Summary
Inhibition of NEDD8 activating enzyme (NAE) has emerged as a highly promising approach to treat cancer through the adenosine sulfamate analogue MLN4924. Here we show that selective pressure results in HCT116 colorectal carcinoma cells with decreased MLN4924 sensitivity and identify a single nucleotide transition that changes alanine 171 to threonine (A171T) of the NAE subunit UBA3. This reduces the enzyme’s affinity for MLN4924 and ATP while increasing NEDD8 activation at physiological ATP concentrations. Expression of UBA3 A171T is sufficient to decrease MLN4924 sensitivity of naive HCT116 cells, indicating it is a dominant suppressor of MLN4924-mediated cell death. Our data suggest the on-target potency of MLN4924 selects for a point mutation in NAE that overcomes the molecule’s inhibitory effects, allowing cancer cell survival.
Introduction
The modification of cullin proteins with NEDD8 (neddylation) regulates the activity of cullin-RING ubiquitin ligases (CRLs) (Bosu and Kipreos, 2008; Pan et al., 2004; Petroski and Deshaies, 2005). Similar to all ubiquitin-like proteins, NEDD8 conjugation relies on an enzymatic cascade involving initial ATP-dependent activation prior to covalent NEDD8 transfer onto substrates (Bohnsack and Haas, 2003; Schulman and Harper, 2009). NEDD8 activating enzyme (NAE) consists of NAE1 (ULA1, APPBP1) and UBA3 (UBE1C), with the latter containing the ATP binding pocket and catalytic cysteine (Gong and Yeh, 1999; Osaka et al., 1998). Neddylation of CRLs triggers structural rearrangements associated with enhanced ubiquitin modification of CRL-bound protein substrates (Duda et al., 2008; Osaka et al., 2000; Podust et al., 2000; Read et al., 2000; Saha and Deshaies, 2008). As cullin neddylation is reversible through the COP9 signalosome (CSN) (Cope et al., 2002; Lyapina et al., 2001), this functions as a highly dynamic mechanism for regulating ubiquitin- and proteasome-dependent protein homeostasis by CRLs.
The anti-cancer effects of the FDA-approved proteasome inhibitor bortezomib (Velcade®) have motivated efforts to develop other molecules that target cellular protein homeostasis mechanisms such as the NAE inhibitor MLN4924 (Cohen and Tcherpakov, 2010; Kane et al., 2003; Kane et al., 2007; Nalepa et al., 2006; Petroski, 2008; Soucy et al., 2009). MLN4924 rapidly eliminates CRL neddylation, leading to CRL substrate accumulation and DNA re-replication prior to cancer cell apoptosis (Soucy et al., 2009). This molecule potently inhibits tumor growth in mouse xenograft studies and appears well tolerated at different doses and treatment regimens (Soucy et al., 2009). These promising pre-clinical studies have motivated Phase I and Phase I/II clinical trials for hematologic and advanced non-hematologic cancers as well as efforts to develop similar inhibitors for other ubiquitin and ubiquitin-like protein activating enzymes (E1s) (Brownell et al., 2010; Chen et al., 2011; Milhollen et al., 2011; Milhollen et al., 2010; Soucy et al., 2009; Swords et al., 2010).
MLN4924 is an adenosine sulfamate analogue that relies on the NAE catalytic cycle to generate the inhibitory NEDD8-MLN4924 covalent adduct in situ (Brownell et al., 2010). This adduct resembles the acyl adenylate intermediate formed between NEDD8 and AMP during NEDD8 activation and functions as a tight binding inhibitor that prevents subsequent ATP and NEDD8 binding (Brownell et al., 2010; Petroski, 2010). Although E1s have conserved catalytic activities and high degrees of sequence similarity (Schulman and Harper, 2009), MLN4924 has remarkable on-target selectivity as it is 300- and 1500-fold more potent against NAE than the E1s for SUMO and ubiquitin respectively (Brownell et al., 2010; Soucy et al., 2009). In contrast, another adenosine sulfamate analogue, Compound 1, functions as a non-selective substrate-assisted inhibitor of canonical E1s in spite of structural features similar to MLN4924 (Brownell et al., 2010; Chen et al., 2011). Thus, these adenosine sulfamate analogues may be fine tuned towards selectively inhibiting a specific E1. It remains a highly significant challenge, however, to understand the complex mechanism of action of these molecules and to determine what underlies E1 selectivity and sensitivity. Here we show that HCT116 colorectal carcinoma cells develop resistance to MLN4924-induced apoptosis and identify a mutation in a previously uncharacterized residue of UBA3 that provides a dominant and transferable decrease in MLN4924 sensitivity.
Results
HCT116 cells develop MLN4924 resistance
Although HCT116 colorectal carcinoma cells transiently treated with MLN4924 undergo DNA re-replication and cell death (Soucy et al., 2009), serially culturing them in the presence of 1 µM of the molecule over 4 weeks resulted in largely unaffected cells. These cells, HMR (HCT116 MLN4924 Resistant) cells, proliferate similarly to untreated HCT116 cells (HMS cells, HCT116 MLN4924 Sensitive) with doubling times of 17.1 hours and 16.6 hours, respectively (Figure 1A). Treatment of HMR cells with 3 µM MLN4924 slows this doubling time to 22.0 hours indicating a partial response to the molecule; however this concentration potently causes HMS cell death.
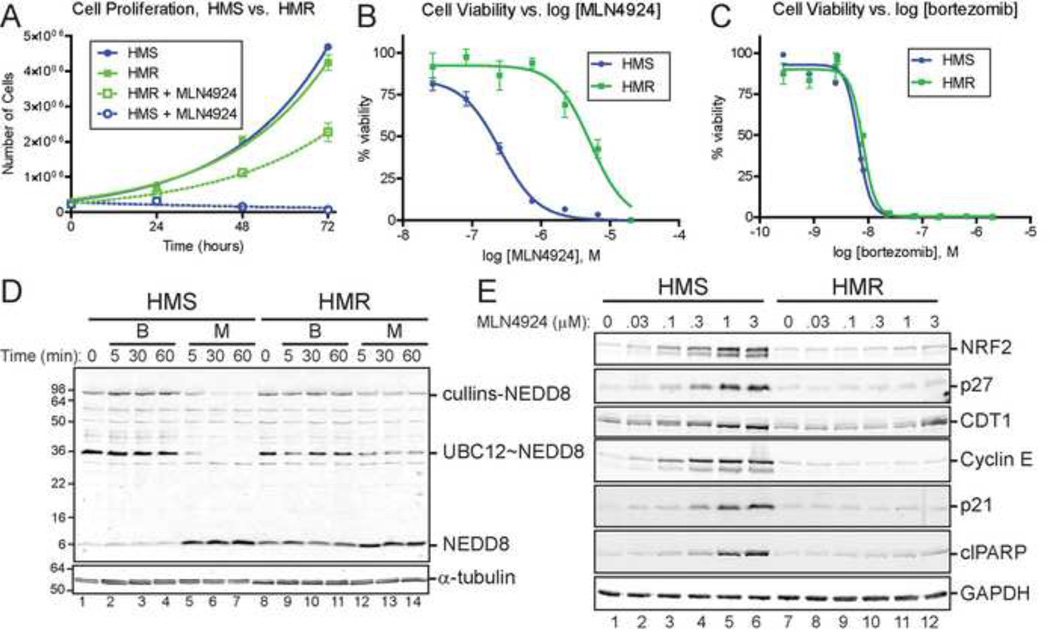
Figure 1. HCT116 cells lose sensitivity to the NEDD8 activating enzyme inhibitor MLN4924 upon prolonged exposure and serial culturing.
(A) Growth of HCT116 cells (HMS cells , HCT116 MLN4924 Sensitive; HMR cells, HCT116 MLN4924 Resistant) in the presence or absence of 3 µM MLN4924. The viability of HMS and HMR cells treated for 48 hours with MLN4924 (B) or bortezomib (C) was assessed by quantifying cellular ATP levels. (D) HMS and HMR cells were treated with 3 µM bortezomib (B) or 3 µM MLN4924 (M) for the indicated times and extracts were analyzed under non-reducing conditions by immunoblotting for NEDD8. NEDD8-modified cullins, NEDD8-activated UBC12, and NEDD8 are indicated. α-tubulin was used as a loading control. See Figure S1 for immunoblotting for ubiquitin. (E) The steady state abundance of CRL substrates NRF2, p27, CDT1, cyclin E, and p21 and the cell death marker cleaved PARP (clPARP)were examined after 24 hours of treatment with the indicated concentration of MLN4924. GAPDH was used as a loading control.
To quantitatively compare the sensitivity of HMS and HMR cells to MLN4924, cell viability assays were performed with increasing concentrations of the molecule (Figure 1B). HMS cells are ∼20-fold less sensitive to MLN4924 than HMR cells (MLN4924 EC50s of 5.2 µM for HMR and 255 nM for HMS). However, both are similarly sensitive to the proteasome inhibitor bortezomib (EC50 6.6 nM and 8.0 nM, respectively), indicating that HMR cells are not generally unresponsive to small molecules.
The decreased sensitivity of HMR cells to MLN4924 could arise through a variety of on- and off-target mechanisms which singly or collectively confer resistance to cell death induced by the molecule. To evaluate if the decreased sensitivity of HMR cells to MLN4924 is due to impaired uptake or retention of the compound or a decrease in its intracellular stability, the steady state levels of MLN4924 in HMS and HMR cells were measured over time upon MLN4924 addition using liquid chromatography tandem mass spectrometry (LC-MS/MS) and no differences were detected (Figure S1A). Additionally, expression profiling did not identify changes in cytochrome P450 superfamily genes (Figure S1B), which are often induced in response to small molecules and are estimated to be responsible for ∼75% of drug metabolism (Guengerich, 2008).
HMR cells maintain neddylated cullins
The effects of MLN4924 and bortezomib on the ubiquitin and neddylation systems in HMR cells were evaluated by immunoblotting extracts for ubiquitin and NEDD8. Bortezomib treatment (3 µM) resulted in the accumulation of high molecular weight polyubiquitinated proteins with no differences between HMS and HMR cells (Figure S1C). Immunoblotting these extracts for NEDD8 under non-reducing conditions revealed that untreated or bortezomib treated HMR cells contain more monomeric NEDD8 than HMS cells and they also maintain activated UBC12 (UBC12∼NEDD8) in the presence of MLN4924 (Figure 1D). As expected, MLN4924 treatment of HMS cells resulted in the rapid loss of NEDD8-modified cullins. While some decrease also occurred in HMR cells, the steady state levels of NEDD8-modified cullins persisted at levels higher than in HMS cells.
Collectively, these studies suggest HMR cells are not completely unresponsive to MLN4924, but are able to maintain sufficient levels of NEDD8-modified cullins. To explore this further, dose response experiments were performed in which HMS and HMR cells were treated for 48 hours with increasing concentrations of MLN4924 and the steady state levels of several well-known CRL substrates were examined by immunoblot (Figure 1E). As expected, these substrates and the apoptosis marker, cleaved PARP, all accumulated in HMS cells in a dose sensitive manner. In contrast, however, similar accumulation was not observed in HMR cells. These observations suggest that the levels of neddylated cullins maintained in the presence of MLN4924 in HMR cells are sufficient to fulfill CRL functions necessary to prevent cell death.
HMR cells contain a UBA3 point mutation
Our data are consistent with an alteration in the NEDD8 system mediating decreased sensitivity of HMR cells to MLN4924. Based on the mechanism of action of MLN4924 (Brownell et al., 2010), we suspected HMR cells contained a mutated form of NAE. As UBA3 contains the nucleotide binding pocket that accommodates MLN4924 and the catalytic cysteine necessary for formation of the inhibitory NEDD8-MLN4924 covalent adduct (Figure 2A), this NAE subunit seemed a likely target for mutations.
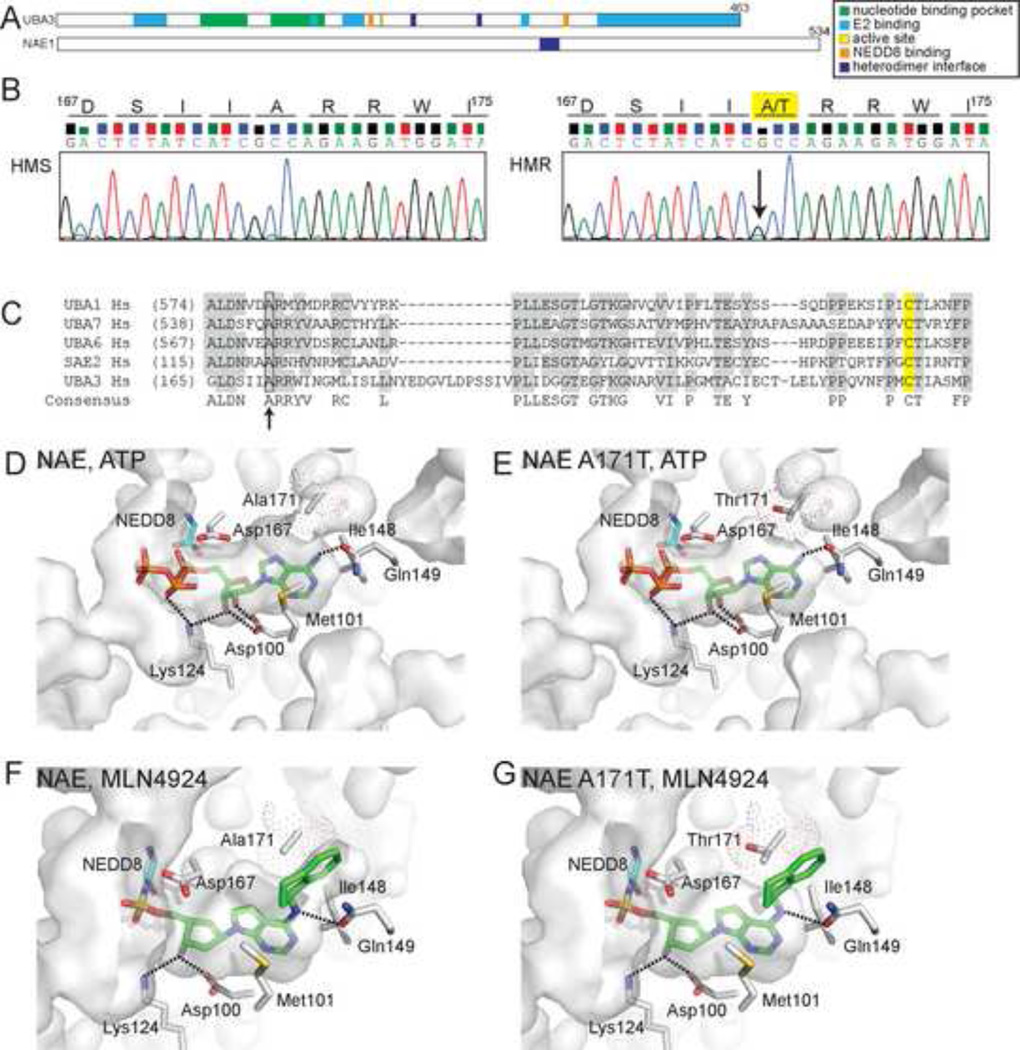
Figure 2. HMR cells are heterozygous for a single nucleotide transition within the coding sequence of the UBA3 nucleotide binding pocket.
(A) Domain structure of NAE subunits, UBA3 (UBE1C) and NAE1 (ULA1, APPBP1). (B) Chromatograms from sequencing reactions analyzing genomic DNA from HMS and HMR cells determined that HMR cells are heterozygous for a G to A transition (indicated by arrow) that alters an amino acid within the UBA3 nucleotide binding pocket. This transition results in the conversion of alanine 171 to threonine (A171T) in the UBA3 open reading frame. See Figure S2 for sequences of cDNAs and extended genomic DNA sequence. (C) A sequence alignment of canonical E1s indicates similarly positioned alanine residues (indicated by arrow) found in other E1s. The catalytic cysteines are highlighted in yellow. Molecular models of the NAE nucleotide binding pocket with ATP bound to UBA3 (D) or UBA3 A171T (E) and MLN4924-NEDD8 bound to UBA3 (F) or UBA3 A171T (G). The solvent accessible nucleotide binding pocket is indicated as are side chains important for MLN4924 binding. Dot surface representation for A171 and computer modeled A171T is shown with only the rotamer with the highest frequency of occurrence in proteins. Computer modeled A171T rotamers in (E) had significant van der Waals overlaps with other residues in UBA3, suggesting it could alter the nucleotide binding pocket. Interestingly, the two major A171T rotamers modeled in (G) had significant van der Waals overlap with the aminoindane of MLN4924, but only minor contacts with other UBA3 residues. These models are based on Protein Data Bank accession codes 1R4N (D, E) and 3GZN (F, G).
Sequencing of cloned UBA3 cDNAs from HMR cells identified a single point mutation at position 511 of the UBA3 open reading frame in 3 out of 6 sequenced cDNAs distinct from those of HMS cells (Figure S2A). This mutation occurs within the coding sequence of the nucleotide binding pocket and represents a purine base transition from guanine (G) to adenine (A). Sequencing of genomic DNA corresponding to this region determined that HMR cells are heterozygous at this nucleotide position which results in either an alanine (A, the wild-type residue, encoded by GCC codon) or threonine (T, generated by a nucleotide transition to ACC codon) encoded at position 171 within the UBA3 open reading frame (Figure 2B and Figure S2B). Alanine 171 represents a previously uncharacterized residue within UBA3 and is found at a similar position in other canonical E1s (Figure 2C).
Molecular models of NAE with bound ATP and NEDD8 from crystal structures (Figure 2D) suggest the residue participates in defining an edge of the solvent accessible nucleotide binding pocket, but does not directly contact ATP. The model of NAE containing NEDD8-MLN4924 (Brownell et al., 2010) indicates this alanine side chain aligns parallel and in close proximity to the aminoindane moiety of MLN4924 (Figure 2F). Computer simulated models of UBA3 containing A171T, in contrast, suggest A171T may not be compatible with MLN4924 binding as this side chain has a steric clash with the aminoindane (Figure 2G). Although it does not appear to directly contact ATP (Figure 2E), the simulated A171T had significant van der Waals overlaps with adjacent residues in UBA3. These suggest a mechanism of aminoindane-dependent binding of MLN4924 through interactions with A171 and that decreased sensitivity to MLN4924 may be conferred by A171T inhibiting aminoindane-dependent MLN4924 binding. Moreover, steric clashes observed in the simulated UBA3 A171T model with ATP suggest this mutation may alter the nucleotide binding pocket.
UBA3 A171T alters NAE properties
To explore the effect of A171T on NAE activity, we generated recombinant NAE complexes containing either UBA3 or UBA3 A171T and examined these in ATP:PPi exchange experiments (Bohnsack and Haas, 2003; Brownell et al., 2010; Bruzzese et al., 2009). This assay measures the rate of radiolabeled ATP formed from 32P-labeled PPi in a NAE- and NEDD8-dependent manner, allowing for kinetic parameters of the enzyme in NEDD8 activation to be directly extracted. In MLN4924 titration experiments using ATP concentrations of either 100 µM (Figure 3A, raw data shown in Figure S3A) or 1 mM (Figure 3B and Figure S3B), NAE (UBA3 A171T) had an IC50 ∼2-fold higher than NAE (192 nM vs. 63 nM, 100 µM ATP; 290 nM vs. 170 nM, 1 mM ATP). These observations are consistent with A171T reducing MLN4924 binding as expected from the modeling described above. Moreover, the differences in IC50s at the two different ATP concentrations for both enzymes reflect MLN4924’s function as an ATP-competitive inhibitor (Soucy et al., 2009; Swords et al., 2010).
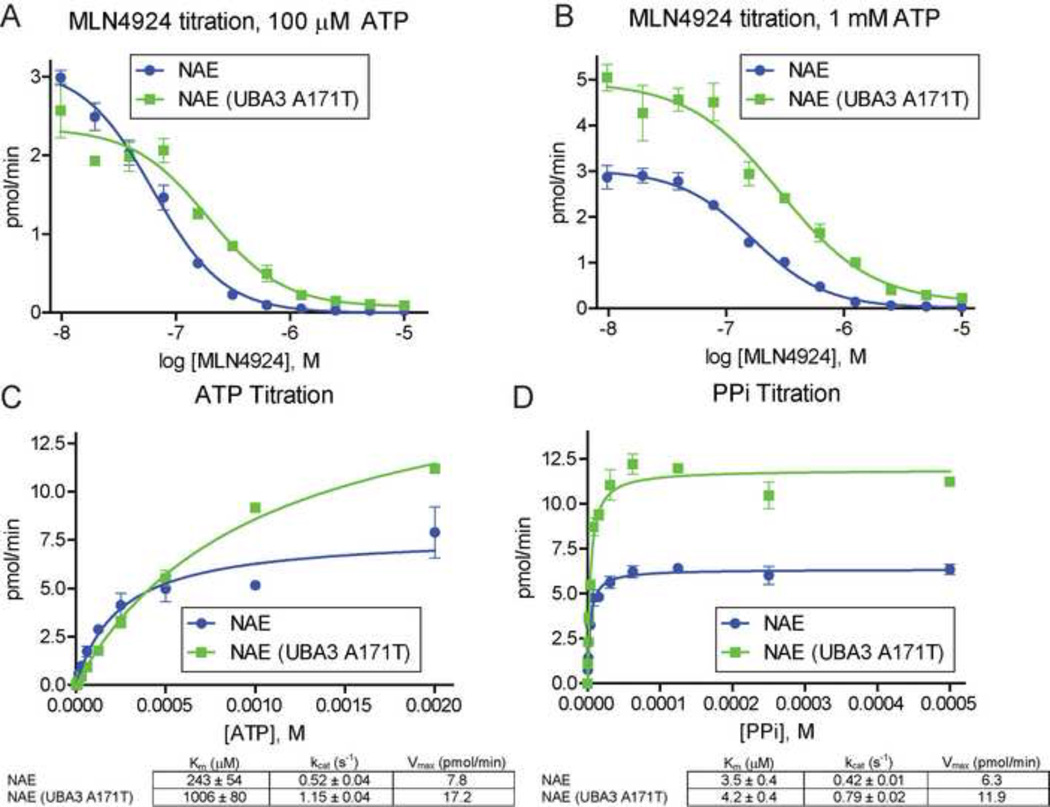
Figure 3. NAE (UBA3 A171T) is less sensitive to MLN4924 and activates NEDD8 more efficiently in vitro.
MLN4924 titrations (2-fold dilutions) in NAE ATP:PPi exchange assay using 100 µM ATP (A) or 1 mM ATP (B). Kinetic measurements of ATP:PPi exchange by NAE and NAE (UBA3 A171T) in ATP (C) and PPi (D) titrations. ATP titrations used 1 mM PPi with the indicated concentrations of ATP. PPi titrations used 2 mM ATP with the indicated concentrations of PPi. All reactions included 50 cpm/pmol of [32P] PPi, 100 µM NEDD8, and 10 nM of NAE or NAE (A171T), and were incubated at 37°C for 30 minutes. See Figure S3 for raw data.
Notably, NAE (UBA3 A171T) had a consistently higher rate of ATP synthesis than NAE at 1 mM ATP with a similar rate at 100 µM ATP. These observations suggest the A171T mutation also alters the nucleotide binding pocket to enhance NEDD8 activation at high ATP concentrations. To explore this hypothesis, we performed titration experiments with ATP (Figure 3C and Figure S3C) and PPi (Figure 3D and Figure S3D). Although NAE and NAE (UBA3 A171T) had similar rates of ATP synthesis at low concentrations of both ATP and PPi, higher concentrations of either substrate resulted in increased rates for NAE (UBA3 A171T).
Both titrations revealed ∼2-fold increases in the turnover number kcat for NAE (UBA3 A171T), with 1.15 vs. 0.52 s−1 for ATP and 0.79 vs. 0.42 s−1 for PPi compared to NAE. We also measured a 4-fold increase in Km for ATP (1006 µM for NAE (UBA3 A171T) and 243 µM for NAE) with a minor effect for PPi (4.2 µM and 3.5 µM). These studies suggest UBA3 A171T decreases MLN4924 sensitivity by altering the nucleotide binding pocket to reduce NAE’s affinity for both MLN4924 and ATP. This mutation also results in correspondingly increased rates of NEDD8 activation at high ATP concentrations. Interestingly, intracellular ATP is typically at 1–10 mM (Beis and Newsholme, 1975), close to the measured Km of NAE (UBA3 A171T).
UBA3 A171T confers MLN4924 resistance
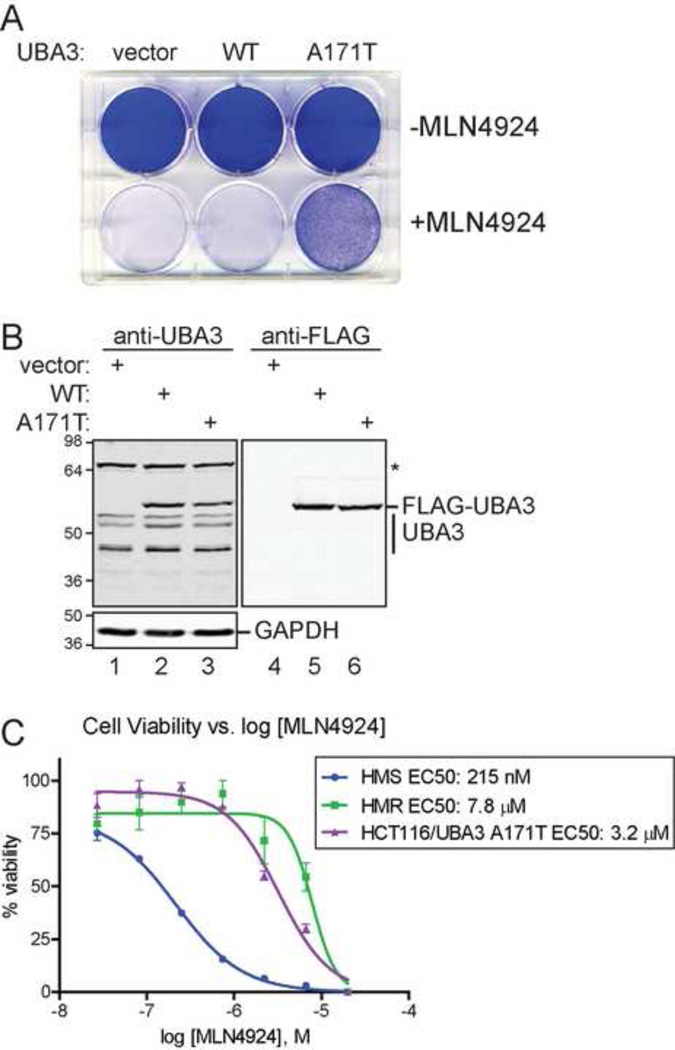
As HMR cells are heterozygous for this mutation in the UBA3 gene, UBA3 A171T should provide a dominant effect on NAE functions to allow cell survival in the presence of MLN4924. To test this hypothesis, HCT116 cells were transfected with plasmids to express either UBA3 (WT) or UBA3 A171T and were treated with either DMSO (-MLN4924) or 3 µM MLN4924 for 72 hours. Although cells transfected with either vector or UBA3 did not survive MLN4924 treatment, those expressing UBA3 A171T did (Figure 4A). This effect is not attributable to UBA3 over-expression as both transfected forms were expressed similarly and at levels comparable to the endogenous UBA3 isoforms detected by immunoblot (Figure 4B).
Figure 4. Expression of UBA3 A171T is sufficient to confer MLN4924 resistance.
(A) HCT116 cells transfected with the indicated plasmids were either treated with DMSO (-MLN4924) or 3 µM MLN4924 for 72 hours prior to crystal violet staining. (B) The expression of FLAG-tagged UBA3 was examined by immunoblot of UBA3 and FLAG in transfected HCT116 cells. GAPDH was used as a loading control. (C) Expression of UBA3 A171T increases HCT116 cells survival in the presence of increasing concentrations of MLN4924. Cell viability measurements for HMS and HMR cells treated with MLN4924 are shown for comparison.
To compare the effects of transient UBA3 A171T expression in HCT116 cells to HMS and HMR cells, viability assays were performed with increasing MLN4924 concentrations (Figure 4C). The measured EC50 for HCT116 cells expressing UBA3 A171T was 3.2 µM which is near the 7.8 µM for HMR cells and 15-fold higher than the 215 nM EC50 for HMS cells. Thus, UBA3 A171T confers MLN4924 resistance to HCT116 cells. Moreover, the underlying single nucleotide transition is sufficient for increased cell survival in the presence of the molecule.
Discussion
We have identified a point mutation in UBA3 that causes decreased sensitivity to the NAE inhibitor MLN4924. This mutation arises through on-target selective pressure and alters the UBA3 nucleotide binding pocket such that MLN4924 binding is impaired through a reduction in the enzyme’s affinity for both ATP and MLN4924. This effectively decreases MLN4924’s function as an ATP competitive inhibitor as the cellular concentration of ATP is in the millimolar range (Beis and Newsholme, 1975)—near the Km for NAE (UBA3 A171T). Additionally, cancer cells typically have elevated ATP levels through increases in aerobic respiration via the Warburg effect (Warburg, 1956). These collectively support sufficient levels of essential NEDD8-dependent CRL functions in the presence of MLN4924 concentrations that typically induce cancer cell death.
In vitro approaches similar to the one described here have successfully identified clinical resistance mechanisms to cancer therapeutics such as the BCR-ABL inhibitor imatinib (Gleevec®) and the epidermal growth factor receptor (EGFR) inhibitors gefitinib (Iressa®), and erlotinib (Tarceva®) (Bell et al., 2005; Cools et al., 2003; Kobayashi et al., 2005; Pao et al., 2005; Shah et al., 2002; Shah et al., 2004; Tamborini et al., 2004; Tamborini et al., 2006; von Bubnoff et al., 2005). EGFR and BCR-ABL contain a “gatekeeper” residue that controls inhibitor access to the ATP binding site while not directly contacting ATP. Mutation of the gatekeeper residue to one that prevents inhibitor binding is a major mechanism of clinical resistance in patients undergoing treatment with EGF receptor and BCR-ABL inhibitors. The observation that A171 comes in close proximity to the aminoindane of MLN4924 (Figure 2F) while not contacting ATP (Figure 2D) suggests it functions in a similar gatekeeper role. It will be interesting to see if mutation of this residue plays a role in clinical resistance to MLN4924 therapy. Conservation of this residue in other canonical E1s (Figure 2C) and the mechanism of action of aminoindane-containing E1 inhibitors (Brownell et al., 2010; Chen et al., 2011), suggest this alanine may play a more general role as a gatekeeper residue for adenosine sulfamate-based E1 inhibitors.
Experimental Procedures
MLN4924 Synthesis
A detailed description of the synthesis is described in Supplementary Methods. Purity and characterization of intermediate compounds and MLN4924 were established by a combination of liquid chromatography-mass spectroscopy (LC-MS) and analytical NMR.
Isolation of MLN4924 resistant HCT116 cells
HCT116 cells (2×105, ATCC) were incubated in 1 µM MLN4924 with the media containing the compound changed every 48 hours. After 17 days, the MLN4924 concentration was increased to 1.5 µM, followed by 2 µM at Day 21 and 3 µM at Day 25. These HMR (HCT116 MLN4924 resistant) cells were maintained in 3 µM MLN4924.
Cell Proliferation and Viability Assays
HMR and HMS cells (105 cells) were incubated in DMSO (0.1% final concentration) or 3 µM MLN4924. After 24, 48, and 72 hours, cells were counted using a TC10 Automated Cell Counter (Bio-Rad). Culture media were replenished every 24 hours. Cell numbers represent the mean (n=3) with error bars representing standard error. See Extended Experimental Procedures for additional details.
For viability measurements, cells were plated at 4000 cells per well and media were added containing 3-fold serial dilutions of MLN4924 from a maximum of 20 µM or bortezomib from 2 µM after 24 hours. After 48 hours, ATP content was measured using Cell Titer-Glo Luminescent Viability Assay (Promega). Extended Experimental Procedures describe analysis methods.
Protein Expression Analyses
Cells (2.8×106 for 24 hour treatments or 5×106 for 60 minute time course experiments) were incubated with DMSO or MLN4924 (0.03 to 3 µM for 24 hour treatments, 3 µM for time courses) after 24 hours. At the indicated times, cells were washed and scraped into cold PBS and resuspended in 25 mM HEPES pH 7.6, 300 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 5 mM MgCl2, 5 mM 1,10-phenanthroline, 1 mM iodoacetamide, and Halt protease inhibitor cocktail (Thermo Scientific). Protein concentrations were determined by Bradford assay prior to analysis of equivalent lysate amounts by SDS-PAGE and immunoblotting. Antibodies used are described in Extended Experimental Procedures .
Molecular Models
All models were rendered in Pymol (Schrodinger, LLC) and used publicly available data from PDB: 1R4N (Walden et al., 2003) and 3GZN (Brownell et al., 2010). The solvent accessible nucleotide binding pocket (rendered using 3 solvent radii for cavity detection and 1 solvent radius for cavity detection cutoff) and side chains of UBA3 important for MLN4924 binding to NAE. Computer modeling of A171T in NAE was performed using the Pymol mutagenesis algorithm in which backbone dependent rotamers of threonine were examined. Only the rotamer with the highest frequency of occurrence in proteins is shown.
Recombinant NAE and NEDD8
ULA1, UBA3 and UBA3 A171T (isoform 1), and NEDD8 open reading frames were prepared as baculoviruses for insect cell expression (ULA1, UBA3, UBA3 A171T) or for E.coli expression (NEDD8). NAE complexes were purified by Ni-NTA chromatography from co-infecting Hi5 cultures (106 cells/ml, 100 ml) with viruses to express ULA1 and hexahistidine tagged UBA3 or UBA3 A171T for 48 hours. NEDD8 was expressed in Rosetta 2 E.coli (Novagen) and purified by SP Sepharose chromatography. Protein concentrations were measured from Coomassie stained gels scanned on a Licor Odyssey, using known amounts of purified BSA to generate a standard curve and the instrument’s analysis software.
ATP:PPi Exchange Assay
The assay is based on a published method (Brownell et al., 2010; Bruzzese et al., 2009). Experiments were performed such that NAE or NAE (UBA3 A171T) was pre-mixed with titrants (MLN4924, ATP, or PPi) and other assay components, including 50 cpm/pmol [32P] PPi. Reactions (50 µl final volume in 25 mM HEPES pH 7.6, 5 mM MgCl2, 25 mM NaCl, 0.5 mM DTT and using NAE and NAE (UBA3 A171T) at 10 nM) were initiated by the addition of NEDD8 to 1 µM, incubated at 37°C for 30 minutes, and terminated with 400 µl 5% TCA, 10 mM PPi. Reactions were transferred to activated charcoal paper by vacuum filtration with each washed four times with 250 µl 2% TCA, 10 mM PPi and the filter washed 5 times with 100 ml over 25 minutes. The filter was exposed to a phosphor screen and imaged on a Fuji FLA-5100. Resulting signals representing NAE generated radiolabeled ATP were quantified using Multi Gauge software and ATP standards. All measurements are shown as the mean (n=3) with error bars representing the SEM. Raw data are shown in Figure S3 as well as descriptions of additional experimental details. Data analyses used GraphPad Prism software and a nonlinear regression algorithm to measure kcat and extract other best-fit values from plots of rates of ATP synthesis vs. substrate concentration.
Transient UBA3 expression
Plasmids expressing FLAG-tagged UBA3 and UBA3 A171T from the CMV promoter were transfected into HCT116 cells using Lipofectamine 2000 (Invitrogen). After 24 hours, protein extracts from a portion of the cells were normalized as described above and immunoblotted for UBA3, FLAG-M2 (Sigma, F1804), and GAPDH. The remaining cells were plated at 4×105 in a 6-well plate. After another 24 hours, 0.1 % DMSO or 3 µM MLN4924 was added and incubated for 72 hours prior to staining with 1% crystal violet in 10% ethanol. Cell viability assays were performed as described above except cells were first transferred to a 10 cm2 plate 24 hours post-transfection and treated for 48 hours with 3 µM MLN4924 prior to seeding into a 96-well plate.
Supplementary Material
Acknowledgements
We thank Ray Deshaies, Dieter Wolf, and Ze’ev Ronai for critically reading the manuscript and helpful suggestions and personnel of the Flow Cytometry, Medicinal Chemistry, Microarray, and Informatics and Data Management Shared Resources at SBMRI for technical assistance. This work was supported by an American Cancer Society Research Scholars Grant (RSG-11-224-01-DMC), institutional start-up funding, and a Scholars Grant from the V Foundation for Cancer Research to M.D.P.
References
- Beis I, Newsholme EA. The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem J. 1975;152:23–32. doi: 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, Haber DA. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Bruzzese FJ, Tsu CA, Ma J, Loke HK, Wu D, Li Z, Tayber O, Dick LR. Development of a charcoal paper adenosine triphosphate:pyrophosphate exchange assay: kinetic characterization of NEDD8 activating enzyme. Anal Biochem. 2009;394:24–29. doi: 10.1016/j.ab.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Tsu CA, Gavin JM, Milhollen MA, Bruzzese FJ, Mallender WD, Sintchak MD, Bump NJ, Yang X, Ma J, et al. Mechanistic studies of substrate-assisted inhibition of ubiquitin activating enzyme by adenosine sulfamate analogues. J Biol Chem. 2011 doi: 10.1074/jbc.M111.279984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- Kane RC, Dagher R, Farrell A, Ko CW, Sridhara R, Justice R, Pazdur R. Bortezomib for the treatment of mantle cell lymphoma. Clin Cancer Res. 2007;13:5291–5294. doi: 10.1158/1078-0432.CCR-07-0871. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9(Suppl 1):S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD. Mechanism-based neddylation inhibitor. Chem Biol. 2010;17:6–8. doi: 10.1016/j.chembiol.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- Tamborini E, Bonadiman L, Greco A, Albertini V, Negri T, Gronchi A, Bertulli R, Colecchia M, Casali PG, Pierotti MA, Pilotti S. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Tamborini E, Pricl S, Negri T, Lagonigro MS, Miselli F, Greco A, Gronchi A, Casali PG, Ferrone M, Fermeglia M, et al. Functional analyses and molecular modeling of two c-Kit mutations responsible for imatinib secondary resistance in GIST patients. Oncogene. 2006;25:6140–6146. doi: 10.1038/sj.onc.1209639. [DOI] [PubMed] [Google Scholar]
- von Bubnoff N, Veach DR, van der Kuip H, Aulitzky WE, Sanger J, Seipel P, Bornmann WG, Peschel C, Clarkson B, Duyster J. A cell-based screen for resistance of Bcr-Abl-positive leukemia identifies the mutation pattern for PD166326, an alternative Abl kinase inhibitor. Blood. 2005;105:1652–1659. doi: 10.1182/blood-2004-06-2445. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr., Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.