Abstract
Over the previous decade, there has been an explosion of interest in network science, in general, and its application to the human brain, in particular. Most brain network investigations to date have used linear correlations (LinCorr) between brain areas to construct and then interpret brain networks. In this study, we applied an entropy-based method to establish functional connectivity between brain areas. This method is sensitive to both nonlinear and linear associations. The LinCorr-based and entropy-based techniques were applied to resting-state functional magnetic resonance imaging data from 10 subjects, and the resulting networks were compared. The networks derived from the entropy-based method exhibited power-law degree distributions. Moreover, the entropy-based networks had a higher clustering coefficient and a shorter path length compared with that of the LinCorr-based networks. While the LinCorr-based networks were assortative, with nodes with similar degrees preferentially connected, the entropy-based networks were disassortative, with high-degree hubs directly connected to low-degree nodes. It is likely that the differences in clustering and assortativity are due to “mega-hubs” in the entropy-based networks. These mega-hubs connect to a large majority of the nodes in the network. This is the first work clearly demonstrating differences between functional brain networks using linear and nonlinear techniques. The key finding is that the nonlinear technique produced networks with scale-free degree distributions. There remains debate among the neuroscience community as to whether human brains are scale free. These data support the argument that at least some aspects of the human brain are perhaps scale free.
Key words: : brain networks, correlation, graph theory, resting state
Introduction
With the advent of the network science field, newly developed network methods have been applied to the most complex of biological systems: the human brain. The connectivity patterns in the human brain have been modeled as networks, from the neuronal level (Bonifazi et al., 2009; Yu et al., 2008) to macroscale brain organization (Achard et al., 2006; Eguiluz et al., 2005; Hagmann et al., 2008; Hayasaka and Laurienti, 2010; Iturria-Medina et al., 2008; Stam, 2004). Functional brain networks are typically derived from linear correlation (LinCorr) of time series from multiple data points in the brain, most commonly from functional magnetic resonance imaging (fMRI) (Achard et al., 2006; Eguiluz et al., 2005; Hayasaka and Laurienti, 2010; van den Heuvel et al., 2008). This work uses nonlinear analyses to establish connections in functional brain networks and demonstrates significant differences compared with networks generated using linear analyses. Networks constructed with linear and nonlinear analyses show differences most notably in the degree distributions with nonlinear networks having nodes with a substantially higher degree. Given that there is no gold standard, it is vital that these differences be further studied. One should not assume that linear outcomes, which are the default in the literature, are correct and that methods producing different outcomes are wrong. This work sets the groundwork for future studies that examine relationships between brain networks and cognition to determine whether these nonlinear findings have behavioral relevance.
In order to create a functional brain network from time series data, the strength of temporal association or coherence needs to be summarized between disparate areas of the brain. Although there are many possible ways to summarize such functional connectivity (Smith et al., 2011), Pearson's correlation coefficient is perhaps most widely used due to its computational ease and familiarity among researchers. However, a correlation coefficient is only sensitive to a linear component of a relationship between two variables. Consequently, a correlation coefficient may miss any nonlinear relationships among brain areas. To overcome this limitation, in recent decades, there has been ongoing interest in combined linearity and nonlinearity in measures of brain connectivity, especially with electroencephalogram (EEG) (Pritchard et al., 1995; Stam et al., 1996, 1998). Mutual information has been used as a nonlinear measure of cross-relation in fMRI studies (Hartman et al., 2011; Hlinka et al., 2011); by a nonlinear measure, we mean one sensitive to both linear and nonlinear cross-relation. This is in contrast to the Pearson's correlation coefficient or LinCorr, which only assesses the former. After Gaussianization of the data, these studies (Hartman et al., 2011; Hlinka et al., 2011) report a significant but small amount of nonlinearity using phase-angle randomized multivariate surrogate data. Using other nonlinear measures along with surrogate data, Xie and coworkers (2008) also reported significant nonlinearity in fMRI data.
For our nonlinear measure of cross-relation, we chose cross-sample entropy (XSampEnt) (Richman and Moorman, 2000). The implementation of XSampEnt is described in the “Materials and Methods” section. XSampEnt is an order-2 measure; in general, such measures are able to probe smaller distances in reconstructed state space than order-1 measures (Xie et al., 2010). XSampEnt also assesses the pattern synchronization between time series rather than just strict nontemporal (zero lag) cross-relation (Richman and Moorman, 2000; Xie et al., 2010). XSampEnt has been successfully applied to the analysis of cardiovascular measures (Yang et al., 2002), renal sympathetic nerve activity (Zhang et al., 2007), fMRI data (Hu and Shi, 2006), EEG data (Vakorin et al., 2010; Xie et al., 2010), and financial data (Liu et al., 2010).
Besides its proven utility in analyzing experimental and observational data as described earlier, XSampEnt has been extensively validated using simulated data with known properties. In their original paper, Richman and Moorman (2000) tested XSampEnt using realizations of the MIX(p) process. MIX(p) starts with N sinusoidal data points, and pN of the original sine-wave points are randomly chosen to be replaced by random numbers, where p is a probability value. Richman and Moorman calculated XSampEnt between MIX(p) realizations of p=0.3 versus p=0.1 and p=0.3 versus p=0.6, and hypothesized that the comparison 0.3 versus 0.1 should show much more (sine wave) pattern synchronization than 0.3 versus 0.6. For a range of the closeness parameter r (see “Materials and Methods” section for the definition), Richman and Moorman were able to confirm their hypothesis (Richman and Moorman, 2000). Similarly, Hu and Shi (2006) generated simulated data consisting of regularly spaced sinusoidal pulses and irregularly spaced sinusoidal pulses. Thus, both time series consisted of the same pattern (identical sinusoidal pulses), but in the first time series the pulses were regularly spaced in time while in the second they were irregularly spaced (some Gaussian noise was added to both). Although LinCorr between the two was very low (0.0032), XSampEnt between the two was virtually the same as the sample entropy (SampEnt) measured for each individually (XSampEnt=2.22 vs. SampEnt regular and irregular=2.24), indicating that XSampEnt successfully detected the identical asynchronous patterning in each (Hu and Shi, 2006). Xie and colleagues (2010) examined XSampEnt using five different simulations, including both deterministic and stochastic models (coupled broadband noises, coupled Lorenz–Lorenz data, coupled Rossler–Rossler data, coupled Rossler–Lorenz data, and a neural mass model) for both noise-free simulations and simulations with added noise. For a wide range of coupling strengths, XSampEnt performed well, although slightly less so than their new measure of cross-fuzzy entropy (Xie et al., 2010).
Thus, in this work, we applied XSampEnt to resting-state fMRI data and examined the resulting connectivity pattern. In particular, we constructed brain functional connectivity networks having the same set of nodes but links defined by different connectivity measures, namely XSampEnt and LinCorr. The topological characteristics of these networks were then examined. Since XSampEnt is sensitive to nonlinear relationships in addition to linear associations identified by LinCorr, we expected that the network organization might differ between the two types of networks.
Materials and Methods
The fMRI data used in this study were acquired from 10 healthy young subjects. These subjects were a part of a larger study (Peiffer et al., 2009), and these are the same set of subjects used in our previous network analysis study (Hayasaka and Laurienti, 2010). All the subjects signed a written informed consent, and the original study was approved by the Institutional Review Board of Wake Forest School of Medicine and conducted in accordance with the Declaration of Helsinki. The fMRI data from each subject consisted of 120 images acquired during 5 min of resting using a gradient echo-planar imaging protocol with TR/TE=2500/40 msec on a 1.5T GE twin-speed LX scanner with a birdcage head coil (GE Medical Systems, Milwaukee, WI). The acquired images were motion corrected, spatially normalized to the MNI (Montreal Neurological Institute) space, and re-sliced to 4×4×5 mm voxel size using an in-house processing script based on SPM99 package (Wellcome Trust Centre for Neuroimaging, London, United Kingdom). In order to avoid artificially introducing local spatial correlation, the resulting images were not smoothed (van den Heuvel et al., 2008). To correct for physiological noises, the spatially normalized fMRI time series for each subject was first band-pass filtered (0.009–0.08 Hz) to reduce respiratory and other physiological noises (Fox et al., 2005; van den Heuvel et al., 2008). Moreover, the mean time courses from the entire brain, the deep white matter, and the ventricles were regressed out from the filtered time series (Fox et al., 2005; van den Heuvel et al., 2008). To account for subject motion, the six rigid-body motion parameters from the motion correction process were also regressed out from the time series. Finally, node time courses were obtained by averaging the voxel time courses in 90 distinct anatomical areas defined by the Anatomical Automatic Labeling atlas (Tzourio-Mazoyer et al., 2002). Further details on fMRI data processing can be found elsewhere (Hayasaka and Laurienti, 2010).
Once the time series data were obtained for each subject, a LinCorr-based brain network was constructed. This was performed by producing a 90×90 cross-correlation matrix, with each element corresponding to LinCorr between the time series data of two brain areas. The resulting correlation matrix was then thresholded in order to characterize high positive correlation values as functional connections. This process yielded a binary matrix known as an adjacency matrix, describing the LinCorr network. A number of thresholds, corresponding to 70–97.5th percentiles of the correlation coefficients in the matrix, were used in this process to form adjacency matrices with strong positive correlations as edges. During the thresholding process, the elements (1s) along the main diagonal were set to zero to eliminate self-loops.
With the same fMRI time series data described earlier, a network based on XSampEnt connectivity was also constructed for each of the subjects. XSampEnt is based on Pincus' order-1 cross-approximate entropy (XApEnt) (Richman and Moorman, 2000). In essence, in an initial delay-time embedding dimension m, this measure first calculates the average number of vectors in reconstructed state space from series 1 that are close to vectors from series 2 (close being defined by the critical distance, or closeness parameter r). The embedding dimension m is then increased by 1, and the average number of close vectors is recomputed. XSampEnt is then the negative log of a ratio, the second average over the first average. XSampEnt represents an improvement over XApEnt in that it only requires one pair of points within the critical distance of each other to yield a value. XSampEnt increases as the number of vectors that remain close decreases, reflecting less pattern synchronization in the data. For our calculation of XSampEnt, we set the m parameter (initial embedding dimension) equal to 1 based on preliminary investigations. Following Pincus (1991), we set the r parameter equal to 0.2 of the average of standard deviation (SD) of each pair of the time series. We used the calculation method by Zhang and associates (2007) to assess XSampEnt in our fMRI data. In brief, let u and v represent two time series: u=[u(1), u(2),…u(N)] and v=[v(1), v(2),…v(N)], where N is the number of data points in each series. With m and r fixed (as defined earlier), we form two vector sequences xm and ym: xm(i)=[u(i), u(i+1),…u(i+m–1)] and ym(i)=[v(i), v(i+1),…v(i+m–1)]. Then, for each i≤N – m, we define
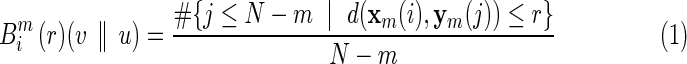 |
where j ranges from 1 to N – m, and d(.) is the distance function. Then, let
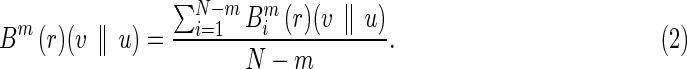 |
In other words, Bm (r)(v || u) is the average value of 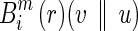 . Similarly, we define
. Similarly, we define 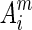 and Am as
and Am as
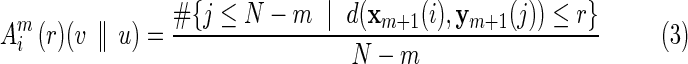 |
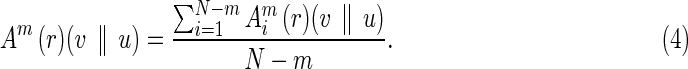 |
Finally, we can calculate XSampEnt as
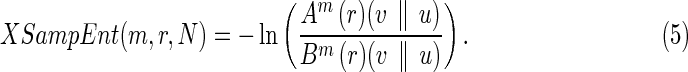 |
With the method described earlier, a 90×90 connectivity matrix was constructed, with each element quantifying XSampEnt between two brain areas. In the resulting matrix, the main diagonal elements were set to zero to eliminate self-loops. A number of thresholds, corresponding to 70–97.5th percentiles of XSampEnt in the matrix, were then applied so that a small proportion of strongest connections were retained as edges. Thus, the edge density of the resulting XSampEnt network was the same as that of the corresponding LinCorr network, facilitating comparisons of characteristics between the two types of networks.
In this study, we hypothesized that XSampEnt might capture brain connectivity which cannot be explained by the LinCorr method. This was examined by comparing various topological characteristics of the XSampEnt and LinCorr networks. Since both types of networks were constructed from the same fMRI data of the same set of subjects, any differences found in this study can be attributed to the difference in the connectivity measures. In this study, we examined topological characteristics often discussed in the brain network literature. First, we examined the node degree, or the number of edges at each node. In particular, we examined the distribution of node degrees from each subject's networks (LinCorr and XSampEnt) by Kolmogorov–Smirnov (KS) tests (Clauset et al., 2009) to compare against known parametric distributions: a power law distribution, an exponentially truncated power law distribution, and an exponential distribution. Furthermore, we also compared the degree distributions directly between the LinCorr and XSampEnt networks for each subject by a KS test. In these analyses, a significant deviation was detected at p<0.05 significance level. A degree distribution is often used to examine heterogeneity of node degrees (Amaral et al., 2000; Clauset et al., 2009). For example, in a class of networks, known as scale-free networks, the degree distribution follows a power-law distribution—or scale-free distribution—indicating the existence of very few extremely high-degree nodes; whereas the vast majority of nodes have only a small number of edges (Barabasi and Albert, 1999; Clauset et al., 2009). In the brain network literature, the degree distribution of a brain network, whether it is functional or structural, is found to be an exponentially-truncated power-law distribution (Achard et al., 2006; Gong et al., 2009; Hayasaka and Laurienti, 2010; He et al., 2007). It is a variant of power-law distributions, with its tail truncated; there are still extremely high-degree nodes, but which are attenuated in magnitude and abundance compared with power-law distributions (Mossa et al., 2002).
In addition to degree distributions, we also examined the clustering coefficient C and the shortest average path length L (Reijneveld et al., 2007; Rubinov and Sporns, 2010). The clustering coefficient C is defined as the probability that a node's neighbors are also neighbors to each other, and it summarizes how tightly nodes are interconnected. Large C is indicative of a network with strong local interconnections. The path length L is the average of shortest distances between any two nodes in a network, in terms of the number of edges separating them. In a network with small L, information can travel from any node to any other nodes just by a few steps. A network with large C and small L is often characterized as a small-world network (Watts and Strogatz, 1998), simultaneously supporting local specializations and efficient global communication (Bullmore and Sporns, 2009; Rubinov and Sporns, 2010; Telesford et al., 2011a). Both C and L have been closely examined in the brain network literature, and brain networks, both anatomical and functional, have been found to be small-world networks (Bullmore and Sporns, 2009; Humphries and Gurney, 2008; Telesford et al., 2011b). To compare C and L between the LinCorr and XSampEnt networks, normalized clustering coefficient and path length were also calculated. While the path length was normalized against that of the equivalent random network with the same degree distribution (Maslov and Sneppen, 2002), the clustering coefficient was normalized against that of the equivalent lattice network with the same degree distribution (Sporns and Zwi, 2004; Telesford., 2011b). In particular, 30 realizations of random and lattice networks were generated from each of the original networks, and the resulting path lengths and clustering coefficients were averaged. This resulted in the average clustering coefficient Clatt from the lattice networks and path length Lrand from the random networks, which were used to normalize the clustering coefficient by C/Clatt and the path length by L/Lrand. These normalized statistics were then compared between the LinCorr and XSampEnt networks by paired t-tests.
We also examined the assortativity of the LinCorr and XSampEnt networks. The assortativity in a network is defined as the tendency for high (or low)-degree nodes being preferentially connected to other high (or low)-degree nodes (Newman, 2002, 2003). The assortative tendency of a network can be quantified by the assortativity coefficient ranging from −1 to 1, calculated as the correlation coefficient between the origin node degree and the terminus node degree of all the edges in the network (Newman, 2002, 2003). A highly negative assortativity coefficient indicates that the network is disassortative, with nodes with dissimilar degrees being preferentially connected, while a highly positive coefficient indicates that the network is assortative. In general, social networks tend to be assortative, whereas technological and biological networks tend to be disassortative (Newman, 2003). However, brain networks have been found to be assortative (Hagmann et al., 2008; Joyce et al., 2010). The assortative or disassortative tendency of a network can also be examined by a 2D connection probability plot. A 2D connection probability plot, also known as the correlation profile, shows the probability that an edge with the origin node degree k is connected to the terminus node with degree j (Newman, 2003). A 2D connectivity plot can be generated from a single network by summarizing the abundance of edges in that network with certain origin and terminus degrees (Maslov and Sneppen, 2002; Maslov et al., 2004; Newman, 2003; Song et al., 2006; Xu et al., 2009). Such plots enable an examination of assortativity in detail, visually summarizing how nodes with different levels of degrees are connected in the network. If nodes with similar degrees are connected in an assortative network, then connection probability is elevated along the leading diagonal. In contrast, if nodes with dissimilar degrees are connected in a disassortative network, connection probability is elevated along the axes instead.
Results
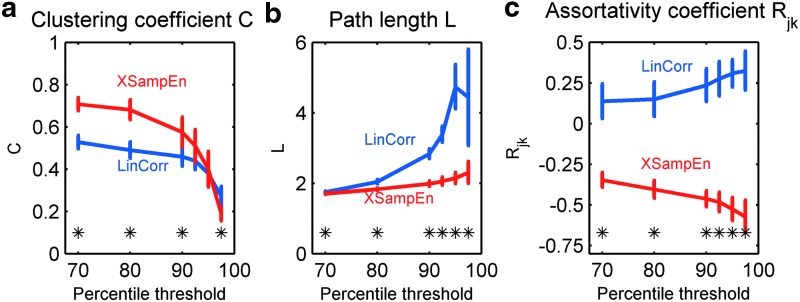
Clustering coefficient C and path length L, metrics associated with small-world characteristics of a network (Telesford et al., 2011b; Watts and Strogatz, 1998), were first compared between the LinCorr and XSampEnt networks. This was done by calculating C and L over a range of percentile thresholds (70–97.5%). The average C and L from all the subjects are plotted in Figure 1. The data clearly demonstrated that C and L significantly differed between the two types of networks, indicated by asterisks for p<0.01 (paired t-test). The clustering coefficient C was significantly larger for the XSampEnt networks for 70–90th percentile thresholds (Fig. 1a), while the path length was significantly shorter for the XSampEnt networks at any threshold (Fig. 1b). In addition to C and L, the assortativity coefficient Rjk was also calculated and compared between the two types of networks (Fig. 1c). Interestingly, Rjk was positive (i.e., assortative) for the LinCorr networks, whereas Rjk was negative (i.e., disassortative) for the XSampEnt networks. The difference was significant (p<0.01, paired t-test) at any threshold. This difference in assortativity may be due to the presence of high-degree mega-hubs in XSampEnt networks that are connected to a large number of low-degree nodes.
FIG. 1.
Clustering coefficient C, path length L, and assortativity coefficient Rjk at different thresholds The averages of clustering coefficient C (a), path length L (b), and assortativity coefficient Rjk (c) were calculated from the linear correlation (LinCorr) and cross-sample entropy (XSampEnt) networks of all the subjects formed at different percentile thresholds (70–97.5%). The error bars show standard deviations (SD). Significant differences (p<0.01) assessed by paired t-tests are indicated by asterisks. Color images available online at www.liebertpub.com/brain
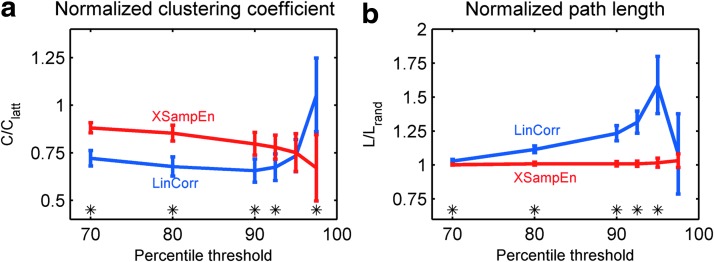
To account for the likely differences in comparable null networks, we compared normalized clustering coefficient (C/Clatt) and path length (L/Lrand) as described in the “Methods” section. Figure 2 shows the normalized clustering coefficients and path lengths. For the XSampEnt networks, the normalized clustering coefficient was significantly higher (p<0.01) than that of the LinCorr networks, except at high thresholds (95th percentile or higher; Fig. 2a). This indicates that relative to comparable lattice networks, the clustering was higher in the XSampEnt networks. On the other hand, the normalized path length was significantly smaller (p<0.01) for the XSampEnt networks compared with the LinCorr networks, except at the highest threshold (Fig. 2b). The results support the observation that the path length is shorter in the XSampEnt networks. This was particularly true as the threshold was increased. These results may be due to the presence of mega-hubs that are connected to a large portion of the network, enabling even relatively isolated nodes to reach the remainder of the network in just a few steps. It should be noted that, according to the formula provided by Laurienti and coworkers (2011), the edge density of a 90-node network should be approximately 7.89×90−0.986=0.093% or 9.3%. Thus, the 90th percentile threshold may be the most appropriate threshold in this study, producing networks with a comparable density to other types of self-organized networks (Laurienti et al., 2011). For this reason, we focus on the results from the networks formed with the 90th percentile threshold in the rest of this article.
FIG. 2.
Normalized clustering coefficient and path length at different thresholds. The normalized clustering coefficient (C/Clatt) (a) and path length (L/Lrand) (b) were calculated at different percentile thresholds (70–97.5%) and averaged across subjects. The plots show the mean of the normalized statistics, along with the SD as error bars. Significant differences (p<0.01) assessed by paired t-tests are indicated by asterisks. Color images available online at www.liebertpub.com/brain
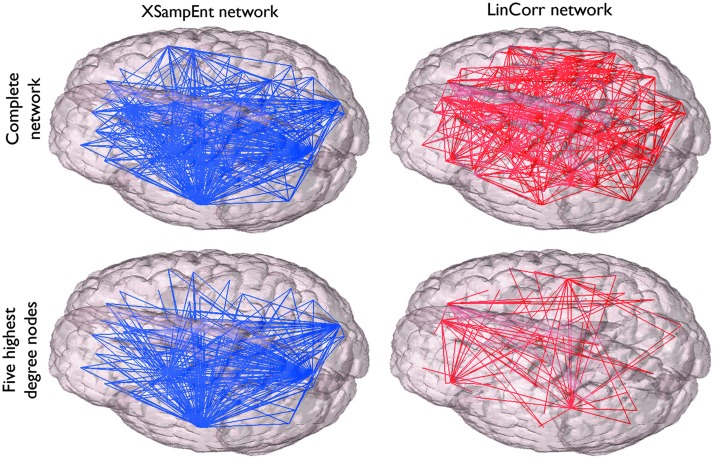
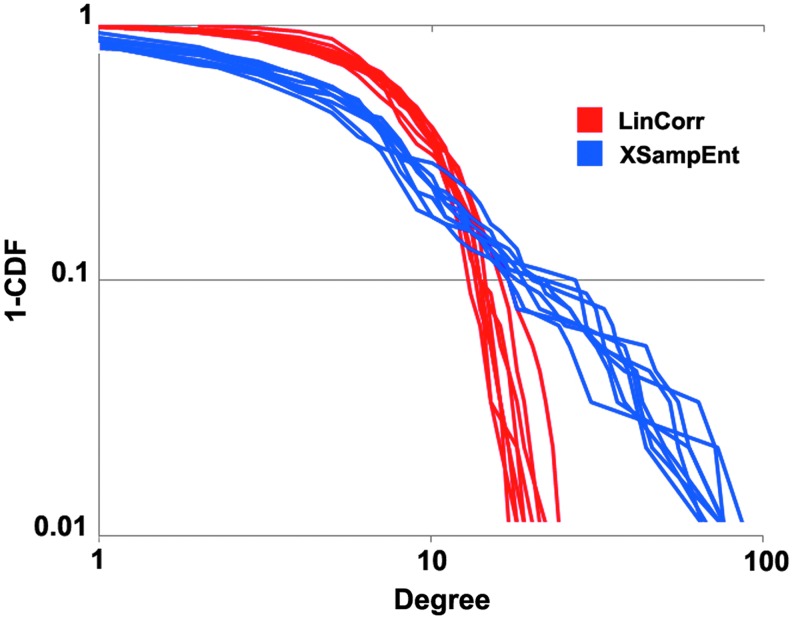
The XSampEnt networks had mega-hubs connected to the vast majority (up to 84%) of the other nodes, while such nodes did not exist in the LinCorr networks. This can be seen in Figure 3, in which all edges originating from the 5 highest-degree nodes are emphasized. In contrast, in the LinCorr networks, there were no nodes with more than 18 connections (20% of nodes). This disparity in connectivity can be seen in the degree distributions for the LinCorr and XSampEnt networks (Fig. 4). While the degree distributions of the XSampEnt networks followed a scale-free distribution, the degree distributions of the LinCorr networks, although generated from the same time series data, resulted in exponentially truncated curves as previously reported (Hayasaka and Laurienti, 2010). KS tests (Clauset et al., 2009) indicated no evidence of a significant departure from a power-law distribution among the degree distributions of the XSampEnt networks (p-value range: 0.12–0.92), while 8 out of 10 LinCorr networks had degree distributions significantly deviating from a power-law distribution (p-value range: 0.0002–0.18). The exponent of the power-law fit for the XSampEnt networks was between −2 and −3 (mean±SD=2.53±0.16).The degree distribution for each network was fit to a power law, and the corresponding R2 was computed. A paired t-test was used to compare the goodness of fit between the LinCorr (average R2=0.66, 0.059 SD) and XSampEnt (average R2=0.91, 0.026 SD). The networks generated using XSampEnt exhibited a significantly better fit (p<0.0001) than those generated using LinCorr based on a two-tailed T-test.
FIG. 3.
A XSampEnt network and a LinCorr network. The XSampEnt network (top left) and the LinCorr network (top right) were generated from the same data for one of the subjects. The nodes are embedded in the brain space according to their anatomical coordinates. To examine the number of connections mediated by hubs, the edges originating from the five highest degree nodes in each of these networks are also shown (bottom). The XSampEnt network shows a few mega-hubs that are connected to the vast majority of nodes (bottom left). On the other hand, although there are some hub nodes mediating a dozen or so connections in the LinCorr network, there are no mega-hubs with a tremendously large degree (bottom right). Color images available online at www.liebertpub.com/brain
FIG. 4.
Degree distributions. The degree distributions of the networks with connectivity based on XSampEnt (blue) and LinCorr (red) are shown. The x-axis denotes the node degree, and the y-axis denotes the complimentary cumulative distribution function (1-CDF) describing the abundance of nodes with a certain degree or higher. The degree distributions from the XSampEnt networks follow a scale-free distribution characterized by a straight line on a log–log plot, indicating that there are a few nodes with an extremely large degree or mega-hubs; while the vast majority of nodes have just a few connections at most. On the other hand, the degree distributions from the LinCorr networks are exponentially truncated. Although high-degree nodes exist in the LinCorr networks, with k≈20, such nodes have far less connections than the mega-hubs observed in the XSampEnt networks. Color images available online at www.liebertpub.com/brain
These are interesting findings, as the vast majority of brain network studies have identified truncated degree distributions rather than scale-free distributions (Bullmore and Sporns, 2009), with just a few exceptions (Eguiluz et al., 2005; van den Heuvel et al., 2008). This degree distribution difference has significant implications for fundamental property differences between the XSampEnt and LinCorr networks. For example, the XSampEnt-derived networks would have shorter connections between nodes and might be more resistant to a random node failure but would be tremendously vulnerable to failure of a hub (Albert et al., 2000). In contrast, the LinCorr networks with truncated degree distributions would be more resistant to hub failure (Achard et al., 2006). Mega-hubs in the XSampEnt networks also suggest that such hubs can exist if both linear and nonlinear dynamics are considered. We also note that the SampEnt (Richman and Moorman, 2000) in our XSampEnt networks was negatively correlated with degree (R=−0.624), indicating that the fMRI time series of mega-hubs are themselves dynamically more regular than that of nonhubs. In some way, regular dynamical activity at a node aids in its forming nonlinear connections with other nodes.
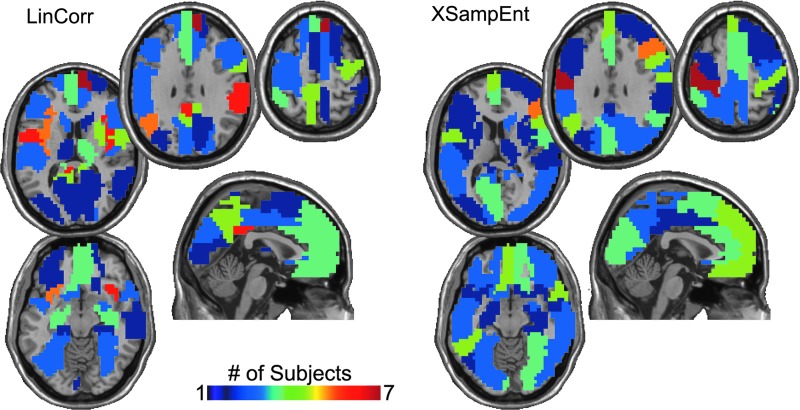
The location of the hubs within the brain was determined by mapping the degree of each node back into brain space. Figure 5 shows the location of the network hubs (defined at the top 20% of nodes). We recognize the arbitrary nature of a threshold such as this, but the purpose of this figure is to simply demonstrate the spatial location of the high-degree nodes. This was done, because simple averages are inappropriate when the degree distribution follows a power law. The hub maps show the number of subjects for which each node was considered a hub. In both the LinCorr and XSampEnt networks, the maximum number of subjects that exhibited a hub in the same location was 7 out of a possible 10. The location of the most consistent hubs was markedly different for the two methods. For LinCorr, the most consistent hub was in the right superior, medial frontal gyrus. Additional areas of relatively high consistency were the bilateral insula, the left Rolandic operculum, left mid cingulum, and the right supramarginal gyrus. For XSampEnt networks, the most consistent hub was located in the left motor cortex. Additional areas of relatively high consistency included the right inferior frontal gyrus, the left superior, medial frontal gyrus, left supramarginal gyrus, left Rolandic operculum, and the left inferior temporal gyrus. On comparing the methods, it is clear that the Rolandic operculum and the supramarginal gyri were consistent hubs. The superior, medial frontal gyrus was a hub based on both methods but it was on the right in LinCorr and on the left in XSampEnt. A noticeable absence of hubs was consistent for the two methods in the left superior middle frontal gyrus and the right superior parietal lobe.
FIG. 5.
Hub maps showing the consistency of the location of the top 20% of all brain hubs. The color bar indicates the number of subjects that had a hub in each brain region. The maximum number of subjects with the same hub location was seven for both the LinCorr and the XSampEnt networks (dark red). The right side of the image is the right side of the brain. Color images available online at www.liebertpub.com/brain
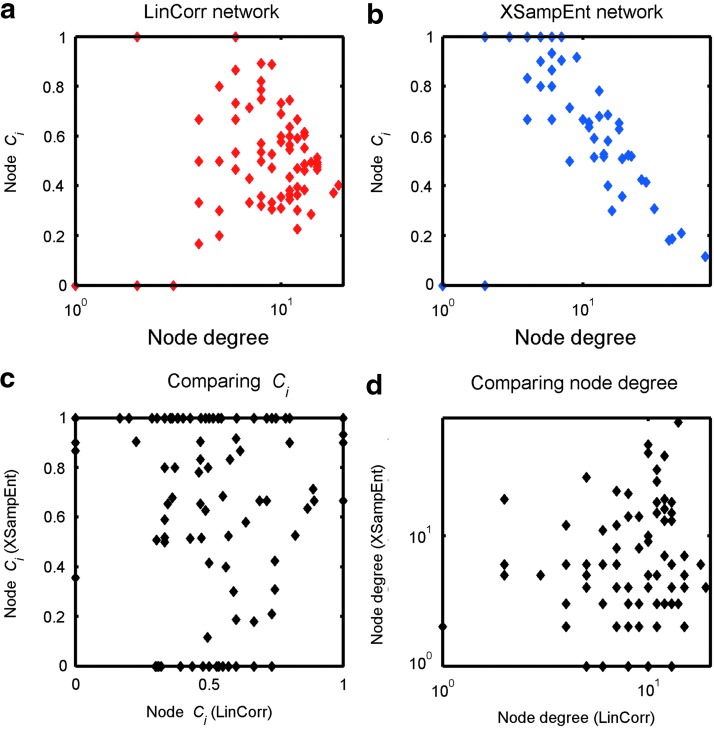
To further understand the relationship between change in clustering and degree, we plotted the nodal clustering coefficient Ci against the node degree. As can be seen in Figure 6a, there is no clear relationship between Ci and node degree in a LinCorr network. On the other hand, Figure 6b shows elevated Ci among low-degree nodes and attenuated Ci among high-degree nodes in the XSampEnt network. While these plots are based on the network data from a single subject, the pattern is consistent across subjects. These results indicate that these two types of networks have different topological organizations. We also compared Ci and node degree between the LinCorr and XSampEnd networks (Fig. 6c and d, respectively), and we found no apparent relationships between the two types of networks.
FIG. 6.
The relationship between node degree and clustering coefficient. The node degree and the node clustering coefficient Ci was plotted from the LinCorr network (a) and the XSampEnt network (b) generated with the top 10% strongest connections from the same subject. While the variability of Ci seems to decrease as the node degree increases in the LinCorr network (a), there is no obvious relationship between Ci and the degree. On the other hand, in the XSampEnt network, Ci is elevated for low-degree nodes but it decreases for nodes with higher degrees (b). When Ci and the node degree are compared between the two types of networks [(c) and (d), respectively], no obvious relationship can be observed. These patterns are consistent among the other subjects. Color images available online at www.liebertpub.com/brain
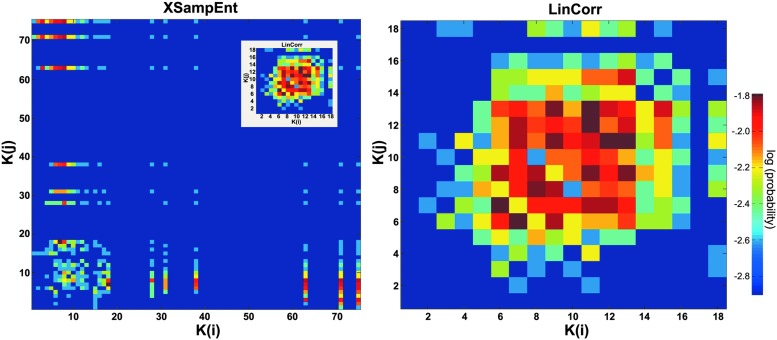
Providing further support to the substantial differences between the XSampEnt and LinCorr networks are the connection probability plots. An example from one of the subjects is shown in Figure 6. These plots show the likelihood of edges connecting a node with degree K(i) at the origin to another node with degree K(j) at the terminus. From Figure 7, we can easily see that the LinCorr network is assortative (assortativity coefficient 0.236±0.100) with elevated connection probability along the main diagonal (Fig. 7, right). On the other hand, the XSampEnt network is disassortative (assortativity coefficient −0.463±0.048), which is indicated by connection probability spanning along the axes (Fig. 7, left). It is interesting to note that the connection probability among low-degree nodes (degree <20) in the XSampEnt network somewhat resembled that of the corresponding LinCorr network. To facilitate the comparison, the connection probability plot for the LinCorr network was comparably scaled down and displayed as an inset in Figure 7, left. Despite this similarity, however, the connection probability attributable to high-degree nodes is concentrated along the axes, effectively lowering the assortativity coefficient to negative.
FIG. 7.
Connection probability plots. Each plot shows the connection probability of a network, representing how likely an edge with a certain origin degree and a terminus degree exists in the network. The connection probability is concentrated along the x- and y-axes in the XSampEnt network from a representative subject (left), indicating that nodes with a variety of degrees are connected to high-degree nodes. This concentration along the axes is a sign of disassortative connectivity in which nodes with disparate degrees tend to be preferentially connected. On the other hand, the LinCorr network from the same subject (right) shows a concentration of probability along the diagonal, resulting from nodes with similar degrees being connected with each other. This is an indication of assortative connectivity in the LinCorr network. The inset shows the connection probability plot of the LinCorr network scaled to the comparable size of that of the XSampEnt network; this is to emphasize the differences in the connection probability plots between the two networks. Color images available online at www.liebertpub.com/brain
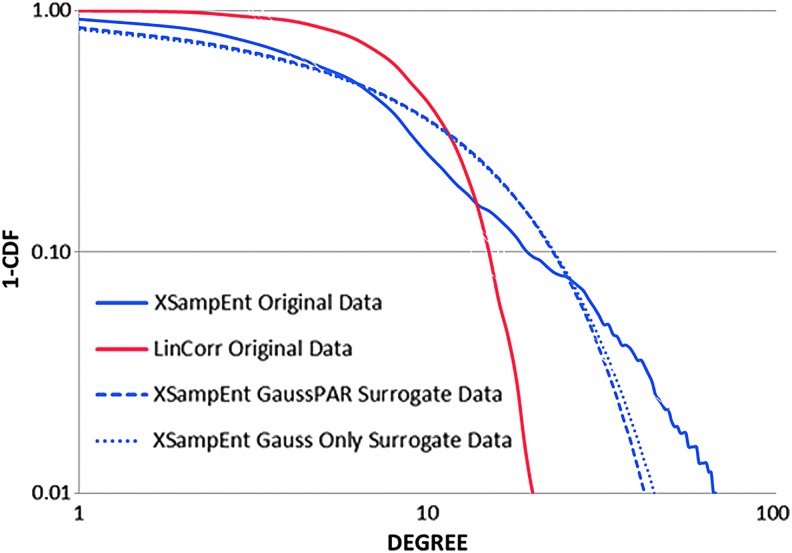
Our results described earlier seem to contradict with the results found by Hartman and colleagues (2011), in which the degree of nonlinearity, although significant, was very small in magnitude. We believe that this discrepancy is due to the Gaussianization process applied to Hartman and colleagues's data. To demonstrate this point, we examined the effects of Gaussianization on the network metrics by Gaussianizing our data and examining the properties of the resulting networks. For our Gaussianization process, rather than sorting perfect Gaussian percentiles into the data rank order, we sorted Gaussian random numbers produced by Gasdev function as described in the Numerical Recipes (Press, 1992). This is because real samples of Gaussian data are not perfectly Gaussian except in the limit approaching infinite data length. For each data set, we produced 99 Gaussian surrogates and XSampEnt networks were constructed based on the Gaussianized fMRI data. We also Gaussianized and then randomized multivariate phase angles (Prichard and Theiler, 1994), and the effect of this process on the XSampEnt network measures was also examined (again, 99 Gaussianized and phase-angle randomized [GaussPAR] surrogates). We found that Gaussianizing the data lowered the clustering coefficient of the XSampEnt networks without affecting the path length. Most notably, Gaussianzing the data markedly degraded the scale-free property of the XSampEnt networks as can be seen in the degree distributions in Figure 8. The additional step of phase-angle randomization produced almost no difference from Gaussianization alone; the cumulative degree distribution from the GaussPAR data closely followed the degree distribution from the Gaussianization only data. We also noted that Gaussianzation (with or without phase-angle randomization) did not completely transform the degree distribution of the XSampEnt network to that of the LinCorr network; we interpret this as the ability of XSampEnt to assess pattern synchronization as well as nonlinearity.
FIG. 8.
Degree distributions of Gaussianized data. Degree distributions (1-CDF) are plotted from XSampEnt networks with Gaussianized data, with and without phase-angle randomization (PAR). As a visual comparison, the degree distributions for the corresponding LinCorr and XSampEnt (without Gaussianization) networks are also shown. Color images available online at www.liebertpub.com/brain
Discussion
In this work, we demonstrated interesting characteristics of brain networks formed by functional connectivity based on XSampEnt compared with the widely used LinCorr. The primary finding was that resulting XSampEnt networks exhibited characteristics of scale-free networks, with mega-hubs connected to the majority of nodes in the network. Such hubs have far more connections than typical hubs found in functional brain networks constructed with LinCorr connectivity. The degree distributions of the XSampEnt networks followed a power-law distribution; to the best of our knowledge, this has not been observed in a brain network of a comparable scale (Bullmore and Sporns, 2009). While the location of the brain hubs differed between the two types of networks, there were consistent hubs in the medial frontal, opercular cortex, and suparmarginal gyrus. The XSampEnt had its major hub in the left motor cortex, while the primary hub in the LinCorr networks was in the right superior frontal gyrus. It is interesting to note that in these data, neither type of network exhibited the primary hub in precuneus, and the location was often reported to be the core of human brain networks (Hagmann et al., 2008). The LinCorr had this regions show up as a relatively consistent hub across subjects, but it was not among the most consistent regions. We can only speculate why hubs in the XSampEnt networks favored areas of motor cortex. Under resting conditions, the participant's sole task is to simply remain as still as possible. Perhaps this leads to numerous inputs and outputs to and from the motor cortex of a nonlinear nature involved in the inhibition of unwanted movement. Further work comparing the location of the mega-hubs from rest to various cognitive and motor tasks provides insights into the role and importance of these hubs. We hypothesize that the location of the mega-hubs will be associated with brain regions that are critical in performing specific tasks. While we view the brain as a distributed system, there is strong evidence for localization of function as well. The mega-hubs may be situated in areas that would be the most activated by the task but connect throughout the brain to enable distributed information processing.
We also discovered that the XSampEnt networks had a higher clustering coefficient and a shorter path length compared with that of the LinCorr networks constructed on the same data. Our examination on assortativity also demonstrated that the XSampEnt brain networks were disassortative as in many other biological networks reported in the literature (Newman, 2003). The changes in clustering and assortativity are most likely attributed to the presence of mega hubs. Such hubs are connected to a very large percentage of all nodes in the network, most of which are low degree. This results in an increase in the clustering in low-degree nodes and transforms the network from assortative to disassortative. It is important to note that the size of the networks used here likely contributed to this outcome. Since the networks were limited to 90 nodes, the mega-hubs (essentially out of necessity in order to, in fact, be mega-hubs) connected a large percentage of all nodes. In a network with many more nodes, this would not likely be the outcome because all the low-degree nodes would not be connected to a common hub. Such a situation would produce disassortative networks with lower clustering rather than with higher clustering.
Our results using XSampEnt also demonstrate its sensitivity to pattern synchronization between time series data (Richman and Moorman, 2000). This is an important distinction from LinCorr, in which the temporal ordering of data points is simply ignored. Mutual information, another method to measure nonlinear association, similarly does not assess temporal relationships in patterns between time series; it only assesses zero-lag relations (Hlinka et al., 2011). In particular, Hlinka and associates (2011) stated:
“When linear correlation is used as a measure of functional connectivity, there are some implicit assumptions made. The first is that the information in the temporal order of the samples can be ignored (both within each time series and the mutual interaction). While the extent of justifiability of this assumption deserves exploration of its own, we keep this interim assumption for the purposes of this paper, not least in order to keep the comparison of linear correlation to nonlinear measures [mutual information] fair.”
As we explored the validity of this assumption in this work, we were able to demonstrate that there are obviously important nonlinear relations in the temporal order. That said, we would like to note that XSampEnt's sensitivity to nonlinear association does not mean it is “better” or “worse” than LinCorr. Rather, our results demonstrate that the functional brain networks constructed with the added nonlinear components result in drastically different networks from those constructed with the more traditional linear components alone and, thus, may provide additional valuable information in future investigations regarding brain function during active processing tasks.
While examining functional connectivity in fMRI data is becoming a popular research topic among neuroimaging researchers (Smith et al., 2011), a limited number of time points in typical fMRI time series data can pose a serious challenge in application of more sophisticated connectivity methods based on information theory, such as XSampEnt. Information theory-based methods are found to be the most useful in EEG and MEG time series data with a large number of temporal data points with a small number of recording sites (Ponten et al., 2007; Rubinov et al., 2009; Stam et al., 2007; Vakorin et al., 2010; Xie et al., 2010). On the other hand, typical fMRI data have a large number of recording sites (or voxels) and a small number of time points. The limited number of time points may pose a challenge in reliably estimating functional connectivity between two brain areas, and a large number of nodes necessitate calculating tremendously large number of connectivity measures. If there are N nodes, then there are N (N−1)/2 possible pairs of nodes. While calculating such a large number of correlation coefficients has been done in multiple studies (Eguiluz et al., 2005; Hayasaka and Laurienti, 2010; van den Heuvel et al., 2008), calculating a large number of nonlinear connectivity measures may be computationally challenging. A comparison of LinCorr and XSampEnt networks formed at a finer regional parcellation would be a next logical step in examining their differences further. We are in the process of implementing methods that enable us to use this methodology in voxel-based networks with ∼20,000 nodes; the results will be presented in our future work.
In summary, our findings demonstrate the differences between assessing both linear and nonlinear connections when building a network based on time series data versus assessing just the linear connections. Although a LinCorr coefficient is widely used as a measure of connectivity in various time series data, due to its simplicity and computational ease, it is only sensitive to linear relationships and will miss other underlying nonlinear connectivity in the data, if such nonlinearities exist. The XSampEnt approach assessing both linear and nonlinear connections produced networks with mega-hubs. These mega-hubs connect to nearly all the nodes in the network, with resultant effects on clustering, pathlength, and assortativity. This is the first work clearly demonstrating differences between functional brain networks using linear and nonlinear techniques. The key finding is that the nonlinear technique produced network with scale-free degree distributions. There remains debate among the neuroscience community as to whether human brains are scale free, and these data support the argument that the human brain is perhaps scale free.
While this work clearly shows differences in the network topology between LinCorr and XSampEnt networks, it is not possible at this time to conclude which method is more correct. Given that there is no gold standard to be used as a comparator, such a conclusion should wait for studies that combine cognitive function with network analyses. Studies will have to be designed to determine whether one or the other method produces networks that better explain cognitive function. Given that the networks exhibited quantitative and qualitative differences, presumably one of the methods will better reflect cognition than the other. This future work is required to determine whether the neuroscience community should shift focus more toward information theory-based connectivity measures that are sensitive to various types of associations, both linear and nonlinear, such as our XSampEnt technique. It should be noted that the correlation-based linear method should not be considered the gold standard just because it came first and is easier to perform, just as the XSampEnt should not be considered a new gold standard. Obviously, we were able to uncover a functional brain network structure previously unobserved by the conventional LinCorr method. While further research is needed to fully characterize the linear and nonlinear interactions between brain regions, this XSampEnt technique provides new evidence of how the brain may work and shows that the self-organized human brain network, while perhaps the most complex of biological systems, may be operationally organized similar to other biological systems.
Acknowledgment
This study was supported by the National Institute of Neurological Disorders and Stroke (NINDS; NS070917).
Author Disclosure Statement
No competing financial interests exist.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. 2006. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci 26:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi AL. 2000. Error and attack tolerance of complex networks. Nature 406:378–382 [DOI] [PubMed] [Google Scholar]
- Amaral LAN, Scala A, Barthelemy M, Stanley HE. 2000. Classes of small-world networks. Proc Natl Acad Sci U S A 97:11149–11152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. 1999. Emergence of scaling in random networks. Science 286:509–512 [DOI] [PubMed] [Google Scholar]
- Bonifazi P, Goldin M, Picardo MA, Jorquera I, Cattani A, Bianconi G, Represa A, Ben-Ari Y, Cossart R. 2009. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science 326:1419–1424 [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10:186–198 [DOI] [PubMed] [Google Scholar]
- Clauset A, Shalizi CR, Newman MEJ. 2009. Power-law distributions in empirical data. SIAM Rev 51:661–703 [Google Scholar]
- Eguiluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. 2005. Scale-free brain functional networks. Phys Rev Lett 94:018102. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C. 2009. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex 19:524–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman D, Hlinka J, Palus M, Mantini D, Corbetta M. 2011. The role of nonlinearity in computing graph-theoretical properties of resting-state functional magnetic resonance imaging brain networks. Chaos 21:013119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Laurienti PJ. 2010. Comparison of characteristics between region-and voxel-based network analyses in resting-state fMRI data. Neuroimage 50:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. 2007. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex 17:2407–2419 [DOI] [PubMed] [Google Scholar]
- Hlinka J, Palus M, Vejmelka M, Mantini D, Corbetta M. 2011. Functional connectivity in resting-state fMRI: is linear correlation sufficient? Neuroimage 54:2218–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shi P. 2006. Interregional functional connectivity via pattern synchrony. The 9th International Conference on Control, Automation, Robotics and Vision, Singapore [Google Scholar]
- Humphries MD, Gurney K. 2008. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS One 3:e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodriguez EJ, Aleman-Gomez Y, Melie-Garcia L. 2008. Studying the human brain anatomical network via diffusion-weighted MRI and Graph Theory. Neuroimage 40:1064–1076 [DOI] [PubMed] [Google Scholar]
- Joyce KE, Laurienti PJ, Burdette JH, Hayasaka S. 2010. A new measure of centrality for brain networks. PLoS One 5:e12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurienti PJ, Joyce KE, Telesford Q, Burdette JH, Hayasaka S. 2011. Universal fractal scaling of self-organized networks. Physica A 390:3608–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-Z, Qian X-Y, Lu H-Y. 2010. Cross-sample entropy of foreign exchange time series. Physica A 389:4785–4792 [Google Scholar]
- Maslov S, Sneppen K. 2002. Specificity and stability in topology of protein networks. Science 296:910–913 [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K, Zaliznyak A. 2004. Detection of topological patterns in complex networks: correlation profile of the internet. Physica A 333:529–540 [Google Scholar]
- Mossa S, Barthelemy M, Stanley HE, Amaral LAN. 2002. Truncation of power law behavior in scale-free network models due to information filtering. Phys Rev Lett 88:138701. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. 2002. Assortative mixing in networks. Phys Rev Lett 89:208701. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. 2003. Mixing patterns in networks. Phys Rev E 67(2 Pt 2):026126. [DOI] [PubMed] [Google Scholar]
- Peiffer AM, Hugenschmidt CE, Maldjian JA, Casanova R, Srikanth R, Hayasaka S, Burdette JH, Kraft RA, Laurienti PJ. 2009. Aging and the interaction of sensory cortical function and structure. Hum Brain Mapp 30:228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus SM. 1991. Approximate entropy as a measure of system complexity. Proc Natl Acad Sci U S A 88:2297–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten SC, Bartolomei F, Stam CJ. 2007. Small-world networks and epilepsy: graph theoretical analysis of intracerebrally recorded mesial temporal lobe seizures. Clin Neurophysiol 118:918–927 [DOI] [PubMed] [Google Scholar]
- Press WH. 1992. Numerical Recipes in FORTRAN: The Art of Scientific Computing. Cambridge, United Kingdom: Cambridge University Press [Google Scholar]
- Prichard D, Theiler J. 1994. Generating surrogate data for time series with several simultaneously measured variables. Phys Rev Lett 73:951–954 [DOI] [PubMed] [Google Scholar]
- Pritchard WS, Duke DW, Krieble KK. 1995. Dimensional analysis of resting human EEG. II: surrogate-data testing indicates nonlinearity but not low-dimensional chaos. Psychophysiology 32:486–491 [DOI] [PubMed] [Google Scholar]
- Reijneveld JC, Ponten SC, Berendse HW, Stam CJ. 2007. The application of graph theoretical analysis to complex networks in the brain. Clin Neurophysiol 118:2317–2331 [DOI] [PubMed] [Google Scholar]
- Richman JS, Moorman JR. 2000. Physiological time-series analysis using approximate entropy and sample entropy. Am J Physiol Heart Circ Physiol 278:H2039–H2049 [DOI] [PubMed] [Google Scholar]
- Rubinov M, Knock SA, Stam CJ, Micheloyannis S, Harris AW, Williams LM, Breakspear M. 2009. Small-world properties of nonlinear brain activity in schizophrenia. Hum Brain Mapp 30:403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52:1059–1069 [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. 2011. Network modelling methods for FMRI. Neuroimage 54:875–891 [DOI] [PubMed] [Google Scholar]
- Song C, Havlin S, Makse HA. 2006. Origins of fractality in the growth of complex networks. Nat Phys 2:275–281 [Google Scholar]
- Sporns O, Zwi JD. 2004. The small world of the cerebral cortex. Neuroinformatics 2:145–162 [DOI] [PubMed] [Google Scholar]
- Stam CJ. 2004. Functional connectivity patterns of human magnetoencephalographic recordings: a ‘small-world’ network? Neurosci Lett 355:25–28 [DOI] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. 2007. Small-world networks and functional connectivity in Alzheimer's disease. Cereb Cortex 17:92–99 [DOI] [PubMed] [Google Scholar]
- Stam CJ, Pijn JPM, Pritchard WS. 1998. Reliable detection of nonlinearity in experimental time series with strong periodic components. Physica D 112:361–380 [Google Scholar]
- Stam CJ, van Woerkom TC, Pritchard WS. 1996. Use of non-linear EEG measures to characterize EEG changes during mental activity. Electroencephalogr Clin Neurophysiol 99:214–224 [DOI] [PubMed] [Google Scholar]
- Telesford Q, Simpson SL, Burdette JH, Hayasaka S, laurienti PJ. 2011a. The brain as a complex system: using network science as a tool for understanding the brain. Brain Connect 1:295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford QK, Joyce KE, Hayasaka S, Burdette JH, Laurienti PJ. 2011b. The ubiquity of small-world networks. Brain Connect 1:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289 [DOI] [PubMed] [Google Scholar]
- Vakorin VA, Ross B, Krakovska O, Bardouille T, Cheyne D, McIntosh AR. 2010. Complexity analysis of source activity underlying the neuromagnetic somatosensory steady-state response. Neuroimage 51:83–90 [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Boersma M, Hulshoff Pol HE. 2008. Small-world and scale-free organization of voxel-based resting-state functional connectivity in the human brain. Neuroimage 43:528–539 [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. 1998. Collective dynamics of ‘small-world’ networks. Nature 393:440–442 [DOI] [PubMed] [Google Scholar]
- Xie HB, Guo JY, Zheng YP. 2010. A comparative study of pattern synchronization detection between neural signals using different cross-entropy measures. Biol Cybern 102:123–135 [DOI] [PubMed] [Google Scholar]
- Xie X, Cao Z, Weng X. 2008. Spatiotemporal nonlinearity in resting-state fMRI of the human brain. Neuroimage 40:1672–1685 [DOI] [PubMed] [Google Scholar]
- Xu X-K, Zhang J, Sun J, Small M. 2009. Revising the simple measures of assortativity in complex networks. Phys Rev E 80:056106. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang T, Coote JH. 2002. Synchrony analysis between blood pressure and sympathetic nerve signal inhibited by atrial receptor stimulation in Wistar rats. Exp Physiol 87:461–468 [DOI] [PubMed] [Google Scholar]
- Yu S, Huang D, Singer W, Nikolic D. 2008. A small world of neuronal synchrony. Cereb Cortex 18:2891–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Yang Z, Coote JH. 2007. Cross-sample entropy statistic as a measure of complexity and regularity of renal sympathetic nerve activity in the rat. Exp Physiol 92:659–669 [DOI] [PubMed] [Google Scholar]