Abstract
True tendon regeneration in human patients remains a vision of musculoskeletal therapies. In comparison to other mesenchymal lineages the biology of tenogenic differentiation is barely understood. Specifically, easy and efficient protocols are lacking that might enable tendon cell and tissue differentiation based on adult (stem) cell sources. In the murine mesenchymal progenitor cell line C3H10T½, overexpression of the growth factor bone morphogenetic protein 2 (BMP2) and a constitutively active transcription factor, Smad8 L+MH2, mediates tendon cell differentiation in vitro and the formation of tendon-like tissue in vivo. We hypothesized that during this differentiation secreted factors involved in extracellular matrix formation exert a major impact on tendon development. Gene expression analyses revealed four genes encoding secreted factors that are notably upregulated: periostin, C-type lectin domain family 3 (member b), RNase A4, and follistatin-like 1. These factors have not previously been implicated in tendon biology. Among these, periostin showed a specific expression in tenocytes of adult mouse Achilles tendon and in chondrocytes within the nonmineralized fibrocartilage zone of the enthesis with the calcaneus. Overexpression of periostin alone or in combination with constitutively active BMP receptor type in human mesenchymal stem cells and subsequent implantation into ectopic sites in mice demonstrated a reproducible moderate tenogenic capacity that has not been described before. Therefore, periostin may belong to the factors contributing to the development of tenogenic tissue.
Introduction
The molecular mechanisms involved in tendon and ligament formation during embryogenesis are quite well characterized both in vertebrate and invertebrate systems [1]. Much less is known about processes that contribute to de novo tendon formation and tendon repair in adult organisms. Tendon regeneration in human patients imposes major clinical problems since, so far, only mechanically inferior fibrotic scar tissue is formed. This causes significant dysfunction and disability leading to re-injury in the long term. Specifically, adult tendons lack a true regeneration capacity that, however, is present in the fetus [2]. Therefore, identification of those specific properties contributing to regeneration during ontogenesis but not previously implicated in tendon formation will be informative and may help to devise biological strategies for tendon regeneration. This may hold true especially since cells in the adult organism are either specialized/differentiated or multipotent stem-cell-like mesenchymal stem cells [also called mesenchymal stromal cells (MSCs)] that seem to lack some factors for achieving such a regeneration. Nevertheless, in contrast to cells from embryonic or fetal stages, MSCs are more readily available, for example, from bone marrow, and therefore identification of novel factors that may contribute to or even direct their differentiation into tendon cells is a therapeutically attractive approach. Among other parameters, secreted soluble proteins and extracellular matrix proteins are such promising candidates. Several growth factors, including members of the transforming growth factor-beta/bone morphogenetic protein (TGF-β/BMP) family, have been attributed a role in tendon repair in adult animals and humans and a variety of preclinical and clinical investigations have been conducted [3]. Unfortunately, none of these approaches—although promising—has reached routine clinical application. Therefore, present research aims to identify further molecular events that may contribute to de novo tendon formation in the adult organism. This research must also include the identification of novel “markers” on the transcript or protein level to distinguish tendon cells from other mesenchymal cell types.
The ordered collagen structure of tendons that accounts for its specific biomechanical properties is the principal difference to other connective tissues. Secreted matrix components contribute to the establishment of this ordered collagen structure. Consequently, these secreted factors may be involved as key regulators during exertion of the tenogenic differentiation program and arrangement of the extracellular matrix. They may also assist in the activation of endogenous regeneration processes. There are several studies that investigated the role of secreted matrix factors for tendon regeneration. A study by Chen et al. used MSCs that were derived from human embryonic stem cells for generation of tendon-like tissue [4]. They observed the formation of fetal matrix component genes (collagen III, XIV, and tenascin-c) and differentiation factors (including TGF-β3, growth and differentiation factor 5, BMP2, and fibroblast growth factor 2), which then activated the endogenous regeneration process in a rat patellar tendon repair model.
Using a cellular model system of murine mesenchymal progenitor cells C3H10T½, we had demonstrated before that combined overexpression of a growth factor, BMP2, and the constitutively active variant of a transcription factor (Smad8 L+MH2, hereafter designated Smad8ca) leads to the formation of tendon-like tissue both in vitro and in vivo or even to the formation of tendon-bone junctions in vivo [5,6]. However, the molecular processes that are triggered by BMP2 and Smad8ca and that result in the formation of tendon-like tissue have not yet been elucidated. We hypothesized that the BMP2/Smad8ca signaling system may trigger the synthesis of secreted factors that in turn support tendon formation. Therefore, we focused our attention on secreted factors that were upregulated during the BMP2/Smad8ca-dependent early tenogenic differentiation in C3H10T½ mesenchymal progenitors.
Microarray gene expression analyses identified the periostin gene (also called OSF-2: osteoblast-specific factor-2) as upregulated in the course of tenogenic C3H10T½ mesenchymal progenitor differentiation. Periostin encodes a matricellular protein first described in a mouse osteoblastic cell line as a putative cell adhesion protein for preosteoblasts and was therefore originally named osteoblast-specific factor [7]. Later, it was renamed based on its preferential expression in periodontal ligament and periosteum [8]. Periostin expression is induced by TGF-β [9] and by BMP2 [10]. It interacts with extracellular matrix molecules both intra- and extracellularly. Through interaction with collagen type I—the major component of tendon extracellular matrix—periostin regulates collagen I fibrillogenesis. Reduced collagen fibril diameters were found in skin dermis of periostin knockout mice [11]. We investigated the potential role of periostin in tendon formation and report that periostin is expressed in tenocytes of adult murine tendon within the vicinity of the enthesis (tendon-bone junction) and in chondrocytes within the nonmineralized fibrocartilage zone of this enthesis. Finally, it was shown that periostin supports the formation of tendon-like tissue in an ectopic transplantation model.
Materials and Methods
Stable C3H10T½ cell lines and HEK293T cells
The stable cell lines C3H10T½-BMP2 and C3H10T½-BMP2/Smad8ca have been described in detail [5]. Human embryonic kidney 293T (HEK293T) cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g/L glucose (Sigma) containing 10% fetal bovine serum (Invitrogen), 100 U/mL penicillin (Sigma), and 100 μg/mL streptomycin (Serva). All cell lines were maintained at 37°C and with 5% CO2.
Ethics statement
Human bone marrow aspirates used in the present study were obtained after approval of the institutional ethical committee of Medical School Hannover. Written informed consent was obtained from all donors. All personal information, including age and gender, was made anonymous. The bone marrow aspirates were harvested from left-over tissue during total hip arthroplasty from otherwise healthy persons.
MSC isolation and propagation
Human bone marrow MSCs were isolated from bone marrow aspirates. Mononuclear cells were isolated by density gradient centrifugation with Biocoll (Biochrom AG). Outgrowing colonies of plastic-adherent cells were harvested by trypsinization before reaching confluence and subcultured at a density of 2,000 to 5,000 cells/cm2 in DMEM (Biochrom)/10% (v/v) fetal bovine serum (Thermo Scientific:Hyclone)/20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Biochrom AG)/100 U/mL penicillin (Sigma)/100 μg/mL streptomycin (Serva)/2 ng/mL human recombinant FGF2 (Peprotech). For all experiments, cells were used between passages 3 and 6. MSC characteristics were confirmed by flow cytometric analysis of cell surface molecules and by in vitro differentiation as detailed by Shahab-Osterloh et al. [6].
Differentiation experiments
C3H10T½-BMP2 or C3H10T½-BMP2/Smad8ca cell lines were seeded at 5,000 cells/cm2. At confluence (arbitrarily termed day 0), medium was supplemented with 50 μg/mL ascorbic acid and 10 mM beta-glycerophosphate. Medium changes were performed every other day. At days 0, 7, 10, and 17 of differentiation, samples were processed for RNA and protein isolation and were used for the microarray analysis (cf. section “Microarrays”). For lentivirally infected MSCs the differentiation was allowed to proceed for 21 days and samples were then processed for protein and RNA analyses.
Western blotting
Cell pellets were lysed in 1% (w/v) Nonidet P-40, 150 mM NaCl, 20 mM Tris (pH 7.5), 2 mM EDTA, 50 mM NaF, and 1 mM Na4P2O7, supplemented with protease inhibitors (Protease Inhibitor Tablets; Roche Applied Science). Protein concentration was determined by the Bradford method, and equal amounts (25 μg/lane) of total protein were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subsequent western blotting. The periostin antibody (Acris, DP2019) was used at a dilution of 1:1,000. The secondary antibody goat anti-rabbit-IgG (H+L), horseradish peroxidase conjugated (Dianova), was used at a dilution of 1:10,000. For detection, ECL reagent (GE Healthcare) was used in conjunction with Hyperfilm ECL (GE Healthcare). Exposed films were scanned with the Epson scanner 1680 Pro.
Isolation of RNA
RNA was isolated from cell cultures using TriReagent® lysates according to the instructions of the manufacturer (Life Technologies). For isolation of RNA from mouse Achilles tendons and muscle, the following protocol was used. The respective tissues were prepared and pooled from nine black/six mice (male, 6-week old). During dissection, extreme care was taken to isolate pure tissues only, that is, tendon without muscle or skeletal attachment sites, respectively. The tissues were washed in ice-cold PBS, frozen in liquid nitrogen, and stored at −70°C after removal of residual fluid. The tissues were transferred to 300 μL buffer RLT containing beta-mercaptoethanol and 4 ng/μL carrier-RNA (RNeasy micro kit; Qiagen) and distributed into tubes containing Lysing Matrix A beads (MP Instruments). Subsequently, the tissue was grinded with a homogenizer (FastPrep®-24 Instrument; MP Biomedicals, 4.5 m/s, 24×2, 4×10 s). Five hundred ninety microliters of water was added per sample and protein digested with 10 μL proteinase K (20 mg/mL; Thermo Scientific) for 10 min at 55°C. After centrifugation for 3 min at 10,000 g, the supernatants were divided into two reaction vessels each. About 0.5 volumes of absolute ethanol were added and the samples were applied to RNeasy microcolumns (Qiagen). RNA was isolated according to the instructions of the manufacturer and eluted with 14 μL of diethylpyrocarbonate-treated water. The entire material (12 μL eluate) was reverse transcribed as detailed in the next section.
cDNA synthesis and reverse transcription–polymerase chain reaction
Five micrograms of total RNA from cell cultures was reverse transcribed with the cDNA synthesis kit (MMLV-RT; Life Technologies) and with oligo-dT(12–18)-primer. For tendon and muscle tissue, the entire material was used for cDNA synthesis, which was carried out after DNase treatment to remove genomic DNA according to the protocol of the supplier (Revert Aid First Strand cDNA synthesis Kit; Fermentas) using random hexamer primers. cDNA aliquots were subjected to polymerase chain reaction (PCR). Quantitative real-time PCR analysis was used to quantify the transcripts of C-type lectin domain family 3, member b (Clec3b; previously called tetranectin) [RefSeq: NM_011606.2] (Mm00495657_m1), collagen 1a1 [RefSeq: NM_007742.3] (Mm00801666_g1), follistatin-like 1 (Fstl1) [RefSeq: NM_008047.5] (Mm00433371_m1), periostin [RefSeq: NM_001198765.1; NM_001198766.1; NM_015784.3] (Mm00450111_m1), RNase A4 [RefSeq: NM_021472.4] (Mm00491347_m1), scleraxis [RefSeq: NM_198885.3] (Mm01205675_m1), sine oculis-related homeobox 2 (Six2) [RefSeq: NM_011380.2] (Mm03808565_m1), and tenomodulin [RefSeq: NM_022322.2]. Hypoxanthine phosphoribosyl transferase (HPRT) [RefSeq: NM_013556.2] (Mm01545399_m1) was used as housekeeping gene. Control reactions were performed with water. The PCR was accomplished in a duplex run with TaqMan Fast Advanced Master Mix using the StepOnePlus Real-Time PCR System (Life Technologies) with the following conditions: 20 s at 95°C and 40 cycles of 1 s at 95°C and 20 s at 60°C. Differences between samples and controls were calculated based on 2−ΔΔCt method. Duplicate experiments were performed for all genes investigated and the standard deviation is indicated.
For the human MSC samples, semiquantitative reverse-transcription PCR (RT-PCR) was performed using GoTaq DNA polymerase according to the instructions of the manufacturer (Promega). The thermocycler conditions consisted of 90 s at 94°C followed by 28–35 cycles of 20 s at 94°C, 20 s at the respective annealing temperature, 30 s at 72°C, and 15 min at 72°C for the final extension. All reactions were performed in the linear range by appropriate choice of cycling conditions. The transcriptional levels of HPRT were used for normalization. The PCR primers and the annealing temperature were as follows: HPRT: 5′-GGTCAGGCAGTATAATCCAAAGA, 5′-AGGCTCATAGTGCAAATAAACAGT (404 bp/52.5°C); collagen 1a1: 5′-ATCCAGCTGACCTTCCTGCG, 5′-TCGAAGCCGAATTCCTGGTCT (323 bp/57.3°C); collagen 3a1: 5′-GTGGACAGATTCTAGTGCTGAG, 5′-ATAGGTAGTCTCACAGCCTTGC (378 bp/56.5°C); biglycan: 5′-GGACTCTGTCACACCCACCT, 5′-AGCTCGGAGATGTCGTTGTT (159 bp/56.5°C), decorin: 5′-GGACCGTTTCAACAGAGAGG, 5′-TCAGAACACTGGACCACTCG (158 bp/56.5°C), lumican: 5′-CTTCAATCAGATAGCCAGACTGC, 5′-AGCCAGTTCGTTGTGAGATAAAC (150 bp/55.8°C), scleraxis: 5′-CCACTCCGGGCCCGCCTTCTTCC, 5′-TCTTTCTGTCGCGGTCCTTGCTCA (161 bp/63.6°C); tenomodulin: 5′-CCATGCTGGATGAGAGAGGT, 5′-CTCGTCCTCCTTGGTAGCAG (123 bp/57.3°C).
Microarrays
cDNA microarrays [12] were generated using ∼20,000 murine cDNA clones (arrayTAG clone collection) from LION Bioscience. In this array, several genes are represented by more than one probe. For example, among the well-annotated differentially expressed probes (DEPs), 2 genes are represented by 5 or 4 DEPs, another 20 genes by 3 DEP each, and 209 genes by 2 DEPs. Labeling and hybridization reactions were performed using the 3DNA Array 50 Expression Array Detection kit (Genisphere) according to the manufacturer's instructions. RNA samples from two experimental series were investigated: tenogenic C3H10T½-BMP2/Smad8ca cells at four time points of differentiation—cellular confluence (day 0), day 7, 10, and 17 after confluence—and RNA samples from nontenogenic C3H10T½-BMP2 at the same four time points of differentiation. For each series, three cohybridizations were performed: control sample (day 0) versus test sample 1 (day 7), test sample 2 (day 10), and test sample 3 (day 17). Briefly, 20 μg of total RNA of control and test sample pairs was reverse transcribed using primers with a specific capture sequence. The two cDNA samples were pooled and hybridized to the microarrays in a humidified chamber at 42°C for 16 h. Dye-swap repeats were performed for each of the three pairs. Thus, six arrays were used per series. Slides were washed at room temperature in 2×SSC/0.2% (w/v) sodium dodecyl sulfate for 10 min, 2×SSC for 10 min, and 0.2×SSC for 10 min. Bound cDNA was visualized by hybridization with 3DNA (dendrimers specific for respective capture sequences conjugated to either Cy3 or Cy5) in a humidified chamber at 42°C for 3 h. Subsequently, slides were washed as before. Image acquisition and data analysis was done according to Gurok et al. [12]. From the hybridization data obtained we established a list of DEPs. Differential gene expression is presented as an estimation of a natural logarithmic ratio at each time point relative to the control at day 0. Therefore a log2 ratio of 0.6931 is considered a fold change of 2. A log2 ratio>0.6931 was considered upregulated (“up”) and a log2 ratio<−0.6931 was considered downregulated (“down”) as compared with the controls (day 0) in at least one of the three differentiation time points (day 7, 10, or 17).
Nonradioactive RNA Northern blot analysis
Five micrograms of RNA was applied per lane. About 300 ng of digoxigenin-labeled RNA probe was applied to RNA blots and processed according to the manufacturer's advice (Roche Diagnostics).
Lentivirus generation
Murine periostin with a C-terminal HA-Tag was cloned from mRNA isolated from C3H10T½-BMP2/Smad8ca cells (day 7 of differentiation) into the lentiviral expression vector pLOX/TwGFP. The integrity of the construct was confirmed by sequencing with the ABI Prism 310 capillary sequencer (BigDye Terminator Cycle Sequencing Ready Reaction Mix v.1.1; Applied Biosystems) and matches periostin variant 1 (NM_015784). Constitutively active human BMP receptor IA (hereafter called caALK3) obtained from Peter ten Dijke (Leiden University Medical Center, Leiden, Netherlands) was integrated into pLOX. For virus production, HEK293T cells were transfected with either pUCL-MIK (tetracycline-dependent transactivator), pLOX-TwGFP, periostin-pLOX, BMP2- pLOX or ca. alk3-pLOX and pMD.G VSV-G (encoding the vesicular stomatitis virus envelope glycoprotein) plus pCMVΔR8.2 (packaging plasmid) by using the calcium phosphate technique. Forty-eight and 72 h after transfection, supernatants were harvested and frozen in aliquots at −70°C for later use. For in vitro analyses, supernatants were chromatographically purified and concentrated with Lenti-X™ Maxi Purification Kit (TaKaRa) according to the instructions of the manufacturer before freezing. Virus titrations were performed with the Lenti-X qRT-PCR Titration Kit (TaKaRa).
Lentiviral infection of MSCs
For infection, MSCs were seeded at 5,000 cells/cm2 in T25 flasks. For ectopic implantations, viral supernatants were used at a multiplicity of infection of 100. For in vitro differentiation kinetics, purified virus was used at a multiplicity of infection of 100. Twenty-four hours after infection, doxycycline was added at a final concentration of 1 μg/mL in standard culture medium to induce gene expression.
Ectopic implantations of MSCs
Ectopic implantations were performed essentially as described by Shahab-Osterloh et al. [6]. Briefly, 5 days after infection, cell-collagen implants seeded with 3×105 modified human MSCs were mounted on individual type I collagen sponges (3×3×3 mm3, No. ID-2205; Duragen, Integra Life Sciences) and transplanted into the thigh muscle or subcutaneously into female nude (nu/nu) mice (6-week old). Three to 5 days before implantation, mice obtained doxycycline in their drinking water (0.2 mg/mL). Four weeks after implantation, mice were sacrificed. Explanted transplants were fixed in 4% formalin solution overnight and were immediately dehydrated and paraffin embedded. These experiments were approved by the local administration of Lower-Saxony (No. 33.11.42502-14-051/07).
Histology
Explanted tissues with engrafted stem cells were immersion fixed in 4% neutral buffered formalin for 24 h, embedded in paraffin, cut at 5 μm using a motorized microtome (Leica Microsystems), and stained with hematoxylin and eosin (HE) following routine procedures. Images were acquired using a Keyence VHX-2000 digital microscope with accompanying software with high dynamic range image option and processed with Adobe Photoshop CE. No specific feature within an image was manipulated.
In situ hybridization
DIG (digoxigenin-11-UTP)–labeled cRNA probes of defined length (antisense and sense) were generated by in vitro transcription of linearized plasmids according to the manufacturer's instructions (Roche Diagnostics) and as detailed by Shahab-Osterloh et al. [6]. Counterstaining was performed with methyl green.
Immunohistology
Immunohistochemical analyses were performed with deparaffinized tissue sections. For periostin staining, proteinase K digestion (Dako Cytomation No. S3004, 1:50) was performed for 30 min at 37°C. Slides were rinsed with Tris-buffered saline (TBS), and endogenous peroxidases were quenched with 3% H2O2 in TBS for 30 min. Slides were washed again and blocked with 10% normal goat serum in TBS for 30 min. The periostin antibody (Abcam; ab14041) was diluted 1:300 and applied overnight. After washing, En Vision goat anti-rabbit secondary antibodies conjugated with peroxidase (Dako No. K4002) were applied for 30 min in the dark. For color development, 3,3′-diaminobenzidine was used. For immunofluorescence analysis, antigenic epitopes were demasked by boiling in citrate buffer. Parallel sections were then blocked with normal goat serum followed by incubation with the primary antibody (rabbit anti-human collagen type III also reacting with mouse collagen III; Abcam) for 30 min. After washing, the slides were incubated with donkey anti-rabbit secondary antibodies conjugated to Cy3 (Dianova) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Green fluorescent protein (GFP) fluorescence was visualized directly. Negative controls were performed without primary antibody.
Results
Microarray analyses identify secreted factors during the onset of tenogenic differentiation in C3H10T½ cells expressing BMP2/Smad8ca
Murine mesenchymal progenitor cells C3H10T½ overexpressing BMP2 and the constitutively active variant of the transcription/signaling factor Smad8 (Smad8 L+MH2, now designated Smad8ca) form tendon-like structures without bony tissue in vitro and in vivo [5]. Following the hypothesis that secreted factors should contribute to the establishment of the ordered structure of tendons, this particular cell line was subjected to microarray analyses to pinpoint secreted factors during the onset of the tenogenic developmental sequence.
Tenogenic C3H10T½-BMP2/Smad8ca cells were compared with C3H10T½-BMP2 cells that do not possess a tenogenic capacity but which efficiently differentiate along the osteogenic, chondrogenic, and adipogenic lineages [13]. Four time points of differentiation were examined [cellular confluence (day 0), day 7, 10, and 17]. To identify genes with a role in the BMP2/Smad8ca-mediated tendon formation, gene expression changes during in vitro differentiation were analyzed, using a 20,000 clone mouse cDNA microarray. From the hybridization data obtained, we established a list of candidate array probes that either showed higher or lower transcript levels as compared with the controls (day 0) in at least one of the three differentiation time points (day 7, 10, or 17) for each of the two data sets (C3H10T½-BMP2 and C3H10T½-BMP2/Smad8ca). Probes that showed an upregulation in one time point and downregulation at another time point of either series are termed “ambiguous.” According to these criteria, 2,966 DEPs were identified in the C3H10T½-BMP2 series and 1,936 DEPs were identified in the C3H10T½-BMP2/Smad8ca series.
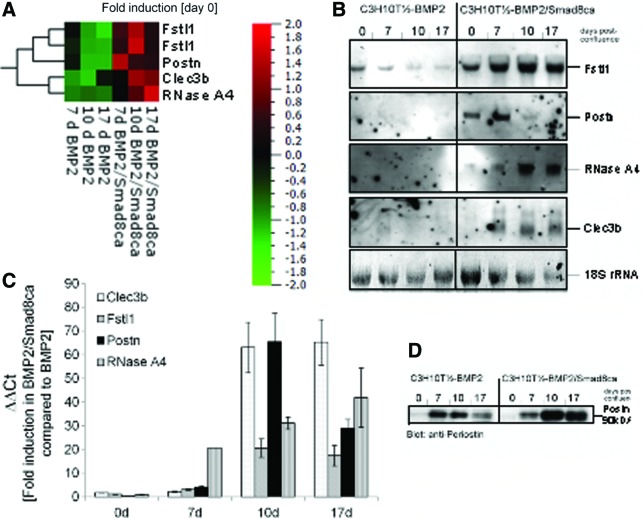
These DEPs were evaluated according to the following criteria: (1) being upregulated during the tenogenic developmental sequence (C3H10T½-BMP2/Smad8ca differentiation), (2) being downregulated during the osteogenic or chondrogenic differentiation (C3H10T½-BMP2), and (3) encoding genes for secreted factors. Four factors were selected for further investigations: Fstl1, Clec3b, RNase A4, and periostin (Fig. 1A). Figure 1A shows that in tenogenic BMP2/Smad8ca cells, the expression of these four factors was considerably higher than in the C3H10T½-BMP2 cells used as a control. Periostin exhibited induction levels up to 28.5-fold at day 7 post-confluence (induction levels estimated by microarray), that is, at an early time during tenogenic development. Fstl1 expression (two probes) remained moderately high at all time points investigated but was maximal at day 10. Gene expression levels of Clec3b and RNase A4 were highest at days 10 and 17 with RNase A4 obtaining a maximum at the late time point. Within the C3H10T½-BMP2 data set, all four genes were downregulated during two or even three (RNase A4) time points (Fig. 1A).
FIG. 1.
Identification of secreted factors during tenogenic differentiation in C3H10T½-BMP2/Smad8ca in comparison with nontenogenic C3H10T½-BMP2 cells in a 20,000 clone mouse cDNA microarray. (A) Heat map of microarray data according to the results of the microarray analyses. Color-coded expression levels of differentially expressed genes in BMP2- or BMP2/Smad8 cell lines are shown. Normalization (day 0 of each cell line) was applied to accommodate for cell-line-specific effects. Qlucore Omics Explorer 2.3 Software was used to generate a heat map. High-expressed genes are depicted in red and low-expressed genes in green. For the Fstl1 gene, two different probes were present on the arrays. The results obtained with both probes are comparable. Periostin (Postn): ID 15044; Clec3b: ID 17142; RNase A4: ID 2987; and Fstl1: ID 807, ID 10435. (B) Northern blot analyses of genes shown in (A). RNA from in-vitro-cultivated C3H10T½ cells was blotted and subjected to nonradioactive hybridization as described in “Materials and Methods” section. Staining for 18S-rRNA was used as loading control. (C) The quantitative real-time PCR data from both cell lines at the four time points are presented as ΔΔCt using the BMP2 cell line as reference. Supplementary Figure S1 shows the corresponding ΔCt values (using the housekeeping gene HPRT as a reference). (D) Periostin expression in cultivated C3H10T½ cells as assessed by western blot analyses. Proteins were isolated as described in “Materials and Methods” section, subjected to sodium dodecyl sulfate gel electrophoresis, and blotted. Periostin was detected with an anti-periostin antibody (Acris, Herford, Germany). Periostin shows a higher expression rate in cells undergoing tenogenic differentiation than in nontenogenic mesenchymal progenitor cells C3H10T½-BMP2. BMP, bone morphogenetic protein; Clec3b, C-type lectin domain family 3, member b (previously called tetranectin); Fstl1, follistatin-like 1; HPRT, hypoxanthine phosphoribosyl transferase; PCR, polymerase chain reaction. Color images available online at www.liebertpub.com/scd
mRNA, quantitative RT-PCR, and protein level analyses confirm upregulation of Fstl1, Clec3b, RNase A4, and periostin in tenogenic mesenchymal progenitors
Northern blots were performed in order to verify the microarray data for the four selected genes (Fig. 1B). By and large, the Northern blots confirmed the expression data obtained by microarray analyses. In BMP2/Smad8ca-expressing C3H10T½ cells, the four selected genes were considerably upregulated. Specifically, no hybridization of the probes for periostin, RNase A4, Clec3b, or just a faint signal for Fstl1 could be detected in BMP2-expressing C3H10T½ cells. To more quantitatively evaluate the induction levels of the genes, quantitative RT-PCR analyses were performed (Fig. 1C and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). Figure 1C compares the results obtained for the four genes in the BMP2/Smad8ca cell line using the BMP2 cell line as a reference (ΔΔCt). Periostin is the gene with the strongest induction (about 66-fold at day 10). Although the temporal pattern predicted by the arrays is not exactly mirrored (quantitative RT-PCR exhibits maximum induction at day 10 whereas microarrays indicated the maximum induction for day 7), the clear induction of the gene in the course of tenogenic differentiation is thereby confirmed. Clec3b exhibits similarly high induction levels especially at days 10 and 17 but its overall expression is low in both cell lines (Supplementary Fig. S1 shows the ΔCt values using the housekeeping gene HPRT as a reference for both cell lines and all four time points.). Fstl1 obtains a maximum at days 10 and 17 of cultivation. RNase A4 gene expression increases from day 0 toward day 17, which is in absolute concordance with the array data.
In the case of periostin, it was confirmed with western blot analyses (Fig. 1D) that periostin is an upregulated protein in BMP2/Smad8ca-expressing C3H10T½ cells with a maximum expression at day 10. A considerably lower expression of periostin is present in the BMP2 cell line declining from day 7 to 17. In conclusion, the protein data support the gene expression analysis that periostin is notably expressed in the tenogenic model cell line.
Gene expression analyses confirm expression of Fstl1, Clec3b, RNase A4, and periostin in adult mouse tendon
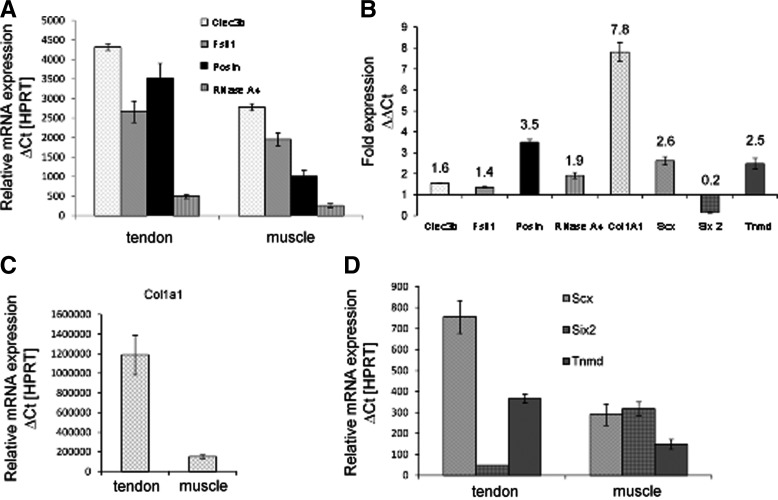
Since the four factors identified by the present study have not been linked to tendon formation, their presence in adult tendon has not been tested previously. We therefore performed mRNA expression analyses in adult mouse tendon in comparison to adult mouse muscle in order to confirm that these factors may have the potential to affect tenogenesis. Quantitative RT-PCR experiments with RNA isolated from pooled Achilles tendon or muscle from adult mice (6 weeks of age) showed that all secreted factors described previously are expressed in tendon (Fig. 2A). Among the four factors selected, periostin showed the highest relative expression in tendon tissue compared to muscle (about 3.5-fold, Fig. 2B) and is even higher than the expression level of tendon-specific genes, such as scleraxis and tenomodulin (see below). This specificity could be verified using patella tendon tissue (not shown). The relative expression levels of Clec3b, Fstl1, and RNase A4 were also higher in tendon than in muscle but these effects were less prominent. Since all four genes were expressed in both tissues, with a higher level in tendon, they may play a role in the formation of the extracellular matrix of connective tissue.
FIG. 2.
Expression rates of secreted factors upregulated in adult murine tendon and muscle tissue. RNA was isolated from murine thigh muscle and Achilles tendon tissue (mouse age: 6 weeks) as described in “Materials and Methods” section. The primers are indicated in “Materials and Methods” section. (A) Relative mRNA expression (ΔCt values normalized to HPRT) for Clec3b (tetranectin), follistatin-like 1 (Fstl1), periostin (Postn), and RNase A4. (B) Fold expression of the same four genes as well as of collagen 1a1 (Col1a1), scleraxis (Scx), sine oculis-related homeobox 2 (Six2), and tenomodulin (Tnmd) in tendon compared to muscle (ΔΔCt). (C) Relative mRNA expression (ΔCt values normalized to HPRT) for Col1a1, and (D) for Scx, Six2, and Tnmd. Apart from collagen 1a1, periostin is the gene with the most prominent expression in tendon compared to muscle.
We additionally analyzed the expression of genes that have been described in the literature as “specific” or “characteristic” for tendon tissue, in order to obtain a more complete picture of gene expression in murine tendon and muscle: collagen 1a1, scleraxis, Six2, and tenomodulin. The expression of collagen 1a1 is about 7.8-fold higher in tendon than in muscle and thereby demonstrates the largest difference in relative expression values between both tissues (Fig. 2B, C). Higher amounts of scleraxis (2.6-fold) and tenomodulin (2.5-fold) expression were seen in Achilles tendon, indeed (Fig. 2B, D). In contrast, Six2 expression was predominantly demonstrated in muscle (Fig. 2B, D). In conclusion, although these genes have been designated as being “tendon specific,” they are also expressed in muscle tissue—some of them like Clec3b at a considerable level (Fig. 2A). Therefore, the relatively high abundance of periostin in tendon tissue is particularly striking. Consequently, our further studies were focused on periostin.
Periostin protein is expressed in adult murine tendons and in the tendon-bone junction
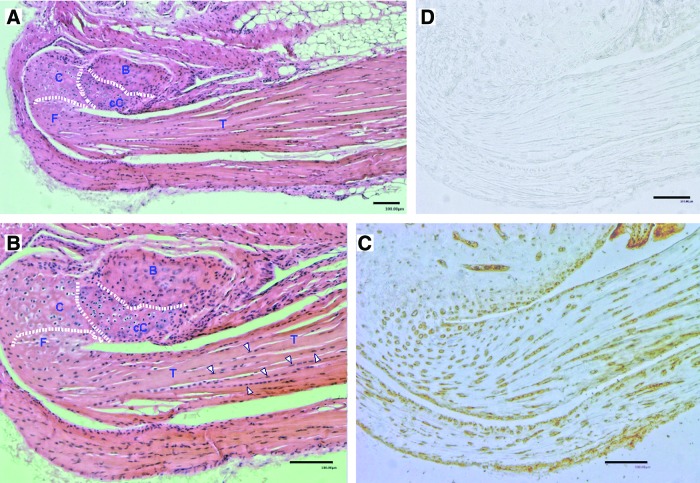
We were interested to know the cellular distribution of periostin in adult murine tendons and in their attachment site to bone, the osteo-tendinous junction or enthesis. We again used Achilles tendon as a study material including its attachment to the calcaneus. Tissue was prepared, decalcified, and paraffin embedded. Figure 3 shows histological and immunohistological results of serial sections. Figure 3A is an overview of the Achilles tendon and its insertion into the bone stained with HE. Figure 3B presents a detailed view of the insertion of the tendon (T) into the calcaneus. Since this is an indirect or fibrocartilaginous enthesis, two zones of fibrocartilage can be clearly distinguished. The tidemark separating nonmineralized cartilage (C) from mineralized cartilage (cC) is indicated in Fig. 3B by a dotted line. The bony portion (B) that is characterized by more intense eosin staining is also indicated. When compared with the control (Fig. 3D), immunohistological staining (Fig. 3C) shows that a distinct periostin protein expression was present in the fibrocartilage portion of the enthesis, in some nonmineralized chondrocytes, but not in the mineralized chondrocyte region and in bony tissue. Tenocytes within the Achilles tendon also stained positive. Similar data were obtained with a different periostin antibody (Acris).
FIG. 3.
Histology and periostin immunohistology of mouse Achilles tendon and its insertion into the calcaneus. The entire anatomical structure was removed from the animal, fixed with 4% paraformaldehyde overnight at 4°C, and subsequently decalcified with 16.8% EDTA for 7 days and then stored in 70% ethanol. After paraffin embedding, 5-μm sections were cut and parallel sections were stained. (A) HE stain, overview. An enlarged view is shown in (B). (B) Detailed view of tendon approaching the calcaneus. Dotted lines delineate bone from mineralized, calcified cartilage (cC) and from nonmineralized cartilage (C). Some representative tenocytes are indicated by white arrowheads. (C) Periostin staining. Positively stained, round cartilage cells and tenocytes are clearly visible. (D) Negative control without primary antibody. B, bone; cC, calcified cartilage; C, nonmineralized (fibro)cartilage; F, fibrocartilage; T, tendon. HE, hematoxylin and eosin. Color images available online at www.liebertpub.com/scd
Overexpression of periostin in MSCs does not result in differentiation of tenocyte-like cells in vitro
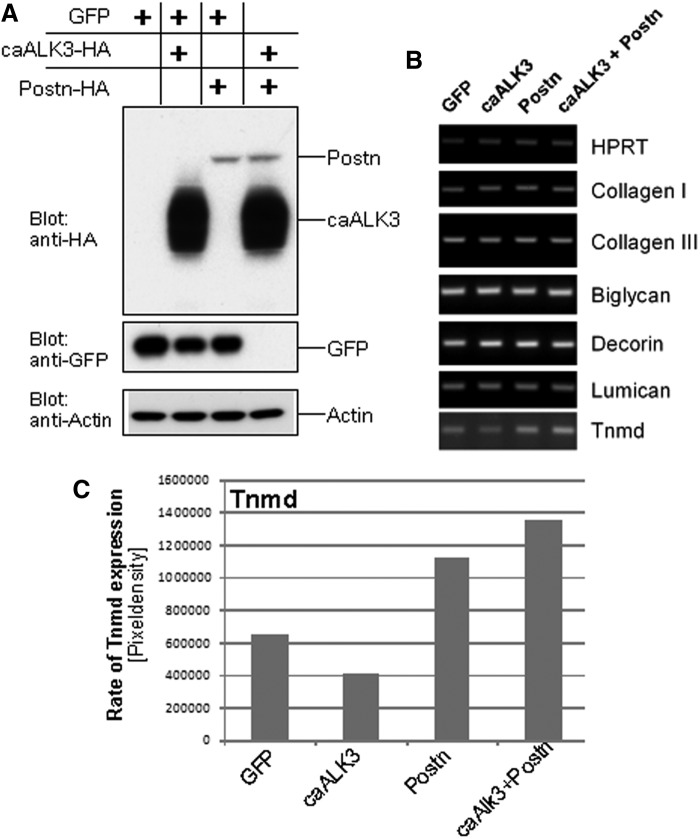
To analyze the presumptive role of periostin in tendon formation by mesenchymal progenitors, the full-length periostin cDNA was incorporated in a tetracycline-inducible lentiviral vector. Human MSCs were modified with infectious lentiviral particles expressing GFP, periostin, and caALK3. GFP-expressing cells were used as a negative control. Western blotting demonstrated the successful overexpression of caALK3 and periostin (Fig. 4A). After reaching confluence, the cells were treated with a medium previously described as suitable for in vitro tenogenic differentiation, that is, containing 50 μg/mL ascorbic acid and 10 mM beta-glycerophosphate, for 3 weeks (cf. “Materials and Methods” section and ref. [5]). Subsequent gene expression analysis (Fig. 4B) for matrix proteins (collagens type I and III) and proteoglycans (biglycan, decorin, and lumican) showed no distinct changes. In particular, the gene expression of the transcription factor scleraxis was negative. However, the type II transmembrane protein tenomodulin was upregulated 1.7-fold in periostin-overexpressing and 2-fold in periostin plus caALK3-overexpressing MSCs indicative for the onset of a tenogenic differentiation program (Fig. 4B, C). In contrast, expression of caALK3 alone was unable to support enhanced tenomodulin expression and even led to a 1.5-fold reduction of tenomodulin mRNA synthesis in MSCs (Fig. 4B, C). A notable upregulation of tenomodulin was also seen in C3H10T½ cells lentivirally infected with periostin as compared with GFP-expressing control cells (data not shown).
FIG. 4.
Overexpression of caALK3, periostin, or caALK3 plus periostin in vitro induces tenomodulin transcription but does not result in formation of tenocyte-like cells. Human MSCs were lentivirally modified to overexpress GFP (control), caALK3, periostin, or caALK3 plus periostin. The cells were cultured under conditions favoring tenogenic differentiation (cf. “Materials and Methods” section) for 21 days after obtaining confluence. (A) Western blot analysis demonstrates expression of caALK3 and periostin with an HA antibody since both constructs are C-terminally HA tagged. Analysis was performed at day 0, that is, upon reaching of confluence and start of tenogenic differentiation. (B) Semiquantitative RT-PCR was performed to analyze expression of genes relevant for tenocytes and fibroblasts. Matrix proteins: collagen type I and collagen type III. Proteoglycans: biglycan, decorin, and lumican. Transmembrane protein: tenomodulin (Tnmd). No amplificates were obtained for the transcription factor scleraxis. (C) Rate of Tnmd upregulation by periostin. The periostin-dependent upregulation of Tnmd transcription was assessed by determining the pixel density of the RT-PCR bands of (B). Evaluation was performed with the ImageJ 1.41o software (NIH). caALK3, constitutively active human BMP receptor IA; GFP, green fluorescent protein; MSCs, mesenchymal stromal cells; RT-PCR, reverse transcription–polymerase chain reaction.
Overexpression of periostin in MSCs supports ectopic tendon formation in vivo
Human MSCs were lentivirally modified to overexpress periostin, caALK3, or periostin plus caALK3. caALK3 was used to simulate BMP2-dependent matrix production without the secretion of biologically active BMP2 into the surrounding tissue. This should avoid the BMP-induced immigration and differentiation of host stem cells and restrict BMP-dependent matrix production and cartilage/bone formation to genetically modified stem cells only. Moreover, the caALK3-mediated BMP-like activity should also enhance a putative tendon-like differentiation potential since, due to the use of this constitutively active receptor variant, BMP2-dependent signaling pathways would become activated. This activity has been found necessary in our original cellular model involving active BMP2 and Smad8ca [5].
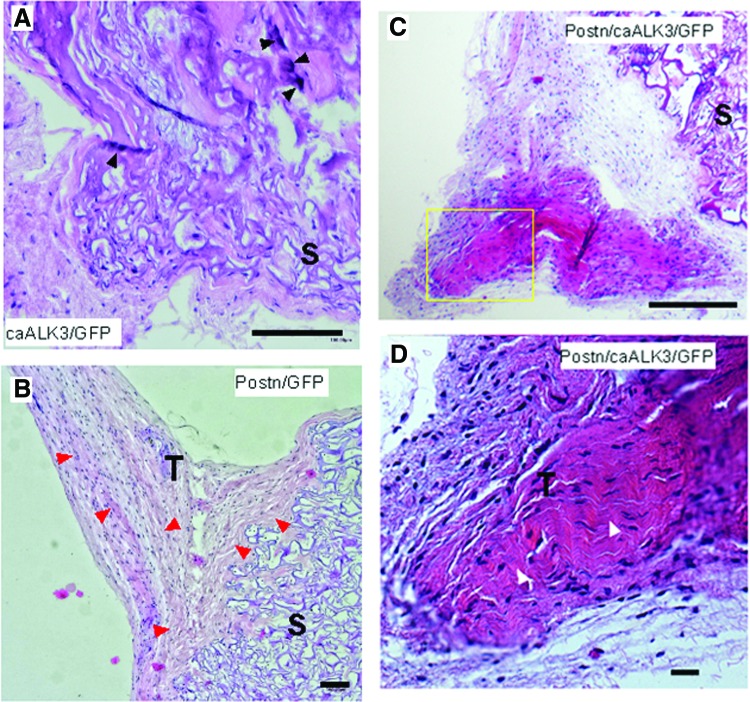
Any particular combination of cellular modifications was implanted ectopically into five nude mice, both intramuscularly and subcutaneously. Tissue formation was analyzed after 4 weeks. We did not observe notable differences in tissue formation in subcutaneous versus intramuscular sites. Therefore, this aspect is not further distinguished here. The lentiviral infection of MSCs with caALK3 alone resulted in the formation of tissue with bony elements as evidenced by intense eosin staining (Fig. 5A). Overexpression of periostin alone resulted in formation of fibroblastic tendon-like tissue (Fig. 5B) with the characteristic wavy crimp pattern. In the combined presence of periostin and caALK3, the explants demonstrated even more pronounced areas with tendon-like tissue; one representative implant (Fig. 5C) and an enhanced view (Fig. 5D) are shown. The production of ordered wavy fibroblastic tissue with elongated cells can be observed indicative for ongoing development of tendon-like structures. These results show that periostin is able to organize fibroblastic matrix into a tendon-like tissue.
FIG. 5.
Periostin-dependent development of ordered collagen structures in human MSCs subjected to ectopic transplantations. Human MSCs were modified with lentiviruses encoding periostin, GFP, and caALK3 as described in “Materials and Methods” section. Modified cells were applied on a Duragen sponge and ectopically subcutaneously or intramuscularly implanted in female nude mice (6 weeks of age). After 4 weeks of implantation, mice were sacrificed and implants were explanted and subjected to histological analyses (HE staining). One representative implant of each group is shown in (A–D). (A) Implantation of human MSCs modified by lentiviral expression of caALK3 and GFP. The formation of bony elements may be assessed by the regions of intense HE staining (black arrowheads). (B) Human MSCs overexpressing periostin and GFP. The expression of periostin leads to the formation of ordered tendon-like tissue in the neighborhood of the implanted sponge and is indicated by the red arrowheads. (C) Overview of an implant harboring periostin- and caALK3-modified MSCs and surrounding tissue. The implantation of caALK3/periostin-modified MSCs favors the development of tendon-like tissue. (D) Detailed view of the yellow rectangle from (C). This magnification documents the ordered tendon-like organization of fibroblastic tissue and is pinpointed by white arrowheads. (A–D): scale bar, 100 μm. S, sponge; T, tendon-like tissue organization. Color images available online at www.liebertpub.com/scd
Modified MSCs are detected in the sponge matrix
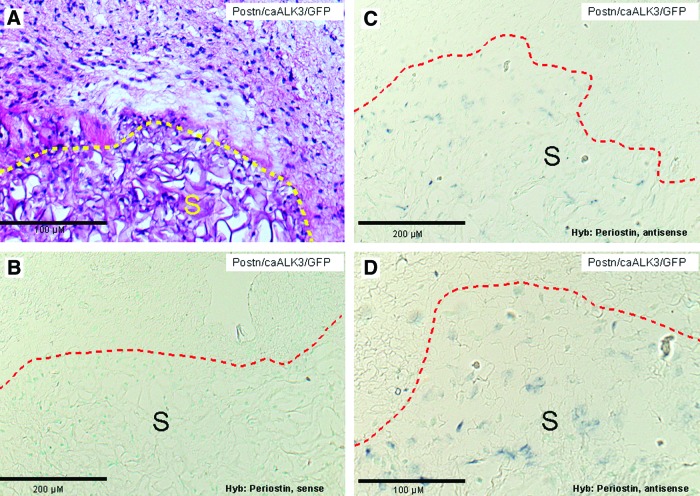
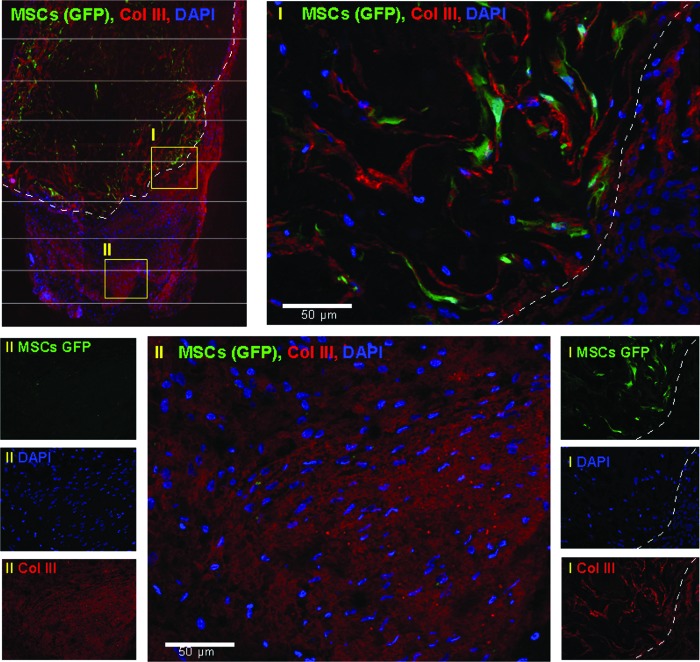
To investigate whether or not periostin/caALK3-modified MSCs are developing along the tenogenic lineage, we performed nonradioactive mRNA in situ hybridizations and fluorescence microscopy for GFP in tissue sections. Periostin-expressing MSCs could be detected with antisense probes in the carrier sponge but not in the fibroblastic cells surrounding the implanted sponge (Fig. 6), indicating that the implanted cells were retained in the carrier. In support of this finding, modified MSCs coexpressing GFP could be detected by the green GFP fluorescence in immunohistochemical stains in the sponge (Fig. 7, region I). We next investigated whether in the tendon-like tissue—documented to be present in Fig. 5C and D—collagen type III can be detected in parallel slides. The latter collagen type is synthesized during early tenogenic differentiation. Indeed, the tendon-like tissue contains collagen type III-rich structures (Fig. 7, region II; region I is positively stained due to the use of a collagen sponge) which is a further substantiation of the periostin-dependent formation of the tendon-like tissue. In conclusion, periostin secreted by the implanted cells seems to be able to mediate formation of organized tissue-like structures in the immediate environment of the implant harboring the genetically modified MSCs (region II) but not in the MSCs themselves.
FIG. 6.
Nonradioactive in situ hybridizations with periostin-specific sense and antisense probes detect periostin-expressing MSCs predominantly in the implanted collagenous sponge. (A) Implant of lentivirally modified human MSCs expressing periostin, caALK3, and GFP. The border of the implant is indicated by the yellow dashed line. HE staining. (B–D) In situ hybridization with nonradioactively labeled periostin probes. Labeling and hybridization was as described in “Materials and Methods” section. The border of the implanted sponge is indicated with a red dashed line. Counter staining was performed with methyl green. (B) Control hybridization with a periostin-specific sense probe does not result in a hybridization signal. (C, D) Hybridization with a periostin-specific sense probe. Periostin-expressing MSCs are detected in the sponge only. S, sponge. Color images available online at www.liebertpub.com/scd
FIG. 7.
Modified MSCs are located in the implanted sponge and the tendon-like tissue surrounding the implant expresses collagen III, an early marker of fibroblastic tendon-like tissue formation. Immunofluorescence analysis was performed as described in “Materials and Methods” section. Red fluorescence: collagen III. Green fluorescence: GFP. Blue fluorescence: 4′,6-diamidino-2-phenylindole (DAPI). Left upper panel: overview of implant and its surrounding tissue. The border of the implant is indicated with a dashed white line. Two yellow rectangles (I, II) indicate regions shown in more detail. Right lower panels: individual immunofluorescence analyses show GFP and collagen III expression and DAPI staining of the implant and its immediate neighborhood (region I). GFP-expressing cells are only found in the implant. Upper right panel: merging of the individual fluorescence analyses (region I) indicates that MSCs remain in the sponge and are not directly involved in tendon differentiation. Left lower panels: individual immunofluorescence analyses do not show GFP-expressing MSCs and indicate collagen III expression and DAPI staining in a tendon-like tissue (region II). Lower middle panel: merging of the individual fluorescence analyses (region II). Tenogenic collagen III is detected in tendon-like tissue. The HE staining of this region is seen in Fig. 5C. Color images available online at www.liebertpub.com/scd
Discussion
Fstl1, Clec3b, RNase A4, and periostin are novel factors identified in adult mouse tendon and the tendon-bone junction
The present study identifies four secreted factors whose gene expression is upregulated in the tenogenic model cell line C3H10T½-BMP2/Smad8ca. These factors have not been linked to tendon formation and their abundance in adult tendon has therefore not been tested previously. Fstl1 encodes a secreted glycoprotein that binds members of the TGF-β superfamily of growth/differentiation factors and is able to interfere with BMP2-induced transcriptional responses in a dose-dependent manner [14]. Its expression might be one of the reasons for the low osteogenic properties of tenogenic C3H10T½-BMP2/Smad8ca cells [5] and for tenogenic tissue in general [15]. Clec3b is a homotrimeric protein that has been suggested to play a role in tissue remodeling, due to its ability to stimulate plasminogen activation and it is expressed in developing tissues, such as bone and muscle. A skeletal phenotype has also been reported for Clec3b-knockout mice [16]. The secreted RNase A4 is located at the same genetic locus as angiogenin, and both genes share the same promoters. Both of these factors are able to induce neurogenesis and neovascularization [17]. All these properties might exert a general impact on tissue formation and homeostasis. Periostin is able to interact with extracellular matrix molecules, in particular, with collagen type I—the major component of tendon extracellular matrix—and to regulate collagen fibrillogenesis and the biomechanical properties of connective tissues [11]. We here confirm that these four factors are indeed expressed in adult mouse tendon and that their expression is higher in tendon as compared to muscle. This is interesting in particular for periostin that exhibited a 3.5-fold higher expression in tendon than in muscle whereas three other genes—scleraxis, Six2, and tenomodulin that are often used to experimentally characterize tendon tissue and tendon cells—are only induced maximally 2.6-fold (Fig. 2B). Only the abundantly expressed collagen 1a1 gene is present at yet higher levels in tendon than in muscle compared with periostin. In conclusion these genes might be attractive candidates for further studies in the tendon field.
The fact that all “tendon-specific” genes are also demonstrated in muscle tissue in our study can be easily explained by the fact that it is well-known that muscle tissue does not only consist of myogenic cells. For example, a considerable proportion of muscle tissue contains intramuscular connective tissue [18,19]. For example, the collagen I gene is expressed by muscle connective tissue cells and not by myogenic cells. Evidence for Six2 as a “tendon marker” comes from several studies using limb tendon embryonic development [20,21]. These and other results that indicate expression of putative tendon marker genes in muscle tissue may either be due to a change of the expression mode of embryonic versus adult tissue or an efficient expression mode of these genes in intramuscular connective tissue.
To our knowledge, periostin expression in a fibrocartilaginous (direct) enthesis and in the nonmineralized chondrocytes of this enthesis has not been described before. Another study reported expression of the periostin-like factor, a splice variant of periostin, in tenocytes of trained but not of untrained tendon tissue [22]. In addition, one study observed periostin expression in a subset of hypertrophic chondrocytes of the growth plate [23].
Tendon-inducing capacity of periostin
Although clearly present, the tendon-formation promoting effect of periostin was not as pronounced and efficient as in the original BMP2/Smad8ca system. This indicates that overexpression of periostin alone and its extracellular secretion even in conjunction with caALK3 is less efficient in promoting a tendon-like phenotype. Also, the separate overexpression of the other three candidate factors in C3H10T½ cells and in human MSCs only partially enhanced the expression of single “tenogenic marker genes” (data not shown). The original BMP2/Smad8ca system more efficiently triggers the necessary molecular events probably due to secretion of a variety of additional extracellular-matrix-forming factors as also identified in the present study. In this context, periostin is upregulated during tendon-like tissue formation and may assist the formation of ordered tendon-like tissue. Undoubtedly, more factors are involved in tendon extracellular matrix formation. For example, in the tendon stem cell niche, biglycan and fibromodulin play a major role [15]. Other major extracellular matrix components of tendon include tenomodulin and tenascin. More secreted mediators of tendon differentiation and tendon tissue organization from adult stem cells remain to be identified. In this context, the additional three secreted factors identified by our study (Fstl1, tetranectin, and RNase A4) that are induced in the BMP2/Smad8ca system might affect the tenogenic program in cooperation with periostin. Although their role is not entirely clear, it may be postulated that these factors contribute to the production of an extracellular matrix that is conducive to further development of a tendon-like matrix.
Periostin and tumorigenesis
Periostin has been implicated as a tumor-related protein. An increase in its expression has been proposed to be a prognostic marker in tumorigenesis [24]. It is frequently dysregulated in various malignant cancers and promotes tumor metastatic growth [25] since it is produced by cancer-associated fibroblasts thereby supporting, for example, growth of gastric cancer [26] or breast-cancer-derived metastases [27]. Therefore, in tumor biology, periostin presents as a possible target within the tumor microenvironment to be eliminated in order to inhibit metastasis. Nevertheless, evidence has also accumulated for a distinct role of periostin in fibrillogenesis and, in addition, periostin is closely involved in mechanosensing [8]. In the context of the work presented here, we, therefore, suggest that periostin is not only expressed in tendons but also that it may influence tendon development and formation. In our studies, we did not observe any indications for tumor or blood vessel formation in our explants, although the time allowed in our ectopic implantations (4 weeks) may be too short for a final conclusion. In contrast, BMP2 clearly stimulates blood vessel formation in ectopic implantations (data not shown).
Use of viral expression systems for genetic modification of stem cells
Lentiviral vectors are used as convenient gene delivery systems because they offer several advantages; they infect nondividing cells, have high transduction efficiencies, and insert stably into the host genome allowing long-time gene expression. However, genome integration by lentiviral vectors may lead to insertional mutations. So, human immunodeficiency virus (HIV)–based vectors may integrate into the growth hormone receptor locus [28]. Also, a potentially oncogenic insertion of HIV integration has been reported in rare T-cell lymphomas [29]. Further, HIV-derived lentiviral vectors may bias analyses by their integration into transcriptional units [30,31]. Recently, it also has been documented that lentiviral vectors cause genome-wide epigenetic changes in the methylation status. Up to 900 genes may be modified by lentiviral infection of CD34+ cells [32]. These problems ask for special safety regards before lentiviral vectors can proceed to the clinic [33] or, for example, the use of nonviral gene delivery systems, such as the “sleeping beauty,” may be advisable [34]. In spite of all these caveats, it has been documented that in vitro and even in vivo lentiviral infection of MSCs did not affect their multilineage differentiation potential [35] and in our case, tenogenic tissue formation of control implantations with GFP-lentiviruses or uninfected MSCs was not observed (data not shown).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge expert technical assistance by Inge Hollatz-Rangosch, Doris Stellfeld (both HZI), Annika Hamm, Peter Erfurt, Mattias Reebmann (all MHH), and Bettina Lipkovitz (MPI). G.G. and U.A.N. were financially supported by the European Community (Key action LSH 1.2.4-3, Integrated project GENOSTEM “Adult MSCs engineering for connective tissue disorders. From the bench to the bed side,” contract no. 503161). A.H. and G.G. additionally acknowledge funding by Deutsche Forschungsgemeinschaft (www.dfg.de) within SFB 599 and DFG HO 2058/4-1 to A.H., who also acknowledges financial support by Förderstiftung MHHplus (Dr. Eckhard Schenke). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Schweitzer R, Zelzer E. and Volk T. (2010). Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137:2807–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Favata M, Beredjiklian PK, Zgonis MH, Beason DP, Crombleholme TM, Jawad AF. and Soslowsky LJ. (2006). Regenerative properties of fetal sheep tendon are not adversely affected by transplantation into an adult environment. J Orthop Res 24:2124–2132 [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann A. and Gross G. (2009). Innovative strategies for treatment of soft tissue injuries in human and animal athletes. Med Sport Sci 54:150–165 [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Song XH, Yin Z, Zou XH, Wang LL, Hu H, Cao T, Zheng M. and Ouyang HW. (2009). Stepwise differentiation of human embryonic stem cells promotes tendon regeneration by secreting fetal tendon matrix and differentiation factors. Stem Cells 27:1276–1287 [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann A, Pelled G, Turgeman G, Eberle P, Zilberman Y, Shinar H, Keinan-Adamsky K, Winkel A, Shahab S, et al. (2006). Neotendon formation induced by manipulation of the Smad8 signalling pathway in mesenchymal stem cells. J Clin Invest 116:940–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahab-Osterloh S, Witte F, Hoffmann A, Winkel A, Laggies S, Neumann B, Seiffart V, Lindenmaier W, Gruber AD, et al. (2010). Mesenchymal stem cell-dependent formation of heterotopic tendon-bone insertions (osteotendinous junctions). Stem Cells 28:1590–1601 [DOI] [PubMed] [Google Scholar]
- 7.Merle B. and Garnero P. (2012). The multiple facets of periostin in bone metabolism. Osteoporos Int 23:1199–1212 [DOI] [PubMed] [Google Scholar]
- 8.Kudo A. (2011). Periostin in fibrillogenesis for tissue regeneration: periostin actions inside and outside the cell. Cell Mol Life Sci 68:3201–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF. and Kudo A. (1999). Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res 14:1239–1249 [DOI] [PubMed] [Google Scholar]
- 10.Inai K, Norris RA, Hoffman S, Markwald RR. and Sugi Y. (2008). BMP-2 induces cell migration and periostin expression during atrioventricular valvulogenesis. Dev Biol 315:383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, et al. (2007). Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem 101:695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurok U, Steinhoff C, Lipkowitz B, Ropers HH, Scharff C. and Nuber UA. (2004). Gene expression changes in the course of neural progenitor cell differentiation. J Neurosci 24:5982–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahrens M, Ankenbauer T, Schröder D, Hollnagel A, Mayer H. and Gross G. (1993). Expression of human bone morphogenetic proteins -2 or -4 in murine mesenchymal progenitor C3H10T½ cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol 12:871–880 [DOI] [PubMed] [Google Scholar]
- 14.Tsuchida K, Arai KY, Kuramoto Y, Yamakawa N, Hasegawa Y. and Sugino H. (2000). Identification and characterization of a novel follistatin-like protein as a binding protein for the TGF-beta family. J Biol Chem 275:40788–40796 [DOI] [PubMed] [Google Scholar]
- 15.Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, et al. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med 13:1219–1227 [DOI] [PubMed] [Google Scholar]
- 16.Iba K, Durkin ME, Johnsen L, Hunziker E, Damgaard-Pedersen K, Zhang H, Engvall E, Albrechtsen R. and Wewer UM. (2001). Mice with a targeted deletion of the tetranectin gene exhibit a spinal deformity. Mol Cell Biol 21:7817–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Sheng J, Hu JK, Yu W, Kishikawa H, Hu MG, Shima K, Wu D, Xu Z, et al. (2012). Ribonuclease 4 protects neuron degeneration by promoting angiogenesis, neurogenesis, and neuronal survival under stress. Angiogenesis 16:387–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvinen TA, Jozsa L, Kannus P, Jarvinen TL. and Jarvinen M. (2002). Organization and distribution of intramuscular connective tissue in normal and immobilized skeletal muscles. An immunohistochemical, polarization and scanning electron microscopic study. J Muscle Res Cell Motil 23:245–254 [DOI] [PubMed] [Google Scholar]
- 19.Mackey AL, Donnelly AE. and Roper HP. (2005). Muscle connective tissue content of endurance-trained and inactive individuals. Scand J Med Sci Sports 15:402–408 [DOI] [PubMed] [Google Scholar]
- 20.Oliver G, Wehr R, Jenkins NA, Copeland NG, Cheyette BN, Hartenstein V, Zipursky SL. and Gruss P. (1995). Homeobox genes and connective tissue patterning. Development 121:693–705 [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto-Shiraishi Y. and Kuroiwa A. (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev Biol 377:363–374 [DOI] [PubMed] [Google Scholar]
- 22.Rani S, Barbe MF, Barr AE. and Litvin J. (2009). Induction of periostin-like factor and periostin in forearm muscle, tendon, and nerve in an animal model of work-related musculoskeletal disorder. J Histochem Cytochem 57:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen KS, Tatarczuch L, Mirams M, Ahmed YA, Pagel CN. and Mackie EJ. (2010). Periostin expression distinguishes between light and dark hypertrophic chondrocytes. Int J Biochem Cell Biol 42:880–889 [DOI] [PubMed] [Google Scholar]
- 24.Tian B, Zhang Y. and Zhang J. (2014). Periostin is a new potential prognostic biomarker for glioma. Tumour Biol [Epub ahead of print]; DOI: 10.1007/s13277-014-1778-3 [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Liu J, Wang Z, Huang Y, Liu W, Zhu X, Cai Y, Fang X, Lin S, Yuan L. and Ouyang G. (2013). Periostin contributes to the acquisition of multipotent stem cell-like properties in human mammary epithelial cells and breast cancer cells. PLoS One 8:e72962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi Y, Kunita A, Iwata C, Komura D, Nishiyama T, Shimazu K, Takeshita K, Shibahara J, Kii I, et al. (2014). The niche component periostin is produced by cancer-associated fibroblasts, supporting growth of gastric cancer through ERK activation. Am J Pathol 184:859–870 [DOI] [PubMed] [Google Scholar]
- 27.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF. and Huelsken J. (2012). Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481:85–89 [DOI] [PubMed] [Google Scholar]
- 28.Bokhoven M, Stephen SL, Knight S, Gevers EF, Robinson IC, Takeuchi Y. and Collins MK. (2009). Insertional gene activation by lentiviral and gammaretroviral vectors. J Virol 83:283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiramizu B, Herndier BG. and McGrath MS. (1994). Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res 54:2069–2072 [PubMed] [Google Scholar]
- 30.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR. and Bushman F. (2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529 [DOI] [PubMed] [Google Scholar]
- 31.Bushman FD. (2003). Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135–138 [DOI] [PubMed] [Google Scholar]
- 32.Yamagata Y, Parietti V, Stockholm D, Corre G, Poinsignon C, Touleimat N, Delafoy D, Besse C, Tost J, Galy A. and Paldi A. (2012). Lentiviral transduction of CD34(+) cells induces genome-wide epigenetic modifications. PLoS One 7:e48943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly JB. (2002). Lentiviruses in gene therapy clinical research. Gene Ther 9:1730–1734 [DOI] [PubMed] [Google Scholar]
- 34.Ivics Z. and Izsvak Z. (2011). Nonviral gene delivery with the sleeping beauty transposon system. Hum Gene Ther 22:1043–1051 [DOI] [PubMed] [Google Scholar]
- 35.Worsham DN, Schuesler T, von KC. and Pan D. (2006). In vivo gene transfer into adult stem cells in unconditioned mice by in situ delivery of a lentiviral vector. Mol Ther 14:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.