithelial sheet is visualized with anti-γ-tubulin antibody, which stains centrosomes (green). (E) GPF expression in the eye of a zebrafish embryo following injection of a DNA construct containing the GFP gene under the control of a heat-shock promoter. The transgene is expressed in only a small subpopulation of cells. (F) A confocal z-series through the eye of a living transgenic zebrafish, carrying a GFP transgene under the control of a rod opsin promoter (Fadool, 2003). Bright expression is present in rod photoreceptor cells (ca. 3 dpf). Scale bar, 50 μm. pe, pigmented epithelium; le, lens. Panel E reprinted from Malicki et al. (2002) with permission from Elsevier.
3. Lipophilic Tracers
Details of cell morphology can also be studied using lipophilic carbocyanine dyes, DiI, DiO, and others, which label cell membranes (Honig and Hume, 1986; 1989). In the retina, these are especially useful in the analysis of ganglion cells. Carbocyanine dyes can be used as anterograde as well as retrograde tracers. When applied to the retina, DiI and DiO allow one to trace the retinotectal projections (Baier et al., 1996). When applied to the optic tectum or the optic tract, they can be used to determine the position of ganglion cell perikarya, and even to study the stratification and branching of ganglion cell dendrites (Burrill and Easter, 1995; Malicki and Driever, 1999; Mangrum et al., 2002). Since DiI and DiO have different emission spectra, they can be used simultaneously to label different cell populations (Baier et al., 1996).
4. Fluorescent Proteins
Fluorescent proteins (hereafter FPs), frequently fused to other polypeptides, offer a very rich source of markers to visualize tissues, cells, and even subcellular structures.
These can be expressed in embryos either transiently or from stably integrated trans-genes. Numerous derivatives of two FPs—green fluorescent protein (GFP, from jellyfish, Aequorea victoria) and red fluorescent protein (RFP, from coral species)—are currently available (reviewed in Shaner et al., 2007) and differ in brightness as well as emission spectra. Many of them have been applied in zebrafish. The uses of FPs can be grouped in at least three categories:
-
Visualization of gene activity. The purpose of these experiments is to determine where and when a gene of interest is transcribed. Although the same goal can be accomplished using in situ hybridization, the use of FP fusions may result in higher sensitivity of detection (see for example a sonic hedgehog study by Neumann and Nuesslein-Volhard, 2000), and, importantly, allow one to create time-lapse images tracking spatial-temporal changes in gene expression. The biggest challenge in this type of study is the need to include all regulatory elements in the transgene to faithfully recapitulate the expression of the endogenous transcript. The best way to accomplish that is to insert an FP coding sequence into the open reading frame of a gene derived from a phage or bacterial artificial chromosome (PAC or BAC). For example, to study the expression of zebrafish green opsin genes, a modified PAC clone of ca. 85 kb was used to generate transgenic lines. To visualize expression, the first exon after the initiation codon was replaced with a GFP sequence in each of these genes (Tsujimura et al., 2007). The use of artificial chromosomes is frequently necessary as distant regulatory elements are likely to affect the expression of a given gene. One has to note, however, that even using an artificial chromosome does not assure that all relevant regulatory elements will be included in the transgene.
In some experiments, when temporal characteristics of expression need to be faithfully reproduced, excessive stability of FP may pose a problem. FPs tend to be stable in the cell’s cytoplasm and may persist for much longer than the transcript of the gene being studied, making it difficult to determine when the gene of interest is turned off. To circumvent this difficulty, FPs characterized by reduced stability, such as dRFP (destabilized RFP) or short half-life GFP, are available (Yeo et al., 2007; Yu et al., 2007). dRFP was used, for example, to study Notch pathway activity in the zebrafish retinal neuroepithelium (Del Bene et al., 2008).
Visualization of the subcellular localization of proteins. In this type of experiment, it is not necessary to recapitulate the tissue distribution of the protein being studied and thus expression can be driven ubiquitously. Consequently, transient expression methods based on mRNA or DNA injection are preferred. Although they usually do not allow for the targeting of expression to particular tissues, they are much less time consuming, compared to using stable transgenic lines. The expression of FP fusions is especially valuable when antibodies are difficult to generate, as has been the case for the Elipsa protein, for example (Omori et al., 2008). This procedure is not without drawbacks, however. First, adding GFP polypeptide to a protein may change its binding properties, and thus cause aberrant localization in the cell. Second, as FP fusions are frequently expressed at a higher level compared to their wild-type counterparts, they may display nonspecific binding. Finally, fusion proteins may be toxic to cells. These problems can be largely, although not entirely, eliminated by placing FP tags in multiple locations and testing whether the resulting fusion proteins can rescue mutant/morphant phenotypes.
Monitoring of fate, differentiation, and cell physiology. In these studies, FP fusions are used solely to mark cells and/or subcellular structures. In the simplest case, this approach can be used to monitor the gross morphology of the cell and its survival. In more sophisticated variants of this technique, one monitors cell division patterns, migration trajectories, or specific aspects of cell morphology, such as the shape of dendritic processes, subcellular distribution of organelles, or intracellular transport. Zebrafish FP transgenic lines have been generated to monitor the differentiation of fine morphological features of various retinal cell classes, including bipolar interneurons (Schroeter et al., 2006), horizontal interneurons (Shields et al., 2007), amacrine interneurons (Godinho et al., 2005; Kay et al., 2004), ganglion cells (Xiao et al., 2005), and Müller glia (Bernardos and Raymond, 2006) (Table II). These transgenic lines allow one to continuously observe fine features of cells, and even follow the entire trajectory of the retinotectal projection, or the phylopodia of differentiating bipolar cell axon terminals. In most studies conducted so far, FP fusions were expressed from stably integrated transgenes, although in some cases the GAL4–VP16-based system (Koster and Fraser, 2001, see below) is used to drive transient expression in retinal interneurons (Mumm et al., 2006; Shields et al., 2007). While generating stable transgenic lines, it is necessary to compare expression patterns from at least two different transgenic lines since the integration of same construct can produce very different expression patterns in different lines, due to position-specific effects. For example, depending on the integration site, a hexamer of the DF4 regulatory element of the Pax6 gene can drive expression either throughout the retina or in subsets of amacrine cells (Godinho et al., 2005; Kay et al., 2004).
In some experimental contexts, FPs can also be used to monitor the behavior of cellular organelles. This is accomplished by generating FPs fused to subcellular localization signals or to entire proteins that display a desirable subcellular localization. The H2A-GFP transgene, for example, allows one not only to visualize cell nuclei but also to distinguish when cells undergo mitosis, and even to determine the orientation of mitotic spindles in the retinal neuroepithelium (Cui et al., 2007; Pauls et al., 2001). Similarly, GFP-centrin can be used to monitor the position of the centrosome in differentiating ganglion cells (Zolessi et al., 2006), and GFP fused to a mitochondrial localization sequence can be applied to observe the distribution of mitochondria (Kim et al., 2008). GFP fused to the 44 C-terminal amino acids of rod opsin is targeted to the photoreceptor outer segment and can be used as a specific marker of this structure (Perkins et al., 2002). FPs can also be applied to mark specific cell membrane domains: PAR-3/EGFP fusion, for example, labels the apical surface of retinal neuroepithelial cells (Zolessi et al., 2006).
Photoconvertible FPs are yet another class of markers that can be used to visualize cell morphology. Kaede and Dronpa have been used most frequently in the zebrafish so far (Aramaki and Hatta, 2006; Hatta et al., 2006; Sato et al., 2006). Kaede is irreversibly converted from green to red fluorescence using UV irradiation, whereas Dronpa green fluorescence can be reversibly activated and deactivated multiple times by irradiating it with blue and UV light, respectively. The advantage of these FPs is that they can be used to reveal morphology of single neurons by selective photocon−version in the cell soma (anterograde labeling) or in cell processes (retrograde labeling). This is particularly useful when appropriate regulatory elements are not available to drive FP expression in specific cell populations. Moreover, one can potentially use photoactivatable FPs to trace the journey of tagged proteins within cells. Although as yet this approach has not been applied in the zebrafish retina, it is potentially useful to analyze protein trafficking in photoreceptor cells.
The number of different FPs and the variety of their applications in zebrafish have been growing at a breathtaking pace. Given the multitude of available promoter sequences, the diversity of spectral variants, and the variety of methods for protein expression in the zebrafish embryo, one is frequently confronted with the task of generating multiple combinations of regulatory elements and FP tags. This is made easier by recombination cloning approaches (Kwan et al., 2007; Villefranc et al., 2007; see the description of the Gateway cloning system on page 241). The use of FPs to monitor the divisions, movements, and differentiation of cells and their organelles has been one of the fastest growing approaches in the studies of zebrafish embryogenesis.
C. Analysis of Cell Movements and Lineage Relationships
The best-established and the most versatile approach to cell labeling in living zebrafish embryos is iontophoresis. This technique was applied in numerous zebra-fish cell fate studies (Collazo et al., 1994; Devoto et al., 1996; Raible et al., 1992). In the context of visual system development, iontophoretic cell labeling was used to determine the developmental origins of the optic primordium (Woo and Fraser, 1995) and later to study cell rearrangements that accompany optic cup morphogenesis (Li et al., 2000b). A potentially very informative variant of cell fate analysis is to perform it in the retinae of mutant animals (Poggi et al., 2005; Varga et al., 1999). Iontophoretic cell labeling has been applied to study cell lineage relationships in the developing retina of Xenopus laevis (Holt et al., 1988; Wetts and Fraser, 1988). Lineage analysis has been performed in the zebrafish retina to a very limited extent, perhaps because of the perception that it would be unlikely to add much to the results previously obtained in higher vertebrates (Holt et al., 1988; Turner and Cepko, 1987; Turner et al., 1990). An alternative to iontophoresis is the activation of caged fluorophores using a laser beam. Caged flourescein (Molecular Probes, Inc.) is particularly popular in this type of experiment, and was applied to study cell fate changes caused by a double knockdown of vax1 and vax2 gene function (Take-uchi et al., 2003). One study of lineage relationships in the zebrafish eye also took advantage of a transgenic line that expresses GFP in retinal progenitor cells (Poggi et al., 2005).
D. Analysis of Cell and Tissue Interactions
Transplantation techniques are used to reveal cell or tissue interactions. The size of a transplant varies from a small group of cells, or even a single cell, to the entire organ. In the case of mutations that affect retinotectal projections, it is important to determine whether defects originate in the eye or in brain tissues. This can be accomplished by transplanting the entire optic lobe at 12 hpf, and allowing the animals to develop until desired stages (Fricke et al., 2001). Smaller size fragments of tissue can be transplanted to document cell–cell signaling events within the optic cup. This approach was used to demonstrate inductive properties of the optic stalk tissue, and to test the presence of cell–cell interactions within the optic cup (Kay et al., 2005; Masai et al., 2000). Transplantation can also be used to study interactions between the lens and the retina. Lens transplantation is performed following a procedure similar to that developed for Astyanax mexicanus (Yamamoto and Jeffery, 2002) and recently applied to zebrafish (Zhang et al., 2009)
Mosaic analysis is a widely used approach that combines genetic and embryological manipulations (Ho and Kane, 1990). The goal of such experiments is to determine the site of the genetic defect responsible for a mutant phenotype. In simple terms, cell-autonomous phenotypes are caused by gene function defects within the affected cells, while cell-nonautonomous phenotypes are caused by defects in other (frequently neighboring) cells. In contrast to approaches used in Drosophila, genetic mosaics in zebrafish are generated via blastomere transplantation, essentially a surgical procedure performed on the early embryo (Ho and Kane, 1990; Westerfield, 2000). As this technique has been widely used in zebrafish, also in the context of eye development, we provide a more extensive description of how it is applied.
In the first step, the donor embryos are labeled at the one- to eight-cell stage with a cell tracer. Dextrans conjugated with biotin or a fluorophore are the most commonly used tracers, and frequently a mix of both is used. The choice of the tracer depends on how it is going to be detected during later stages of the experiment, when the fate of donor-derived cells is analyzed. Within a few minutes after injection into the yolk, tracers diffuse throughout the embryo, labeling all blastomeres. Subsequently, starting at about 3 hpf, blastomeres are transplanted from tracer-labeled donor embryos to unlabeled host embryos using a glass needle. The number of transplanted blastomeres usually varies from a few to hundreds, depending on the experimental context. One donor embryo is frequently sufficient to supply blastomeres for several hosts. The transplanted blastomeres become incorporated into the host embryo and randomly contribute to various tissues, including those of experimental interest. To increase the frequency of donor-derived cells in the retina, blastomeres should be transplanted into the animal pole of a host embryo (Moens and Fritz, 1999). Cells in that region will later contribute to eye and brain structures (Woo and Fraser, 1995). Embryos that contain descendants of donor blastomeres in the eye are identified using UV illumination between 24 and 30 hpf, when the retina is only weakly pigmented and contains large radially oriented neuroepithelial cells (Fig. 4C and D). An elegant way to control cell autonomy tests is to transplant cells from two donor embryos—one wild type, one mutant—into a single host (Ho and Kane, 1990). In such a case, each of the donors has to be labeled with a different tracer.
Tracer purity and the quality of the transplantation needle are two important technical aspects of mosaic analysis. To increase the survival rate of donor embryos and transplanted cells, it is important to purify dextran by filtering it through a spin column several times (Microcon YM-3, Millipore Inc.). This procedure removes small molecular weight contaminants that are toxic for cells. The preparation of transplantation needle requires considerable manual dexterity, and is fairly time consuming. A good transplantation needle has several features: (1) a smooth opening with a diameter that is slightly larger than blastomeres at the “high” stage (Kimmel et al., 1995); (2) a fairly constant width near the tip; (3) lumen free of glass debris, which frequently accumulate when the needle is beveled; and (4) a sharp glass spike at the very tip, to help in penetrating the embryo. Needle preparation requires two instruments: a beveler and a microforge, available from WPI and Narishige, respectively. Useful technical details of needle preparation and other aspects of blastomere transplantation protocol are provided in The Zebrafish Book (Westerfield, 2000).
Following successful transplantations, the analysis of donor-derived cells in mosaic embryos can proceed in several ways. In the simplest case, the donor-derived cells are labeled with a fluorescent tracer or a transgene and directly analyzed in whole embryos using conventional or confocal miscroscopy (Zolessi et al., 2006). Such analysis is sufficient to provide information about the position and sometimes the morphology of donor-derived cells. When more detailed analysis is necessary, the donor-derived cells can be further analyzed on frozen or plastic sections (Avanesov et al., 2005). In such cases, the donor blastomeres are usually labeled with both fluorophore- and biotin-conjugated dextrans. The fluorophore-conjugated tracer is used to distinguish which embryos contain donor-derived cells in the desired tissue as described above. The biotin-conjugated dextran, on the other hand, is used in detailed analysis at later developmental stages. The HRP-conjugated streptavidin version of the ABC kit (Vector Laboratories Inc.) or fluorophore-conjugated avidin (Jackson ImmunoRe−search Inc., Molecular Probes, Inc.) can be used to detect biotinylated dextran (Fig. 4C and D, respectively). HRP detection can be performed on whole mounts and analyzed on plastic sections, as described above for histological analysis. In contrast to that, fluorophore-conjugated avidin is preferably used after sectioning of the frozen tissue, owing to degradation of some flurophores during embedding of specimen in plastic. In these experiments, cryosections are prepared as described for antibody staining above. In some experiments, it is desirable to analyze the donor-derived cells for the expression of molecular markers (see Fig. 4D for an example). On frozen sections, avidin detection of donor-derived cells can be combined with antibody staining. Another way to visualize donor-derived cells and analyze expression at the same time is to combine HRP detection of donor-derived cells with in situ hybridization or antibody staining (Halpern et al., 1993; Schier et al., 1997). When HRP is used for the detection of donor-derived cells, the resulting reaction product inhibits the detection of the in situ probe with AP (Schier et al., 1997). Because of this, the opposite sequence of enzymatic detection reactions is preferred: in situ probe detection first and HRP staining second.
When mosaic analysis is performed in the zebrafish retina at 3 dpf or later, the dilution of a donor-cell tracer can make the interpretation of the results difficult. This is because the descendants of a single transplanted blastomere divide a variable number of times. Thus in the donor-derived cells which undergo the highest number of divisions the label may be diluted so much that it is no longer detectable. In mosaic animals, such a situation can lead to the appearance of a mutant phenotype or to the rescue of a mutant phenotype in places seemingly not associated with the presence of donor cells and complicate the interpretation of experimental results. Increasing the concentration of the tracer or, in the case of whole-mount experiments, improving the penetration of staining reagents can sometimes alleviate this problem. Alternatively, collagenase treatment of fixed embryos improves reagent penetration during the detection of donor-derived cells (Doerre and Malicki, 2001). The amount of injected dextran should be increased carefully as excessively high concentrations are lethal for labeled cells.
If the dilution of tracer cannot be circumvented, an excellent alternative is the use of transgenes. An ideal transgene to mark donor cells in mosaic analysis would drive the expression of FP at a high level in all cells throughout development. In the context of the retina, the mCFP Q01 line largely meets these requirements, although its expression becomes somewhat dimmer as development advances (Godinho et al., 2005). This line has been used, for example, to study photoreceptor and glia defects in ale oko mutant retinae (Jing and Malicki, 2009). An additional advantage of using transgenic FP tracers is that they eliminate the need for tracer injections into the donors, which decreases mechanical damage to embryos. Lastly, FP are relatively nontoxic, which increases the survival of donor-derived cells further. A disadvantage of transgene use in this context is that it takes one generation to in-cross an FP transgene into a mutant line. In summary, mosaic analysis is an important approach that has been widely used to study zebrafish retinal mutants (Avanesov et al., 2005; Cerveny et al., 2010; Doerre and Malicki, 2001; 2002; Goldsmith et al., 2003; Jensen et al., 2001; Jing and Malicki, 2009; Krock et al., 2007; Link et al., 2000; Malicki and Driever, 1999; Malicki et al., 2003; Pujic and Malicki, 2001; Wei and Malicki, 2002; Yamaguchi et al., 2010).
E. Analysis of Cell Proliferation
Several techniques are available to study cell proliferation in the retina. The amount of cell proliferation, the timing of cell cycle exit (birth date), and cell cycle length can be evaluated by H3-thymidine labeling (Nawrocki, 1985) or via bromodeoxyuridine (BrdU) injections into the embryo (Hu and Easter, 1999). Such studies can be very informative in mutant animals (Kay et al., 2001; Link et al., 2000; Yamaguchi et al., 2008). To identify the population of cells that exit the cell cycle in a particular window of time, BrdU labeling can be combined with iododeoxyuridine (IdU) (Del Bene et al., 2008). Finally, another useful technique that can be used to test for cell cycle defects in mutant strains is fluorescence activated cell sorting (FACS) of dissociated retinal cells (Plaster et al., 2006; Yamaguchi et al., 2008).
F. Behavioral Studies
Several vision-dependent behavioral responses have been described in zebrafish larvae and adults: the optomotor response (Clark, 1981), the optokinetic response (Clark, 1981; Easter and Nicola, 1996), the startle response (Easter and Nicola, 1996), the phototaxis (Brockerhoff et al., 1995), the escape response (Li and Dowling, 1997), and the dorsal light reflex (Nicolson et al., 1998). Not surprisingly, larval feeding efficiency also depends on vision (Clark, 1981). While some of these behaviors are already present by 72 hpf, others have been described in adult fish only (for a review see Neuhauss, 2003). The vision-dependent behaviors of zebrafish proved to be very useful in genetic screening (see Phenotype Detection Methods on page 244). The optokinetic response appears to be the most robust and versatile. It is useful both in quick tests of vision and in quantitative estimates of visual acuity. In addition to genetic screens, behavioral tests have been used to study the function of the zebrafish optic tectum (Roeser and Baier, 2003).
G. Electrophysiological Analysis of Retinal Function
In addition to behavioral tests, measurements of electrical activity in the eye are another, more precise way to evaluate retinal function. Electrical responses of the zebrafish retina can be evaluated by electroretinography (ERG) already by 4 dpf (for example, Avanesov et al., 2005). Similar to other vertebrates, the zebrafish ERG response contains two main waves: a small negative a-wave, originating from the photoreceptor cells, and a large positive b-wave, which reflects the function of the INL (Dowling, 1987; Makhankov et al., 2004). The goal of an ERG study in zebrafish is no different from that of a similar procedure performed on the human eye. ERG can be used to evaluate the site of retinal defects in mutant animals. Ganglion cell defects do not affect the ERG response (Gnuegge et al., 2001), whereas the absence of the a-wave or the b-wave suggests a defect in photoreceptors or in the INL, respectively. The a-wave appears small in ERG measurements because of an overlap with the b-wave. To measure the a-wave amplitude, the b-wave has to be blocked pharmacologically (Kainz et al., 2003). An additional ERG wave, the d-wave, is produced when longer (ca. 1 s) flashes of light are used. Referred to as the OFF response, the d-wave is thought to reflect the activity of OFF-bipolar cells and photoreceptors (Kainz et al., 2003; Makhankov et al., 2004).
Retinal responses are usually elicited using a series of light stimuli that vary by several orders of magnitude in intensity (Allwardt et al., 2001; Kainz et al., 2003). This allows the evaluation of the visual response threshold, a parameter that is sometimes abnormal in mutant animals (Li and Dowling, 1997). Another important variable in ERG measurements is the level of background illumination. ERG measurements can be performed on light-adapted retinae using background illumination of a constant intensity, or on dark-adapted retinae, which are maintained in total darkness for at least 20 min prior to measurements (Kainz et al., 2003). Most frequently recordings are performed on intact anesthetized animals (Makhankov et al., 2004). Alternatively, eyes may be gently removed from larvae and bathed in an oxygenated buffer solution. The latter ensures the oxygen supply to the retina in the absence of blood circulation (Kainz et al., 2003). ERG recordings have become a standard assay when evaluating zebrafish eye mutants (Allwardt et al., 2001; Avanesov et al., 2005; Biehlmaier et al., 2007; Brockerhoff et al., 1998; Kainz et al., 2003; Makhankov et al., 2004; Morris et al., 2005).
In addition to ERG, other more technically sophisticated electrophysiological measurements can be used to evaluate zebrafish (mutant) retinae. The ganglion cell function, for example, can be evaluated by recording action potentials from the optic nerve (Emran et al., 2007). Such measurements revealed ganglion cell defects in the retinae of nbb and mao mutants (Gnuegge et al., 2001; Li and Dowling, 2000). Similarly, photoreceptor function has been evaluated by measuring outer segment currents in isolated cells (Brockerhoff et al., 2003).
H. Biochemical Approaches
Genetic experiments in animal models are frequently supplemented with studies of protein–protein interactions. Although this type of analysis has not been traditionally a strength of the zebrafish model, zebrafish embryos can be used to analyze binding interactions. In the context of the visual system, biochemical analysis has been largely applied to study the intraflagellar transport (IFT) in photoreceptor outer segment formation. As IFT occurs in many tissues, it can be studied via co-immunoprecipitation from embryonic or larval extracts (Krock and Perkins, 2008). Alternatively, extracts from the retinae of adult animals can be used in this type of experiment (Insinna et al., 2008). A clear advantage of using larvae is that one can apply biochemical methods to analyze mutant phenotypes. As most zebrafish mutants are lethal at embryonic or larval stages, adult retinae are not suitable for this purpose. In addition to immunopre−cipitation experiments, a more sophisticated but also more laborious and technically demanding approach is to identify binding partners by tandem affinity purification (TAP) (reviewed in Collins and Choudhary, 2008). The TAP tag procedure involves attaching a peptide tag to the protein of interest, and expressing it in zebrafish embryos. Following the preparation of embryonic extract, the peptide tag is used to purify the bait protein along with its binding partners using appropriate affinity columns. The identities of the binding partners are established using mass spectrometry. The TAP tag approach was applied in the zebrafish to identify the binding partners of Elipsa, a determinant of outer segment differentiation (Omori et al., 2008). It is a relatively demanding technique, as it requires the expression of the bait protein in thousands of embryos. As more efficient affinity purification tags are engineered (Burckstummer et al., 2006), TAP is likely to become easier to apply in the zebrafish.
I. Chemical Screens
Another approach that is gaining popularity in the zebrafish model is the screening of chemical libraries for compounds that affect developmental processes. The characteristics that render the zebrafish embryo suitable for genetic experiments—small size, rapid development, and transparency—also make it exceptionally useful for small-molecule screening (Kokel et al., 2010; Peterson et al., 2000; Tran et al., 2007; Zon and Peterson, 2005). In this type of experiment, hundreds or even thousands of small batches of embryos are each exposed to a different chemical compound, and analyzed for developmental or behavioral changes. Such an approach has been applied either to wild-type embryos or to carriers of genetic defects (Cao et al., 2009; North et al., 2007, 2009; Peterson et al., 2004). In the latter case, compounds that rescue a mutant phenotype can be screened for. When mutations that resemble human abnormalities are used, this approach can be a powerful way to identify chemicals of potential therapeutic importance (Cao et al., 2009; Hong et al., 2006; Peterson et al., 2004).
Chemical compound libraries ranging in size from hundreds to tens of thousands of molecules are commercially available. Phenotype detection approaches in a small-molecule screen are potentially as varied as in a genetic screen (Kaufman et al., 2009; Phenotype Detection Methods on page 244). Gross evaluation of morphological features is the simplest option. Transgenic lines that express FPs in target tissues make it possible to detect subtle phenotypes. In a recent experiment, for example, an flk-GFP transgenic line was used to screen ca. 2000 small molecules for their effects on retinal vasculature (Kitambi et al., 2009). Although little precedent exists at this time for small-molecule screens focusing on retinal development, this approach has been successful in the analysis of several zebrafish organs and behaviors (Hong et al., 2006; Kokel et al., 2010; North et al., 2007; Sachidanandan et al., 2008), and thus is also likely to find its way into the studies of the visual system.
IV. Analysis of Gene Function in the Zebrafish Retina
A. Reverse Genetic Approaches
A series of mutant alleles of varying severity is arguably the most informative tool of gene function analysis. Although a great variety of mutant lines have been identified in forward genetic sceens, for many loci chemically induced mutant alleles are not yet available. In these cases, other approaches must be applied to study gene function. In this section, we briefly discuss advantages and disadvantages of different loss-of−function and gain-of-function approaches in the context of the zebrafish visual system, and provide references to more comprehensive discussions of each.
1. Loss-of-Function Analysis
In the absence of loss-of-function mutations, antisense-based interference is by far the most common way to obtain information about gene function in the zebrafish embryo (Nasevicius and Ekker, 2000). The reasons for this popularity are low cost and low labor expense involved in their use. Although antisense morpholino-modified oligonucleotides have been shown to reproduce mutant phenotypes quite well, their use suffers from two main disadvantages. First, they become progressively less effective as development proceeds, presumably because of degradation. Second, some morpholinos produce nonspecific toxicity, which must be distinguished from specific features of a morpholino-induced phenotype. Morpholino oligos can be used to interfere either with translation initiation or with splicing. Importantly, the efficiency of splice-site morpholinos can be monitored by reverse transcription polymerase chain reaction (RT-PCR) (Draper et al., 2001; Tsujikawa and Malicki, 2004b). In general, splice-site morpholinos reduce wild-type transcript expression below the level of RT−PCR detection throughout the first 36 h of development, although some have been reported to remain active until 3 or even 5 dpf (Tsujikawa and Malicki, 2004b). Most morpholinos are thus sufficient to interfere with genetic pathways involved in retinal neurogenesis but not to study later differentiation events or retinal function. Some help in designing morpholinos can be obtained from their manufacturer (Gene Tools LLC). Detailed protocols for the use of morpholinos, including their target site homology requirements, injection protocols, and methods to control for specificity, are available in literature (reviewed by Eisen and Smith, 2008; Malicki et al., 2002).
A powerful alternative to the use of morpholinos in loss-of-function studies is TILLING (targeted induced local lesions in genomes) (Colbert et al., 2001; McCal−lum et al., 2000; Wienholds et al., 2002). This approach combines chemical mutagen−esis with a PCR-based protocol for detecting mutations in a locus of choice, and yields a series of mutant alleles that vary in strength. Its main disadvantage is the vast amount of preparation that needs to be done to initiate these experiments. One particularly labor-intensive step is the collection of thousands of sperm and DNA samples from F1 males. Because of this limitation, TILLING experiments are frequently performed by core facilities, which serve a group of laboratories, or the entire research community.
A recent addition to mutagenesis approaches in zebrafish is the use of zinc finger nucleases (ZFNs) to induce lesions in desired genes. ZFNs consist of a DNA recognition module, essentially a tandem array of two to four zinc finger-type DNA binding domains, and a catalytic module, which is usually derived from the Fok I restriction endonuclease (reviewed in Porteus and Carroll, 2005). ZFN binding to its target sequence induces double-stranded DNA breaks, which results in heritable defects because of improper repair. Needless to say, DNA binding specificity is critical for the application of ZFNs in animal models. The ability to manipulate binding is based on several findings: individual zinc fingers primarily interact with a single triplet of the DNA sequence; this interaction involves a significant degree of sequence specificity; and multiple zinc fingers can be assembled together to recognize longer target sequences (Porteus and Carroll, 2005). In zebrafish, pilot studies confirmed that ZFNs can be used to induce mutations in desired genes with good efficiency and specificity (Doyon et al., 2008; Meng et al., 2008). Nonetheless, the engineering of zinc finger binding domains of predetermined specificities remains laborious as it requires lengthy in vitro and/or in vivo selection procedures (Doyon et al., 2008; Meng et al., 2008). Detailed protocols for the selection of zinc finger combinations that will efficiently target predetermined DNA sequences and for the subsequent generation of mutant zebrafish have been described (Foley et al., 2009; Wright et al., 2006).
2. Approaches to Gene Overexpression
To obtain a comprehensive understanding of gene function, one often needs to supplement loss-of-function analysis with overexpression data. In the simplest scenario, this can be accomplished in zebrafish by RNA or DNA injections into the embryo. Several variants of this procedure exist, each with unique advantages and drawbacks (reviewed in Malicki et al., 2002). The main disadvantage of injecting RNA into embryos is its limited stability. The injection of DNA constructs, on the other hand, produces expression for a much longer period of time but only in a small number of cells. The fraction of cells that express a gene of interest following the injection of a DNA construct into the embryo can be increased by placing the gene to be studied under the control of UAS (Upstream Activating Sequence, multiple copies are used in tandem) and driving its expression using GAL4–VP16 fusion protein expressed from either a ubiquitous or a tissue-specific promoter (Koster and Fraser, 2001). Alternatively, transgene integration efficiency (estimated as the fraction of cells that express DNA construct) can be greatly improved by using Tol2 transposon-based vectors (Kawakami, 2004). These are injected into one- to two-cell embryos (the earlier the better) along with transposase mRNA (Kawakami, 2004; Kwan et al., 2007). The integration of these constructs into the genome relies on terminal transpo−son sequences, including the terminal inverted repeats (TIRs). The Tol2-derived sequences can be as short as 150–200 bp, but tend to be longer in older vectors, such as T2KXIG (Kawakami, 2004; Urasaki et al., 2006). In addition to transposon terminal sequences, these vectors contain an FP marker that helps to follow the pattern of transgene inheritance in embryonic tissues. Genes of interest can also be placed in these vectors under the control of appropriate regulatory elements. The heat-shock promoter has been used, for example, to drive the expression of a crumbs gene from a Tol2-based vector in the zebrafish retinal neuroepithelium. This approach produced expression in nearly half of neuroepithelial cells (Omori et al., 2008).
Overexpression phenotypes can also be studied in stable transgenic lines, provided that the resulting dominant phenotype is viable or can be conditionally induced. Several efficient methods for generating transgenic zebrafish are available. To develop a good understanding of its function, a gene under investigation may have to be expressed under the control of several regulatory elements and/or as a fusion with more than one tag (FP tags with different spectral characteristics and/or a myc tag, for example). As generating appropriate expression constructs using traditional cloning approaches is laborious, recombination cloning-based strategies have been specifically tailored for use in zebrafish (Kwan et al., 2007; Villefranc et al., 2007). These methods utilize a set of bacteriophage λ recombination enzymes to transfer DNA fragments from so-called entry vectors into so-called destination vectors, and are referred to as Gateway cloning (Hartley et al., 2000). One of the most obvious advantages of the Gateway system is that it allows one to combine several different DNA elements relatively efficiently in a single enzymatic reaction. In one example of how this method can be applied, three entry clones were assembled in the correct configuration into a Tol2-based zebrafish destination vector in a single step (Kwan et al., 2007). The use of the Gateway system requires some preparatory work. Recombination sites need to be added to generate entry vectors, and, similarly, the destination vectors have to be prepared by inserting recombination sites and selection markers. These procedures are nonetheless straightforward, and most standard laboratory vectors can be fairly easily converted into destination vectors. To make this approach even more attractive, several destination vectors are already available for use in the zebrafish (Kwan et al., 2007; Villefranc et al., 2007).
A frequent limitation of overexpression studies is the pleiotropy of mutant phenotypes: for many loci, early embryonic phenotypes are so severe that they preclude the analysis of late developmental processes, such as retinal neurogenesis. Several experimental tools are available to overcome this problem, including the use of heat-shock promoters, the GAL4–UAS overexpression system, and caged nucleic acids. Similar to invertebrate model systems, the use of heat-shock-induced expression in zebrafish relies on the hsp70 promoter (Halloran et al., 2000). An interesting variant of this protocol involves the activation of a heat-shock promoter-driven transgene in a small group of cells in a living embryo by heating them gently with a laser beam, which provides both temporal and spatial control of overexpression pattern (Halloran et al., 2000).
GAL4–UAS system is another method to achieve spatial control of gene expression. Modeled after Drosophila (Brand and Perrimon, 1993), the GAL4–UAS overexpres−sion approach takes advantage of two transgenic strains. The activator strain expresses the GAL4 transcriptional activator in a desired subset of tissues, while the effector strain carries the gene of interest under the control of a GAL4 responsive promoter (UAS, upstream activating sequence). The effector transgene is activated by crossing its carrier strain to a line that carries the activator transgene (Scheer et al., 2002). One variant of this system involves a fusion of the Gal4 DNA binding domain to the viral VP16 activation domain and uses a multimer of 14 UAS sites in the reporter construct (Koster and Fraser, 2001; see also comments above). The GAL4–UAS system was initially used in the zebrafish eye to study notch function (Scheer et al., 2001), and since then has gained popularity (Del Bene et al., 2008; Godinho et al., 2005; Mumm et al., 2006; Yeo et al., 2007). Importantly, enhancer trap screens have generated hundreds of transgenic strains that express the Gal4 activator in a variety of patterns and can be used to drive the expression of UAS effector transgenes in many organs, including the eye (Asakawa and Kawakami, 2008; Scott et al., 2007). Finally, an interesting method to control gene overexpression patterns is the use of Bhc-caged nucleic acids (Ando et al., 2001). In this approach, embryos are injected with an inactive form of an overexpression construct, which is then later activated in a selected tissue using UV illumination. Both RNA and DNA templates can be used to produce overexpression in this approach (Ando and Okamoto, 2003).
B. Forward Genetics
The use of zebrafish in genetic studies offers several obvious advantages. The most important of these is the possibility of performing efficient forward genetic screens. Genetic screening is feasible because adult zebrafish are highly fecund and are easily maintained in large numbers in a fairly small laboratory space. Screens performed in on the zebrafish so far identified hundreds of visual system mutants (Baier et al., 1996; Fadool et al., 1997; Malicki et al., 1996; Muto et al., 2005; Neuhauss et al., 1999). While designing a genetic screen, one has to consider three important variables: the type of mutagen to be used, the design of the breeding scheme, and mutant defect recognition criteria. Each of these is discussed below.
1. Mutagenesis Approaches
The majority of screens performed in zebrafish so far involved the use of N-ethyl-N−nitrosourea (ENU) (Mullins et al., 1994; Solnica-Krezel et al., 1994). This mutagenesis approach is very effective as evidenced by the fact that the vast majority of mutations isolated so far are ENU induced. A powerful alternative to chemical mutagenesis is insertional retroviral mutagenesis. Although the efficiency of this mutagenesis approach is still lower than that of chemical methods, an obvious advantage of a retroviral mutagen is that it provides means for very rapid identification of mutant genes (Amster−dam et al., 1999; Golling et al., 2002). Retroviral mutagenesis has also been applied on a large scale to identify hundreds of mutant strains (Golling et al., 2002). The photo−receptor mutant nrf is an example of a retinal defect induced using this approach (Becker et al., 1998). More recently, a rescreen of 250 retrovirus-induced mutants led to the identification of defects in several aspects of eye development (Gross et al., 2005).
In addition to chemical mutagens and retroviral vectors, transposons provide another option for effective mutagenesis. Transposable elements of the Tc-1/mariner (Sleeping beauty) and hAT (Tol2) families integrate into the zebrafish genome in a transposase−dependent manner (Fadool et al., 1998; Kawakami et al., 2000; Raz et al., 1998). Although initial efforts to induce mutations using transposon-based vectors were unsuccessful (Balciunas et al., 2004; Kawakami et al., 2004), recent experiments that rely on improved vector design generate mutants with high efficiency (Nagayoshi et al., 2008; Sivasubbu et al., 2006). Both Tol2- and Sleeping beauty-based constructs were used in these efforts. Transposon-based mutagenesis is an attractive alternative to retrovirus-mediated one because transposon-based vectors efficiently integrate into the zebrafish genome, and their mutagenicity (measured as the fraction of genome insertion events that lead to mutant phenotypes in homozygous animals) already exceeds that of retroviral mutagenesis (Nagayoshi et al., 2008; Sivasubbu et al., 2006). The use of transposons does not require technically difficult packaging of DNA into viral particles, and also appears to pose few safety concerns. An added bonus of using transposons is that they can be remobilized from preexisting lines to generate additional insertions (Kondrychyn et al., 2009). One has to bear in mind, however, that just like in the case of viral insertions, transposon integration is not entirely random (Kondrychyn et al., 2009). As the efficiency of transposable element-mediated muta−genesis is gradually improving, future genetic screens are likely to be performed with the help of transposons.
Transposon-mediated mutagenesis is usually performed using enhancer or gene trap vectors, which carry FP reporter genes (reviewed in Balciunas et al., 2004; Nagayoshi et al., 2008). Such a design is important for several reasons. First, it allows one to visually detect integration events that occur in the vicinity of genes because the nearby regulatory elements frequently drive FP reporter expression. Such integrations are much more likely to produce phenotypic defects, compared to insertions into non-transcribed regions of the genome. Second, as different integration events tend to produce different expression patterns, at least in some cases one can distinguish them from each other via simple inspection of living embryos. Consequently, potentially mutagenic insertions can be driven to homozygocity already in the F2 generation of a screen (Nagayoshi et al., 2008). Moreover, as gene/enhancer trap expression patterns suggest the function for genes in which insertions have occurred, they may allow one to focus a genetic screen on a specific developmental or physiological process. Finally, trap-induced mutant alleles are easier to maintain as their presence can be selected for in heterozygotes based on expression pattern. Although retroviral mutagenesis vectors can also be engineered to function as traps (Ellingsen et al., 2005), mutants generated using retroviral trap vectors have not been reported in zebrafish so far.
2. Breeding Schemes
The second important consideration is the type of breeding scheme that will carry genetic defects from mutagenized animals (G0) to the generation in which the screening for mutant phenotypes is performed. The most straightforward option, but also the most space- and time-consuming one, is screening for recessive defects in F3 generation embryos. This procedure was used in early large-scale genetic screens (Amster−dam et al., 1999; Driever et al., 1996; Haffter et al., 1996). Its main disadvantage is that it requires a very large number of tanks to raise the F2 generation to adulthood. As the majority of laboratories do not have access to several thousands of fish tanks, more space-efficient procedures are frequently required. In this regard, the zebrafish offers some possibilities not available in other genetically studied vertebrates—haploid and early pressure screens (for a review see Malicki, 2000). The major asset of these screening strategies is that one generation of animals is omitted and consequently time and the amount of laboratory space required is dramatically reduced. Although there are obvious advantages, these two screening strategies also suffer from some limitations. The most significant disadvantage of haploids is that their development does not proceed in the same way as wild-type embryogenesis. Haploid embryos do not survive beyond 5 dpf, and even at earlier stages of development they display obvious defects. Although the eyes of haploid zebrafish appear fairly normal at least until 3 dpf, the architecture of their retinae tends to be disorganized. By 5 dpf, haploid embryos are markedly smaller than the wild type and display numerous abnormalities. In the context of the visual system, haploid screens appear useful to search for early patterning defects prior to the onset of neurogenesis.
Screening of embryos generated via the application of early pressure (Streisinger et al., 1981) is another strategy that can be used to save both time and space. Similar to haploidization, this technique also allows one to screen for recessive defects in F2 generation embryos. The early pressure technique also involves some shortcomings. Embryos produced via this method display a high background of developmental abnormalities, which complicate the detection of mutant phenotypes, especially at early developmental stages. Another limitation of early pressure screens is that the fraction of homozygous mutant animals in a clutch of early pressure-generated embryos depends on the distance of a mutant locus from the centromere. For centro−meric loci, the fraction of mutant embryos approaches 50%, whereas for telomeric genes it decreases below 10% (Streisinger et al., 1986). In other types of screens, mutant phenotypes can be distinguished from non-genetic developmental abnormalities based on their frequencies (25% in the case of screens on F3 embryos). Clearly, this criterion cannot be used in early pressure screens. Despite these limitations, early pressure screens are useful, especially in small-scale endeavors. The experimental techniques involved in haploid and early pressure screens have been previously reviewed in depth (Beattie et al., 1999; Walker, 1999).
While the approaches discussed above are used to identify recessive mutant phenotypes, an entirely different breeding scheme is used in searches for dominant defects. These can already be detected in embryos, larvae, or adults of the F1 generation. Although this category of screens requires just a single generation and consequently a very small amount of laboratory space, few experiments focusing on dominant defects have been performed in zebrafish so far (van Eeden et al., 1999). An example of a search for dominant defects of the visual system is provided by a small behavioral screen of adult animals for defects of visual perception, which identified a late-onset photoreceptor degeneration phenotype (Li and Dowling, 1997).
3. Phenotype Detection Methods
The third important consideration while designing a genetic screen is the mutant phenotype detection method. This aspect of screening allows for substantial creativity. Phenotype detection criteria range from very simple to very sophisticated. Ideally, the mutant phenotype recognition strategy should fulfill the following requirements: (1) involve minimal effort, (2) detect gross abnormalities as well as subtle changes, and (3) exclude phenotypes irrelevant to the targeted process. One class of irrelevant phenotypes are nonspecific defects. In large-scale mutagenesis screens performed so far, more than two-thirds of all phenotypes were classified as nonspecific (Driever et al., 1996; Golling et al., 2002; Haffter et al., 1996). The most frequent nonspecific phenotypes in zebrafish are early degeneration spreading across the entire embryo, and developmental retardation affecting brain, eyes, fins, and jaw. The latter class of mutants affects tissues that display robust proliferation between 3 and 5 dpf. Nonspecific phenotypes are not necessarily without value, but are usually considered uninteresting because they are likely to be produced by defects in a broad range of housekeeping mechanisms (such as metabolic pathways or DNA replication machinery; see for example Allende et al., 1996; Plaster et al., 2006). Another category of irrelevant phenotypes includes specific defects that are of no interest to investigators performing the screen. Such phenotypes are isolated when a screening procedure detects mutations affecting multiple organs, only one of which is of interest. A good example of such a situation is provided by behavioral screens involving the optomotor response. Lack of the optomotor response may be due to defects of photoreceptor neurons or skeletal muscles. These two cell types are seldom interesting to the same group of investigators. It is one of the virtues of a well-designed screen that irrelevant phenotypes are efficiently selected against.
The simplest way to screen for mutant phenotypes is by visual inspection. The most significant disadvantage of this method is that it detects changes only in structures easily recognizable using a microscope (preferably a dissecting scope). Thus visual inspection screens are suitable to search for defects in trunk blood vessels (which are easy to see in larvae), but would not detect a loss of a small population of neurons hidden in the depths of the retina or the brain. Visual inspection criteria work well when the aim of a screen is to detect gross morphological changes. Within the eye, such changes may reflect specific defects in a single neuronal lamina. In many mutants, the changes of eye size are caused by a degeneration of photoreceptor cells (Doerre and Malicki, 2002; Jing and Malicki, 2009; Malicki et al., 1996). In this case, the affected cell population is numerous enough to cause a major change of morphology. Most likely, a morphological screen would not detect abnormalities in a less numerous cell class.
Changes confined to small populations of cells cannot usually be identified in a visual inspection screen. To detect these changes, the target cell population must somehow be made accessible to inspection. Several options exist in this regard: analysis of histological sections, whole-mount antibody staining, in situ hybridization, retrograde or anterograde labeling of neurons, and cell class-specific FP transgenes. One technically simple but rather laborious approach is to embed zebrafish larvae in paraffin and prepare histological sections. This approach was used to screen more than 2000 individuals from ca. 50 clutches of F2 early pressure-generated mutagenized larvae and led to the identification of two photoreceptor mutants (Mohideen et al., 2003). In addition to histological analysis, individual cell populations can be visualized in mutgenized animals using antibody staining or in situ hybridization. In one screening endeavor, staining of 700 early pressure-generated egg clutches with anti-tyrosine hydroxylase antibody led to the isolation of two retinal mutants (Guo et al., 1999).
An excellent example of a genetic screen that involves labeling of a specific neuronal population has been performed to uncover defects of the retinotectal projection (Baier et al., 1996; Karlstrom et al., 1996; Trowe et al., 1996). In this screen, two subpopulations of retinal ganglion cells were labeled with the carbocyanine tracers, DiI and DiO. Labeling procedures usually make screening much more laborious. To reduce the workload in this screen, DiI and DiO labeling were highly automated. For tracer injection, fish larvae were mounted in a standardized fashion in a temperature-controlled mounting apparatus. After filling the apparatus with liquid agarose and mounting the larvae, the temperature was lowered allowing the agarose to solidify. Subsequently, the blocks of agarose containing mounted larvae were transferred into the injection setup. Upon injection, the larvae were stored overnight at room temperature to allow for the diffusion of the injected tracer, and then transferred to a microscope stage for phenotypic analysis. The authors of this experiment estimate that using this highly automated screening procedure allowed them to inspect over 2000 larvae per day and to reduce the time spent on the analysis of a single individual to less than 1 min (Baier et al., 1996). Other labeling procedures can also be scaled up to process many clutches of embryos in a single experiment. Antibody or in situ protocols, for example, involve multiple changes of staining and washing solutions. To perform these protocols on many embryos in parallel, one can use multiwell staining dishes with stainless steel mesh at the bottom. Such staining dishes can be quickly transferred from one solution to another. Since many labeling procedures are time consuming, it is essential that during a screen they are performed in parallel on many embryos.
Recent advances provide an additional way to label specific cell populations in a much less labor-intensive way by using FP transgenes, such as the ones described earlier in this chapter. Transgenic FP lines can be either directly mutagenized or crossed to mutagenized males. Then the resulting progeny is used to search for defects in fine features of retinal cell populations. In contrast to other cell labeling procedures, the use of FP transgenes requires very little additional effort, compared to simple morphological observations of the external phenotype.
Behavioral tests are yet another screening alternative. Several screens based on behavioral criteria have been performed in recent years, leading to the isolation of interesting developmental defects (Brockerhoff et al., 1997; 2003; Li and Dowling, 1997; Muto et al., 2005; Neuhauss et al., 1999). Behavioral screens allow one to detect subtle functional defects of the retina that might evade other search criteria. They can be used to search for both recessive and dominant defects in larvae as well as in adult fish (Li and Dowling, 1997). Similar to many labeling procedures, however, behavioral screens tend to be laborious. In one instance of a screen involving the optokinetic response, the authors estimate that screening of a single zebrafish larva took, on average, 1 min. (Brockerhoff et al., 1995). Since optomotor tests can be performed on populations of animals, they tend to be less time consuming, compared to optokinetic response tests. They do, however, produce more false-positive hits (Muto et al., 2005). In addition, since behavioral responses usually involve the cooperation of many cell classes, screens of this type tend to detect a wide range of defects. The optokinetic response screens, for example, may lead to the isolation of defects in the differentiation of lens cells, the specification of the retinal neurons or glia, the formation of synaptic connections, the mechanisms of neurotransmitter release, or the development of ocular muscles. Additional tests are necessary to assure that the isolated mutants belong to the desired category. To be useful for screening, the behavioral response should be robust and reproducible, and should involve the simplest possible neuronal circuitry. In light of these criteria, the optokinetic response appears to be superior to other behaviors; both optomotor and startle responses require functional optic tecta while the optokinetic response does not (Clark, 1981; Easter and Nicola, 1996). The optokinetic response also appears to be more robust than the optomotor response and phototaxis (Brockerhoff et al., 1995; Clark, 1981). The most extensive visual behavior-based screen conducted so far relied on two tests conducted in parallel: optokinetic and optomotor responses (Muto et al., 2005). Although the results of this experiment are quite informative, they also illustrate inconsistencies associated with the use of behavioral tests as a screening tool. First, the initial round of screening was characterized by a very high false-positive rate (> 90% for the optomotor test). Second, surprisingly, the two behavioral tests used in this study uncovered largely non-overlapping sets of mutants. Following retests it turned out, however, that all mutants display both optomotor and optokinetic defects to varying degrees. Finally, as pointed out above, a broad range of phenotypic abnormalities in different cell classes were found in this experiment.
4. Positional and Candidate Cloning
Molecular characterization of defective loci is usually a crucial step that follows the isolation of mutant lines. The development of positional and candidate gene cloning strategies is one of the most significant advances in the field of zebrafish genetics within the last decade. These approaches are currently well established and have played a key role in many important contributions to the understanding of eye development and function. The positional cloning strategy involves a standard set of steps, such as mapping, chromosomal walking, transcript identification, and the delivery of a proof that the correct gene has been cloned. These steps are largely the same, regardless of the nature of a mutant phenotype. An example of a positional cloning strategy, laborious but eventually successful, is the cloning of the nagie oko locus (Wei and Malicki, 2002).
5. Mutant Strains Available
Large and small mutagenesis screens identified numerous genetic defects of retinal development in zebrafish. Mutant phenotypes affect a broad range of developmental stages, starting with the specification of the eye primordia, through optic lobe morphogenesis, the specification of neuronal identities, and include the final steps of differentiation, such as outer segment development in photoreceptor cells. Lists of mutant lines, excluding those that produce nonspecific degeneration of the entire retina, have been provided previously (Avanesov and Malicki, 2004; Malicki, 1999). Although these are still useful, many new mutants have been generated in recent years. The descriptions of these are available in the Zebrafish Model Organism Database (ZFIN, http://zfin.org).
V. Summary
Relative simplicity, rapid development, and accessibility to genetic analysis make the zebrafish retina an excellent model system for the studies of neurogenesis in the vertebrate CNS. Numerous genetic screens have led to isolation of many mutants affecting the retina and the retinotectal projection in zebrafish. Mutant phenotypes are being studied using a rich variety of markers: antibodies, RNA probes, retrograde and anterograde tracers, as well as transgenic lines. A particularly impressive progress has been made in the characterization of the zebrafish genome. Consequently, positional and candidate cloning of mutant loci are now fairly easy in zebrafish. Many mutant genes have been cloned, and their analysis has provided insights into genetic circuitries that regulate retinal pattern formation, and the differentiation of retinal neurons and glia. Genetic screens for visual system defects will continue in the future, and progressively more sophisticated screening approaches will make it possible to detect an increasingly broad and varied assortment of mutant phenotypes. The remarkable evolutionary conservation of the vertebrate eye provides the basis for the use of the zebrafish retina as a model of human-inherited eye defects. As new techniques are being introduced and rapidly improved, the zebrafish will continue to be an important organism for the studies of the vertebrate visual system.
Acknowledgments
The authors are grateful to Brian Perkins, Ichiro Masai, and Brian Link for critical reading of earlier versions of this manuscript and helpful comments. The authors’ research on the retina is supported by grants from the National Eye Institute to JM (R01 EY018176 and R01EY016859).
References
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- Allwardt BA, Lall AB, Brockerhoff SE, Dowling JE. Synapse formation is arrested in retinal photoreceptors of the zebrafish nrc mutant. J Neurosci. 2001;21:2330–2342. doi: 10.1523/JNEUROSCI.21-07-02330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Turner D, Cepko C. Specification of cell type in the vertebrate retina. In: Lam D, Shatz C, editors. Development of the Visual System. The MIT Press; Cambridge, MA: 1991. pp. 37–58. [Google Scholar]
- Alvarez Y, Cederlund ML, Cottell DC, Bill BR, Ekker SC, Torres-Vazquez J, Weinstein BM, Hyde DR, Vihtelic TS, Kennedy BN. Genetic determinants of hyaloid and retinal vasculature in zebrafish. BMC Dev Biol. 2007;7:114. doi: 10.1186/1471-213X-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- Ando H, Okamoto H. Practical procedures for ectopic induction of gene expression in zebrafish embryos using Bhc-diazo-caged mRNA. Methods Cell Sci. 2003;25:25–31. doi: 10.1023/B:MICS.0000006846.13226.38. [DOI] [PubMed] [Google Scholar]
- Aramaki S, Hatta K. Visualizing neurons one-by-one in vivo: optical dissection and reconstruction of neural networks with reversible fluorescent proteins. Dev Dyn. 2006;235:2192–2199. doi: 10.1002/dvdy.20826. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K. Targeted gene expression by the Gal4–UAS system in zebrafish. Dev Growth Differ. 2008;50:391–399. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Dahm R, Sewell WF, Malicki JJ. Mutations that affect the survival of selected amacrine cell subpopulations define a new class of genetic defects in the vertebrate retina. Dev Biol. 2005;285:138–155. doi: 10.1016/j.ydbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Malicki J. Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- Bahadori R, Rinner O, Schonthaler HB, Biehlmaier O, Makhankov YV, Rao P, Jagadeeswaran P, Neuhauss SC. The zebrafish fade out mutant: a novel genetic model for Hermansky–Pudlak syndrome. Invest Ophthalmol Vis Sci. 2006;47:4523–4531. doi: 10.1167/iovs.05-1596. [DOI] [PubMed] [Google Scholar]
- Baier H, Klostermann S, Trowe T, Karlstrom RO, Nusslein-Volhard C, Bonhoeffer F. Genetic dissection of the retinotectal projection. Development. 1996;123:415–425. doi: 10.1242/dev.123.1.415. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the sleeping beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel LK, Raymond PA. Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. doi: 10.1177/38.9.2201738. [DOI] [PubMed] [Google Scholar]
- Barthel LK, Raymond PA. Subcellular localization of alpha-tubulin and opsin mRNA in the goldfish retina using digoxigenin-labeled cRNA probes detected by alkaline phosphatase and HRP histochemistry. J Neurosci Methods. 1993;50:145–152. doi: 10.1016/0165-0270(93)90002-9. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie CE, Raible DW, Henion PD, Eisen JS. Early pressure screens. Methods Cell Biol. 1999;60:71–86. [PubMed] [Google Scholar]
- Becker TS, Burgess SM, Amsterdam AH, Allende ML, Hopkins N. Not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor-1 and avian initiation binding repressor. Development. 1998;125:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Biehlmaier O, Makhankov Y, Neuhauss SC. Impaired retinal differentiation and maintenance in zebrafish laminin mutants. Invest Ophthalmol Vis Sci. 2007;48:2887–2894. doi: 10.1167/iovs.06-1212. [DOI] [PubMed] [Google Scholar]
- Biehlmaier O, Neuhauss SC, Kohler K. Synaptic plasticity and functionality at the cone terminal of the developing zebrafish retina. J Neurobiol. 2003;56:222–236. doi: 10.1002/neu.10243. [DOI] [PubMed] [Google Scholar]
- Bodick N, Levinthal C. Growing optic nerve fibers follow neighbors during embryogenesis. Proc Natl Acad Sci U S A. 1980;77:4374–4378. doi: 10.1073/pnas.77.7.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchek T, Bremiller R. The development of photoreceptors in the zebrafish, Brachydanio rerio. I Structure. J Comp Neurol. 1984;224:107–115. doi: 10.1002/cne.902240109. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brennan C, Monschau B, Lindberg R, Guthrie B, Drescher U, Bonhoeffer F, Holder N. Two Eph receptor tyrosine kinase ligands control axon growth and may be involved in the creation of the retinotectal map in the zebrafish. Development. 1997;124:655–664. doi: 10.1242/dev.124.3.655. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Dowling JE, Hurley JB. Zebrafish retinal mutants. Vision Res. 1998;38:1335–1339. doi: 10.1016/s0042-6989(97)00227-7. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, Dowling JE. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc Natl Acad Sci U S A. 1995;92:10545–10549. doi: 10.1073/pnas.92.23.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff SE, Hurley JB, Niemi GA, Dowling JE. A new form of inherited red-blindness identified in zebrafish. J Neurosci. 1997;17:4236–4242. doi: 10.1523/JNEUROSCI.17-11-04236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff SE, Rieke F, Matthews HR, Taylor MR, Kennedy B, Ankoudinova I, Niemi GA, Tucker CL, Xiao M, Cilluffo MC, Fain GL, Hurley JB. Light stimulates a transducin-independent increase of cytoplasmic Ca2+ and suppression of current in cones from the zebrafish mutant nof. J Neurosci. 2003;23:470–480. doi: 10.1523/JNEUROSCI.23-02-00470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Burrill JD, Easter SS., Jr Development of the retinofugal projections in the embryonic and larval zebrafish (Brachydanio rerio) J Comp Neurol. 1994;346:583–600. doi: 10.1002/cne.903460410. [DOI] [PubMed] [Google Scholar]
- Burrill J, Easter S. The first retinal axons and their microenvironment in zebrafish cryptic pioneers and the pretract. J Neurosci. 1995;15:2935–2947. doi: 10.1523/JNEUROSCI.15-04-02935.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. La retine des vertebres. Cellule. 1893;9:17–257. [Google Scholar]
- Cameron DA, Carney LH. Cell mosaic patterns in the native and regenerated inner retina of zebrafish: implications for retinal assembly. J Comp Neurol. 2000;416:356–367. [PubMed] [Google Scholar]
- Cao Y, Semanchik N, Lee SH, Somlo S, Barbano PE, Coifman R, Sun Z. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc Natl Acad Sci U S A. 2009;106:21819–21824. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development. 2003;130:2329–2339. doi: 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Whitmore AV, Jeffery G, Raff M. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J Neurosci. 2001;21:5643–5651. doi: 10.1523/JNEUROSCI.21-15-05643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedrone C, Culasso F, Cesareo M, Zapelloni A, Cedrone P, Cerulli L. Prevalence of glaucoma in Ponza, Italy: a comparison with other studies. Ophthalmic Epidemiol. 1997;4:59–72. doi: 10.3109/09286589709057098. [DOI] [PubMed] [Google Scholar]
- Cerveny KL, Cavodeassi F, Turner KJ, de Jong-Curtain TA, Heath JK, Wilson SW. The zebrafish flotte lotte mutant reveals that the local retinal environment promotes the differentiation of proliferating precursors emerging from their stem cell niche. Development. 2010;137:2107–2115. doi: 10.1242/dev.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen A, Hamaoka T, Yamada Y, Kawamura S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics. 2003;163:663–675. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Clark T. Ph D dissertation. University of Oregon; Eugene, OR: 1981. Visual responses in developing zebrafish (Brachydanio rerio) [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo A, Fraser SE, Mabee PM. A dual embryonic origin for vertebrate mechanorecep−tors. Science. 1994;264:426–430. doi: 10.1126/science.8153631. [DOI] [PubMed] [Google Scholar]
- Collins MO, Choudhary JS. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr Opin Biotechnol. 2008;19:324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Connaughton VP, Behar TN, Liu WL, Massey SC. Immunocytochemical localization of excitatory and inhibitory neurotransmitters in the zebrafish retina. Vis Neurosci. 1999;16:483–490. doi: 10.1017/s0952523899163090. [DOI] [PubMed] [Google Scholar]
- Cui S, Otten C, Rohr S, Abdelilah-Seyfried S, Link BA. Analysis of aPKCλ and aPKCζ reveals multiple and redundant functions during vertebrate retinogenesis. Mol Cell Neurosci. 2007;34:431–444. doi: 10.1016/j.mcn.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R, Schonthaler HB, Soehn AS, van Marle J, Vrensen GF. Development and adult morphology of the eye lens in the zebrafish. Exp Eye Res. 2007;85:74–89. doi: 10.1016/j.exer.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Dapson RW. Glyoxal fixation: how it works and why it only occasionally needs antigen retrieval. Biotech Histochem. 2007;82:161–166. doi: 10.1080/10520290701488113. [DOI] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Doerre G, Malicki J. A mutation of early photoreceptor development, mikre oko, reveals cell–cell interactions involved in the survival and differentiation of zebrafish photoreceptors. J Neurosci. 2001;21:6745–6757. doi: 10.1523/JNEUROSCI.21-17-06745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerre G, Malicki J. Genetic analysis of photoreceptor cell development in the zebrafish retina. Mech Dev. 2002;110:125–138. doi: 10.1016/s0925-4773(01)00571-8. [DOI] [PubMed] [Google Scholar]
- Dowling J. The Retina. Harvard University Press; Cambridge, MA: 1987. [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Drescher U, Bonhoeffer F, Muller BK. The Eph family in retinal axon guidance. Curr Opin Neurobiol. 1997;7:75–80. doi: 10.1016/s0959-4388(97)80123-7. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Dryja T, Li T. Molecular genetics of retinitis pigmentosa. Hum Mol Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS, Jr, Malicki JJ. The zebrafish eye: developmental and genetic analysis. Result Probl Cell Differ. 2002;40:346–370. doi: 10.1007/978-3-540-46041-1_17. [DOI] [PubMed] [Google Scholar]
- Easter S, Nicola G. The development of vision in the zebrafish (Danio rerio) Dev Biol. 1996;180:646–663. doi: 10.1006/dbio.1996.0335. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Ellingsen S, Laplante MA, Konig M, Kikuta H, Furmanek T, Hoivik EA, Becker TS. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- Emran F, Rihel J, Adolph AR, Dowling JE. Zebrafish larvae lose vision at night. Proc Natl Acad Sci U S A. 2010;107:6034–6039. doi: 10.1073/pnas.0914718107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran F, Rihel J, Adolph AR, Wong KY, Kraves S, Dowling JE. OFF ganglion cells cannot drive the optokinetic reflex in zebrafish. Proc Natl Acad Sci U S A. 2007;104:19126–19131. doi: 10.1073/pnas.0709337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev Biol. 2003;258:277–290. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Brockerhoff SE, Hyatt GA, Dowling JE. Mutations affecting eye morphology in the developing zebrafish (Danio rerio) Dev Genet. 1997;20:288–295. doi: 10.1002/(SICI)1520-6408(1997)20:3<288::AID-DVG11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Hartl DL, Dowling JE. Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc Natl Acad Sci U S A. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fashena D, Westerfield M. Secondary motoneuron axons localize DM-GRASP on their fasciculated segments. J Comp Neurol. 1999;406:415–424. doi: 10.1002/(sici)1096-9861(19990412)406:3<415::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser S. Patterning of retinotectal connections in the vertebrate visual system. Curr Oppin Neurobiol. 1992;2:83–87. doi: 10.1016/0959-4388(92)90167-j. [DOI] [PubMed] [Google Scholar]
- Fricke C, Lee JS, Geiger-Rudolph S, Bonhoeffer F, Chien CB. Astray, a zebrafish roundabout homolog required for retinal axon guidance. Science. 2001;292:507–510. doi: 10.1126/science.1059496. [DOI] [PubMed] [Google Scholar]
- Gnuegge L, Schmid S, Neuhauss SC. Analysis of the activity-deprived zebrafish mutant macho reveals an essential requirement of neuronal activity for the development of a fine-grained visuotopic map. J Neurosci. 2001;21:3542–3548. doi: 10.1523/JNEUROSCI.21-10-03542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- Goldsmith P, Baier H, Harris WA. Two zebrafish mutants, ebony and ivory, uncover benefits of neighborhood on photoreceptor survival. J Neurobiol. 2003;57:235–245. doi: 10.1002/neu.10274. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin S-Y, Nissen RM, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Gray MP, Smith RS, Soules KA, John SW, Link BA. The aqueous humor outflow pathway of zebrafish. Invest Ophthalmol Vis Sci. 2009;50:1515–21. doi: 10.1167/iovs.08-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JM, Perkins BD. Zebrafish mutants as models for congenital ocular disorders in humans. Mol Reprod Dev. 2008;75:547–555. doi: 10.1002/mrd.20831. [DOI] [PubMed] [Google Scholar]
- Gross JM, Perkins BD, Amsterdam A, Egana A, Darland T, Matsui JI, Sciascia S, Hopkins N, Dowling JE. Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics. 2005;170:245–261. doi: 10.1534/genetics.104.039727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev Biol. 1999;208:473–487. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Halpern M, Ho R, Walker C, Kimmel C. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell. 1993;75:99–111. [PubMed] [Google Scholar]
- Hanker JS. Osmiophilic reagents in electronmicroscopic histocytochemistry. Prog Histochem Cytochem. 1979;12:1–85. doi: 10.1016/s0079-6336(79)80002-9. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Hinds J, Hinds P. Early ganglion cell differentiation in the mouse retina: an electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- Hisatomi O, Satoh T, Barthel LK, Stenkamp DL, Raymond PA, Tokunaga F. Molecular cloning and characterization of the putative ultraviolet-sensitive visual pigment of goldfish. Vision Res. 1996;36:933–939. doi: 10.1016/0042-6989(95)00189-1. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, VanDeRyt JT, Wilson SW. Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Holt C, Bertsch T, Ellis H, Harris W. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Hong CC, Peterson QP, Hong JY, Peterson RT. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr Biol. 2006;16:1366–1372. doi: 10.1016/j.cub.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Hume RI. Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term cultures. J Cell Biol. 1986;103:171–187. doi: 10.1083/jcb.103.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Hume RI. Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends Neurosci. 1989;12(333–335):340–341. [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Hudak LM, Lunt S, Chang CH, Winkler E, Flammer H, Lindsey M, Perkins BD. The intraflagellar transport protein ift80 is essential for photoreceptor survival in a zebrafish model of jeune asphyxiating thoracic dystrophy. Invest Ophthalmol Vis Sci. 2010;51:3792–3799. doi: 10.1167/iovs.09-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt GA, Schmitt EA, Marsh-Armstrong N, McCaffery P, Drager UC, Dowling JE. Retinoic acid establishes ventral retinal characteristics. Development. 1996;122:195–204. doi: 10.1242/dev.122.1.195. [DOI] [PubMed] [Google Scholar]
- Humphrey C, Pittman F. A simple methylene blue-azure II-basic fuchsin stain for epoxy-embedded tissue sections. Stain Technol. 1974;49:9–14. doi: 10.3109/10520297409116929. [DOI] [PubMed] [Google Scholar]
- Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. Developmental Neurobiology. Plenum Press; New York: 1991. [Google Scholar]
- Jensen AM, Walker C, Westerfield M. Mosaic eyes: a zebrafish gene required in pigmented epithelium for apical localization of retinal cell division and lamination. Development. 2001;128:95–105. doi: 10.1242/dev.128.1.95. [DOI] [PubMed] [Google Scholar]
- Jing X, Malicki J. Zebrafish ale oko, an essential determinant of sensory neuron survival and the polarity of retinal radial glia, encodes the p50 subunit of dynactin. Development. 2009;136:2955–2964. doi: 10.1242/dev.037739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T. Double in situ hybridization techniques in zebrafish. Methods. 2001;23:345–358. doi: 10.1006/meth.2000.1147. [DOI] [PubMed] [Google Scholar]
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Kainz PM, Adolph AR, Wong KY, Dowling JE. Lazy eyes zebrafish mutation affects Müller glial cells, compromising photoreceptor function and causing partial blindness. J Comp Neurol. 2003;463:265–280. doi: 10.1002/cne.10763. [DOI] [PubMed] [Google Scholar]
- Karlsson J, von Hofsten J, Olsson PE. Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Mar Biotechnol (NY) 2001;3:522–527. doi: 10.1007/s1012601-0053-4. [DOI] [PubMed] [Google Scholar]
- Karlstrom RO, Trowe T, Klostermann S, Baier H, Brand M, Crawford AD, Grunewald B, Haffter P, Hoffmann H, Meyer SU, et al. Zebrafish mutations affecting retinotectal axon pathfinding. Development. 1996;123:427–438. doi: 10.1242/dev.123.1.427. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4:1422–1432. doi: 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara A, Chien CB, Dawid IB. The homeobox gene mbx is involved in eye and tectum development. Dev Biol. 2002;248:107–117. doi: 10.1006/dbio.2002.0709. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a zebrafish atonal homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kay JN, Link BA, Baier H. Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development. 2005;132:2573–2585. doi: 10.1242/dev.01831. [DOI] [PubMed] [Google Scholar]
- Kay JN, Roeser T, Mumm JS, Godinho L, Mrejeru A, Wong RO, Baier H. Transient requirement for ganglion cells during assembly of retinal synaptic layers. Development. 2004;131:1331–1342. doi: 10.1242/dev.01040. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Segawa H, Tokumoto M, Tsubokawa T, Hotta Y, Uyemura K, Okamoto H. Ocular and cerebellar defects in zebrafish induced by overexpression of the LIM domains of the islet-3 LIM/homeodomain protein. Neuron. 1997;18:369–382. doi: 10.1016/s0896-6273(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Kang KH, Kim CH, Choi SY. Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. Biotechniques. 2008;45:331–334. doi: 10.2144/000112909. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Sessions SK, Kimmel RJ. Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron. J Comp Neurol. 1981;198:101–120. doi: 10.1002/cne.901980110. [DOI] [PubMed] [Google Scholar]
- Kitambi SS, McCulloch KJ, Peterson RT, Malicki JJ. Small molecule screen for compounds that affect vascular development in the zebrafish retina. Mech Dev. 2009;126:464–477. doi: 10.1016/j.mod.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitambi S, Peterson R, Malicki J. Small molecule screen for compounds that affect vascular development in the zebrafish retina. Mech Dev. 2008;126:464–477. doi: 10.1016/j.mod.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D, Bryan J, Laggner C, White R, Cheung CY, Mateus R, Healey D, Kim S, Werdich AA, Haggarty SJ, Macrae CA, Shoichet B, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6:231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Toida K, Dubreuil V, Alexandre P, Schenk J, Kiyokage E, Attardo A, Mora-Bermudez F, Arii T, Clarke JD, Huttner WB. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008;27:3151–3163. doi: 10.1038/emboj.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Krock BL, Bilotta J, Perkins BD. Noncell-autonomous photoreceptor degeneration in a zebrafish model of choroideremia. Proc Natl Acad Sci U S A. 2007;104:4600–4605. doi: 10.1073/pnas.0605818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krock BL, Perkins BD. The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT-particle-kinesin-II dissociation in vertebrate photoreceptors. J Cell Sci. 2008;121:1907–1915. doi: 10.1242/jcs.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Laessing U, Giordano S, Stecher B, Lottspeich F, Stuermer CA. Molecular characterization of fish neurolin: a growth-associated cell surface protein and member of the immunoglobulin superfamily in the fish retinotectal system with similarities to chick protein DM-GRASP/SC-1/BEN. Differentiation. 1994;56:21–29. doi: 10.1046/j.1432-0436.1994.56120021.x. [DOI] [PubMed] [Google Scholar]
- Laessing U, Stuermer CA. Spatiotemporal pattern of retinal ganglion cell differentiation revealed by the expression of neurolin in embryonic zebrafish. J Neurobiol. 1996;29:65–74. doi: 10.1002/(SICI)1097-4695(199601)29:1<65::AID-NEU5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Larison K, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:567–576. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Li L, Dowling JE. A dominant form of inherited retinal degeneration caused by a non-photoreceptor cell-specific mutation. Proc Natl Acad Sci U S A. 1997;94:11645–11650. doi: 10.1073/pnas.94.21.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dowling JE. Disruption of the olfactoretinal centrifugal pathway may relate to the visual system defect in night blindness b mutant zebrafish. J Neurosci. 2000;20:1883–1892. doi: 10.1523/JNEUROSCI.20-05-01883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS., Jr Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio) Dev Dyn. 2000a;219:391–401. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1063>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Li Z, Joseph NM, Easter SS., Jr The morphogenesis of the zebrafish eye, including a fate map of the optic vesicle. Dev Dyn. 2000b;218:175–188. doi: 10.1002/(SICI)1097-0177(200005)218:1<175::AID-DVDY15>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Link BA, Fadool JM, Malicki J, Dowling JE. The zebrafish young mutation acts non−cell-autonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127:2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D, Kalnins VI, McInnes RR. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mech Dev. 2004;121:1189–1197. doi: 10.1016/j.mod.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Macdonald R, Barth KA, Xu Q, Holder N, Mikkola I, Wilson SW. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development. 1995;121:3267–3278. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- Macdonald R, Scholes J, Strahle U, Brennan C, Holder N, Brand M, Wilson SW. The Pax protein Noi is required for commissural axon pathway formation in the rostral forebrain. Development. 1997;124:2397–2408. doi: 10.1242/dev.124.12.2397. [DOI] [PubMed] [Google Scholar]
- Macdonald R, Wilson S. Distribution of Pax6 protein during eye development suggests discrete roles in proliferative and differentiated visual cells. Dev Genes Evol. 1997;206:363–369. doi: 10.1007/s004270050065. [DOI] [PubMed] [Google Scholar]
- Mack AF, Fernald RD. New rods move before differentiating in adult teleost retina. Dev Biol. 1995;170:136–141. doi: 10.1006/dbio.1995.1202. [DOI] [PubMed] [Google Scholar]
- Makhankov YV, Rinner O, Neuhauss SC. An inexpensive device for non-invasive electroretinography in small aquatic vertebrates. J Neurosci Methods. 2004;135:205–210. doi: 10.1016/j.jneumeth.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Malicki J. Development of the retina. Methods Cell Biol. 1999;59:273–299. doi: 10.1016/s0091-679x(08)61830-0. [DOI] [PubMed] [Google Scholar]
- Malicki J. Harnessing the power of forward genetics—Analysis of neuronal diversity and patterning in the zebrafish retina. Trends Neurosci. 2000;23:531–541. doi: 10.1016/s0166-2236(00)01655-6. [DOI] [PubMed] [Google Scholar]
- Malicki J, Driever W. Oko meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell–cell interactions of the retinal neuroepithelial sheet. Development. 1999;126:1235–1246. doi: 10.1242/dev.126.6.1235. [DOI] [PubMed] [Google Scholar]
- Malicki J, Jo H, Pujic Z. Zebrafish N-cadherin, encoded by the glass onion locus, plays an essential role in retinal patterning. Dev Biol. 2003;259:95–108. doi: 10.1016/s0012-1606(03)00181-7. [DOI] [PubMed] [Google Scholar]
- Malicki J, Jo H, Wei X, Hsiung M, Pujic Z. Analysis of gene function in the zebrafish retina. Methods. 2002;28:427–438. doi: 10.1016/s1046-2023(02)00262-1. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Mangrum WI, Dowling JE, Cohen ED. A morphological classification of ganglion cells in the zebrafish retina. Vis Neurosci. 2002;19:767–779. doi: 10.1017/s0952523802196076. [DOI] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, Easter SS., Jr Neurogenesis in the visual system of embryonic and adult zebrafish (Danio rerio) Vis Neurosci. 1999;16:417–424. doi: 10.1017/s095252389916303x. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Del Bene F, Nica G, Hammerschmidt M, Bovolenta P, Wittbrodt J. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev Cell. 2005;8:565–574. doi: 10.1016/j.devcel.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Masai I, Lele Z, Yamaguchi M, Komori A, Nakata A, Nishiwaki Y, Wada H, Tanaka H, Nojima Y, Hammerschmidt M, Wilson SW, Okamoto H. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development. 2003;130:2479–2494. doi: 10.1242/dev.00465. [DOI] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Masai I, Yamaguchi M, Tonou-Fujimori N, Komori A, Okamoto H. The hedgehog–PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development. 2005;132:1539–1553. doi: 10.1242/dev.01714. [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe WK. Sensory neuron growth cones comigrate with posterior lateral line primordial cells in zebrafish. J Comp Neurol. 1985;238:218–224. doi: 10.1002/cne.902380208. [DOI] [PubMed] [Google Scholar]
- Metcalfe WK, Myers P, Trevarrow B, Bass M, Kimmel C. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Moens CB, Fritz A. Techniques in neural development. Methods Cell Biol. 1999;59:253–272. doi: 10.1016/s0091-679x(08)61829-4. [DOI] [PubMed] [Google Scholar]
- Mohideen MA, Beckwith LG, Tsao-Wu GS, Moore JL, Wong AC, Chinoy MR, Cheng KC. Histology-based screen for zebrafish mutants with abnormal cell differentiation. Dev Dyn. 2003;228:414–423. doi: 10.1002/dvdy.10407. [DOI] [PubMed] [Google Scholar]
- Morris AC, Schroeter EH, Bilotta J, Wong RO, Fadool JM. Cone survival despite rod degeneration in XOPS-mCFP transgenic zebrafish. Invest Ophthalmol Vis Sci. 2005;46:4762–4771. doi: 10.1167/iovs.05-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. Anatomisch-physiologische untersuchungen uber die Retina bei Menschen und Wirbelthieren. Z Wiss Zool. 1857;8:1–122. [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien CB, Baier H, Wong RO. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron. 2006;52:609–621. doi: 10.1016/j.neuron.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, Gahtan E, Xiao T, Nevin LM, Gosse NJ, Staub W, Finger-Baier K, et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, Horikawa K, Ikeo K, Takeda H, Kawakami K. Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: Tcf7 and synembryn-like. Development. 2008;135:159–169. doi: 10.1242/dev.009050. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nawrocki W. Ph D dissertation. University of Oregon; Eugine, OR: 1985. Development of the neural retina in the zebrafish (Brachydanio rerio) [Google Scholar]
- Neuhauss SC. Behavioral genetic approaches to visual system development and function in zebrafish. J Neurobiol. 2003;54:148–160. doi: 10.1002/neu.10165. [DOI] [PubMed] [Google Scholar]
- Neuhauss SC, Biehlmaier O, Seeliger MW, Das T, Kohler K, Harris WA, Baier H. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J Neurosci. 1999;19:8603–8615. doi: 10.1523/JNEUROSCI.19-19-08603.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Nicolson T, Rusch A, Friedrich RW, Granato M, Ruppersberg JP, Nusslein-Volhard C. Genetic analysis of vertebrate sensory hair cell mechanosensation: the zebrafish circler mutants. Neuron. 1998;20:271–283. doi: 10.1016/s0896-6273(00)80455-9. [DOI] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nornes HO, Dressler GR, Knapik EW, Deutsch U, Gruss P. Spatially and temporally restricted expression of Pax2 during murine neurogenesis. Development. 1990;109:797–809. doi: 10.1242/dev.109.4.797. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak AE, Ribera AB. Immunocytochemistry as a tool for zebrafish developmental neurobiology. Methods Cell Sci. 2003;25:79–83. doi: 10.1023/B:MICS.0000006894.43940.b1. [DOI] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nature Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Levine EM, Canger AK, Raymond PA, Schechter N. Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J Comp Neurol. 1997;388:495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Pathak N, Obara T, Mangos S, Liu Y, Drummond IA. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z: GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211:603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Perkins BD, Kainz PM, O’Malley DM, Dowling JE. Transgenic expression of a GFP−rhodopsin COOH-terminal fusion protein in zebrafish rod photoreceptors. Vis Neurosci. 2002;19:257–264. doi: 10.1017/s0952523802192030. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Fadool JM, McClintock J, Linser PJ. Müller cell differentiation in the zebrafish neural retina: evidence of distinct early and late stages in cell maturation. J Comp Neurol. 2001;429:530–540. doi: 10.1002/1096-9861(20010122)429:4<530::aid-cne2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Tu C, Linser PJ. Isolation and characterization of a carbonic anhydrase homologue from the zebrafish (Danio rerio) J Mol Evol. 1997;44:432–439. doi: 10.1007/pl00006163. [DOI] [PubMed] [Google Scholar]
- Plaster N, Sonntag C, Busse CE, Hammerschmidt M. p53 deficiency rescues apoptosis and differentiation of multiple cell types in zebrafish flathead mutants deficient for zygotic DNA polymerase delta1. Cell Death Differ. 2006;13:223–235. doi: 10.1038/sj.cdd.4401747. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Pujic Z, Malicki J. Mutation of the zebrafish glass onion locus causes early cell−nonautonomous loss of neuroepithelial integrity followed by severe neuronal patterning defects in the retina. Dev Biol. 2001;234:454–469. doi: 10.1006/dbio.2001.0251. [DOI] [PubMed] [Google Scholar]
- Pujic Z, Malicki J. Retinal pattern and the genetic basis of its formation in zebrafish. Semin Cell Dev Biol. 2004;15:105–114. doi: 10.1016/j.semcdb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Pujic Z, Omori Y, Tsujikawa M, Thisse B, Thisse C, Malicki J. Reverse genetic analysis of neurogenesis in the zebrafish retina. Dev Biol. 2006;293:330–347. doi: 10.1016/j.ydbio.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Raymond P, Barthel L, Curran G. Developmental patterning of rod and cone photoreceptors in embryonic zebrafish. J Comp Neurol. 1995;359:537–550. doi: 10.1002/cne.903590403. [DOI] [PubMed] [Google Scholar]
- Raymond P, Barthel L, Rounsifer M, Sullivan S, Knight J. Expression of rod and cone visual pigments in godfish and zebrafish: a rhodopsin-like gene is expressed in cones. Neuron. 1993;10:1161–1174. doi: 10.1016/0896-6273(93)90064-x. [DOI] [PubMed] [Google Scholar]
- Raz E, van Luenen HG, Schaerringer B, Plasterk RH, Driever W. Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- Rembold M, Loosli F, Adams RJ, Wittbrodt J. Individual cell migration serves as the driving force for optic vesicle evagination. Science. 2006;313:1130–1134. doi: 10.1126/science.1127144. [DOI] [PubMed] [Google Scholar]
- Ren JQ, McCarthy WR, Zhang H, Adolph AR, Li L. Behavioral visual responses of wild-type and hypopigmented zebrafish. Vision Res. 2002;42:293–299. doi: 10.1016/s0042-6989(01)00284-x. [DOI] [PubMed] [Google Scholar]
- Riley BB, Chiang M, Farmer L, Heck R. The deltaA gene of zebrafish mediates lateral inhibition of hair cells in the inner ear and is regulated by pax2.1. Development. 1999;126:5669–78. doi: 10.1242/dev.126.24.5669. [DOI] [PubMed] [Google Scholar]
- Robinson J, Schmitt E, Dowling J. Temporal and spatial patterns of opsin gene expression in zebrafish (Danio rerio) Vis Neurosci. 1995;12:895–906. doi: 10.1017/s0952523800009457. [DOI] [PubMed] [Google Scholar]
- Rodieck RW. The Vertebrate Retina: Principles of Structure and Function. W. H. Freeman & Co; San Francisco, CA: 1973. [Google Scholar]
- Roeser T, Baier H. Visuomotor behaviors in larval zebrafish after GFP-guided laser ablation of the optic tectum. J Neurosci. 2003;23:3726–3734. doi: 10.1523/JNEUROSCI.23-09-03726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandan C, Yeh J, Peteerson Q, Peteerson R. Identification of a novel retinoid by small molecule screening with zebrafish embryos. PLoS One. 2008;3:1–9. doi: 10.1371/journal.pone.0001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell J, Martin S, Heinrich G. The development of GABA immunoreactivity in the retina of the zebrafish. J Comp Neurol. 1994;345:596–601. doi: 10.1002/cne.903450409. [DOI] [PubMed] [Google Scholar]
- Sanes JR. Topographic maps and molecular gradients. Curr Opin Neurobiol. 1993;3:67–74. doi: 10.1016/0959-4388(93)90037-y. [DOI] [PubMed] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4–UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev. 2002;112:9–14. doi: 10.1016/s0925-4773(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Dowling J. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J Comp Neurol. 1994;344:532–542. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Comparison of topographical patterns of ganglion and photo−receptor cell differentiation in the retina of the zebrafish, Danio rerio. J Comp Neurol. 1996;371:222–234. doi: 10.1002/(SICI)1096-9861(19960722)371:2<222::AID-CNE3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J Comp Neurol. 1999;404:515–536. [PubMed] [Google Scholar]
- Schmitz B, Papan C, Campos-Ortega J. Neurulation in the anterior trunk of the zebrafish Brachydanio rerio. Roux’s Arch Dev Biol. 1993;202:250–259. doi: 10.1007/BF00363214. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Wong RO, Gregg RG. In vivo development of retinal ON-bipolar cell axonal terminals visualized in nyx: MYFP transgenic zebrafish. Vis Neurosci. 2006;23:833–843. doi: 10.1017/S0952523806230219. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Seddon J. Age-relatred macular degeneration: epidemiology. In: Albert B, Jakobiec F, editors. Principles and Practice of Ophthalmology. Philadelphia: 1994. pp. 1266–1274. [Google Scholar]
- Seo HC, Drivenes O, Ellingsen S, Fjose A. Expression of two zebrafish homologues of the murine Six3 gene demarcates the initial eye primordia. Mech Dev. 1998;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shields CR, Klooster J, Claassen Y, Ul-Hussain M, Zoidl G, Dermietzel R, Kamermans M. Retinal horizontal cell-specific promoter activity and protein expression of zebrafish connexin 52.6 and connexin 55.5. J Comp Neurol. 2007;501:765–779. doi: 10.1002/cne.21282. [DOI] [PubMed] [Google Scholar]
- Sivasubbu S, Balciunas D, Davidson AE, Pickart MA, Hermanson SB, Wangensteen KJ, Wolbrink DC, Ekker SC. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev. 2006;123:513–529. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Schier A, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1–20. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soules KA, Link BA. Morphogenesis of the anterior segment in the zebrafish eye. BMC Dev Biol. 2005;5:12. doi: 10.1186/1471-213X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle U, Blader P, Adam J, Ingham PW. A simple and efficient procedure for non-isotopic in situ hybridization to sectioned material. Trends Genet. 1994;10:75–76. doi: 10.1016/0168-9525(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Streisinger G, Singer F, Walker C, Knauber D, Dower N. Segregation analyses and gene– centromere distances in zebrafish. Genetics. 1986;112:311–319. doi: 10.1093/genetics/112.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- Stuermer CA. Retinotopic organization of the developing retinotectal projections in the zebrafish embryo. J Neurosci. 1988;12:4513–4530. doi: 10.1523/JNEUROSCI.08-12-04513.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Take-uchi M, Clarke JD, Wilson SW. Hedgehog signalling maintains the optic stalk–retinal interface through the regulation of Vax gene activity. Development. 2003;130:955–968. doi: 10.1242/dev.00305. [DOI] [PubMed] [Google Scholar]
- Takechi M, Hamaoka T, Kawamura S. Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 2003;553:90–94. doi: 10.1016/s0014-5793(03)00977-3. [DOI] [PubMed] [Google Scholar]
- Takechi M, Kawamura S. Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J Exp Biol. 2005;208:1337–1345. doi: 10.1242/jeb.01532. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Hurley JB, Van Epps HA, Brockerhoff SE. A zebrafish model for pyruvate dehydrogenase deficiency: rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc Natl Acad Sci U S A. 2004;101:4584–4589. doi: 10.1073/pnas.0307074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, Baranowski TC, Rubinstein AL, Doan TN, Dingledine R, Sandberg EM. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67:11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- Trowe T, Klostermann S, Baier H, Granato M, Crawford AD, Grunewald B, Hoffmann H, Karlstrom RO, Meyer SU, Muller B, Richter S, Nüsslein-Volhard C, et al. Mutations disrupting the ordering and topographic mapping of axons in the retinotectal projection of the zebrafish, Danio rerio. Development. 1996;123:439–450. doi: 10.1242/dev.123.1.439. [DOI] [PubMed] [Google Scholar]
- Tsujimura T, Chinen A, Kawamura S. Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc Natl Acad Sci U S A. 2007;104:12813–12818. doi: 10.1073/pnas.0704061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Genetics of photoreceptor development and function in zebrafish. Int J Dev Biol. 2004a;48:925–934. doi: 10.1387/ijdb.041890mt. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron. 2004b;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- Turner D, Cepko C. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner D, Snyder E, Cepko C. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Odenthal J, Haffter P. Developmental mutant screens in the zebrafish. Methods Cell Biol. 1999;60:21–41. doi: 10.1016/s0091-679x(08)61892-0. [DOI] [PubMed] [Google Scholar]
- Varga ZM, Wegner J, Westerfield M. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops. Development. 1999;126:5533–5546. doi: 10.1242/dev.126.24.5533. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Doro CJ, Hyde DR. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Raff M. Retinal astrocytes are immigrants from the optic nerve. Nature. 1988;332:834–837. doi: 10.1038/332834a0. [DOI] [PubMed] [Google Scholar]
- Wei X, Malicki J. Nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene: 2000. The Zebrafish Book. [Google Scholar]
- Wetts R, Fraser S. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Wise G, Dollery C, Henkind P. The Retinal Circulation. Harper & Row; New York: 1971. [Google Scholar]
- Woo K, Fraser SE. Order and coherence in the fate map of the zebrafish nervous system. Development. 1995;121:2595–2609. doi: 10.1242/dev.121.8.2595. [DOI] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–2967. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Fujimori-Tonou N, Yoshimura Y, Kishi T, Okamoto H, Masai I. Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53. Development. 2008;135:1247–1257. doi: 10.1242/dev.011015. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Imai F, Tonou-Fujimori N, Masai I. Mutations in N-cadherin and a Stardust homolog, Nagie oko, affect cell-cycle exit in zebrafish retina. Mech Dev. 2010;127:247–264. doi: 10.1016/j.mod.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Jeffery WR. Probing teleost eye development by lens transplantation. Methods. 2002;28:420–426. doi: 10.1016/s1046-2023(02)00261-x. [DOI] [PubMed] [Google Scholar]
- Yazulla S, Studholme KM. Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. J Neurocytol. 2001;30:551–592. doi: 10.1023/a:1016512617484. [DOI] [PubMed] [Google Scholar]
- Yeo SY, Kim M, Kim HS, Huh TL, Chitnis AB. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Yu CJ, Gao Y, Willis CL, Li P, Tiano JP, Nakamura PA, Hyde DR, Li L. Mitogen−associated protein kinase- and protein kinase A-dependent regulation of rhodopsin promoter expression in zebrafish rod photoreceptor cells. J Neurosci Res. 2007;85:488–496. doi: 10.1002/jnr.21157. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCulloch K, Malicki J. Lens transplantation in zebrafish and its application in the analysis of eye mutants. J Vis Exp. 2009 doi: 10.3791/1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XC, Yee RW, Norcom E, Burgess H, Avanesov AS, Barrish JP, Malicki J. The zebrafish cornea: structure and development. Invest Ophthalmol Vis Sci. 2006;47:4341–4348. doi: 10.1167/iovs.05-1611. [DOI] [PubMed] [Google Scholar]
- Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]