Summary
Bacterial cell shapes are manifestations of programs carried out by multi-protein machines that synthesize and remodel the peptidoglycan (PG) mesh and other polymers surrounding cells. GpsB protein is conserved in low-GC Gram-positive bacteria and is not essential in rod-shaped Bacillus subtilis, where it plays a role in shuttling penicillin binding proteins (PBPs) between septal side-wall sites of PG synthesis. In contrast, we report here that GpsB is essential in ellipsoid-shaped, ovococcal Streptococcus pneumoniae (pneumococcus), and depletion of GpsB leads to formation of elongated, enlarged cells containing unsegregated nucleoids and multiple, unconstricted rings of fluorescent-vancomycin staining, and eventual lysis. These phenotypes are similar to those caused by selective inhibition of Pbp2x by methicillin that prevents septal PG synthesis. Dual-protein 2D and 3D-SIM (structured illumination) immunofluorescence microscopy (IFM) showed that GpsB and FtsZ have overlapping, but not identical, patterns of localization during cell division and that multiple, unconstricted rings of division proteins FtsZ, Pbp2x, Pbp1a, and MreC are in elongated cells depleted of GpsB. These patterns suggest that GpsB, like Pbp2x, mediates septal ring closure. This first dual-protein 3D-SIM IFM analysis also revealed separate positioning of Pbp2x and Pbp1a in constricting septa, consistent with two separable PG synthesis machines.
Keywords: GpsB, Pbp2x, FtsZ ring closure, Pbp1a, graphical user interface
Introduction
The shape, size, and chaining of bacterial cells are fundamental, evolved properties that allow bacteria to survive and compete in their ecological niches (Young, 2007, Young, 2010). These morphological characteristics are largely determined by the resilient peptidoglycan (PG) exoskeleton that surrounds bacterial cell membranes and are manifestations of the mechanisms that carry out and regulate PG synthesis during bacterial cell growth and division (reviewed in (Egan & Vollmer, 2013, Lovering et al., 2012, Sun & Jiang, 2011, Typas et al., 2012)). Extensive work in rod-shaped model bacterial systems has revealed that PG synthesis is carried out by large, dynamic, molecular machines that extend from cell membranes into the extracellular space. The identities and structures of many component proteins and their interactions in the two machines that carry out lateral (sidewall) and septal PG synthesis are reviewed in (Egan & Vollmer, 2013, Typas et al., 2012, White & Gober, 2012). In rod-shaped bacteria, lateral and septal PG synthesis are organized by the cytoskeletal proteins, MreB and FtsZ, respectively, along with other organizer proteins that regulate the flippases and penicillin binding proteins (PBPs) that carry out PG synthesis (see (Egan & Vollmer, 2013, Lovering et al., 2012, Sun & Jiang, 2011, Typas et al., 2012)).
Much less is known about PG synthesis in cocci (spherical cells) or ovococci (prolate ellipsoids), which lack MreB homologues. Early ultrastructure studies suggested that ellipsoid-shaped bacteria, such as Streptococcus pneumoniae (pneumococcus), synthesize their PG by a combination of peripheral (sidewall-like) and septal synthesis that occurs in the midcell regions of dividing cells (Higgins & Shockman, 1976, Massidda et al., 2013, Pinho et al., 2013, Zapun et al., 2008). These two modes are thought to be analogous to the lateral sidewall and septal PG synthesis that occur at different locations in rod-shaped bacteria (Massidda et al., 2013, Sham et al., 2012, Zapun et al., 2008). Current speculative models for the composition of the peripheral and septal machines assume that pneumococcal PG synthesis proteins will have analogous functions to their homologues in rod-shaped bacteria (see (Massidda et al., 2013)). The idea that ovococci, but not spherical cocci, have two modes of PG synthesis is supported by mutagenesis, drug treatment, and biochemical studies of several cocci and ovococci species (Berg et al., 2013, Land & Winkler, 2011, Lleo et al., 1990, Perez-Nunez et al., 2011, Raz et al., 2012, Zapun et al., 2012). Similarities and important differences between PG synthesis in ovococci and rod-shaped bacteria have emerged from these initial studies (see Massidda et al., 2013).
The PG synthesis machines of S. pneumoniae contain three class A bifunctional transglycosylase (TG) - transpeptidase (TP) PBPs (Pbp1a, Pbp1b, and Pbp2a), two class B monofunctional transpeptidases (TPs) (Pbp2x and Pbp2b), and one low-molecular-weight D,D-carboxypeptidase PBP (Pbp3 or DacA) (Fig. 1a) (Barendt et al., 2011, Hoskins et al., 1999, Kocaoglu et al., 2012, Massidda et al., 2013, Paik et al., 1999, Zapun et al., 2008). Pneumococcal Pbp2x and Pbp2b are individually essential and are the homologues of essential PBP3 and PBP2 involved in septal and lateral PG synthesis, respectively, in E. coli (Berg et al., 2013, Zapun et al., 2008). Evidence that Pbp2x functions in septal PG synthesis in ovococci was provided by the demonstration that inhibition of Pbp2x by the β-lactam antibiotic methicillin leads to the formation of elongated cells in Lactococcus lactis (Perez-Nunez et al., 2011). Unexpectedly, depletion of Pbp2x in S. pneumoniae laboratory strain R6 was recently reported to lead to the formation of enlarged, sometimes elongated, cells with oddly pointed ends (Berg et al., 2013), indicative of a complicated division defect. Evidence that Pbp2b functions in peripheral PG synthesis in ovococci was provided by the demonstration that lack of Pbp2b, which is not essential in L. lactis, causes the formation of spherical cells (Perez-Nunez et al., 2011). Depletion of Pbp2b in S. pneumoniae R6 was reported to cause the appearance of compressed, more rounded cells in chains (Berg et al., 2013). Although different from the phenotype in L. lactis, this result is consistent with a role of Pbp2b in peripheral PG synthesis in S. pneumoniae. The initial interpretation from 2D immunofluorescence microscopy (IFM) that pneumococcal Pbp2x and Pbp2b localize exclusively to division septa and daughter cell equators, respectively (Morlot et al., 2003), was later modified to localization of Pbp2x and Pbp2b to midcell regions of dividing cells (Zapun et al., 2008).
Fig. 1.
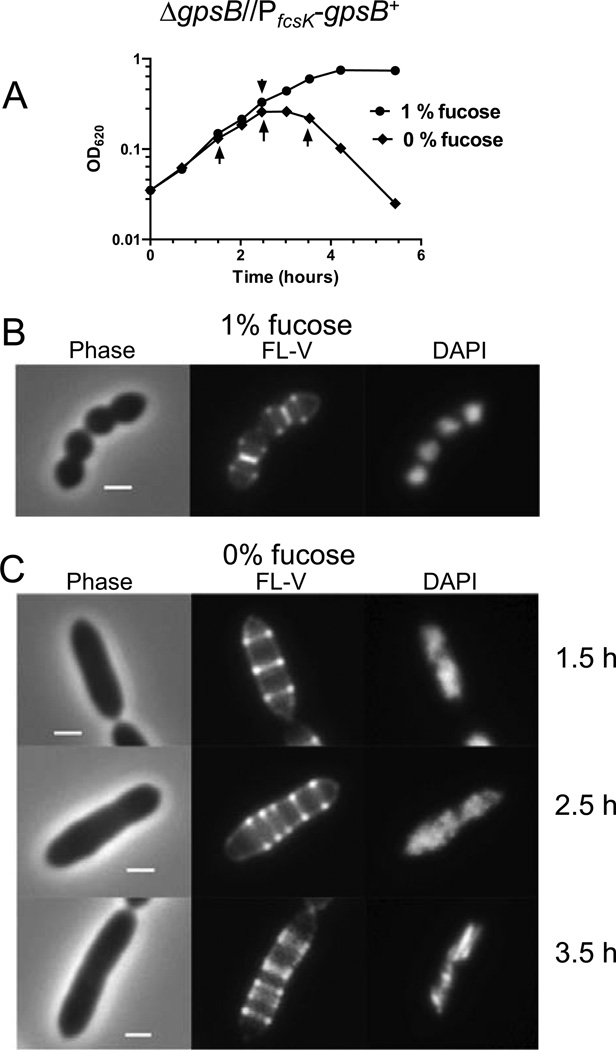
Elongated cell morphology, multiple rings of FL-V staining, and unsegregated nucleoids caused by depletion of GpsB. Strain IU4888 (ΔgpsB//PfcsK-gpsB+) was grown exponentially in BHI broth containing 1.0% (wt/vol) fucose to OD620 ≈ 0.1, washed, and resuspended to OD620 ≈ 0.03 in BHI broth containing 0 or 1% (wt/vol) fucose. (A) Growth curves after resuspension in BHI broth containing or lacking fucose. Arrows indicate when samples were taken for staining and microscopy. (B) Representative images 2.5 h after resuspension in BHI broth containing fucose. Cells were stained with FL-V and DAPI and observed with phase-contrast (Phase) and epifluorescence microscopy as described in Experimental procedures. (C) Representative images of IU4888 after resuspension in BHI broth lacking fucose for 1.5, 2.5, and 3.5 h. All images are shown at the same magnification (scale bar = 1 µm). The experiment was performed two times with similar results.
Unlike the class B PBPs, class A Pbp1a, Pbp2a, and Pbp1b are individually not essential in S. pneumoniae, although the Δpbp1a Δpbp2a combination is synthetically lethal (Hoskins et al., 1999, Land & Winkler, 2011, Paik et al., 1999). However, Pbp1a and Pbp2a are not simply functionally redundant, because the essentiality of MreCD in S. pneumoniae is suppressed in Δpbp1a but not in Δpbp2a mutants in the serotype 2 D39 genetic background (Land & Winkler, 2011). Notably, Δpbp1a mutants showed reduced cell lengths and widths compared to their parent strain, which was an unencapsulated derivative of strain D39 used to reduce masking of cell-shape phenotypes by capsule (Barendt et al., 2009, Land & Winkler, 2011). Depletion of pneumococcal MreCD resulted in formation of spherical cells, consistent with a role of MreCD in peripheral PG synthesis (Land & Winkler, 2011). Taken together, these results suggest that MreCD regulates the location, activity, or both of Pbp1a in peripheral PG synthesis in S. pneumoniae, although this interpretation does not exclude a role of Pbp1a in septal PG synthesis (Land & Winkler, 2011).
This article provides the first characterization of the pneumococcal GpsB cell cycle protein during division. The gpsB gene (for guiding PBP1 shuttling) was identified by a synthetic lethal screen with ezrA in Bacillus subtilis (Claessen et al., 2008). ΔgpsB ΔezrA double mutants of B. subtilis grew poorly, and ΔgpsB mutants only showed growth phenotypes under salt stress (Claessen et al., 2008). GpsB (called YpsB) was identified independently based on the high similarity of its N-terminal amino acid sequence to that of the DivIVA division protein (Tavares et al., 2008). In that study, gpsB showed a synthetic lethal phenotype with ftsA, but not with ezrA (Tavares et al., 2008). GpsB is conserved in a range of Gram-positive bacteria (Claessen et al., 2008, Tavares et al., 2008). Bacterial two-hybrid assays showed strong self-interaction of B. subtilis GpsB, and interaction of GpsB with PBP1, MreC, and EzrA (Claessen et al., 2008). GpsB-GFP localized as discrete spots on lateral walls during B. subtilis cell elongation (Claessen et al., 2008) and joined divisomes in the middle of their assembly, before the start of cell constriction (Gamba et al., 2009, Tavares et al., 2008). PBP1 distribution was not altered in a ΔgpsB mutant compared to its gpsB+ parent, but PBP1 was localized to cell poles in a strain expressing a DivIVA-GpsB fusion protein (Claessen et al., 2008). PBP1 was mislocalized in a B. subtilis ΔezrA ΔgpsB double mutant, and it was proposed that GpsB plays a role in shuttling PBP1 between the septal and lateral PG synthesis machines (Claessen et al., 2008). GpsB was reported to localize at septa in the coccus, Staphylococcus aureus, self-associate, interact with EzrA, and mislocalize when EzrA was depleted, but the essentiality and depletion phenotypes of GpsB in S. aureus were not reported (Steele et al., 2011).
We report here that GpsB is essential in S. pneumoniae and that depletion of GpsB causes S. pneumoniae cells to elongate greatly and increase in size, similar to the morphological defect caused by specific inhibition of Pbp2x, which blocks septal closure. Dual-protein 2D and 3D-SIM (three-dimensional structured illumination microscopy) IFM was used to localize GpsB relative to FtsZ, Pbp2x, Pbp1a (which is the homologue of B. subtilis PBP1), MreC, and nucleoids during different division stages in wild-type and GpsB-depleted cells. To record and analyze patterns of localization in 2D IFM images, a versatile, simple-to-use graphical user interface was developed. We show that GpsB follows a different, but overlapping, pattern of localization compared to FtsZ at different stages of cell division and that the FtsZ and GpsB localization patterns differ from those of Pbp2x and Pbp1a. The super-high-resolution 3D-SIM IFM used in this study provided fundamental new information showing separate positions of Pbp2x and Pbp1a in contracting septa, non-uniformity of FtsZ and other division protein rings, and concurrent constriction and nucleoid separation in dividing S. pneumoniae cells. Finally, we found that elongated GpsB-depleted cells contain multiple, non-constricted rings of FtsZ and other division proteins, suggesting that GpsB may be required for an early step in septal ring closure in S. pneumoniae.
Results
Depletion of GpsB results in enlarged, elongated cells containing unsegregated nucleoids and multiple bands of fluorescent-vancomycin (FL-V) staining
A homologue of B. subtilis gpsB was identified in S. pneumoniae (spd_0339 in serotype 2 strain D39) (Claessen et al., 2008, Tavares et al., 2008). Earlier studies suggested that gpsB may not be essential in some pneumococcal strains (see Massidda et al., 2013); however recent Tn-Seq experiments identified gpsB(gene sp0372) as essential in serotype 4 strain TIGR4 (van Opijnen et al., 2009). To test the essentiality of gpsB and minimize the accumulation of suppressor mutations, we determined primary phenotypes caused by GpsB depletion using merodiploid strain IU4888 (ΔgpsB//PfcsK-gpsB+) (Table S1). IU4888 was constructed by transforming a ΔgpsB<>aad9 amplicon into merodiploid strain IU4846 (gpsB+//PfcsK-gpsB+), which contains an ectopic copy of gpsB+ under control of an inducible fucose promoter (see (Land & Winkler, 2011, Sham et al., 2011)). Transformation mixtures added to plates containing 0.8% (wt/vol) fucose yielded >500 colonies after 20 h of incubation, compared to <20 colonies of diverse sizes on plates lacking fucose in >24 h. As expected, growth of the resulting strain IU4888 (ΔgpsB//PfcsK-gpsB+) was fucose dependent (Fig. 1A). Direct transformation of the ΔgpsB<>aad9 amplicon into the parent gpsB+ strain (IU1945) resulted in the appearance of ≈20 colonies after prolonged incubation. Besides the ΔgpsB replacement mutation, these strains contained additional mutations that likely act as suppressors (data not shown). These combined results support the conclusion that gpsB is essential for growth of serotype 2-derived pneumococcal strains.
IU4888 was grown exponentially in BHI broth containing fucose, washed, and resuspended in fresh BHI broth containing or lacking fucose (Fig. 1A). Both cultures grew similarly for ≈2 h, after which the culture lacking fucose stopped growing and started autolysis (Fig. 1A). At various times after the medium change, cells were stained with fluorescent vancomycin (FL-V) and DAPI to label regions of active PG synthesis (Daniel & Errington, 2003, Ng et al., 2004, Pinho & Errington, 2005, Wheeler et al., 2011) and nucleoids, respectively (Fig. 1B and 1C). In the presence of fucose, cells retained wild-type patterns of septal and equatorial staining with FL-V and had separated, defined nucleoids (Fig. 1B). In contrast, cells depleted for GpsB formed enlarged, elongated cells containing multiple bands of FL-V staining and diffuse nucleoids that were often unsegregated (Fig. 1C). Cells continued to elongate as slightly irregular rods until ≈3 h, with median lengths and widths increasing by ≈3.7x and ≈1.2x, respectively (Fig. 1C; Fig. S1B and S1C). Interestingly, the number of FL-V rings per cell increased linearly with cell length, and plots of fractional ring location normalized to cell length confirmed that the rings were placed non-randomly in cells (data not shown). As cells started to lyse at ≈3 h, cells with pointed ends and bulges started to appear (Fig. S1B).
Control experiments showed that independently constructed ΔgpsB//PfcsK-gpsB+ merodiploid strains IU5846 and IU5847 (Table S1) had the same phenotypes as IU4888 when depleted for GpsB (data not shown). In addition, similar cell morphology defects were observed when IU4888 was depleted for GpsB by a different protocol of fucose limitation (Fig. S2). Inactivation of class A PBP1 of B. subtilis relieved the bulge formation phenotype exhibited by ΔezrA ΔgpsB double mutants (Claessen et al., 2008). We tested whether deletion of pbp1a (which encodes the pneumococcal homologue of B. subtilis PBP1), pbp1b, or pbp2a alleviated lethality associated with depletion of GpsB in strain IU4888 (Fig. S3); but, none of these deletion mutations in pneumococcal class A pbp genes restored growth and viability of bacteria depleted for GpsB.
Selective inhibition of PBP2x results in elongated cells containing unsegregated nucleoids and multiple bands of FL-V staining similar to GpsB depletion
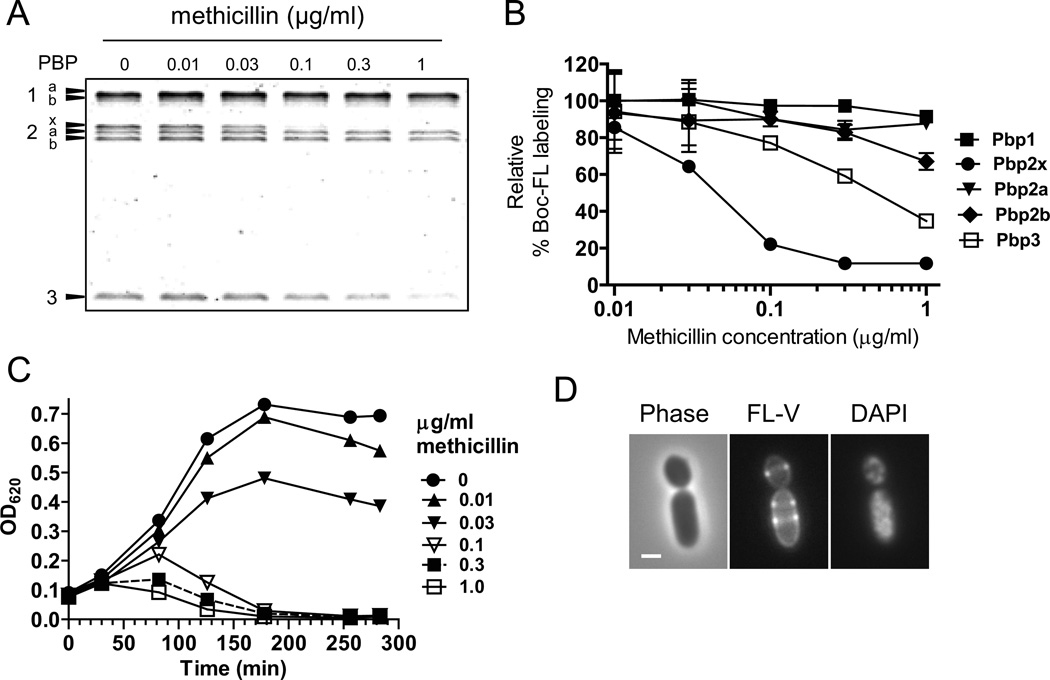
Treatment of several ovococci species with intermediate concentrations of methicillin has resulted in the formation of rod-shaped or filamentous cells; however, the dose-response curve for inhibition of individual PBPs by methicillin was not included in these previous studies (Lleo et al., 1990, Perez-Nunez et al., 2011, Raz et al., 2012). We assayed the specificity of methicillin adduct formation with pneumococcal PBPs (see Introduction) by secondary labeling with fluorescent bocillin (Boc-FL) (Kocaoglu et al., 2012, Zhao et al., 1999). Parent strain IU1945 grown in BHI broth was treated with concentrations of methicillin from 0.01 to 1.0 µg/ml and then labeled with Boc-FL (Fig. 2; Experimental procedures). Treatment with 0.1 µg/ml of methicillin resulted in ≈80% or ≈20% reduction in Boc-FL labeling of Pbp2x or Pbp3, respectively, without significantly (<10%) reducing the labeling of Pbp1a/1b, Pbp2a, and Pbp2b (Fig. 2A and 2B). Inhibition of specific PBPs was not detected for other β-lactam antibiotics, including cefotaxime and piperacillin (Fig. S4).
Fig. 2.
Inhibition of Pbp2x activity with methicillin leads to elongation of S. pneumoniae cells. (A) Representative SDS-PAGE titration profile of Boc-FL binding to PBPs of strain IU1945 (D39 Δcps parent) treated with the indicated amounts of methicillin for 30 min as described in Experimental procedures. (B) Inhibition of Boc-FL labeling of individual PBPs by different doses of methicillin. Means and standard errors from two independent experiments, including panel A, are shown. (C) Inhibition of growth and lysis of IU1945 at various times after addition of the indicated amounts of methicillin at t = 0 (see Experimental procedures). The experiment was performed 3 times with similar results. (D) Representative images of IU1945 after addition of methicillin at 0.1 µg/mL for 50 min. Cells were stained with FL-V and DAPI and observed by phase-contrast (Phase) or epifluorescence microscopy (FL-V, DAPI) as described in Experimental procedures. Scale bar = 1 µm. The experiment was performed 3 times with similar results. See Fig. S5 for related results.
The dose-response of methicillin binding to Pbp2x was correlated with the inhibition of bacterial growth at ≈0.1 µg/ml methicillin (Fig. 2B and 2C). Since 0.1 µg/ml methicillin primarily inhibited Pbp2x and to a lesser extent Pbp3, we examined cell morphologies before cell lysis. Between 40–60 min after methicillin addition, most cells assumed an enlarged, rod shape that contained multiple rings of FL-V staining and dispersed, often unsegregated nucleoids (Fig. 2 and S5B-S5D). At treatment times >60 min, large, oval-shaped cells appeared in addition to rod-shaped cells as cultures started to lyse (Fig. 2C; data not shown). Cell elongation at this dose of methicillin likely reflects Pbp2x inhibition, because lack of Pbp3 (DacA) causes cells to become spherical without lysing (see Barendt et al., 2011). Cells with pointed, conical ends reported for Pbp2x depletion (Berg et al., 2013) were not observed for methicillin treatment. The formation of large, elongated rod-shaped cells upon inhibition of Pbp2x supports the interpretation that Pbp2x functions primarily in septal PG synthesis that closes septal rings.
Criteria used for strains expressing epitope-tagged division and PG synthesis proteins used in IFM
In this study we used an unencapsulated derivative (IU1945) of encapsulated virulent strain D39, because the presence of capsule promotes chain formation and can mask or change phenotypes caused by pneumococcal division mutants (Barendt et al., 2009). This unencapsulated D39 strain is different from laboratory strain R6, which although originally derived from strain D39, contains numerous mutations that change the PG composition, the essentiality of genes that regulate PG synthesis, and central metabolism compared to its D39 progenitor (Barendt et al., 2009, Carvalho et al., 2013, Land & Winkler, 2011). Rigorous criteria were set for strains containing epitope-tagged division and PG synthesis proteins used for IFM, and numerous constructs were vetted to find ones that meet these criteria. Each tagged fusion protein was expressed from its native locus in the chromosome under normal control and was the sole source of that protein in the cell (Table S1). No cell morphology or division phenotypes or growth defects were detected in strains expressing one or two epitope-tagged proteins (see micrographs throughout and Fig. S6). Western blots probed for the epitope tags showed a single band of each fusion protein and no overt signs of instability or tag separation (Fig. S6). Finally, Boc-FL staining showed nearly wild-type levels for Pbp2x-FLAG3, Pbp1a-FLAG, and Pbp1a-L-FLAG3 (Fig. S6H and S6K.). The one exception was Pbp2x-HA, which labeled to ≈60% of the wild-type level with Boc-FL (Fig. S6K). Nevertheless, strains expressing the Pbp2x-HA construct grew similarly to the wild-type strain (Fig. S6I), and Pbp2x-HA localization in cells was indistinguishable from that of Pbp2x-FLAG3 (data not shown). Numerous specificity controls for IFM were performed and are described in Experimental procedures.
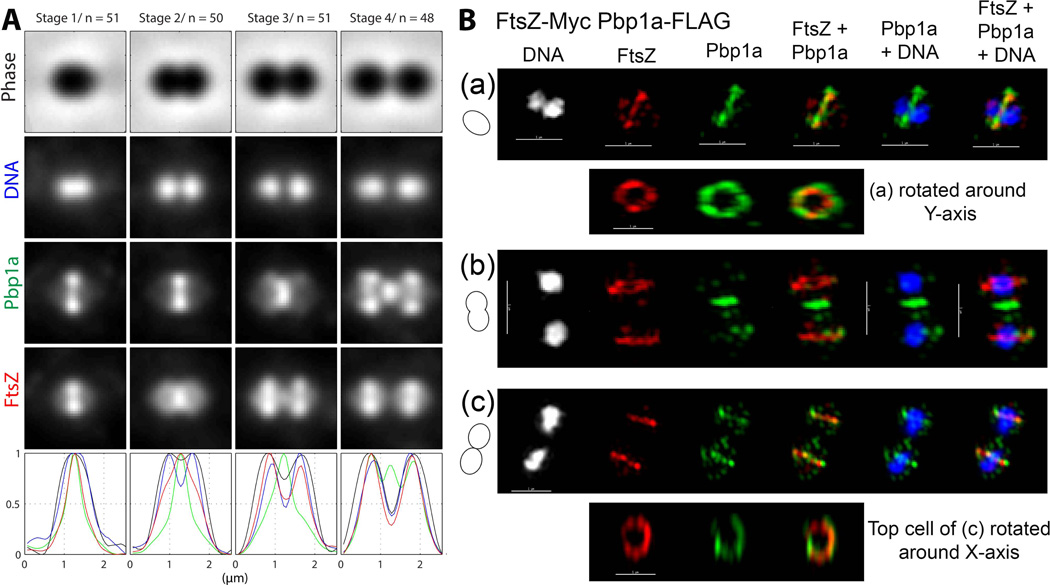
GpsB and FtsZ have different, but overlapping, localization patterns at each stage of cell division
We constructed strain IU6964 that expresses FtsZ-Myc and GpsB-FLAG in the same cell (Table S1). Standard-resolution dual-protein 2D IFM was performed as described in Experimental procedures, including staining nucleoids with DAPI (Fig. S7A-C). The planes of pneumococcal division are perpendicular to the long cell axis (Fig. 3A), and cells in different stages of division can be binned retroactively from phase-contrast images of single and diplococcal cells (Morlot et al., 2003, Tsui et al., 2011). To record and quantitate 2D IFM images, we developed an easy-to-use graphical user interface program (Experimental methods). This program allows rapid manual selection of cells, binning to division stage based on phase contrast images and localization of nucleoids, and averaging of multiple images of cells at each division stage to show shape, nucleoid position, and protein locations (Fig. 3A). The increase in overall cell length of dividing ovococcus pneumococcal cells is not strictly correlated with division stage (Fig. S7D) (see (Wheeler et al., 2011)); hence, manual sorting, rather than automated sorting based on length, was used to bin cells by division stage. This program can be used for bacterial cells of other shapes and is available upon request from the corresponding authors.
Fig. 3.
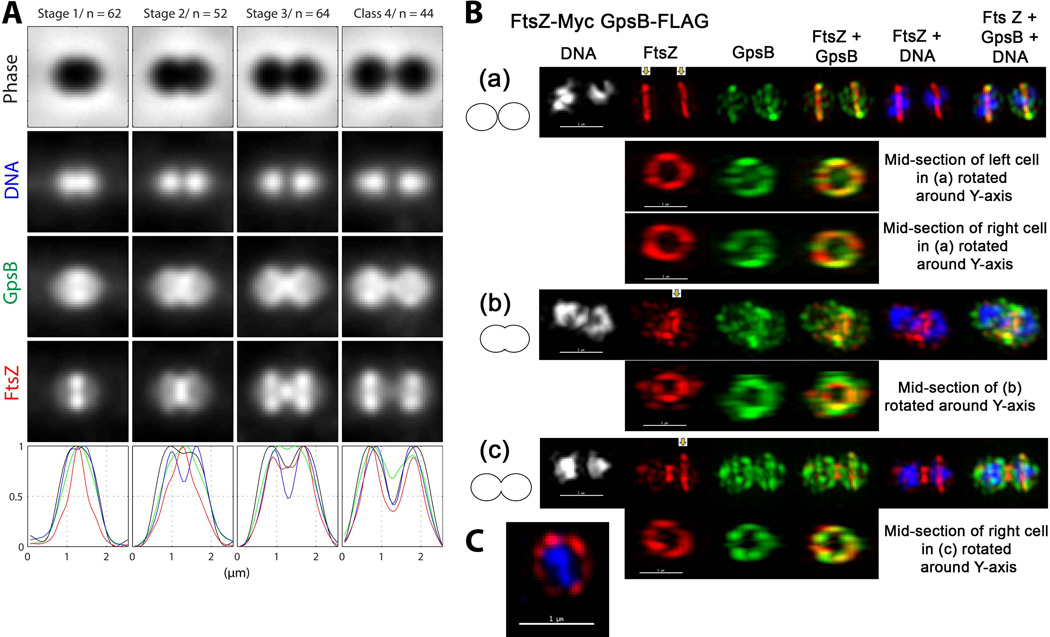
GpsB and FtsZ have different, but overlapping, localization patterns at each stage of cell division. (A) Averaged images and fluorescence intensity traces of strain IU6964 (ftsZ-Myc gpsB-FLAG) grown to mid-exponential phase in BHI broth and processed for dual-protein 2D IFM and DAPI labeling as described in Experimental procedures. Cells were binned into division Stages 1–4, and images from the indicated number of cells (n) from two independent biological replicates were quantified using the graphical user interface program (GUI) described in Experimental procedures. Row 1, cell shapes from phase-contrast images; row 2, nucleoid locations from DAPI labeling; row 3, GpsB locations from IFM; row 4, FtsZ locations from IFM; row 5, normalized average fluorescence intensity distributions along the horizontal cell axis for each channel (black, phase image; blue, DNA; green, GpsB; red, FtsZ). (B) Representative 3D-SIM IFM images of strain IU6964 at different division stages (scale bar = 1 µm). DNA (DAPI stained image) are false-colored white or blue in column(s) 1 or 5 and 6, respectively. FtsZ and GpsB are pseudo-colored as red and green, respectively, and overlapping FtsZ and GpsB signal is represented by yellow. Cartoons at the left of images depict stage of cell division. (a) First row; transverse section of separated pre-divisional cells, corresponding to stage 1 in A. Second and third rows, mid-sections containing FtsZ and GpsB of the left and the right cells were selected (arrows) with the softWorx program and rotated around the Y-axis to show distributions and overlap of FtsZ and GpsB in midcell rings (see Movie S1). (b) First row; mid-divisional cell corresponding to Stage 2 in A with segregating DNA still in the septum of a contracting FtsZ ring. Bottom row; mid-section of the constricting septum (arrow) rotated around the Y-axis. (c) Top row; transverse section of a late-divisional cell corresponding to Stage 4 in A. Bottom row; mid-section of right daughter cell (arrow) rotated around the Y-axis showing that although GpsB is diffusely distributed throughout the daughter cells, a distinct equatorial ring of GpsB has formed where FtsZ is located. Images shown are representative of >30 examined cells in different division stages from two biological replicates. (C) 3D-SIM image of a vertically standing cell expressing FtsZ-Myc (pseudo-colored red) in a non-uniform “bead-like” ring around a nucleoid core (pseudo-colored blue) (scale bar = 1 µm).
As reported in previous studies (Morlot et al., 2003, Tsui et al., 2011), FtsZ localizes to the midcell of pre-divisional cells (Stage 1. Fig. 3A), remains at constricting septa in mid-divisional cells (Stage 2, Fig. 3A), and dynamically relocates to the equators of daughter cells in later-divisional cells (Stages 3 and 4, Fig 3A). In contrast, GpsB follows a different pattern of localization that overlaps that of FtsZ at each stage of cell division (Fig. 3A). GpsB forms a broader band than FtsZ at the midcells of pre-divisional cells, and GpsB is more dispersed than FtsZ throughout cells at this stage (Stage 1, Fig. 3A). Unlike FtsZ, GpsB extends from septa along the inner hemispheres of mid-divisional cells to form a “chi” pattern (Stages 2 and 3, Fig. 3A). In late-divisional cells, GpsB remains at septal regions, while FtsZ has largely relocated to division equators, and GpsB is more dispersed throughout cells in contrast to FtsZ (Stage 4, Fig. 3A).
We performed high-resolution 3D-SIM IFM to gain additional information about the relative localization of GpsB and FtsZ (Fig. 3B). In all pre-divisional cells analyzed that contained unsegregated nucleoids (n = 22), GpsB and FtsZ co-localized as non-uniform rings at midcells (Fig. 3B (a); Movie S1). FtsZ was concentrated to these rings, whereas GpsB was also dispersed to the facing hemispheres formed in the previous division. In mid-divisional cells, both FtsZ and GpsB remained in non-uniform septal rings, but FtsZ and GpsB had started to migrate to the regions between the future equators of the daughter cells (Fig. 3B (b)). In late-divisional cells, FtsZ remained at septa and had started to re-organize at equators (arrow, Fig. 3B (c)). In contrast, GpsB was at the septum, co-localized as a non-uniform ring with FtsZ at equators (arrow), and was also dispersed throughout both daughter cells (Fig. 3B (c)), consistent with the averaged 2D images (Stage 4, Fig. 3A). Two observations indicate that the GpsB localization pattern is unlikely an artifact of the IFM approach (Fig. 3A). The same GpsB localization pattern (Fig. 3A) was obtained for cells containing the GpsB-FLAG fusion alone or containing a different GpsB-Myc fusion (strain IU7199, Table S1; data not shown). In addition, the GpsB localization pattern was non-random, and no degradation or tag loss was detected for the GpsB-FLAG or GpsB-Myc fusions (Fig. S6B; data not shown). We conclude that GpsB localization in S. pneumoniae is dynamic and coincident with localization of FtsZ, although GpsB can also be found in intracellular regions not occupied by FtsZ.
Separate positions of Pbp2x and Pbp1a in constricting septa of wild-type S. pneumoniae cells
To better understand GpsB function, we wanted to localize Pbp2x and Pbp1a in pneumococcal cells depleted for GpsB (Fig. 1). We focused on Pbp2x because of its essential role in septum closure (Fig. 2) and Pbp1a, because it is the homologue of the B. subtilis PBP1, which is thought to be shuttled by GpsB (Claessen et al., 2008). Before performing these studies, we needed to determine the localization patterns of Pbp2x and Pbp1a in wild-type pneumococcal cells relative to FtsZ and to each other.
To perform these experiments, we constructed strains IU6978, which expresses FtsZ-Myc and Pbp2x-FLAG3, and IU6976, which expresses FtsZ-Myc and Pbp1a-FLAG (Tables S1). Pbp2x and Pbp1a followed similar overall patterns of localization (Fig. 4 and 5) that are different from those of FtsZ and GpsB (Fig. 3). Based on 2D IFM, Pbp2x and Pbp1a located with FtsZ at the midcell of pre-divisional cells (Stage 1, Fig. 4A and 5A). In mid-divisional cells, Pbp2x and Pbp1a remained at division septa even after most FtsZ appeared to migrate to equators of daughter cells (Stage 3, Fig. 4A and 5A). In late-divisional cells, Pbp2x and Pbp1a remained at a region between the newly divided cells, corresponding to a highly constricted septum or new poles, while FtsZ had moved completely to equators (Stage 4, Fig. 4A and 5A). This apparent co-localization of Pbp2x and Pbp1a separate from FtsZ confirms IFM results reported previously (Morlot et al., 2003).
Fig. 4.
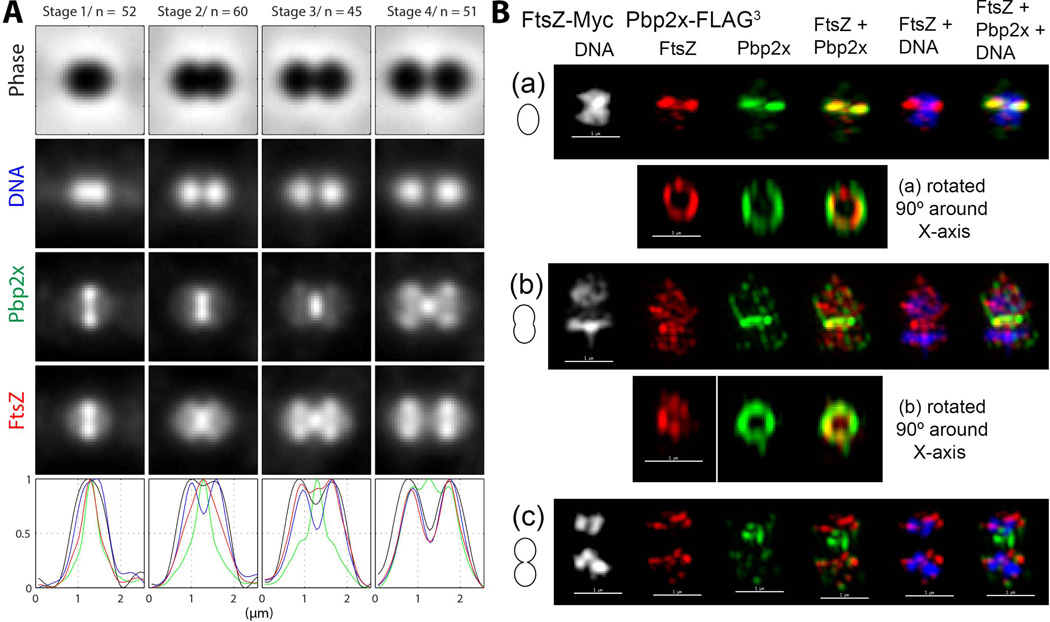
Pbp2x remains at division septa after most FtsZ has migrated to equators of daughter cells. (A) Averaged images and fluorescence intensity traces of strain IU6978 (ftsZ-Myc pbp2x-FLAG3) grown to mid-exponential phase in BHI broth and processed for dual-protein 2D IFM and DAPI labeling (see Experimental procedures). Cells from two independent biological replicates were sorted into division stages and images were averaged and intensity traces graphed as described for Figure 3A and in Experimental procedures, except that row 3 and green color correspond to Pbp2x. (B) Representative 3D-SIM IFM and DAPI images of strain IU6978 at different division stages (scale bar = 1 µm). Pseudo-coloring is similar to Figure 3B, except green corresponds to Pbp2x. Cartoons at the left of images again depict stage of cell division. (a) First row; transverse section of a pre-divisional cell corresponding to Stage 1 in A showing co-localization of FtsZ and Pbp2x. Second row; a section from the midcell containing FtsZ and Pbp2x was selected and rotated around the X-axis to show that the FtsZ ring is smaller and concentric with the Pbp2x ring (see text and Movie S2). (b) First, row; mid-divisional cells corresponding to Stage 3 in A. Second row, a section from the midcell region containing the Pbp2x ring was selected and rotated around the X-axis to show a nearly constricted FtsZ ring. (c) Traverse section of a late-divisional cell corresponding to Stage 4 in A showing that Pbp2x remains at septa after FtsZ has migrated to equators of daughter cells. Images shown are representative of >30 examined cells in different division stages from two biological replicates.
Fig. 5.
Pbp1a localization resembles that of Pbp2x and remains at division septa after most FtsZ has migrated to equators of daughter cells. (A) Averaged images and fluorescence intensity traces of strain IU6976 (ftsZ-Myc pbp1a-FLAG) grown to mid-exponential phase in BHI broth and processed for dual-protein 2D IFM and DAPI labeling (see Experimental procedures). Cells from two independent biological replicates were binned into division stages and images were averaged and intensity traces graphed as described for Figure 3A and in Experimental procedures, except that row 3 and green color correspond to Pbp1a. (B) Representative 3D-SIM IFM and DAPI images of strain IU6976 at different division stages (scale bar = 1 µm). Pseudo-coloring is similar to Figure 3B, except green corresponds to Pbp1a. Cartoons at the left of images again depict stage of cell division. (a) First row; transverse section of a pre-divisional cell corresponding to Stage 1 in A showing co-localization of FtsZ and Pbp1a. Second row; a section in the midcell containing FtsZ and Pbp1a was selected and rotated around the Y-axis to show that Pbp1a, like Pbp2x (Fig. 4B (a)), forms a larger concentric ring around the FtsZ ring (see text and Movie S3). (b) Traverse section of late-divisional cell corresponding to Stage 4 in A showing that Pbp1a, like Pbp2x (Fig. 4B(c), remains at septa after FtsZ has migrated to equators of daughter cells. Initial migration of Pbp1a to equators can also be seen. (c) First row, transverse section of newly divided cells corresponding to Stage 1 in A showing co-localization of FtsZ and most of Pbp1a at equators of daughter cells. Second row, section of the midcell of upper daughter cell rotated around the X-axis. Images shown are representative of >30 examined cells at various division stages obtained from two biological replicates.
3D-SIM IFM extended the results of the 2D IFM (Fig. 4B and 5B). In pre-divisional cells, FtsZ formed a concentric smaller ring than Pbp2x and Pbp1a (see Movies S2 and S3), consistent with the intracellular and extracellular locations of FtsZ and the tagged functional domains of Pbp2x and Pbp1a, respectively (Egan & Vollmer, 2013, Massidda et al., 2013, Typas et al., 2012). A smaller diameter of FtsZ rings compared to Pbp2x and Pbp1a rings was indicated in the averaged 2D images (Stage 1, Fig. 4A and 5A), further validating this conclusion. 3D-SIM IFM images showed conclusively that Pbp2x and Pbp1a remain largely at the septa of mid-to-late divisional cells in which FtsZ has migrated to the equators of the daughter cells (Fig. 4B (b) and (c); Fig. 5B (b)). At a late stage in division, Pbp2x and Pbp1a join FtsZ at equators (Fig. 4B (c) and Fig. 5B (c)).
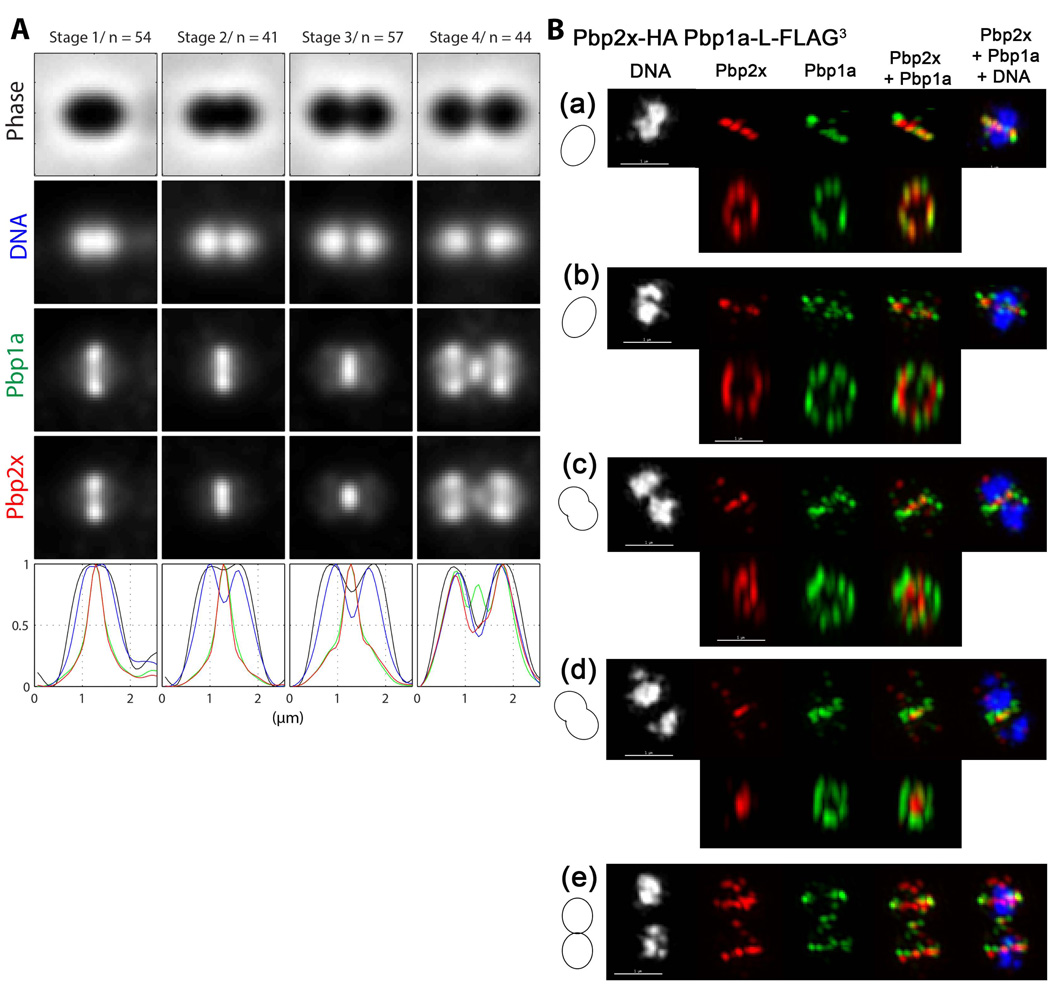
Although the pattern of Pbp2x and Pbp1a localization is the same at different stages of pneumococcal cell division (Fig. 4 and 5) (Morlot et al., 2003), their positions in division septa are separate (Fig. 6). We constructed strain IU7365, which expresses Pbp2x-HA and Pbp1a-L-FLAG3 in the same cell (Table S1). Analysis of 2D IFM images confirmed that Pbp2x and Pbp1a follow the same overall localization patterns (Fig. 6A). The diameters of the Pbp1a and Pbp2x rings were similar in pre-divisional cells (Stage 1, Fig. 6A). However, in mid-divisional cells, the average diameter of Pbp1a rings was larger than that of Pbp2x rings (Stages 2 and 3, Fig 6A). Separate positioning of Pbp1a and Pbp2x in septa was verified in 3D-SIM IFM images (Fig. 6B). In pre-divisional cells, Pbp1a and Pbp2x co-localized to the same midcell ring (Fig. 6B (a)). At later division stages, Pbp2x contracted within septa, whereas Pbp1a remained in larger rings, to the point that in mid-to-late divisional cells, Pbp2x formed a solid disk at the center of septa surrounded by a distinct ring of Pbp1a (Fig. 6B (d); Movie S4). We conclude from both the 2D and 3D-SIM results that Pbp1a and Pbp2x occupy different positions in constricting division septa.
Fig. 6.
Separate positioning of Pbp1a and Pbp2x in septa of dividing cells. (A) Averaged images and fluorescence intensity traces of strain IU7365 (pbp2x-HA pbp1a-FLAG) grown to mid-exponential phase in BHI broth and processed for dual-protein 2D IFM and DAPI labeling (see Experimental procedures). Cells from two independent biological replicates were binned into division stages and images were averaged and intensity traces graphed as described for Figure 3A and in Experimental procedures, except that row 3 (green color) and row 4 (red color) now correspond to Pbp1a and Pbp2x, respectively. Average ring diameters viewed from traverse images indicate that the Pbp1a ring is larger than the Pbp2x ring in cells at division Stages 2 and 3 (see text). (B) Representative 3D-SIM IFM and DAPI images of strain IU7365 at different division stages (scale bar = 1 µm). Pseudo-coloring is similar to Figure 3B, except red and green correspond to Pbp2x and Pbp1a, respectively. Cartoons at the left of images again depict stage of cell division. Upper rows in (a)-(d) and row (e) show traverse sections, whereas lower rows in (a)-(d) show midsections from the upper images rotated 90° to show ring organization. Cells in (a)-(d) progress through the stages in cell division corresponding to A. In pre-divisional cells (a), Pbp2x and Pbp1a co-localize, whereas in mid-divisional cells, Pbp2x condenses in the middle of the septum and eventually forms a disk surrounded by a distinct ring of Pbp1a (d) (see Movie S4). In newly separated cells (e), Pbp 2x and Pbp1a again co-localize at equators of daughter cells. Images shown are representative of >30 examined cells in different division stages from two biological replicates.
Non-uniformity of FtsZ and division rings and lack of septal occlusion of nucleoids in wild-type S. pneumoniae cells
This first 3D-SIM IFM analysis of pneumococcal division proteins revealed fundamental new information about the localization patterns of FtsZ and nucleoids in wild-type pneumococcal cells. In pre-divisional and late-divisional cells, rings of FtsZ and other division proteins appeared non-uniform with some regions of discontinuities (Fig. 3B (a) and (c); Fig. 4B (a); Fig. 5B (a) and (c)). These pneumococcal FtsZ structures closely resemble the heterogeneous, bead-like FtsZ rings reported recently in 3D-SIM IFM and FtsZ-GFP images of B. subtilis and S. aureus cells (Strauss et al., 2012). In 3D-SIM, there is a difference between lateral (XY) and axial (Z) resolution that can lead to an appearance of heterogeneity (see (Strauss et al., 2012)). To examine whether this resolution distortion could account for the apparent non-uniformity of pneumococcal FtsZ rings, we compared 5 pairs of FtsZ rings from separated cells that were each captured in images where the optical path was perpendicular to the long axis of the cells (e.g., see Fig. 3B (a); data not shown). We reasoned that if FtsZ ring non-uniformity was caused solely by resolution distortion, then pairs of FtsZ rings from the same images would show similar patterns of non-uniformity. This was not the case for pairs of FtsZ rings or for rings of other division proteins (e.g., see Fig. 3B (a)). In addition, we searched 3D-SIM images for rare cells that were “standing on their heads,” so that we could view the FtsZ ring with uniform resolution in the XY plane. The vertically standing cell in Figure 3C has a non-uniform FtsZ ring containing an odd number of asymmetrically placed “beads.” We conclude that pneumococcal FtsZ and other division proteins form non-uniform rings during division.
Another remarkable feature shown by these 3D-SIM images is the lack of nucleoid occlusion during septation of wild-type pneumococcal cells. In pre-divisional cells, FtsZ rings surround nucleoids to form Saturn-like structures (Fig. 3B (a); Fig. 4B (a); Fig. 5B (a) and (c)). Of >70 pre-divisional cells examined in different strains by 3D-SIM, all contained FtsZ rings surrounding nucleoids that had just started to separate (Fig. 3–5; data not shown). In 42 cells at later division stages that were examined in these strains by 3D-SIM, ≈40% contained segregating nucleoids that were still connected, but extended through constricting FtsZ rings (e.g., see Fig. 3B (b); data not shown). The remaining 60% of cells from later divisional stages contained fully separated nucleoids. Together, these results confirm the lack of nucleoid occlusion in pre-divisional cells and show that complete separation of nucleoids occurs as a later step in pneumococcal cell division concurrent with the start of FtsZ ring constriction.
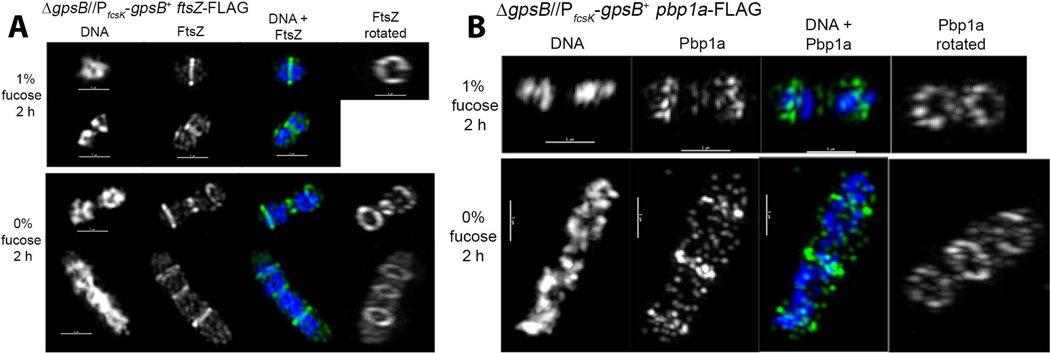
Elongated cells depleted of GpsB contain multiple non-constricted rings of FtsZ and other division proteins
GpsB depletion led to the formation of elongated cells containing multiple bands stained with FL-V (Fig. 1C and S2C). Since FL-V indicates regions of PG pentapeptide concentration associated with active PG synthesis (Daniel & Errington, 2003, Ng et al., 2004, Pinho & Errington, 2005, Wheeler et al., 2011), we anticipated that FtsZ would be present in multiple bands in GpsB-depleted cells. To test this idea, we moved the FtsZ-FLAG or FtsZ-Myc fusion into the IU4888 GpsB depletion strain to give strains IU6944 and IU6946, respectively (Table S1). In BHI medium containing fucose, IU6944 and IU6946 cells appeared similar to wild-type cells (Fig. 7A; data not shown). GpsB depletion caused the same cessation of growth, formation of elongated, enlarged cells, and eventual lysis as for strain IU4888 (Fig. 7A and S8; data not shown). After 2 h of GpsB depletion, 3 FtsZ rings were often observed in the elongated cells (bottom rows, Fig. 7A; middle rows, S8B), and similar to the FL-V bands, the number of FtsZ rings increased to >3 in many cells after 3 h of GpsB depletion (arrowheads, Fig. S8). After 2 h of GpsB depletion, partially constricted FtsZ rings were sometimes observed in 3D-SIM images (e.g., row 3, Fig. 7A). These partially constricted rings may reflect residual GpsB amounts in some cells. However, most FtsZ rings appeared to be unconstricted and surrounded diffuse, unsegregated nucleoids (Fig. 7A and S8). The increase in the number of FtsZ and FL-V rings with time of GpsB depletion suggests that non-constricted rings continued to form and were not simply carried over from before the start of GpsB depletion.
Fig. 7.
Presence of multiple FtsZ and PBp1a rings in elongated cells depleted of GpsB. Strains IU6944 (ΔgpsB//PfcsK-gpsB+ ftsZ-FLAG) (A) and IU5980 (ΔgpsB//PfcsK-gpsB+ pbp1a-FLAG) (B) were depleted for GpsB for 2 h as described for Fig. 1 and in Experimental procedures. DAPI staining and 3D-SIM IFM of cells expressing GpsB (1% fucose) and cells depleted of GpsB (0% fucose) were performed as described in Experimental procedures. (A) Representative FtsZ localization upon GpsB depletion. DNA is pseudo-colored white or blue in column 1 or 3, respectively. FtsZ is pseudo-colored white or green in columns 2 and 4 or 3, respectively. Images are rotated in column 4 to display FtsZ rings (see Movie S5). (B) Representative Pbp1a localization upon GpsB depletion. The color scheme is as in A, except Pbp1a is now pseudo-colored white or green in column(s) 2 and 4 or 3, respectively (see Movie S6). Scale bar = 1 µm. Images shown are representative of 8 cells examined by 3D-SIM and hundreds of cells observed by 2D IFM (see Fig. S8 and S10).
In addition, we depleted GpsB in strains containing Pbp2x-FLAG3 (IU6980), Pbp1a-FLAG (IU5980), and MreC-L-FLAG3 (IU4976) fusions (Table S1). MreC was included in this study because it has been implicated in peripheral PG synthesis in S. pneumoniae (Land & Winkler, 2011), and we predicted that rings of MreC would underlie the elongation and enlargement of cells depleted for GpsB. Multiple non-constricted rings of Pbp2x, Pbp1a, and MreC were present at locations similar to those of FtsZ rings in enlarged, elongated cells depleted of GpsB for 2 or 3 h (Fig. 7B, and S9-S11). As for the FtsZ and FL-V rings, the number of Pbp2x, Pbp1a, and MreC rings per cell increased with time of GpsB depletion (arrowheads, Fig. S9-S11), consistent with new ring formation. We conclude that GpsB depletion blocks the closure of FtsZ and other division-protein rings in S. pneumoniae.
Discussion
In this study, we demonstrate major differences between GpsB of ovococcus S. pneumoniae and its homologue in rod-shaped B. subtilis, with respect to essentiality, localization, function, and order of appearance during cell division. First, B. subtilis ΔgpsB ΔezrA mutants are synthetically sick; but ΔgpsB mutants exhibit aberrant cell morphology only under high-salt stress conditions (Claessen et al., 2008). In addition, ΔgpsB mutants of B. subtilis show a synthetic lethal phenotype when early division protein FtsA is depleted (Tavares et al., 2008). In contrast, GpsB is essential for normal growth of serotype 2-derived strains of S. pneumoniae, and depletion of GpsB causes cessation of growth, substantial cell elongation and enlargement, and eventual lysis (Fig. 1 and S1-S3; Results). Consistent with this finding, the essentiality of pneumococcal gpsB was indicated by its non-recovery in a Tn-seq screen (van Opijnen et al., 2009). Second, GpsB localizes at both the lateral walls and the divisional septa of B. subtilis cells (Claessen et al., 2008), and GpsB is among a second group of division proteins that assemble after ZapA and EzrA at FtsZ-rings in B. subtilis (Gamba et al., 2009, Tavares et al., 2008). In contrast, GpsB localization in S. pneumoniae is dynamic and coincides with localization of FtsZ at different division stages; but, GpsB localizes in a different pattern from FtsZ and can be found at locations not occupied by FtsZ (Fig. 3). Third, GpsB and EzrA were postulated to play roles in the localization cycle of PBP1 in B. subtilis (Claessen et al., 2008). In contrast, pneumococcal GpsB is not required for the formation of rings of Pbp1a, which is the PBP1 homologue, FtsZ, Pbp2x, or MreC in cells depleted of GpsB (Fig. 7 and S8-S11). Taken together, these results suggest that GpsB homologues play different roles in ovococci compared to rod-shaped Gram-positive bacteria.
The striking formation of elongated, enlarged, aseptate cells that occurs upon depletion of pneumococcal GpsB is indicative of a defect in septal PG synthesis. We showed that an analogous elongation occurs when S. pneumoniae was treated with a limited amount of methicillin to specifically inhibit Pbp2x activity (Fig. 2 and S5). Methicillin has been shown to inhibit septum formation in several ovococcus species (Lleo et al., 1990, Perez-Nunez et al., 2011, Raz et al., 2012), and resistance was used to link Pbp2x activity with methicillin-induced filamentation of L. lactis (Perez-Nunez et al (2011)). The elongation caused by Pbp2x inhibition supports an important tenet of the model that pneumococcal PG synthesis is carried out by a septal machine that contains Pbp2x and a peripheral machine, which likely contains Pbp2b and MreCD, that continues to function out-of-balance when the septal machine is inhibited (Massidda et al., 2013, Sham et al., 2012, Zapun et al., 2008). Re-balancing between two PG synthesis machines has been suggested by phenotypes of new mutants in S. pneumoniae. For example, abnormal cell shapes caused by resistance-conferring mutations in pbp2b were corrected by compensatory mutations in pbp2x and pbp1a (Albarracin Orio et al., 2011).
A recent paper reported that depletion of Pbp2x in laboratory strain R6 results in the formation of enlarged, sometimes elongated, pointed cells (Berg et al., 2013) that are shaped differently from the elongated cells caused by methicillin treatment (Fig. 2 and S5). We have confirmed the pointed-cell phenotype for depletion of Pbp2x in D39-derived strains (data not shown). This notable difference in appearance suggests that inhibition of Pbp2x activity inhibits septal ring closure, whereas the absence of Pbp2x may result in a more severe disruption of the division apparatus. The multiple, normal-looking FtsZ, Pbp2x, Pbp1a, and MreC rings in elongated, aseptate cells depleted for GpsB suggests that GpsB functions downstream of ring formation. We postulate that GpsB plays a direct role in the control of FtsZ ring closure in pre-divisional cells, when FtsZ and GpsB co-localize at equators that will become septa (Fig. 3B (a)). At this stage, GpsB may act as a critical connector between the constricting FtsZ ring and other components involved in septal ring closure, such as Pbp2x, StkP/PhpP (Beilharz et al., 2012, Fleurie et al., 2012, Morlot et al., 2013), SepF (YlmF, (Fadda et al., 2003, Hamoen et al., 2006)), FtsL (Kawai & Ogasawara, 2006, Noirclerc-Savoye et al., 2005), and DivIC (Strauss et al., 2012). GpsB could also be required continuously during later stages of septal ring closure. Additional information about the interactions and functions of GpsB in S. pneumoniae is needed to test these hypotheses.
Evidence for separable PG synthesis machines was provided by 2D- and 3D-SIM IFM of Pbp2x and Pbp1a in wild-type cells of S. pneumoniae (Fig. 6). Pbp2x mediates septal PG synthesis (see above), whereas Pbp1a has been strongly implicated in peripheral PG synthesis, although a second role in septal PG synthesis was not ruled out (Land & Winkler, 2011). Paradoxical to these separate functions, Pbp2x and Pbp1a follow the same overall pattern of localization at different stages of pneumococcal cell division (Fig. 3 and 4) (Morlot et al., 2003, Zapun et al., 2008). Pbp2x and Pbp1a localize as concentric rings with FtsZ and GpsB at equators of pre-divisional cells (Fig. 3-6). Remarkably, as nucleoid separation and septal constriction commence, Pbp1a increasingly surrounds Pbp2x until Pbp1a forms a ring around a disk of Pbp2x in the center of septa of mid-to-late divisional cells (Stage 3, Fig. 6A; Fig. 6B (d)). At these later stages of division, FtsZ and GpsB have disassembled from septa and started to form rings at equators of daughter dells (Fig. 3 and 4). Thus, Pbp2x and Pbp1a are at separate locations in mature septa, consistent with their functions in different PG machines. Nevertheless, the juxtaposition of Pbp2x and Pbp1a in septal structures (Fig. 6B) could account for antibiotic resistance phenotypes that suggest interactions (Zerfass et al., 2009).
Some Gram-positive bacteria, like B. subtilis, synthesize a complete wall between daughter cells before the start of cell separation, whereas Gram-negative bacteria use a constrictive mode of cell division, in which septation and cell separation occur simultaneously (reviewed in (Egan & Vollmer, 2013)). Results from 3D-SIM IFM in this study show that FtsZ, Pbp2x, and Pbp1a ring closure, separation of nucleoids, and cell constriction at septa are concurrent during pneumococcal cell division (Fig. 3–5). Therefore, S. pneumoniae follows a constrictive, rather than a separated mode, of cell division. Constrictive division is consistent with results from a previous study in which 3D-SIM was used to determine the dimensions of dividing pneumococcal cells labeled with FL-V (Wheeler et al., 2011). This study concluded that in S. pneumoniae the length of peripheral PG synthesis is extremely short (≈ 300 nm) before the start of measurable septal constriction, especially compared to other ovococcus species, which have an extended period of peripheral elongation before septal constriction begins (Wheeler et al., 2011). Our results indicate that the PG synthesis machine marked by Pbp2x rings is present in all pre-divisional cells observed (Fig. 4), indicating that septal PG synthesis could even begin simultaneously with peripheral PG synthesis, before physical constriction can be detected.
Lack of nucleoid occlusion in pre-divisional cells is particularly apparent in 3D-SIM IFM images of wild-type S. pneumoniae cells (Fig. 3–5). In addition, nucleoids are not completely separated in some cells containing constricting FtsZ rings (see Results), indicating that completion of nucleoid separation occurs after the start of FtsZ ring constriction in S. pneumoniae. Lack of nucleoid occlusion was noted as likely in previous 2D images of pneumococcal cells (Morlot et al., 2003). Unsegregated nucleoids extending through rings of division proteins were also observed in aseptate, elongated cells caused by inhibition of Pbp2x with methicillin or by GpsB depletion (Figs. 1, 2, and 7) and previously in some pbp2b or stkP mutants (Albarracin Orio et al., 2011, Fleurie et al., 2012). The Min system is not present in S. pneumoniae, and the mechanism of pneumococcal FtsZ placement and chromosome segregation are largely unknown. The pneumococcal ParB protein is a homologue of the Noc protein that mediates nucleoid occlusion in B. subtilis (Wu & Errington, 2012). However, pneumococcal ΔparB mutants are viable and accumulate only a low number (<5%) of anucleate cells (Minnen et al., 2011). Likewise, pneumococcal Δsmc mutants, which lack a nucleoid condensin homologue, and double Δsmc ΔparB mutants show mild nucleoid segregation defects (Minnen et al., 2011). Another candidate that likely plays an important role in nucleoid segregation in S. pneumoniae is FtsK (SpoIIIE), which is a divisome component that belongs to a super-family of DNA pumps (see (Wang & Lutkenhaus, 1998, Demarre et al., 2013)). The pneumococcal ftsK (spoIIIE) gene (sp0878) is essential in serotype 4 strain TIGR4 (van Opijnen et al., 2009), but the role of FtsK (SpoIIIE) in pneumococcal nucleoid separation remains to be determined (Massidda et al., 2013, Pinho et al., 2013).
Finally, the appearance of FtsZ rings in S. pneumoniae in 3D-SIM IFM images are remarkably similar to those reported recently for FtsZ rings in B. subtilis and S. aureus (Strauss et al., 2012). Notably, FtsZ rings in pre-divisional pneumococcal cells are non-uniform (Fig. 3–5), and this feature extends to rings of other pneumococcal division proteins that associate with FtsZ rings, including GpsB, Pbp2x, and Pbp1a (Fig. 3–5) (Strauss et al., 2012). Vertically standing pneumococcal cells display bead-like rings of FtsZ surrounding nucleoids (Fig. 3C). Additional control experiments indicated that the apparent non-uniformity of pneumococcal FtsZ rings could not be accounted for solely by differences in resolution distortion in the 3D-SIM technique (see Results). Cryotomographic images of Caulobacter crescentus cells showed sparse, short, non-overlapping FtsZ protofilaments that do not form a continuous ring structure at midcells (Li et al., 2007). Protofilament distributions have been postulated to account for the non-uniform “bead-like” distribution of FtsZ in B. subtilis and S. aureus (Fu et al., 2010, Strauss et al., 2012). The results presented here suggest that this type of mechanism operates for FtsZ organization in S. pneumoniae D39 cells as well.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains used in this study were derived from strain IU1945, an unencapsulated derivative of serotype 2 S. pneumoniae strain D39 (Lanie et al., 2007) (Table S1). Strains containing antibiotic markers were constructed by transforming linear DNA amplicons synthesized by overlapping fusion PCR into competent pneumococcal cells as described previously (Tsui et al., 2010). Primers used for the generation of amplicons are listed in Table S2. All constructs were confirmed by DNA sequencing of chromosomal regions corresponding to the amplicon region used for transformation. Bacteria were grown on plates containing trypticase soy agar II (modified; Becton-Dickinson) and 5% (vol/vol) defibrinated sheep blood (TSAII-BA). Plates were incubated at 37°C in an atmosphere of 5% CO2. For antibiotic selections, TSAII BA plates contained 250 µg kanamycin per ml, 150 µg spectinomycin per ml, 0.3 µg erythromycin per ml, or 250 µg streptomycin per ml. Strains were cultured statically in Becton-Dickinson brain heart infusion (BHI) broth at 37°C in an atmosphere of 5% CO2, and growth was monitored by OD620 using a Spectronic 20 spectrophotometer fitted for measurement of capped tubes (outer diameter, 16 mm). Bacteria were inoculated into BHI broth from frozen cultures or colonies, serially diluted into the same medium, and propagated overnight. For most growth experiments, overnight cultures that were still in exponential phase (OD620 = 0.1 to 0.4) were diluted back to OD620 ≈ 0.002 to start final cultures, which lacked antibiotics.
Transformations of fusion ΔgpsB<>aad9 amplicon into IU4846 (gpsB+//PfcsK-gpsB+) were carried out in the presence of 0.5% (wt/vol) L-fucose (Sigma, F2252 or Arcos, 225880250) in the transformation mix and 0.8% (wt/vol) L-fucose in all subsequent steps. For the transformation of ftsZ-FLAG, ftsZ-Myc, pbp2x-FLAG3, pbp1a-FLAG, and mreC-L-FLAG3 amplicons into IU4888 (ΔgpsB//PfcsK-gpsB+), all media contained 1.0% (wt/vol) L-fucose.
To deplete cells of GpsB, strain IU4888 (ΔgpsB//PfcsK-gpsB+) was first grown overnight in BHI broth containing 0.8 or 1.0% (wt/vol) L-fucose. In one depletion protocol (see Fig. 1 and S1), overnight cultures were diluted to OD620 ≈ 0.005 in BHI broth containing 0.8 to 1% (wt/vol) L-fucose, and cultures were grown to OD620 ≈ 0.1. Cells were collected by centrifugation (5 min at 16,000 × g at 25 °C), washed once in BHI broth lacking L-fucose, and suspended to OD620 ≈ 0.03 in BHI broth lacking or containing 0.8% or 1.0% (wt/vol) L-fucose. Growth was monitored by OD620, and microscopy was performed at the times after resuspension indicated in the figures. In a second protocol (see Fig. S2), cells from overnight cultures were collected by centrifugation (10 min at 16,000 × g at 25°C), washed once in BHI broth lacking L-fucose, and suspended to OD620 ≈ 0.005 in BHI broth containing 0.1% to 1.0% (wt/vol) L-fucose or no fucose.
Western blotting
Strains were grown exponentially in BHI broth to OD620 ≈ 0.13. Lysates were prepared as described previously (Wayne et al., 2012). FLAG-tagged proteins or Pbp2x-HA were detected by Western blotting using a 1:1000 dilution of primary anti-FLAG polyclonal antibody (Sigma, F7425), or anti-HA rabbit polyclonal antibody (Invitrogen, 71-5500) at 1 µg/ml, and a 1:10,000 dilution of HRP-conjugated donkey anti-rabbit antibody (GE Healthcare, NA934), followed by ECL detection reagent. Western blots of FtsZ-Myc were performed with a 1:1000 dilution of primary anti-Myc monoclonal antibody (Life Technologies, R950) and a 1:10,000 dilution of HRP-conjugated donkey anti-mouse antibody (Thermal Scientific, SA1-100). Chemiluminescent signal in protein bands was detected and quantitated by using an IVIS imaging system as described in (Wayne et al., 2010).
Epifluorescence microscopy of DAPI and FL-V stained cells and cell length and width measurement
50 µL samples were taken from exponentially growing BHI broth cultures (OD620 ≈0.15), methicillin-treated cultures, or from cultures of cells depleted of GpsB for indicated times after resuspension in BHI broth lacking fucose. Cells were stained without fixation by adding 4,6-diamidino-2-phenylindole (DAPI), and a 1:1 mixture of vancomycin and Bodipy-FL-conjugated vancomycin (FL-V) (Molecular Probes) to final concentrations of 0.4 µg per ml and 2 µg per ml, respectively, as described previously (Barendt et al., 2009). Cells were viewed using a wide-field epifluorescence microscope, and images were recorded as described before (Barendt et al., 2009, Tsui et al., 2011). In control experiments, cells were examined without staining to ensure that morphology defects observed were not caused by the concentrations of vancomycin used for staining (data not shown). The lengths and widths of separated, pre-divisional (Stage 1, see below) cells were determined for parent strain IU1945 or strain IU4888 growing in BHI containing fucose and compared with the dimensions of separated cells of IU4888 depleted of GpsB for indicated times or IU1945 treated with methicillin (see below). Cell lengths and widths were measured from phase-contrast images by using Nikon NIS-Element AR software as described before (Barendt et al., 2009).
Growth inhibition of cultures and Boc-FL labeling of PBPs in cells following β-lactam antibiotic treatments
Growth inhibition of cultures after antibiotic treatments was monitored by OD620. Cell dimensions were determined from phase-contrast images as described above for cultures of strain IU1945 treated with 0.1 µg methicillin per ml for times indicated in the figures. Binding of β-lactam antibiotics to cellular PBPs was determined by secondary labeling with fluorescent bocillin (Boc-FL) (Molecular Probes), which labels all pneumococcal PBPs (Kocaoglu et al., 2012, Zhao et al., 1999). The following final concentrations of antibiotics were added to 5 ml cultures of parent strain IU1945 growing exponentially (OD620 ≈ 0.09) in BHI broth: methicillin (Sigma), 0.01 to 1.0 µg per ml; cefotaxime (Sigma), 0.001 to 1.0 µg per ml; or piperacillin (Sigma), 0.001 to 1.0 µg per ml. After 30 min incubation at 37°C, 1.5 ml samples were centrifuged (16,100 × g for 2 min at 25°C), and pellets were washed with 1.0 ml of phosphate buffered saline (PBS, pH 7.4) and suspended in 50 µl of PBS containing 5 µg Boc-FL per ml diluted from a stock of 100 µg Boc-FL per ml in DMSO. After 20 min at 25°C, cells were collected by centrifugation, washed with 1.0 ml of PBS, and suspended in 100 µl of PBS containing 1 mg lysozyme per ml. After 30 min at 37°C, cells were further lysed by sonication (Branson Sonifier 250; power setting 3; 30% duty cycle for 4 × 6 s intervals). Membrane proteomes were isolated by centrifugation (16,100 × g for 20 min at 4°C). Pellets were suspended in 100 µl of PBS and homogenized by brief sonication (power setting 1; 10% duty cycle for ≈1 sec). Protein concentrations were estimated by OD280 using a NanoDrop 1000 Spectrophotometer, and samples were diluted to 2 mg protein per ml in PBS. 10 µL of 4 × SDS-PAGE loading buffer was added to 30 µL of each proteome sample, and samples were heated for 5 min at 95°C and cooled to 25°C. 10 µL of samples were separated by 10% SDS-PAGE, and fluorescently labeled proteins were visualized in gels by using a Typhoon 9210 gel scanner (Amersham Biosciences) with a 526 nm short-pass filter. Integrated density values were obtained from scans by using ImageJ software (NIH) for gel band quantitation, and Boc-FL labeling of each PBP in antibiotic-treated samples were expressed relative to samples not treated with antibiotics. Similar results were obtained for methicillin, cefotaxime, or piperacillin treatments of cells resuspended in PBS at 25°C instead of in BHI broth at 37°C (data not shown).
2D IFM
Localization of FLAG-, Myc- and HA-tagged proteins by IFM was performed on exponentially growing cells as described before (Tsui et al., 2011). For-single-protein labeling experiments, primary antibodies used were anti-FLAG rabbit polyclonal antibody (Sigma, F7425, 1:200 dilution for ftsZ-FLAG strains and 1:100 for other strains), anti-c-Myc rabbit polyclonal antibody (Sigma, C3956, 1:100 dilution), or anti-HA rabbit polyclonal antibody (Invitrogen, 71-5500, 1:100), and secondary antibodies used were Alexa Fluor 488 or Alexa Fluor 568 highly cross-absorbed goat anti-rabbit IgG (Life Technologies, A11034, A11036, 1:100 dilution). For dual-protein labeling of strains IU6964 (ftsZ-Myc gpsB-FLAG), IU6976 (ftsZ-Myc pbp1a-FLAG), and U6978 (ftsZ-Myc pbp2x-FLAG3) the following mixtures were used: primary antibodies (1:200 dilution of anti-FLAG rabbit polyclonal antibody and 1:100 dilution of mouse monoclonal antibody against c-Myc (Life Technologies, R950-25)) and secondary antibodies ((1:100 dilution of Alexa Fluor 488 highly cross-absorbed goat anti-rabbit and Alexa Fluor 568 highly cross-absorbed goat anti-mouse (Life Technologies, A11031)). For dual-protein labeling of strain IU7365 (pbp1a-L-FLAG3 pbp2x-HA) the following mixtures were used: (1:100 dilution of anti-HA rabbit polyclonal antibody and 1:100 dilution of anti-FLAG mouse monoclonal M2 antibody (Sigma, F1804) and secondary antibodies for FLAG- and Myc- double tagged strains listed above. Primary and secondary labeling of IU7365 was performed at 37°C for 2 h and 45 min, respectively, instead of at 25°C for 1h for both steps.
Control experiments showed similar localization patterns for each protein when tracked in single or dual-protein labeling experiments (data not shown). Similar localization patterns were observed for proteins fused to different epitope tags or when combinations of tags or antibodies were swapped in dual-protein experiments (data not shown). Control experiments showed no detectable labeling in cells not expressing tagged proteins (IU1945) with the sets of antibodies used (data not shown). Labeling of single-tagged strains with the double labeling procedure containing both sets of primary and secondary antibodies produced signal only in the expected fluorescence channel (data not shown). Together, these controls indicate that there was no detectable non-specific binding of the primary or secondary antibodies to untagged proteins, that non-specific binding of secondary antibodies did not occur to the primary antibodies of a different animal, and that excitation and emission wavelength cutoffs of the fluorescence filter blocks allowed detection of fluorescence signals from only Alexa 488 or Alexa 568, depending on the setting. DNA in nucleoids was stained with DAPI diluted in the secondary antibody incubation mix, as described previously (Wayne et al., 2010), or by using mounting media SlowFade gold antifade reagent with DAPI (Life Technologies, S36936) in place of antifade reagent with no DAPI. Both DAPI staining procedures gave similar results (data not shown). For 2D IFM, cells were examined using a Nikon E-400 epifluorescence phase-contrast microscope equipped with a mercury lamp, a 100× Nikon Plan Apo oil-immersion objective (numerical aperture, 1.40), and filter blocks for fluorescence (DAPI, EX 330 to 380, DM 400, and BA 435 to 485; Alexa 488, EX 460 to 500, DM 505, and BA 510 to 560; Alexa 568, EX 532–587, DM 595, and BA 608 to 683). Images were captured using a CoolSNAP HQ2 charge-coupled device (CCD) camera (Photometrics) and processed with NIS-Elements AR imaging software (Nikon).
3D-SIM IFM
Samples were prepared for 3D-SIM IFM the same way as for 2D IFM (see above), except that fixed cells were deposited on coverslips instead of slides and all subsequent staining and washing steps were performed on coverslips, which were then mounted on slides. 3D-SIM was performed using the OMX 3D-SIM super-resolution system located in the Indiana University Bloomington Light Microscopy Imaging Center (http://www.indiana.edu/~lmic/microscopes/OMX.html). This system consists of an OMX v.3 system (Applied Precision) with structured illumination mode that provides super-high resolution imaging (X, Y resolution ≈100 nm; Z resolution ≈300 nm). Images were acquired with a z-distance of 0.125 µm with a 100x, 1.4 NA objective, using 1.514 immersion oil. Lasers lines used were 405, 488, and 561 and emission filters used were 419–465, 500–550 and 609–705 for examination of samples stained with DAPI, Alexa 488, and Alexa 568, respectively. The system used is equipped with four Photometrics Cascade II EMCCD cameras that allow simultaneous 4 color imaging, and is controlled by DV-OMX software, with image processed by Applied Precision softWoRx 5.0.0 imaging software. Exposure times and %T settings for DAPI, Alexa 488, and Alexa 568 images were 50 to 200 ms and 100%, 50 ms and 10%, and 100 ms and 50%, respectively.
Image analysis of 2D IFM images
Initial qualitative analysis of protein localization at different stages of pneumococcal cell division was performed as described previously (Tsui et al., 2011). Briefly, dozens of cells from multiple fields and independent experiments were binned retrospectively from phase-contrast images into division stages, and then fluorescence images were superimposed on phase-contrast images to locate proteins and nucleoids using Photoshop software. Representative images from each division stage were then determined by visual inspection of image panels (see Fig. S7A-C).
To store and quantitate 2D images, a versatile, easy-to-use graphical user interface (GUI) was organized using MATLAB (The Mathworks) scripts. The general method provides a means to align, classify, and compare cell populations to test hypotheses related to the average relative position of immunofluorescently labeled proteins. Image sets comprising phase contrast, DAPI staining (nucleoid/DNA), and secondary immunofluorescence of 2 different protein targets from the same field of view were imported into the GUI. Individual user selected cells were copied from the raw image and displayed as four separate image channels in the GUI. After manual assignment of cell pole positions, the images were centered and aligned horizontally to a reference crosshair in the image frame by translation and rotation using GUI controls. Images from all four imaging modes were rotated and translated coordinately with no independent rotation or translation of any channel. Final images were taken from the original data file using a single bicubic interpolation for rotation.
Cell length alone was not correlated strongly enough with cell division stage to provide a basis for automated sorting (see Results). Therefore, all separated cells within phase-contrast image fields were selected manually, which could be done rapidly using the GUI. Cells were assigned visually to 1 of 4 division stages based on cell shape in phase-contrast images and position of nucleoids stained with DAPI (see Fig. 3A–6A). Stage 1 contains pre-divisional, ovoid cells with connected nucleoids. Stage 2 contains mid-divisional cells with slight constrictions at midcell and two nucleoids that may be partially connected or fully separated. Stage 3 contains mid-divisional cells with constricted midcell regions that are ≈30–70 % of the width of the new daughter cells and nucleoids that are completely separated. Stage 4 contains late-divisional, separated diplococcal cells with minimal remaining connections and totally separated nucleoids. Stage 4 cells are at the very last stages of septum closure and may include daughter cells with newly formed poles. Approximately 90–95% of selected cells showed consistent staining of proteins throughout cells and were included in intensity analyses. The remaining 5–10% of cells appeared under-stained or uniformly over-stained compared to the vast majority of other cells in the binned stages and were not analyzed further. Data from multiple fields on the same microscope slide or from different experiments were imported into a second GUI for building aggregate data sets. The final averaged images of each strain were obtained from at least two fields of two independent biological replicates.
To determine and compare the average intensity distribution in the fluorescence channels, a mean intensity value was calculated at each pixel position using all of the aligned images of a given class for each channel. Graphical representations of fluorescence distribution along the axis of the cell between poles were created for each channel by obtaining the average intensity of the central 20 pixels vertically through the cell, capturing the cell width, at each pixel position across the horizontally aligned cell. The averaged fluorescence intensity trace was then normalized by subtracting the minimum intensity value and dividing by the resulting maximum intensity value. Intensity traces from the phase contrast image were inverted prior to normalization. Traces represent the normalized average intensity distribution along the horizontal cell axis for each channel for the stage-averaged cell populations.
Supplementary Material
Acknowledgements
We thank Krys Kazmierczak, Michael Boersma, Kevin Bruce, Dan Kearns, and Yves Brun for helpful discussions. We thank the Indiana University Light Microscopy Imaging Center (3D-SIM supported by NIH S10RR028697-01) and Jim Powers for technical support and discussions. This work was supported by NIAID grant AI097289 (M.E.W.), by funds from the Indiana University Bloomington METACyt Initiative, funded in part by a major grant from the Lilly Endowment (M.E.W), by NIH grant DP2OD008592 (E.E.C), by a Pew Biomedical Scholar Award (E.E.C.), and by NSF grant MCB-1157982 (S.L.S.). A.D.L. was a predoctoral trainee on training grant T32GM007757 from the National Institutes of Health. The contents of this paper are solely the responsibility of the authors and do not necessarily represent official views of the funding agencies.
References
- 1.Albarracin Orio AG, Pinas GE, Cortes PR, Cian MB, Echenique J. Compensatory evolution of pbp mutations restores the fitness cost imposed by β-lactam resistance in Streptococcus pneumoniae. PLoS Pathogens. 2011;7:e1002000. doi: 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barendt SM, Land AD, Sham LT, Ng WL, Tsui HC, Arnold RJ, Winkler ME. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J Bacteriol. 2009;191:3024–3040. doi: 10.1128/JB.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barendt SM, Sham LT, Winkler ME. Characterization of mutants deficient in the L,D-carboxypeptidase (DacB) and WalRK (VicRK) regulon, involved in peptidoglycan maturation of Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2011;193:2290–2300. doi: 10.1128/JB.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beilharz K, Novakova L, Fadda D, Branny P, Massidda O, Veening JW. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Nat Acad Sci U.S.A. 2012;109:E905–E913. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg KH, Stamsas GA, Straume D, Havarstein LS. The effect of low Pbp2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol. 2013 Jul 19; doi: 10.1128/JB.00184-13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho SM, Kuipers OP, Neves AR. Environmental and nutritional factors that affect growth and metabolism of the pneumococcal serotype 2 strain D39 and its nonencapsulated derivative strain R6. PloS One. 2013;8:e58492. doi: 10.1371/journal.pone.0058492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Molec Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- 8.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 9.Demarre G, Galli E, Barre FX. The FtsK family of DNA pumps. Adv Exper Med Biology. 2013;767:245–262. doi: 10.1007/978-1-4614-5037-5_12. [DOI] [PubMed] [Google Scholar]
- 10.Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann New York Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 11.Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol. 2003;185:6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleurie A, Cluzel C, Guiral S, Freton C, Galisson F, Zanella-Cleon I, Di Guilmi AM, Grangeasse C. Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Molec Microbiol. 2012;83:746–758. doi: 10.1111/j.1365-2958.2011.07962.x. [DOI] [PubMed] [Google Scholar]
- 13.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PloS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Molec Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins ML, Shockman GD. Study of cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstructions of thin sections of cells. J Bacteriol. 1976;127:1346–1358. doi: 10.1128/jb.127.3.1346-1358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoskins J, Matsushima P, Mullen DL, Tang J, Zhao G, Meier TI, Nicas TI, Jaskunas SR. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J Bacteriol. 1999;181:6552–6555. doi: 10.1128/jb.181.20.6552-6555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai Y, Ogasawara N. Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division. Microbiol. 2006;152:1129–1141. doi: 10.1099/mic.0.28497-0. [DOI] [PubMed] [Google Scholar]
- 19.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol. 2012;7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. The EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lleo MM, Canepari P, Satta G. Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J Bacteriol. 1990;172:3758–3771. doi: 10.1128/jb.172.7.3758-3771.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Ann Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 25.Massidda O, Novakova L, Vollmer W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol. 2013 Jun 18; doi: 10.1111/1462-2920.12189. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Molec Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- 27.Morlot C, Bayle L, Jacq M, Fleurie A, Tourcier G, Galisson F, Vernet T, Grangeasse C, Di Guilmi AM. Interaction of penicillin-binding protein 2x and Ser/Thr protein kinase StkP, two key players in Streptococcus pneumoniae R6 morphogenesis. Molec Microbiol. 2013 Jul 31; doi: 10.1111/mmi.12348. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Morlot C, Zapun A, Dideberg O, Vernet T. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Molec Microbiol. 2003;50:845–855. doi: 10.1046/j.1365-2958.2003.03767.x. [DOI] [PubMed] [Google Scholar]
- 29.Ng WL, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Molec Microbiol. 2004;53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- 30.Noirclerc-Savoye M, Le Gouellec A, Morlot C, Dideberg O, Vernet T, Zapun A. In vitro reconstitution of a trimeric complex of DivIB, DivIC and FtsL, and their transient co-localization at the division site in Streptococcus pneumoniae. Molec Microbiol. 2005;55:413–424. doi: 10.1111/j.1365-2958.2004.04408.x. [DOI] [PubMed] [Google Scholar]
- 31.Paik J, Kern I, Lurz R, Hakenbeck R. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J Bbacteriol. 1999;181:3852–3856. doi: 10.1128/jb.181.12.3852-3856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Nunez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-Lopez R, Guedon E. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Molec Microbiol. 2011;79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 33.Pinho MG, Errington J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Molec Microbiology. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 34.Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 35.Raz A, Talay SR, Fischetti VA. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes. Molec Microbiol. 2012;84:631–647. doi: 10.1111/j.1365-2958.2012.08047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sham LT, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Nat Acad Sci U.S.A. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol. 2012;15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steele VR, Bottomley AL, Garcia-Lara J, Kasturiarachchi J, Foster SJ. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Molec Mmicrobiol. 2011;80:542–555. doi: 10.1111/j.1365-2958.2011.07591.x. [DOI] [PubMed] [Google Scholar]
- 39.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Bbiology. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun SX, Jiang H. Physics of bacterial morphogenesis. Microbiol and Molec Biol Rev: MMBR. 2011;75:543–565. doi: 10.1128/MMBR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavares JR, de Souza RF, Meira GL, Gueiros-Filho FJ. Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome. J Bacteriol. 2008;190:7096–7107. doi: 10.1128/JB.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsui HC, Keen SK, Sham LT, Wayne KJ, Winkler ME. Dynamic Distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. mBio. 2011;2(5):pii, e00202–e00211. doi: 10.1128/mBio.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsui HC, Mukherjee D, Ray VA, Sham LT, Feig AL, Winkler ME. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2010;192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Meth. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Molec Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 47.Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Molec Microbiol. 2012;86:645–660. doi: 10.1111/mmi.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wayne KJ, Sham LT, Tsui HC, Gutu AD, Barendt SM, Keen SK, Winkler ME. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J Bacteriol. 2010;192:4388–4394. doi: 10.1128/JB.00578-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Molec Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 50.White CL, Gober JW. MreB: pilot or passenger of cell wall synthesis? Trends Microbiol. 2012;20:74–79. doi: 10.1016/j.tim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Wu LJ, Errington J. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol. 2012;10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 52.Young KD. Bacterial morphology: why have different shapes? Curr Opin Microbiol. 2007;10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young KD. Bacterial shape: two-dimensional questions and possibilities. Ann Rev Microbiol. 2010;64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zapun A, Noirclerc-Savoye M, Helassa N, Vernet T. Peptidoglycan assembly machines: the biochemical evidence. Microb Drug Resist. 2012;18:256–260. doi: 10.1089/mdr.2011.0236. [DOI] [PubMed] [Google Scholar]
- 55.Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 56.Zerfass I, Hakenbeck R, Denapaite D. An important site in PBP2x of penicillin-resistant clinical isolates of Streptococcus pneumoniae: mutational analysis of Thr338. Antimicrob Agent Chemother. 2009;53:1107–1115. doi: 10.1128/AAC.01107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agent Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.