Abstract
Gene transfer into primary human CD4 T lymphocytes is a critical tool in studying the mechanism of T cell-dependent immune responses and human immunodeficiency virus-1 (HIV-1) infection. Nucleofection® is an electroporation technique that allows efficient gene transfer into primary human CD4 T cells that are notoriously resistant to traditional electroporation. Despite its popularity in immunological research, careful characterization of its impact on the physiology of CD4 T cells has not been documented. Herein, using freshly-isolated primary human CD4 T cells, we examine the effects of Nucleofection® on CD4 T cell morphology, intracellular calcium levels, cell surface activation markers, and transcriptional activity. We find that immediately after Nucleofection®, CD4 T cells undergo dramatic morphological changes characterized by wrinkled and dilated plasma membranes before recovering 1 hour later. The intracellular calcium level also increases after Nucleofection®, peaking after 1 hour before recovering 8 hours post transfection. Moreover, Nucleofection® leads to increased expression of T cell activation markers, CD154 and CD69, for more than 24 hours, and enhances the activation effects of phytohemagglutinin (PHA) stimulation. In addition, transcriptional activity is increased in the first 24 hours after Nucleofection®, even in the absence of exogenous stimuli. Therefore, Nucleofection® significantly alters the activation state of primary human CD4 T cells. The effect of transferred gene products on CD4 T cell function by Nucleofection® should be assessed after sufficient resting time post transfection or analyzed in light of the activation caveats mentioned above.
Keywords: Nucleofection®, gene transfer, T cell activation, calcium, HIV-1, plasmid
1. Introduction
Efficient gene transfer into living cells is a key step in studying gene function and gene therapy. Introduction of exogenous DNA or RNA species into cells is accomplished generally through either viral vectors or chemical/physical methods. While advances in genetically engineered viral vectors have allowed efficient gene transfer with minimal cell damage, these viral approaches are expensive, time consuming, and unable to deliver large DNA fragments that exceed virus packing capacity. Potential contamination of wild-type viruses also limits its use in gene delivery to humans during gene therapy. In comparison, physical approaches, especially electroporation, although accompanied by ing more cell death, offer much easier, faster, and safer ways of gene transfer.
Electroporation works by creating a transient and reversible permeable structure on the cell membrane by applying short and high-voltage pulses to generate an external electric field that surpasses the capacitance of the cell membrane (Rols, 2006). Experimental studies and computer simulations suggest that the electric field leads to disorganization of lipid molecules, entry of water molecules into the lipid bilayer structure, and eventually the formation of pores (Rols, 2006; Tieleman, 2006). The large water permeable pores allow delivery into the cells of not only DNA or RNA molecules, but also small ions, dyes, drugs, and antibodies or other proteins. On the other hand, the disruption of cell integrity caused by the loss of transmembrane potential and leakage of cell contents may lead to cell death.
Gene transfer into primary human CD4 T cells is critical for studying gene functions of the human immune system, mechanisms of human immunodeficiency virus-1 (HIV-1) infection, and endowing cells with new functional attributes for immunotherapy. Unmanipulated resting primary human CD4 T cells are notoriously resistant to transfection with conventional electroporation techniques. However, it was found that CD4 T cells stimulated with sub-optimal doses of phytohemagglutinin (PHA) or concanvalin A (ConA) could be transiently transfected if electroporation was done within a narrow window of time post stimulation (Chrivia et al., 1990; Schubert et al., 1995; Hughes and Pober, 1996; Cron et al., 1997). The transfection efficiency, however, was low (<5%), and although T cell proliferation and IL-2 production were not found after sub-optimal stimulation (Chrivia et al., 1990; Hughes and Pober, 1996), undetected changes in cell cycle status and activation-related gene transcription may undermine the value of these cells in studying the function of naïve T cells.
A more recent advance in gene transfer is an electroporation-based Nucleofection® technique by Lonza (Basel, Switzerland; previously, Amaxa Biosystems). Aided by the unique electrical parameters and buffer solutions customized for each specific cell type, the technique is able to deliver genes of interest directly into the nuclei of non-dividing cells, therefore achieving a very high level of transfection efficiency. It has been widely used to transfer DNA or RNA into dozens of primary cell types of different species, including human primary CD4 T cells. More than 100 research papers have been published using Nucleofection® for gene transfer into primary human T cells (Tahvanainen et al., 2006; Magg et al., 2009; Torgerson et al., 2009) on a range of topics, including T cell activation (Finney et al., 2004; Zhao et al., 2005; Stallwood et al., 2006), signal transduction (Kovacs et al., 2005; Methi et al., 2005; Wabnitz et al., 2006), transcriptional regulation (Grant et al., 2006; Mantel et al., 2006; Mehta et al., 2010), and HIV-1 infection (Chiu et al., 2005; Trushin et al., 2005; Selliah et al., 2008). More recently, this technology has been popularized for selectively knocking down, or silencing, gene expression in primary human CD4 T cells (Zhang et al., 2012; Freeley and Long, 2013). Thus, Nucleofection® has fast become a powerful research tool for scientists studying primary human CD4 T cell physiology.
Despite the popularity of this technique, there has not been a comprehensive study on the effect of the Nucleofection® procedure on the physiology and activation status of primary human CD4 T cells. How Nucleofection® may affect CD4 T cell activation status has important implications in interpreting data from experiments designed to address issues innate to resting primary human CD4 T cells, as opposed to pre-activated CD4 T cells or transformed cell lines. Herein, we examine the changes of primary CD4 T cell activation status by monitoring morphology, intracellular calcium flux, and cell surface activation markers. We also test the impact of Nucleofection® on T cell function by measuring CD4 T cell transcriptional responses with and without stimulation.
2. Materials and Methods
2.1 Plasmids, reagents, and antibodies
Enhanced green fluorescent protein (EGFP) expressing reporter plasmids, pEGFP-N1, pCMS-EGFP, and pIRES2-EGFP, were purchased from Clontech (Mountain View, CA). The empty control expression vector, pRc/CMV, is from Invitrogen (Carlsbad, CA). Luciferase reporter plasmids for NFAT-, NFκB-, and HIV-1 LTR-driven transcriptional activities were described previously (Petrak et al., 1994; Dolganov et al., 1996). Anti-CD25-PE (R-phycoerythrin; catalogue #341009, clone 2A3), anti-CD154-PE (catalogue #555700, clone TRAP1), and anti-HLA-DR-FITC (fluoroscein isothiocyanate; catalogue #555811, clone G46-6) antibodies were purchased from Becton Dickinson Pharmingen (San Jose, CA). Anti-CD69-PE (catalogue #MHCD6904, clone CH/4) was purchased from Caltag (Burlingame, CA). PHA, phorbol myristate acetate (PMA), and the calcium ionophore, ionomycin, were purchased from Sigma (St. Louis, MO). RPMI 1640 (Sigma-Aldrich, St. Louis, MO; catalogue #R8758) was supplemented with 10% fetal calf serum (Gibco/Life Technologies, Grand Island, NY; catalogue #16000036), 100 units/ml penicillin, 100 μg/ml streptomycin, 0.29 mg/ml L-glutamine, non-essential amino acids, 1 mM sodium pyruvate, and 55 μm 2-mercaptoethanol. Antibody-coated beads were made by linking anti-CD28 antibody, 9.3, and anti-CD3 antibody, OKT3, (Ortho Biotech, Bridgewater, NJ) to tosylactivated Dynabeads M-450 (Dynal Biotech, Oslo, Norway).
2.2 Isolation and culture of primary human CD4 T Cells
Adult human CD4 T cells were isolated from peripheral blood of healthy donors by negative selection using a proprietary antibody mix (StemCell Technologies, Vancouver, BC) and red blood cell rosetting as previously described (Cron, 2003). More than 98% of purified cells were CD4-expressing T cells by flow cytometry (see below). CD4 T cells were cultured in complete RPMI1640 medium in the presence of 10 units/ml rhIL-2 R&D Systems, Minneapolis, MN; catalogue #202-IL). Institutional Review Board approval was obtained for these studies.
2.3 Nucleofection®
5x106 primary human CD4 T cells were resuspended in 100 μl of Nucleofection® buffer solution for human primary T cells (Human T cell kit, Lonza) containing plasmid DNA, and the cells were transfected using Program U-014 of the Nucleofector II. Immediately after Nucleofection®, cells were transferred into 500 μl of pre-warmed media.
2.4 Reporter gene activity assay
After resting for varying periods of time, the transfected CD4 T cells were either not stimulated or stimulated with ionomycin (1.5 μM), or PMA (25 ng/ml) and ionomycin, for 6 hours at 37°C. Cells were lysed with Passive Lysis Buffer (Promega, Madison, WI), and firefly luciferase reporter gene activity was determined using a Dual Luciferase Assay Kit (Promega) on a Lumat LB 9507 luminometer (EE and G Berthold, Bad Wildbad, Germany) as previously detailed (Behre et al., 1999; Zhang et al., 2004). The assay was done in duplicate and corrected for transfection efficiency based on co-transfected Renilla luciferase activity as previously described (Cron et al., 2006). For Figure 5, the 2 hour value with no stimulation was assigned as the reference luciferase activity (100%), and the other conditions were calculated according to the formula: (test value/2 hour no stimulation value) x 100%.
Fig. 5.
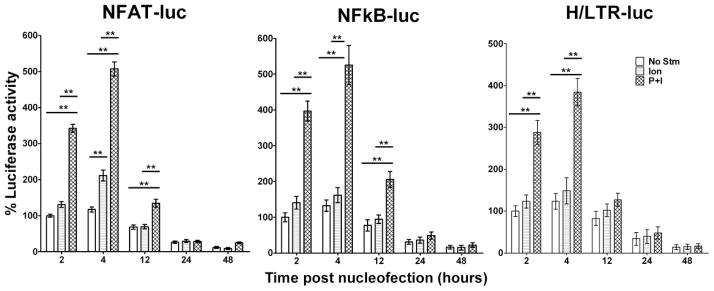
CD4 T cell responses to stimulation as measured by transcriptional activities of luciferase reporter genes introduced by Nucleofection®. 5x106 primary human CD4 T cells underwent Nucleofection® with 2.5 μg of DNA constructs containing luciferase reporter genes driven by NFAT (left), NFκB (middle), or the HIV-1 LTR/promoter (right). pRenilla-null was also transfected as an internal transfection efficiency control. Cells were rested for 2 hours or 4 hours, or cultured in the presence of rhIL-2 (20 units/ml) for 12 hours, 24 hours, or 48 hours prior to stimulation with, ionomycin alone (Ion), or PMA and ionomycin (P+I) for 6 hours. Luciferase activities were assayed after stimulation and percent luciferase levels were calculated as detailed in the Materials and Methods. Results are a summary of three similar experiments with means ± SD plotted. Two-way ANOVA analysis revealed significant differences between the paired groups (**, p<0.01).
2.5 Microscopy and flow cytometry
For intracellular calcium imaging, 5x106 CD4 T cells were washed twice with Dulbecco’s PBS buffer (Gibco/Life Technologies, catalogue #14190-250) and pulsed with 5 μM Fura-2-AM (Invitrogen, Carlsbad, CA) for 30 minutes at room temperature before Nucleofection®. To determine intracellular calcium levels by ratio imaging, 510 nm emissions excited by 380 nm and 340 nm wave lengths were captured using time-lapse imaging with 63x water objectives on a Zeiss Axioplan2 Microscope (Jena, Germany) at 25°C. Data were collected and analyzed with SlideBook software (Intelligent Imaging Innovations, Denver, CO). For flow cytometry, cells were washed twice and blocked in staining buffer (PBS containing 1% BSA and 0.02% sodium azide) for 30 minutes at room temperature, and stained with fluorescently conjugated antibodies for 30 minutes on ice. Stained cells were analyzed using a FACScalibur flow cytometer (Becton Dickinson) and CellQuest (Becton Dickinson) or FlowJo (Ashland, OR) software.
3. Results and Conclusions
3.1 Effective gene transfer into primary human CD4 T cells by Nucleofection®
Freshly isolated resting primary human CD4 T cells underwent Nucleofection® (using the Human T cell kit and the U-014 program setting recommended by the manufacturer) with 3 different EGFP expression vectors [all driven by the immediate-early cytomegalovirus (CMV) promoter] at varying doses. As shown in Fig. 1A, after 24 hours, all three vectors led to GFP expression in transfected cells in a dose-dependent manner. pEGFP-N1 gave the highest detectable transfection rate, with 75% of FCM-gated live cells expressing EGFP after transfection with 1 μg of vector/106 cells. In comparison, cells transfected with pCMS-EGFP or pIRES2-EGFP had approximately five-fold lower detectable transfection rates, possibly due to transcriptional promoter positioning (pCMS-EGFP) or transcript editing (pIRES2-EGFP) in these vectors. While increased amounts of vectors helped improve detectable transfection rates, it also caused substantially more cell death (Fig. 1B). Less than 20% of the cells transfected with 1 μg of vector/106 cells survived the Nucleofection® after 24 hours. The strong impact of the Nucleofection® procedure on cell survival prompted us to investigate how other aspects of T cell physiology, especially T cell activation status, might also be affected.
Fig. 1.
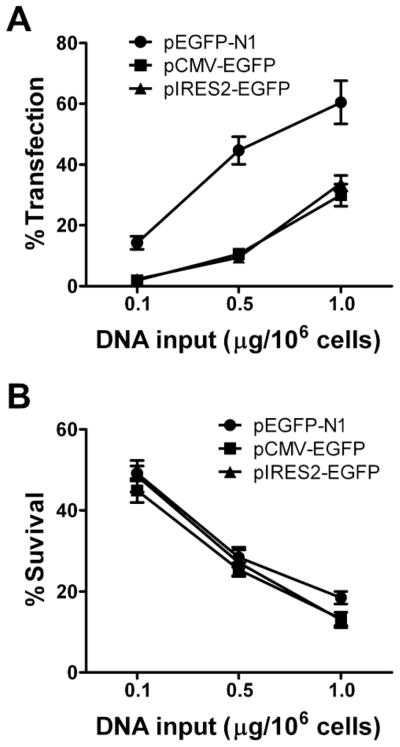
Highly efficient gene transfer into primary human CD4 T cells by Nucleofection®. Freshly isolated human CD4 T cells underwent Nucleofection® with GFP expression vectors, pEGFP-N1, pCMS-EGFP, or pIRES2-EGFP, at 3 different doses. GFP expression (A) and cell viability (B) were assayed 24 hours post transfection and are represented as percent positive live cells and percent survival, respectively. Results are shown as means ± SD for three similar experiments. Two-way ANOVA analysis was performed: GFP expression in pEGFP-N1 transfected cells was significantly higher than in either pCMV-EGFP or pIRES2-EGFP transfected cells, p<0.01; there was no statistically significant difference in cell viability among the groups of transfected cells, p>0.05). Linear regression analysis revealed GFP expression is directly related to transfected DNA dosage for all plasmids studied (p<0.01): pEGFP-N1 (R2=0.8286), pCMV-EGFP (R2=0.9091), and pIRES2-EGFP (R2=0.9085).
3.2 CD4 T cell morphological changes induced by Nucleofection®
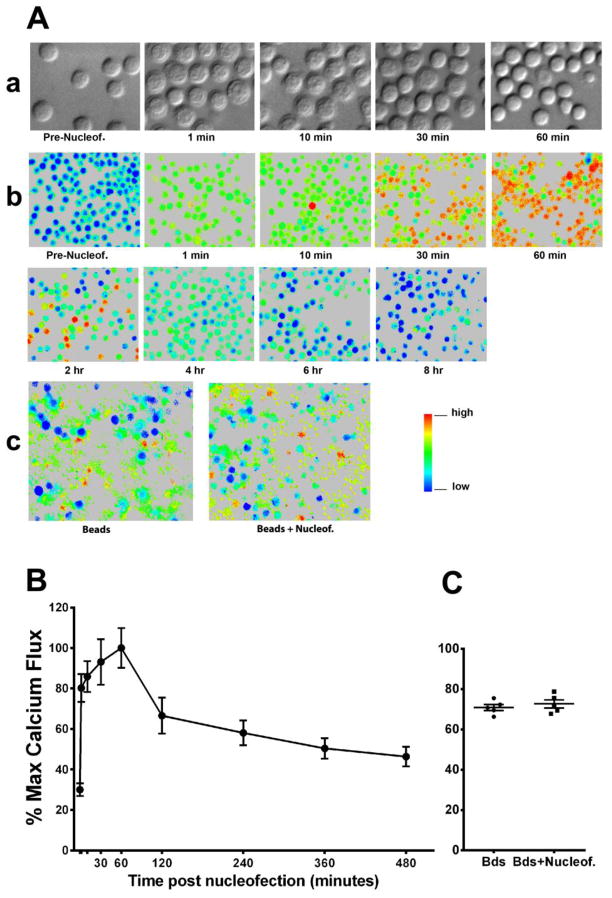
Immediately after Nucleofection® (U-014) with an empty expression vector control, pRc/CMV, CD4 T cells displayed pronounced morphological changes as observed using Nomarski differential interference contrast (DIC) microcopy (Fig. 2A,a). Similar morphologic changes were obtained using alternative manufacture recommend CD4 T cell programs (data not shown). The majority of the CD4 T cells had dramatic expansions of plasma membrane, leading to marked increases in cell size. Since the size of the nucleus appeared to remain unchanged, the expansion of the plasma membrane suggests a volume increase of the previously almost invisible cytoplasm and dilution of its contents by an influx of media through pores in the plasma membrane. The nucleus appeared much rougher in texture than prior to transfection, although it is not certain whether this reflects real structural changes or the appearance was caused by altered optical characteristics of expanded plasma membrane. These changes gradually recovered over time. After an hour, the majority of the cells appeared identical to non-transfected CD4 T cells. Although it is unclear whether and how these early morphological changes might directly influence later T cell activation status and function, such significant alterations of cell morphology by Nucleofection® suggest the process may dramatically impact T cell physiology.
Fig. 2.
The impact of Nucleofection® on CD4 T cell morphology and intracellular Ca2+ levels. 5x106 CD4 T cells were immediately transferred to cell culture medium after Nucleofection® with 2.5 μg of the empty control vector, Rc/CMV. (A) Representative imaging shows (a) morphological changes over time after transfection as evaluated by Nomarski DIC microscopy, (b) changes of intracellular Ca2+ levels after transfection over time (the level of Ca2+ is indicated by the 340/380 ratio shown in pseudocolor), and (c) calcium flux of non-transfected CD4 T cells and CD4 T cells 8 hours post Nucleofection® in response to beads conjugated with anti-CD3 and anti-CD28 antibodies. Results are representative of three similar experiments. (B) The degree of calcium flux before and after Nucleofection® was analyzed by quantifying the 340/380 ratios reflected by color changes; calcium flux percentage was further calculated by normalizing all values of other time points to the value of the one hour time point sample (maximal value). The graph shows a summary of 3 experiments from different donors plotted as means ± SD. (C) A summary of calcium flux percentages obtained in non-transfected CD4 T cells and CD4 T cells eight hours post Nucleofection® in response to anti-CD3/CD28 beads from 5 separate donors. The data is plotted as means ± SD.
3.3 Sustained elevation of CD4 T cell intracellular calcium levels by Nucleofection®
The Ca2+ concentration in the extracellular environment in vivo or in culture medium (~1 mM) is much higher than that inside of T cells (< 1 μM). The ability of Nucleofection® to permeabilize cell membranes to the extent that it allows the entrance of large DNA molecules suggests that it should also permit the passage of small calcium ions. Indeed, a homogeneous increase of intracellular Ca2+ was seen immediately after transfection in the presence (Fig. 2A,b and 2B) or absence of plasmid DNA (data not shown). Interestingly, only limited elevation was seen initially, but the Ca2+ level continued to increase over time and peaked at 60 minutes post Nucleofection®, before gradually declining to normal levels after 8 hours. The peak Ca2+ level was similar to that induced by stimulation with anti-CD3 and anti-CD28 coated beads (Fig. 2A,c and 2C). According to the manufacturer, the transfection buffer does not contain Ca2+. Therefore, the increase was likely a result of gradual Ca2+ influx from the medium. From the kinetics of the Ca2+ level change, it can be speculated that Nucleofection® caused large pores permeable for large DNA molecules very transiently. The large pores were likely closed before the cells were transferred to media containing Ca2+, therefore an immediate high level increase of intracellular Ca2+ was not observed right after the cells were transferred into medium. However, small defects on the membrane may have remained allowing Ca2+ to gradually leak into the cells. During the first hour, this likely could not be balanced by calcium pumps (Ca2+-ATPases) on the membrane that drive Ca2+ out of the cell. Perhaps, Nucleofection® led to compromised calcium pump function, thus resulting in continued increases of intracellular Ca2+ levels. After one hour, with decreasing Ca2+ leakage and/or increasing calcium pump function, intracellular Ca2+ was gradually cleared. Although the direct connection is unknown, it is interesting to note that the time it took for cell morphology to recover and for effective Ca2+ clearance to start were both one hour.
Elevation of intracellular calcium level is an important step in the T cell activation process (Quintana et al., 2005). Signals from the T cell receptor generate inositol-1,4,5-triphosphate (IP3) that induces release of calcium from the endoplasmic reticulum (ER). Depletion of calcium from the ER triggers calcium influx from the extracellular environment through the calcium regulated activation channel (CRAC) (Hogan et al., 2010). Through the phosphatase, calcineurin, the resultant elevation of intracellular calcium level enables the activation and translocation of nuclear factor of activated T cells (NFAT) into the nucleus (Crabtree and Olson, 2002). Ca2+ can also activate nuclear factor kappa B (NF-κB) in a protein kinase C (PKC)-dependent manner (Li and Verma, 2002), and through Ca2+ -dependent kinases, modulate the activity of AP-1 transcription factors (Rao et al., 1997). NFAT, NF-κB, and activation protein-1 (AP-1) are three critical transcription factors in T cells, controlling the transcription of new genes necessary for T cell activation, proliferation, and function. In fact, 75% of all activation-regulated genes show a dependence on Ca2+ influx through the plasma membrane via CRAC channels (Feske et al., 2001). Therefore, when designing experiments aimed at studying the function of primary T cells, the possible impact of sustained elevation of intracellular Ca2+ levels caused by Nucleofection® on T cell activation status should be taken into consideration.
3.4 Changes in CD4 T cell surface activation markers after Nucleofection®
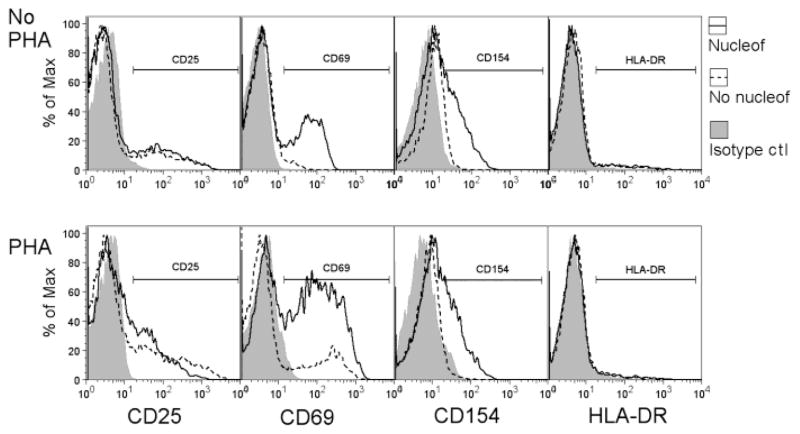
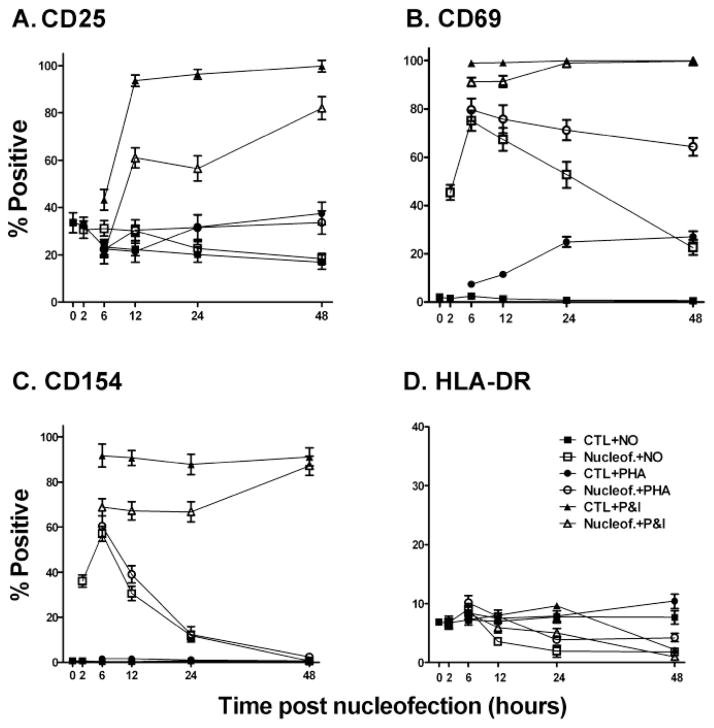
Although Nucleofection® (U-014) did not lead to CD4 T cell proliferation or cytokine production (data not shown), changes in the expression of classical CD4 T cell surface activation markers were observed. Increased expression of CD69 and CD154 was seen on CD4 T cells 24 hours post transfection, although the expression of CD25 and HLA-DR remained largely unchanged (Fig. 3). Following surface marker expression over time revealed that increased expression of CD69 and CD154 started as early as 2 hours and peaked at around 6 hours before declining gradually (Fig. 4). After 48 hours, while CD4 T cells stopped expressing CD154, there were still 25% of CD4 T cells expressing CD69. Similar results in cell surface activation markers were obtained with alternative Nucleofection® protocols (data not shown). It is likely that the expression of CD69 and CD154 are directly linked to the elevated intracellular Ca2+ levels (Jullien et al., 2003), and such sustained expression of surface activation markers suggests altered CD4 T cell activation status.
Fig. 3.
The impact of Nucleofection® on the expression of CD4 T cell surface activation markers. 5x106 primary human CD4 T cells were transfected with 2.5 μg of the empty control vector, Rc/CMV, and rested for 2 hours. Transfected cells (Amaxa, solid line) were then cultured with or without 2.5 μg/ml of PHA for 24 hours before being stained for CD25, CD69, CD154, and HLA-DR expression. As controls, non-transfected CD4 T cells (dashed line) were cultured for 24 hours with or without PHA before being stained for surface markers. Results are representative of three similar experiments.
Fig. 4.
The impact of Nucleofection® on the expression of CD4 T cell surface activation markers over time. 5x106 primary human CD4 T cells were transfected with 2.5 μg of the empty control vector, Rc/CMV, and rested for 2 hours. Transfected cells were then stimulated with 2.5 μg/ml PHA, 10 ng/ml PMA plus 0.5 μM ionomycin, or nothing. Cells were then cultured until the subsequent time points when they were collected and stained for CD25 (A), CD69 (B), CD154 (C), and HLA-DR (D) expression. Non-transfected CD4 T cells were treated the same as controls. Expression of surface markers at 2 hours was monitored for unstimulated CD4 T cells. Values at the 0 hour time point indicate expression levels of non-transfected and unstimulated CD4 T cells. Results are shown as means ± SD for three similar experiments. Two-way ANOVA analysis was performed. For CD25 expression, for either no stimulation of PHA stimulation, there were no differences between control cells and cells undergoing nucleofection (p>0.05). For CD69 expression, transfected cells that were untreated or PHA stimulated had significantly higher expression than controls (p<0.01); PMA and ionomycin (P&I) treated cells undergoing nucleofection had significantly less expression than controls at the 6 and 12 hour time points (p<0.01) but were not different at the later time points. Similar results were noted for CD154 with the exception that only the 48 hour time point was not lower for the transfected cells (p>0.05). There were no significant differences in HLA-DR expression at any time point for the transfected versus the control cells for all 3 conditions.
3.5 Impact of Nucleofection® on subsequent CD4 T cell activation
Despite the sustained elevation of Ca2+ following Nucleofection®, once recovered, CD4 T cells fluxed calcium similarly to non-transfected cells in response to stimulation by beads coated with anti-CD3 and anti-CD28 antibodies (Fig. 2C). This suggests membrane calcium pumps and CRAC channels controlling calcium influx resumed their normal function. To look at surface activation marker expression, CD4 T cells were stimulated with PHA or PMA plus ionomycin two hours post transfection and the cell surface activation markers were monitored at different time points. As shown in Fig. 4A, increased CD25 expression induced by PHA or PMA plus ionomycin was minimally affected by Nucleofection®. PHA induced similar levels of CD25 expression on transfected and non-transfected control cells. CD25 expression was somewhat subdued in transfected CD4 T cells in response to PMA plus ionomycin. This is not unexpected as Nucleofection® itself did not alter CD25 expression. However, for CD69, whose expression was significantly increased by the Nucleofection® procedure, further stimulation with PHA led to sustained expression in over 60% of the CD4 T cells for more than 48 hours (Fig. 4B). PHA did not further increase the expression of CD154 induced by Nucleofection® (Fig. 4C). The expression of CD69 and CD154 induced by PMA and ionomycin was both somewhat subdued in transfected CD4 T cells. Therefore, Nucleofection® appears to enhance the expression of CD69 induced by PHA but subdue the expression of CD25, CD69, and CD154 induced by PMA and ionomycin.
Reporter genes driven by particular transcriptional promoters are frequently introduced into T cells for testing transfection factor activity or identifying activators or inhibitors of transcriptional regulation. To test how suitable Nucleofection® is for introducing reporter genes into primary human CD4 T cells, CD4 T cells were transfected with luciferase genes driven by NFAT- or NFκB-responsive reporter plasmids. After resting for different time periods, transfected CD4 T cells were either not stimulated or stimulated with ionomycin, or PMA and ionomycin, for 6 hours prior to luciferase activity measurement. As shown in Fig. 5, reporter genes measuring NFAT and NFκB activity were notably elevated without additional stimulation within 12 hours of transfection compared to background luciferase control values (routinely <1,000 arbitrary light units). This transcriptional activity was only minimally augmented by the addition of ionomycin alone (Fig. 5). These results likely reflect the fact that Nucleofection® alone results in substantial intracellular calcium levels (Fig. 2A,b) and thus resembles calcium ionophore (ionomycin) treatment. By contrast, substantially higher activities were induced by PMA and ionomycin (P+I) within 12 hours of transfection likely reflecting PMA stimulation of PKC and other kinases. The diminished reporter gene activities at later time points were likely due in part to increased cell death, but other factors are likely at play since there was only a modest decrease in the co-transfected Renilla luciferase tranfection efficiency control plasmids at these later time points.
Similar to the NFAT and NFκB reporter constructs, the HIV-1 LTR-driven reporter gene construct was active within 12 hours of transfection and was only minimally augmented by ionomycin (Fig. 5). These results contrast with prior reports where ionomycin stimulation alone more than doubled HIV-1 LTR transcriptional activity (Cron et al., 2000) when using a more traditional transient transfection protocol (Cron et al., 1997). Nevertheless, addition of PMA notably increased HIV-1 transcription above that of ionomycin alone, particularly at 2 and 4 hours post Nucleofection® (Fig. 5). Because of the importance of studying HIV-1 regulation in primary human CD4 T cells, one must be aware of the calcium flux induced by Nucleofection® when interpreting results such as these.
In summary, Nucleofection® is a highly effective method for delivering DNA constructs into primary human CD4 T cells that are moderately resistant to other means of gene transfer. A relatively small amount of DNA was sufficient for transfecting a large number of CD4 T cells and lead to the expression of the gene of interest in up to 75% of the cell population with an acceptable level of cell death. On the other hand, Nucleofection® caused significant changes in T cell morphology, pronounced elevation of intracellular calcium levels, and sustained increases of certain activation markers on the cell surface of CD4 T cells. Nucleofection® also affected subsequent activation of CD4 T cells, especially in terms of CD69 expression, and transcriptional activity. Therefore, to minimize the impact of the Nucleofection® procedure on CD4 T cell physiology, appropriate resting time post transfection should be considered for CD4 T cells to recover from the changes caused by the procedure and for artificially induced CD4 T cell activities to subside. Unless the significance of these changes in the activation status and the functions of CD4 T cells are clear, care should be taken when interpreting data from cells following Nucleofection®, as they may not represent truly resting primary human CD4 T cells.
Highlights.
Nucleofection® of primary human CD4 T cells results in sustained intracellular calcium levels for up to 8 hours post transfection.
Nucleofection® of primary human CD4 T cells leads to cell surface expression of T cell activation markers for up to 24 hours post transfection.
Nucleofection® of primary human CD4 T cells results in T cell transcriptional activity for at least 24 hours post transfection.
Acknowledgments
This work was supported by NIH grants, R01-AR-48257, R21-AR-49335, and R21-AI-68574 (all to RQC).
Abbreviations
- AP-1
activation protein 1
- ConA
concanvalin A
- CRAC
calcium release activated channel
- CMV
cytomegalovirus
- DIC
differential interference contrast
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HIV-1
human immunodeficiency virus-1
- IP3
inositol-1,4,5-triphosphate
- NFAT
nuclear factor of activated T cells
- NFκB
nuclear factor kappa B
- PHA
phytohemagglutinin
- PMA
phorbol myristate acetate
- PKC
protein kinase C
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mingce Zhang, Email: mzhang@peds.uab.edu.
Zhengyu Ma, Email: zma@nemours.org.
Nithianandan Selliah, Email: nithiselliah@gmail.com.
Greta Weiss, Email: weiss@burnet.edu.au.
Anna Genin, Email: agenin@peds.uab.edu.
Terri H. Finkel, Email: tfinkel@nemours.org.
Randy Q. Cron, Email: rcron@peds.uab.edu.
References
- Behre G, Smith LT, Tenen DG. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. Biotechniques. 1999;26:24–6. 28. doi: 10.2144/99261bm03. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–14. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Wedrychowicz T, Young HA, Hardy KJ. A model of human cytokine regulation based on transfection of gamma interferon gene fragments directly into isolated peripheral blood T lymphocytes. J Exp Med. 1990;172:661–4. doi: 10.1084/jem.172.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Cron RQ. CD154 transcriptional regulation in primary human CD4 T cells. Immunol Res. 2003;27:185–202. doi: 10.1385/IR:27:2-3:185. [DOI] [PubMed] [Google Scholar]
- Cron RQ, Bandyopadhyay R, Genin A, Brunner M, Kersh GJ, Yin J, Finkel TH, Crow MK. Early growth response-1 is required for CD154 transcription. J Immunol. 2006;176:811–8. doi: 10.4049/jimmunol.176.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin Immunol. 2000;94:179–91. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- Cron RQ, Schubert LA, Lewis DB, Hughes CC. Consistent transient transfection of DNA into non-transformed human and murine T-lymphocytes. J Immunol Methods. 1997;205:145–50. doi: 10.1016/s0022-1759(97)00065-3. [DOI] [PubMed] [Google Scholar]
- Dolganov G, Bort S, Lovett M, Burr J, Schubert L, Short D, McGurn M, Gibson C, Lewis DB. Coexpression of the interleukin-13 and interleukin-4 genes correlates with their physical linkage in the cytokine gene cluster on human chromosome 5q23-31. Blood. 1996;87:3316–26. [PubMed] [Google Scholar]
- Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat Immunol. 2001;2:316–24. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- Finney HM, Akbar AN, Lawson AD. Activation of resting human primary T cells with chimeric receptors: costimulation from CD28, inducible costimulator, CD134, and CD137 in series with signals from the TCR zeta chain. J Immunol. 2004;172:104–13. doi: 10.4049/jimmunol.172.1.104. [DOI] [PubMed] [Google Scholar]
- Freeley M, Long A. Advances in siRNA delivery to T-cells: potential clinical applications for inflammatory disease, cancer and infection. Biochem J. 2013;455:133–47. doi: 10.1042/BJ20130950. [DOI] [PubMed] [Google Scholar]
- Grant C, Oh U, Fugo K, Takenouchi N, Griffith C, Yao K, Newhook TE, Ratner L, Jacobson S. Foxp3 represses retroviral transcription by targeting both NF-kappaB and CREB pathways. PLoS Pathog. 2006;2:e33. doi: 10.1371/journal.ppat.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan PG, Lewis RS, Rao A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol. 2010;28:491–533. doi: 10.1146/annurev.immunol.021908.132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CC, Pober JS. Transcriptional regulation of the interleukin-2 gene in normal human peripheral blood T cells. Convergence of costimulatory signals and differences from transformed T cells. J Biol Chem. 1996;271:5369–77. doi: 10.1074/jbc.271.10.5369. [DOI] [PubMed] [Google Scholar]
- Jullien P, Cron RQ, Dabbagh K, Cleary A, Chen L, Tran P, Stepick-Biek P, Lewis DB. Decreased CD154 expression by neonatal CD4+ T cells is due to limitations in both proximal and distal events of T cell activation. Int Immunol. 2003;15:1461–72. doi: 10.1093/intimm/dxg145. [DOI] [PubMed] [Google Scholar]
- Kovacs B, Parry RV, Ma Z, Fan E, Shivers DK, Freiberg BA, Thomas AK, Rutherford R, Rumbley CA, Riley JL, Finkel TH. Ligation of CD28 by its natural ligand CD86 in the absence of TCR stimulation induces lipid raft polarization in human CD4 T cells. J Immunol. 2005;175:7848–54. doi: 10.4049/jimmunol.175.12.7848. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Magg T, Hartrampf S, Albert MH. Stable nonviral gene transfer into primary human T cells. Hum Gene Ther. 2009;20:989–98. doi: 10.1089/hum.2008.180. [DOI] [PubMed] [Google Scholar]
- Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, Schmidt-Weber CB. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- Mehta J, Genin A, Brunner M, Scalzi LV, Mishra N, Beukelman T, Cron RQ. Prolonged expression of CD154 on CD4 T cells from pediatric lupus patients correlates with increased CD154 transcription, increased nuclear factor of activated T cell activity, and glomerulonephritis. Arthritis Rheum. 2010;62:2499–509. doi: 10.1002/art.27554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methi T, Ngai J, Mahic M, Amarzguioui M, Vang T, Tasken K. Short-interfering RNA-mediated Lck knockdown results in augmented downstream T cell responses. Journal of Immunology. 2005;175:7398–7406. doi: 10.4049/jimmunol.175.11.7398. [DOI] [PubMed] [Google Scholar]
- Petrak D, Memon SA, Birrer MJ, Ashwell JD, Zacharchuk CM. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J Immunol. 1994;153:2046–51. [PubMed] [Google Scholar]
- Quintana A, Griesemer D, Schwarz EC, Hoth M. Calcium-dependent activation of T-lymphocytes. Pflugers Arch. 2005;450:1–12. doi: 10.1007/s00424-004-1364-4. [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Rols MP. Electropermeabilization, a physical method for the delivery of therapeutic molecules into cells. Biochim Biophys Acta. 2006;1758:423–8. doi: 10.1016/j.bbamem.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Schubert LA, King G, Cron RQ, Lewis DB, Aruffo A, Hollenbaugh D. The human gp39 promoter. Two distinct nuclear factors of activated T cell protein-binding elements contribute independently to transcriptional activation. J Biol Chem. 1995;270:29624–7. doi: 10.1074/jbc.270.50.29624. [DOI] [PubMed] [Google Scholar]
- Selliah N, Zhang M, White S, Zoltick P, Sawaya BE, Finkel TH, Cron RQ. FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology. 2008;381:161–7. doi: 10.1016/j.virol.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallwood Y, Briend E, Ray KM, Ward GA, Smith BJ, Nye E, Champion BR, McKenzie GJ. Small interfering RNA-mediated knockdown of notch ligands in primary CD4(+) T cells and dendritic cells enhances cytokine production. Journal of Immunology. 2006;177:885–895. doi: 10.4049/jimmunol.177.2.885. [DOI] [PubMed] [Google Scholar]
- Tahvanainen J, Pykalainen M, Kallonen T, Lahteenmaki H, Rasool O, Lahesmaa R. Enrichment of nucleofected primary human CD4+ T cells: a novel and efficient method for studying gene function and role in human primary T helper cell differentiation. J Immunol Methods. 2006;310:30–9. doi: 10.1016/j.jim.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Tieleman DP. Computer simulations of transport through membranes: passive diffusion, pores, channels and transporters. Clin Exp Pharmacol Physiol. 2006;33:893–903. doi: 10.1111/j.1440-1681.2006.04461.x. [DOI] [PubMed] [Google Scholar]
- Torgerson TR, Genin A, Chen C, Zhang M, Zhou B, Anover-Sombke S, Frank MB, Dozmorov I, Ocheltree E, Kulmala P, Centola M, Ochs HD, Wells AD, Cron RQ. FOXP3 inhibits activation-induced NFAT2 expression in T cells thereby limiting effector cytokine expression. J Immunol. 2009;183:907–15. doi: 10.4049/jimmunol.0800216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushin SA, Bren GD, Asin S, Pennington KN, Paya CV, Badley AD. Human immunodeficiency virus reactivation by phorbol esters or T-cell receptor ligation requires both PKCalpha and PKCtheta. J Virol. 2005;79:9821–30. doi: 10.1128/JVI.79.15.9821-9830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabnitz GH, Nebl G, Klemke M, Schroder AJ, Samstag Y. Phosphatidylinositol 3-kinase functions as a Ras effector in the signaling cascade that regulates dephosphorylation of the actin-remodeling protein cofilin after costimulation of untransformed human T lymphocytes. Journal of Immunology. 2006;176:1668–1674. doi: 10.4049/jimmunol.176.3.1668. [DOI] [PubMed] [Google Scholar]
- Zhang M, Clausell A, Robinson T, Yin J, Chen E, Johnson L, Weiss G, Sabbaj S, Lowe RM, Wagner FH, Goepfert PA, Kutsch O, Cron RQ. Host factor transcriptional regulation contributes to preferential expression of HIV type 1 in IL-4-producing CD4 T cells. J Immunol. 2012;189:2746–57. doi: 10.4049/jimmunol.1103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Genin A, Cron RQ. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology. 2004;321:323–31. doi: 10.1016/j.virol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Zhao HR, Li CC, Pardo J, Chu PC, Liao CX, Huang JN, Dong JG, Zhou XL, Huang Q, Huang B, Bennett MK, Molineaux SM, Lu H, Daniel-Issakani S, Payan DG, Masuda ES. A novel E3 ubiquitin ligase TRAC-1 positively regulates T cell activation. Journal of Immunology. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]