Abstract
The activation of innate immune cells triggers numerous intracellular signaling pathways, which require tight control to mount an adequate immune response. The PI3K signaling pathway is intricately involved in innate immunity, and its activation dampens the expression and release of proinflammatory cytokines in myeloid cells. These signaling processes are strictly regulated by the PI3K antagonist, the lipid phosphatase, PTEN, a known tumor suppressor. Importantly, PTEN is responsible for the elevated production of cytokines such as IL-6 in response to TLR agonists, and deletion of PTEN results in diminished inflammatory responses. However, the mechanisms by which PI3K negatively regulates TLR signaling are only partially resolved. We observed that Arginase I expression and secretion were markedly induced by PTEN deletion, suggesting PTEN−/− macrophages were alternatively activated. This was mediated by increased expression and activation of the transcription factors C/EBPβ and STAT3. Genetic and pharmacologic experimental approaches in vitro, as well as in vivo autoimmunity models, provide convincing evidence that PI3K/PTEN-regulated extracellular Arginase I acts as a paracrine regulator of inflammation and immunity.
Introduction
The PI3K/PTEN signaling pathway is a thoroughly described signaling pathway, relevant for numerous physiologically important processes such as cell survival, proliferation, migration, and metabolism (1). Particularly in tumor biology and cancerogenesis, several factors within the activated PI3K signaling axis are pro-oncogenic, and cancer treatment strategies that target this pathway are currently under clinical investigation. In addition, it is clear that the PI3K signaling pathway possesses controversial immunologic properties in myeloid cells. PI3K activation, as well as the efficient regulation of the PI3K pathway by its counteracting phosphatase PTEN, is indispensable for the proper guidance of immune cells to sites of infection or inflammation and is thus considered to be proinflammatory (2, 3). Paradoxically, PI3K activation upon pattern recognition receptor binding of pathogen-associated molecular patterns (PAMPs), such as LPS, is crucial for dampening proinflammatory signals (such as MAPK activation), ultimately exerting anti-inflammatory and tissue-protective effects (4–8). Obviously, this is an intracellular, anti-inflammatory process that protects the organism from inflammation-induced tissue damage (9). Notably, the molecular mechanisms dedicated to controlling this beneficial phenomenon are incompletely understood. Analysis of PTEN-deficient myeloid cells stimulated by a variety of PAMPs revealed differential expression of several relevant immunomodulatory genes, such as IL-10 and DUSP1 (7, 9). Significantly, in that screen, we also identified Arginase I as a differentially expressed gene upon PTEN deficiency. In this study, we aimed to elucidate the molecular basis of the PI3K/PTEN signaling axis regulating Arginase I expression and the potential role of Arginase I in modulating innate immune responses mediated by cell-type–specific PTEN deficiency in cells of monocytic/macrophage origin.
Arginase I is a key element of the urea cycle, which converts arginine to urea, and is predominately active in the liver. However, besides its role in hepatocytes and the detoxification of ammonia, this housekeeping enzyme fulfills a number of important immunologic functions in myeloid cells (10, 11). Innate immune cells, expressing Arginase I, are capable of depleting arginine from the environment under inflammatory conditions. T cells, for instance, are clearly dependent on the semiessential amino acid arginine. Thus, Arginase I–expressing myeloid cells contribute to T cell anergy and prevent Th cell functions (12, 13). Arginase I expression in myeloid cells, at least in the murine situation, is regulated by Th2 cytokines IL-4/IL-13 (14, 15) and TLR agonists such as bacillus Calmette–Guerin (BCG) and LPS (16). Still, it is debatable whether Arginase I expression in the monocytic/macrophage/dendritic cell (DC) lineage is relevant for human pathology, because Arginase I expression seems to be confined to neutrophil granulocytes in humans (17). However, recent findings argue for the presence of Arginase I in macrophages in mycobacterial granulomas of human and nonhuman primate origin (18). In mice, Arginase I is expressed by cells of monocytic origin. It has long been appreciated that Arginase I is a signature molecule for the activation state of macrophages. Arginase I–expressing macrophages are considered to be alternatively activated or M2 macrophages, involved in tissue regeneration and repair, but also required in the immune defense against multicellular pathogens and parasites (19, 20).
Analysis of macrophages derived from PTENfl/fl LysM cre mice revealed a massive increase in Arginase I expression, even in naive macrophages without further activation. On a molecular level, the transcription factors (TFs) C/EBPβ and STAT3 contributed to this PI3K-mediated upregulation. Surprisingly, the in vitro analysis of supernatants (SNs) collected from PTEN−/− macrophages disclosed an increased release and accumulation of extracellular Arginase I, independent of increased apoptosis or cytotoxicity. Transfer experiments of conditioned media derived from naive PTEN−/− macrophages, containing high amounts of Arginase I, as well as the addition of recombinant human pegylated Arginase I (recArgI), which is already in clinical use for the treatment of hepatocellular carcinoma (21), supported the notion that extracellular Arginase I exerts potent anti-inflammatory effects on myeloid cells. This finding was further corroborated by experiments with recArgI present in MLRs using LPS-stimulated, OVA-loaded DCs together with OT-II T cells and by an in vivo autoimmune model, namely, experimental autoimmune encephalomyelitis (EAE), which is clearly dependent on Th1 and Th17 T cell polarization by DCs (22).
In summary, our data support the notion that PTEN contributes to cell fate decisions of macrophages exemplified by increased Arginase I expression. As a functional consequence, sustained Arginase I expression and release lead to a hypoinflammatory environment dampening T cell–mediated pathophysiologic effects in vitro and in vivo. Our findings might have important clinical implications for the treatment of autoimmune disorders, especially in light of the recently approved clinical use of recArgI.
Materials and Methods
Mice
All animals were bred and housed in a specific pathogen-free facility of the Medical University of Vienna with a 12-h/12-h day/night cycle and constant temperature. Floxed PTEN mice were kindly provided by Tak W. Mak (23). LysM cre mice were originally described by Clausen et al. (24). 2D2 myelin oligodendrocyte glycoprotein (MOG)35–55–specific TCR transgenic (tg) mice on a C57BL/6 background were purchased from Jackson Laboratories. Littermate-controlled experiments were performed with 8- to 12-wk-old wild-type (WT), floxed PTEN cre+ and cre− mice on a C57BL/6 background, which were backcrossed for at least 10 generations. Mice of both sexes were used and no gender-specific differences were found. In vivo experiments comply with institutional guidelines for animal experimentation; an ethical approval was obtained by the Federal Ministry for Science and Research, Vienna, Austria (BMWF-66.009/0103-C/GT/2007 and BMWF-66.009/0241-II/3b/2011).
Genotyping
Mice were earmarked 3–4 wk after birth. DNA from lysed (proteinase K lysis buffer) ear tissue was subjected to direct PCR using GoTaq Polymerase (Promega). Specific PCRs were performed with following primers: PTEN primer: forward, 5′-CTCCTCTACTCCATTCTTCCC-3′, reverse, 5′-ACTCCCACCAATGAACAAAC-3′; cre primer: forward, 5′-TCGCGATTATCTTCTATATCTTCAG-3′, reverse, 5′-GCTCGACCAGTTTAGTTACCC-3′.
Preparation and cultivation of primary macrophages
Thioglycollate-elicited peritoneal macrophages (tPMs) were generated by injection of 2 ml 4% thioglycollate (Sigma) into the peritoneal cavity followed by peritoneal lavage with RPMI 1640 (Invitrogen) medium 3 d later. Isolated macrophages were seeded at a concentration of 1 × 106 cells/ml in RPMI 1640 supplemented with 10% FCS, 1% penicillin/streptomycin/Fungizone, 1% l-glutamine, and cultured at 37°C in a 5% CO2 atmosphere. Cells were allowed to recover overnight, and in vitro stimulations were carried out by supplementing the macrophage culture with the following agents and cytokines: 100 ng/ml ultrapure E. coli O111:B4 LPS (Invivogen), 100 nM wortmannin (Sigma), 10 or 200 μM N-hydroxy-l-arginine (Calbiochem), or 5 ng/ml recombinant mouse IL-4/IL-13 (R&D Systems). Stimulations were carried out for 3, 8, or 24 h for RNA extraction, avidin–biotin complex DNA (ABCD) assay, or protein isolation and cytokine measurement, respectively.
Preparation and cultivation of bone marrow–derived DCs
Bone marrow cells were flushed from femurs and tibias of indicated mice and cultivated in complete RPMI medium supplemented with 20 ng/ml recombinant mouse GM-CSF (R&D Systems) at 37°C in a 5% CO2 atmosphere. Half of the medium was replaced with fresh medium supplemented with 20 ng/ml GM-CSF at days 3 and 6 after isolation. DCs were harvested and activated at day 7, and the maturation status was determined by flow cytometry using fluorescent-conjugated Abs against CD80 (PE-Cy5) and MHC class II (PerCP-eFluor 710; eBioscience). Samples were incubated with the Ab mix for 15 min at 4°C and subsequently acquired on a LSRII Flow Cytometer (Becton Dickinson). Data were analyzed using FlowJo Software 10.0 (Tree Star) software.
Measurement of urea
Peritoneal lavage SNs, spleen and lung homogenates, and plasma samples of PTEN-deficient mice or their WT littermates were collected and urea levels were determined colorimetrically (Abnova KA1652) according to the manufacturer’s protocol. The biological samples were assayed in duplicates (using 96-well plates), and concentrations were analyzed using a plate reader (Synergy 2; Biotek) at absorbance 520 nm (Normalization BCA).
Immunoblotting
Cells were homogenized and lysed in Laemmli buffer. Proteins were separated by SDS-PAGE on a 10% denaturing polyacrylamide gel, which was stained with Coomassie brilliant blue (Thermo Scientific Pierce) after electrophoresis. Proteins were blotted onto a polyvinylidene difluoride membrane (Millipore) and, after blocking with 5% dry milk/0.1% Tween 20, incubated overnight with primary Ab. The following Abs were used: chicken anti–Arginase 1 (kindly provided by Dr. Morris), rabbit Abs against inducible NO synthase (iNOS; NEB), PTEN, STAT6, pSTAT6 (Thr641), pSTAT3 (Thr705) (Cell Signaling Technology), C/EBPβ, STAT3 (Santa Cruz Biotechnology), or β-actin, GAPDH (Sigma). After incubation for 2 h at room temperature with the respective peroxidase-conjugated secondary Ab and after development with SuperSignal West Femto (Pierce), signals were detected using chemiluminescence (FluorChem HD2 chemiluminescence imager; Alpha Innotech). Bands were analyzed according to their m.w.
ABCD assay
The ABCD assay was performed as described previously (25). In brief, cells were lysed with lysis buffer (10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA pH 8.0, 10% glycerol, 0.5% Nonidet P-40, 1 mM DTT, protease inhibitor), sonicated (6 impulses for 2 s), centrifuged, and SNs were incubated with buffer H (20 mM HEPES pH 7.9, 50 mM KCl, 20% glycerol, 1 mM DTT, 0.1% Nonidet P-40), 2 μg 5′-biotinylated oligo, and 20 μg herring sperm DNA for 5 min at 37°C followed by incubation on ice for 1 h. For the competitor control (comp), a 10-fold excess of nonbiotinylated oligo was added. The negative control contained cell lysate, herring sperm DNA, and buffer H. Buffer H equilibrated streptavidin-agarose beads (Novagen) were added to pull-downs and controls, and then incubated for 30 min at 4°C on a rotator. The beads were centrifuged, washed several times with buffer H, boiled in Laemmli buffer, and separated by SDS-PAGE. C/EBPβ was detected by Western blot (Santa Cruz Biotech). 5′-Biotinylated and nonbiotinylated oligos were ordered from MicroSynth: C/EBPβ_for: TAT TAG CCA ATA TTA GCC AAT ATT AGC CAA TAT TAG CCA, C/EBPβ_rev: TGG CTA ATA TTG GCT AAT ATT GGC TAA TAT TGG CTA ATA. The oligos were designed to resemble the binding site of C/EBPβ in the promotor site upstream of Arg1 (26).
Total RNA isolation, reverse transcription, and quantitative RT-PCR
Cells were homogenized and isolated with Trifast Reagent (PEQLAB Biotechnology GmbH) following the manufacturer’s instructions. cDNAs were transcribed using the High Capacity cDNA Reverse Transcription kit (Fermentas) as indicated in the instruction manual. Expression of mRNA was quantified by real-time PCR using Fast SYBR Green Master Mix (Applied Biosystems) with the StepOne Real-Time PCR System (Applied Biosystems) and primers. Samples were assayed in duplicates dependent on the quality of their melting curves. Levels of target genes were normalized to HPRT or GAPDH and described as fold induction of unstimulated cells. Target primer sequences were as follows: PTEN: forward, 5′-ACA CCG CCA AAT TTA ACT GC-3′, reverse, 5′-TAC ACC AGT CCG TCC CTT TC-3′; Arginase 1: forward, 5′-GTG AAG AAC CCA CGG TCT GT-3′, reverse, 5′-CTG GTT GTC AGG GGA GTG TT-3′; Ym-1: forward, 5′-TTT CTC CAG TGT AGC CAT CCT T-3′, reverse, 5′-TCT GGG TAC AAG ATC CCT GAA-3′; Fizz-1: forward, 5′-CTG GAT TGG CAA GAA GTT CC-3′, reverse, 5′-CCC TTC TCA TCT GCA TCT CC-3′; Stabilin-1: forward, 5′-CCC TCC TTC TGC TCT GTG TC-3′, reverse, 5′-CAA ACT TGG TGT GGA TGT CG-3′; HPRT: forward, 5′-CGC AGT CCC AGC GTC GTG-3′, reverse, 5′-CCA TCT CCT TCA TGA CAT CTC GAG-3′; and GAPDH: forward, 5′-TCG TCC CGT AGA CAA AAT GG-3′, reverse, 5′-TTG AGG TCA ATG AAG GGG TC-3′.
ELISA
SN levels of selected cytokines secreted by lymphocytes or macrophages were measured by using commercially available ELISA for the quantification of IL-2, IFN-γ, IL-6, IL-12/23 (p40/common subunit), and IL-17A (all from eBioscience), and IL-10 and G-CSF (R&D Systems), according to the manufacturer’s protocol. IL-6 and IL-12/23 were analyzed in the SNs of macrophages 24 h after stimulation with LPS. IL-2, IFN-γ, and IL-17A were measured in SNs collected at day 4 of allogeneic MLRs and of restimulated splenocytes and lymphocytes of MOG-immunized mice. In brief, plates were coated with capture Ab, blocked, and diluted samples and standards were loaded for overnight incubation. Next, plates were incubated with detection Ab and then with Streptavidin-HRP (R&D Systems). For development, TMB 2–Component Microwell Peroxidase Substrate Kit (KPL) was used. The reactions were stopped by the addition of 0.5 M H2SO4 in each well. Absorbance values were measured on an ELISA plate reader at 450 nm. Concentrations were calculated from standard curves using purified recombinant cytokine.
Determination of lactate dehydrogenase levels
Lactate dehydrogenase (LDH) levels were measured in the SNs of unstimulated tPMs of WT or myeloid PTEN−/− mice 24 h after incubation, or in the peritoneal lavage using the LDH Cytotoxicity assay kit supplied by Thermo Scientific Pierce. The assay was performed following the manufacturer’s instructions.
Induction of EAE and ex vivo restimulation
Mice were assigned to four groups each consisting of four to eight mice (see study design in Table I). In brief, for immunization, mice were injected with 150 μl of an emulsion containing equal parts of MOG35–55 (1 mg/ml; Charite Berlin) and IFA (Sigma) supplemented with 10 mg/ml Mycobacterium tuberculosis H37Ra (Difco). At time of immunization and second day after immunization, 200 ng pertussis toxin (Calbiochem) was administered by the i.p. route. For in vivo administration of recArgI, mice were i.v. injected with either 10 mg/kg body weight at days −4 and −2 before immunization, or with 10 or 1 mg/kg body weight at days −4 and −2 before and days 5 and 7 after immunization. Mice were observed daily for clinical signs. Progression of EAE was divided into four clinical stages: grade 0: no signs; grade 1: complete floppy tail; grade 2: severe paraparesis; grade 3: tetraparesis; and grade 4: moribund condition. For ex vivo stimulation, lymphocytes and splenocytes were isolated and stimulated with 30 μg/ml of the cognate MOG peptide for 3 d for proliferation analysis and cytokine measurements.
Table I. Experimental design of EAE study.
| Days Before or After Immunization |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −4 | −2 | 0a | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| Control | No treatment | |||||||||||||||||||
| Group 1: recArgI 10 mg/kg | +b | + | + | + | ||||||||||||||||
| Group 2: recArgI 1 mg/kg | + | + | + | + | ||||||||||||||||
| Group 3: recArgI pre 10 mg/kg | + | + | ||||||||||||||||||
Pertussis toxin was injected i.p. at days 0 and 2.
Day 0 (red) indicates time of immunization.
Plus signs (+) indicate day of recArgI administration.
Adoptive transfer EAE
Bone marrow–derived DCs (BMDCs) were generated as described earlier from WT mice. DCs were loaded with MOG35–55 peptide (30 μg/ml) for 24 h in the presence or absence of recArgI (30 μg/ml) at day 6 of differentiation. Transfer of DCs was performed on day 7: after boosting the cells with LPS (100 ng/ml) for 4 h, they were washed thoroughly and transferred s.c. into 2D2 tg mice (106 cells/mouse). In a second experimental approach, the BMDCs were loaded with 50 μg/ml MOG35–55 peptide instead of 30 μg/ml and the immunization of the 2D2 mice was additionally boosted by the administration of CFA and pertussis toxin.
Mixed leukocyte reaction
WT DCs were generated as described. One part of the cells was stimulated with 30 μg/ml recArg I for 24 h at day 6 of differentiation, and the other part acted as unstimulated control. MLR was performed on day 7 of differentiation in absence of recArgI, which was removed by accurate washing. In brief, all cells were activated and loaded with LPS (100 ng/ml; Invitrogen) and OVA (50 μg/ml; Sigma) for 4 h, washed, and cocultivated with OT-II cells. Responder T cells were isolated from spleens of OT-II mice via positive magnetic cell sorting using a pan T cell isolation kit (Miltenyi Biotec), labeled with 7 μM CFSE/1× 107 cells according to the manufacturer’s instructions (Sigma). DCs (100,000 cells) were washed and plated on a 48-well along with responder T cells (500,000 cells) in 500 μl total volume. On day 3 of the coculture, cells were harvested, fixed, and subjected to intracellular staining. For proliferation, cells were harvested in TruCount tubes and analyzed for CFSE dilution.
[3H]Thymidine proliferation assay
For quantification of T cell proliferation after in vitro stimulation, cells were labeled with [3H]thymidine (1 μCi/well) and incubated for 20 h. Incorporation of [3H]thymidine was measured to assess proliferation.
CD4+ T cell phenotyping by flow cytometry
Cells were isolated from MLR in vitro cultures on day 3 and restimulated with PMA/Ionomycin (Sigma) together with GolgiStop (BD Biosciences) and analyzed for intracellular cytokines. The following Abs were used for cytokine staining: anti-mouse CD4-PerCP (clone RM4-5; BD Pharmingen), CD25-PE-Cy7 (clone PC61.5), IL-2–eFlour 450 (clone JES6-5H4), IL-10–Alexa Flour 647 (clone JES5-16E3), IL-17A–PE-Cy7 (clone 17B7), IFN-γ–PE (clone XMG1.2; all from eBioscience). Cell acquisition and data analysis were performed on an LSR 2 flow cytometer (BD Biosciences) and FlowJo software Version 10.0 (Tree Star).
Determination of apoptotic and necrotic cells by flow cytometry
Thioglycolate-elicited macrophages were isolated as mentioned earlier. Early and late apoptosis was determined by staining the cells with FITC-labeled Annexin V (BMS 306 FI/300; eBioscience) in Annexin V binding buffer (422201; Biolegend) and 7-aminoactinomycin D–PerCP–Cy5.5 (Fluka 06648), respectively, for 20 min at room temperature in the dark. Afterward, the cells were fixed with 1% PFA and flow cytometry analysis was performed using BD Accuri C6 cytometer with BD Accuri C6 software (Accuri cytometers, Version 1.0.264.21).
Statistics
Statistical significance of data was calculated by use of an unpaired two-tailed Student t test. Two-way ANOVA analyses were used to analyze two groups over time. Statistical analysis was performed using GraphPad Prism software (GraphPad Software, La Jolla, CA). Results are presented as the mean ± SD. The p values <0.05 were considered statistically significant (p values were expressed as follows: *p < 0.05, **p < 0.01, ***p < 0.001).
Results
LPS upregulates Arginase I in peritoneal macrophages
First, we analyzed the expression levels of macrophage Arginase I in our experimental settings. tPMs were induced by E. coli O111:B4 LPS for 8 and 24 h. Arginase I expression was increased on mRNA level within 8 h (Fig. 1H) and on protein level within 24 h after LPS induction (Fig. 1I). Therefore, we chose these time points for further analysis of LPS-mediated Arginase I expression in dependence of the PI3K/PTEN signaling pathway in macrophages.
FIGURE 1.
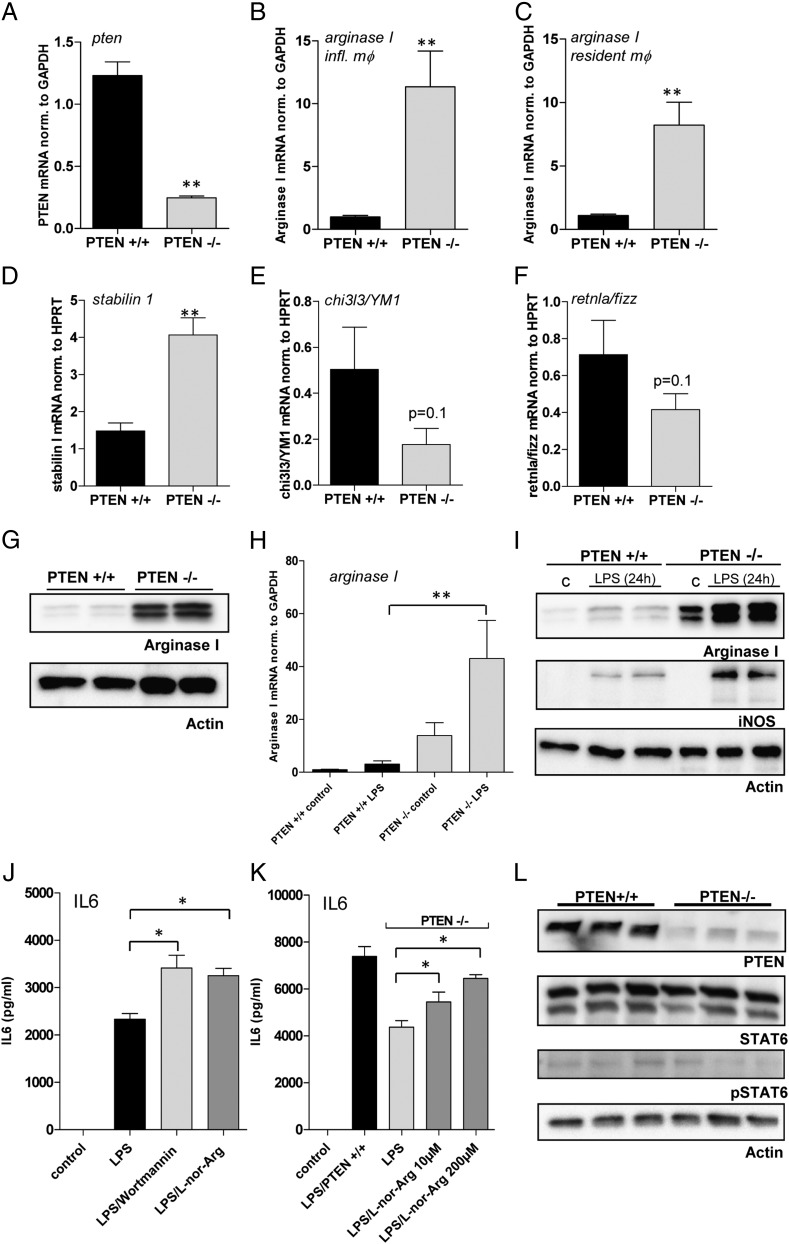
Loss of PTEN leads to a significant increase in Arginase I expression. (A–F) Quantitative real-time analysis of unstimulated PTEN−/− and WT control macrophages. RNA was isolated from tPMs or resident macrophages, and quantitative PCR was performed with (A) PTEN, (B and C) Arginase I, (D) Stabilin1, (E) YM1, and (F) Fizz-specific primers. Target genes were normalized to GAPDH or HPRT as indicated. (G) Western blot analysis of unstimulated PTEN−/− and PTEN+/+ macrophages with Abs specific for Arginase I and β-actin. (H) Quantitative real-time analysis of Arginase I levels in PTEN−/− and PTEN+/+ macrophages 8 h after stimulation with LPS (100 ng/ml). (I) Western blot analysis of PTEN−/− and PTEN+/+ macrophages 24 h poststimulation with LPS. Blots were probed with Abs specific for Arginase I, iNOS, and β-actin. (J and K) tPMs of WT or PTEN−/− mice were stimulated with LPS and treated with the PI3K inhibitor wortmannin (100 nM) or the Arginase inhibitor N-hydroxy-nor-l-arginine (l-nor-Arg) at the indicated concentrations overnight. IL-6 levels were measured by means of ELISA. (L) Naive tPMs of PTEN−/− or WT control mice were subjected to immunoblotting. Levels of PTEN, STAT6, phosphorylated STAT6, and β-actin, as loading control, were determined by Western blot analysis. *p < 0.05, **p < 0.01. Data are representative of three individual experiments (error bars represent SD).
PTEN deletion in macrophages leads to a remarkable increase in Arginase I expression, thereby reducing IL-6 secretion
Previously, we have been using conditional PTEN-deficient mice to investigate the role of PI3K in innate immune responses, in particular, Ag presentation and T cell priming (S. Blüml and G. Schabbauer, submitted for publication). PTEN-deficient macrophages display an immunosuppressed state with decreased TNF-α production and concomitant increased IL-10 secretion when stimulated with PAMPs. We suggested that significantly increased MAPK phosphatase (DUSP1) expression might be one explanation for this phenomenon (7). However, the anti-inflammatory phenotype of PTEN deficiency, leading to altered DUSP expression and reduced MAPK signaling, is still not comprehensively understood. We found increased mRNA transcription of Arginase I in a transcriptome-wide screen in PTEN-deficient DCs and littermate-derived control cells (S. Blüml and G. Schabbauer, unpublished observations). To confirm these findings, we performed transcriptional analyses and found marked upregulation of Arginase I mRNA in unstimulated, naive, PTEN–deficient peritoneal macrophages, in which we observed an efficient gene deletion of >90% each (Fig. 1A and unpublished observations). For these experiments, we isolated resident peritoneal macrophages as well as sterile inflammation-induced peritoneal macrophages (Fig. 1B, 1C). In line with upregulated Arginase I mRNA expression, we found enhanced mRNA transcription for another M2 marker, stabilin 1, in unstimulated PTEN−/− tPMs (Fig. 1D). Although these data indicate an alternative activation, we even found a trend (p = 0.1) for reduced expression of other prominent M2 markers, YM1 and FIZZ (Fig. 1E, 1F).
In addition, we analyzed the protein expression of tPMs. In line with the mRNA data, Arginase I protein was markedly induced by the PTEN deletion (Fig. 1G). Because we have previously investigated the role of PTEN in LPS-induced signaling events, we analyzed Arginase I expression in response to LPS, where similar to baseline, it was also induced (Fig. 1H). Notably, the expression of the prominent M1 marker iNOS is also enhanced in PTEN-deficient macrophages, albeit only in response to LPS.
Arginase I possesses immunomodulatory functions in myeloid cells (10, 13, 16, 17, 27), and a role for Arginase/urea cycle–derived polyamines in the regulation of ILs has been described (28). We stimulated WT macrophages with LPS or LPS in combination with either the fungal pan-PI3K inhibitor wortmannin, which has been reported to enhance cytokine synthesis upon LPS induction in vitro and in vivo (29, 30), or the Arginase inhibitor, N-hydroxy-nor-l-arginine (l-nor-Arg). Inhibition of Arginase I significantly enhanced IL-6 production in macrophages (Fig. 1J). Next, we evaluated the effects of the Arginase-specific inhibitor on the diminished cytokine production of PTEN−/− macrophages as compared with WT macrophages. Indeed, we found a significant, but only partial, restoration of the WT IL-6 production upon treatment with l-nor-Arg in PTEN-deficient tPMs (Fig. 1K). Thus, the pharmacological inhibition of Arginase and the ablation of the anti-inflammatory phenotype in PTEN-deficient Arginase-overexpressing tPMs support the notion that Arginase I contributes to the PI3K-mediated modulation of inflammatory responses. To elucidate the molecular mechanism responsible for the PTEN-mediated gene regulation of Arginase I, we examined several potential candidate TFs. The most prominent regulator of Arginase I is STAT6, which is activated by IL-4 and/or IL-13 in macrophages (20). Unstimulated tPMs, either PTEN-deficient cells or WT cells derived from littermate control animals, did not show any overt changes in STAT6 total protein content or activated STAT6 as measured by phospho-specific STAT6 Abs (Fig. 1L).
PTEN deletion in macrophages upregulates the TF C/EBPβ and activates STAT3, which is crucial for Arginase I promotor activation
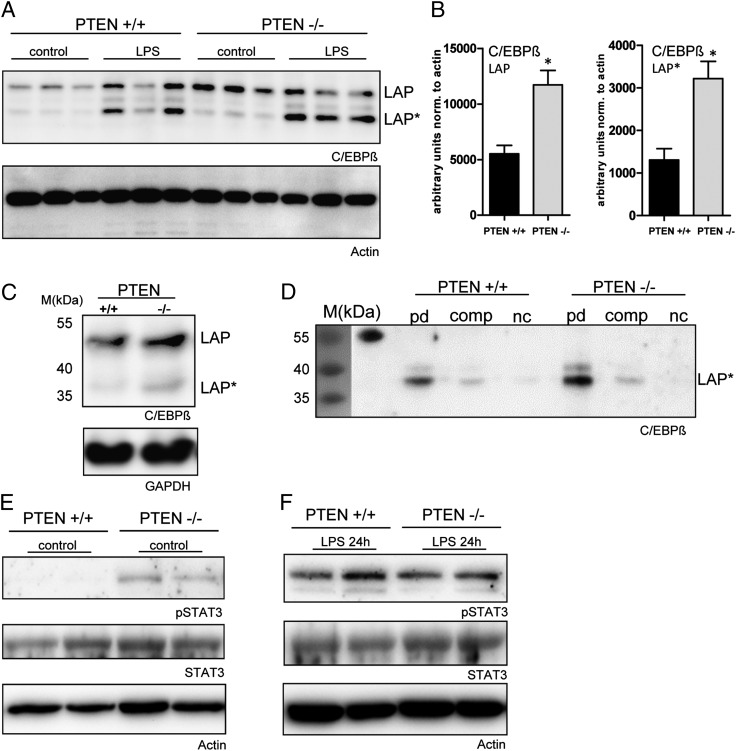
To study the contribution of other candidate TFs, besides STAT6, we analyzed the involvement of C/EBPβ, which has been shown to contribute to pattern recognition receptor–mediated regulation of Arginase I (16). Therefore, we tested the possibility that PTEN regulates C/EBPβ, which, in turn, mediates the constitutive upregulation of Arginase I. Indeed, we found an upregulation of C/EBPβ protein in PTEN-deficient tPMs (Fig. 2A). This was true for two isoforms of C/EBPβ, LAP and LAP* (Fig. 2B). Further induction of the TF was observed within 8 h after LPS induction. However, we could not identify an additional upregulation of C/EBPβ by PTEN deletion upon TLR4 activation, at least at the time point we have evaluated (8 h). Because maximal Arginase I expression is observed 24 h after LPS induction, we cannot exclude that C/EBPβ differential expression might occur earlier in activated PTEN-deficient macrophages. Because of the fact that C/EBPβ is a TF acting on specific DNA elements, we wanted to investigate binding of C/EBPβ to the Arginase enhancer element containing a number of different TF consensus binding sites. We introduced an ABCD binding assay to evaluate C/EBPβ DNA binding properties using biotinylated oligos spanning the C/EBPβ consensus site within the Arginase enhancer 3.8 kb upstream of the transcription start site. First, we tested the suitability of this experimental system in HEK cells overexpressing C/EBPβ. We found that overexpressed C/EBPβ efficiently bound the oligo, which could be precipitated together with the TF, in particular the LAP* isoform, bound to it (Supplemental Fig. 1A). Next, we analyzed effects of PTEN deficiency on C/EBPβ DNA binding in macrophages. First, we determined the presence of C/EBPβ in the lysates we used for the ABCD binding assays. Furthermore, we determined equal input for the ABCD assay by reprobing against GAPDH (Fig. 2C). Analysis of TF binding to the Arginase oligo suggests that in addition to increased protein levels, enhanced binding of C/EBPβ occurred. Again, we found LAP* to be the dominant isoform binding to the Arginase enhancer oligo (Fig. 2D).
FIGURE 2.
C/EBPβ is upregulated and shows higher Arginase I enhancer binding activity in PTEN-deficient macrophages. (A) tPMs of PTEN−/− or WT control mice were stimulated with LPS (100 ng/ml) for 8 h. Levels of C/EBPβ and Actin were determined by Western blot analysis. (B) Quantification of immunoblots of unstimulated PTEN−/− or WT tPMs is presented and normalized to β-actin. (C) Lysates of naive tPMs used for the ABCD were tested for C/EBPβ expression. GAPDH was used to confirm equal input. (D) ABCD pull-down (PD) of C/EBPβ with biotinylated Arginase I enhancer oligos from lysates of tPMs obtained from PTEN−/− and WT littermate control mice: PD, unconjugated competitor oligos (comp) and negative controls (nc). The prestained marker (35–55 kDa) is presented on the left. The first lane of the immunoblot indicates the marker and the cross-reactivity to the 55-kDa marker protein. (E and F) Naive or LPS-stimulated (24 h) tPMs of PTEN−/− or WT control mice were subjected to immunoblotting. Levels of STAT3, phosphorylated STAT3, and Actin, as loading control, were determined by Western blot analysis. *p < 0.05. Data are representative of experiments with three mice per group (A and B) (error bars represent SD).
In addition, we explored the potential contribution of STAT3. We have previously shown that PTEN deficiency in naive macrophages leads to a constitutive IL-10 release (7). Furthermore, Qualls et al. (31) reported autocrine activation via IL-6, IL-10, and G-CSF, promoting Arginase I expression. Analysis of STAT3 activation revealed that indeed phosphorylation at position Thr705 of STAT3 was clearly enhanced in PTEN-deficient macrophages as compared with WT cells derived from littermate mice (Fig. 2E). The activation of unstimulated macrophages is probably due to IL-10, because expression of IL-6, as well as G-CSF, on protein level could not be detected in unstimulated cells. Analysis of LPS-induced macrophages 24 h postinduction did not show overt differences in the STAT3 phosphorylation status. A reason for this is probably simultaneous LPS-induced production of STAT3 inducers such as IL-6, IL-10, and G-CSF. Notably, we found a marked LPS-induced increase of G-CSF release (data not shown) and a reduction for IL-6 release (Fig. 1K) in PTEN-deficient peritoneal macrophages. We conclude that PI3K/PTEN regulates Arginase I expression via C/EBPβ and STAT3.
Arginase I not only acts in the cytosol, but its release into the extracellular space is promoted by sustained PI3K activation
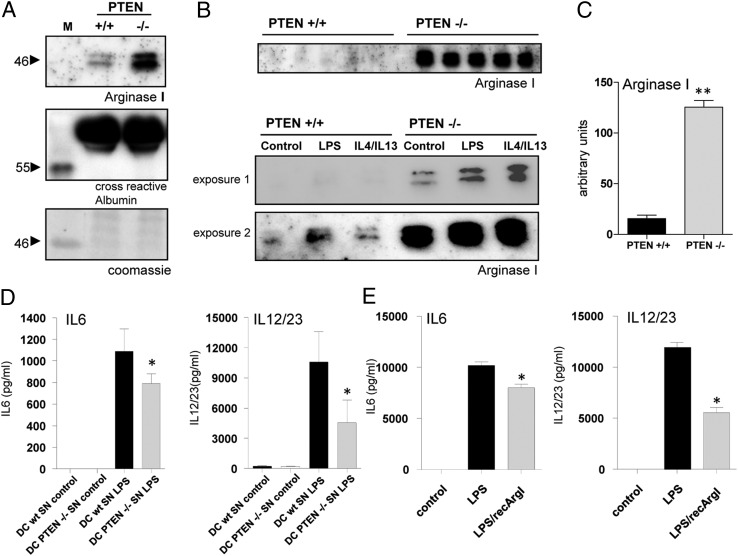
We have previously shown an anti-inflammatory phenotype of PTEN-deficient macrophages and attributed some of the observed effects to reduced MAPK activation and differential DUSP regulation. However, still some of the anti-inflammatory mechanisms in place are not fully understood. Previous studies suggested extracellular properties of Arginase I (10). When we analyzed the SNs of peritoneal macrophages, we surprisingly found that Arginase I is indeed released to the extracellular space. Deficiency of PTEN promoted that effect, which was observed even in unstimulated tPMs (Fig. 3A, top panel). This finding is not related to increased apoptosis as measured by markers of early and late apoptosis, Annexin V and 7-aminoactinomycin D, or cytotoxicity as measured by LDH release (Supplemental Fig. 2A, 2B). Because we did not have markers available to normalize Arginase I immunoblotting, we chose cross-reactive albumin to show equal SN input and total protein content by Coomassie blue stainings in the region of interest around 46 kDa (Fig. 3A, bottom panel). To quantify these unexpected findings, we analyzed macrophages derived from 5 littermate WT and knockout animals. Indeed, we found a >10-fold increase in extracellular Arginase I when PTEN was ablated in macrophages (Fig. 3B, top panel, 3C). Furthermore, we analyzed Arginase I secretion in response to LPS or combined activation by IL-4 and IL-13. Minimal secretion of Arginase I was observed in WT cells. Importantly, we observed increased secretion in PTEN-deficient compared with WT macrophages (Fig. 3B, bottom panel). This finding indicates that Arginase I in the extracellular environment of macrophages might exhibit anti-inflammatory properties in a paracrine fashion, although we were not able to detect significant differences in spleens, lungs, plasma, and peritoneal lavage harvested from unchallenged myeloid PTEN-deficient mice and littermate controls (data not shown).
FIGURE 3.
Constitutively active PI3K promotes Arginase I expression and release into the extracellular space. (A) SNs of unstimulated tPMs of WT and PTEN-deficient mice immunoblotted against Arginase I. Equal loading is presented by Coomassie blue staining and cross-reactive albumin. (B, top panel, and C) Western blot analysis and quantification of Arginase I in SNs of tPMs of WT and PTEN-deficient mice (n = 5). (B, bottom panel) Arginase I immunoblot analysis of tPMs activated with LPS (100 ng/ml) or IL-4/IL-13 (5 ng/ml) for 24 h. (D) Bone marrow DCs incubated with conditioned medium of WT or PTEN−/− tPMs and subsequently activated with LPS. Cytokine secretion was assessed after stimulation for 24 h. (E) Cytokine production of BMDCs preincubated with recArgI (30 μg/ml) and subsequently activated with LPS for 24 h. *p < 0.05, **p < 0.01 (error bars represent SD).
We next examined cytokine production and the potential of APCs to polarize T cells, by transferring conditioned media (Fig. 3A–C) to WT bone marrow–derived GM-CSF differentiated DCs and determining their inflammatory response to LPS. Interestingly, we found protein levels of T cell polarizing cytokines IL-6 and IL-12 p40 (Fig. 3D), the common subunit of IL-12 and IL-23 (denoted as IL-12/23), to be reduced. Next, we attempted to mimic the situation present in the Arginase I high/conditioned media from PTEN-deficient macrophages using recArgI in combination with LPS treatment. Fortunately for us, our study benefits from recent developments using recombinant Arginase in the area of cancer treatment. Administration of recArgI (stabilized by pegylation) proved to be promising therapeutically in combination therapies in the treatment of hepatocellular carcinoma (21). In concordance with the data from conditioned media, we detected reduced expression levels of IL-6 and IL-12/23. Interestingly, this effect we observed was more pronounced for IL-12/23 compared with IL-6 (Fig. 3E).
Free Arginase I potently inhibits T cell polarization
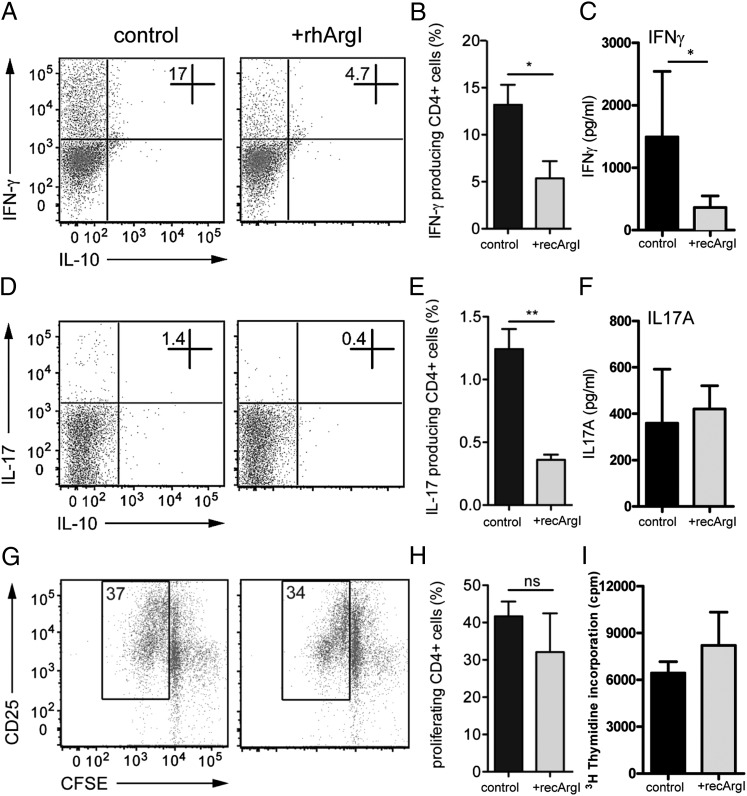
Because we could provide clear evidence for the secretion of Arginase I into the extracellular space, although the mechanism responsible for that is currently unknown, we made use of recArgI to mimic the situation in PTEN-deficient macrophages that release high amounts of Arginase I. To analyze effects on Ag presentation and T cell polarizing properties, we preconditioned bone marrow–derived GM-CSF differentiated WT DCs with recArgI overnight before loading them with OVA and stimulating with LPS. To avoid potential effects on T cells, we efficiently removed recArgI from the DCs by multiple washing steps before cultivating them together with isolated OT-II T cells in a 1:5 ratio. Treatment of DCs with recArgI in a concentration of 30 μg/ml did not alter the surface expression of prototypic DC activation markers CD80 and MHCII, as measured by flow cytometry (Supplemental Fig. 3A, 3B). Importantly, we observed a significant reduction of Th1 and Th17 signature cytokines IFN-γ (Fig. 4A, 4B) and IL-17A (Fig. 4D, 4E) expressing CD4+ T cells. Notably, IL-17–producing cells were present, but in low numbers. When we analyzed the T cell cytokines that were secreted during the MLR after LPS/OVA priming, we found significant differences for the release of IFN-γ, whereas IL-17A was not significantly different (Fig. 4C, 4F). Proliferation as measured by CFSE dilution in dividing T cells was evaluated. However, we did not find significant changes in the presence of recArgI (Fig. 4G, 4H). In line with these data, we did not observe an increased [3H]thymidine incorporation in proliferating OT-II T cells in cocultures (Fig. 4I) of DCs and T cells. Therefore, we assume that extracellular Arginase I in the presence of APCs ameliorates T cell priming, thereby reducing the capacity to polarize preferentially in Th1 cells. To verify that, we decided to test the ability of recArgI in vivo in a clinically relevant model for T cell–mediated autoimmune disease.
FIGURE 4.
Arginase I potently inhibits T cell polarization. (A and D) CD4+-specific cytokine staining for expression of IFN-γ and IL-17 in OT-II T cells stimulated for 3 d with OVA-loaded (50 μg/ml), LPS-pulsed (100 ng/ml), and recArgI (30 μg/ml) pretreated or untreated (control) bone marrow DCs, followed by PMA/Ionomycin T cell stimulation. (B and E) Relative abundance of IFN-γ+ and IL17+ cell populations among CD4+ T cells, expressed as percentage. (G and H) Representative FACS analysis and quantified proliferation of CFSE-stained CD4+ T cells 3 d after coculture with control or recArgI (30 μg/ml) pretreated DCs. (C and F) SNs of cocultures were assessed by ELISA for IFN-γ and IL-17A production by responder T cells. (I) [3H]Thymidine incorporation in proliferating CD4+ T cells was measured. *p < 0.05, **p < 0.01. ns, not significant. Data are representative of experiments with at least three mice per group (error bars represent SD).
Treatment with recombinant Arginase I at early stages of disease development protects from development of EAE
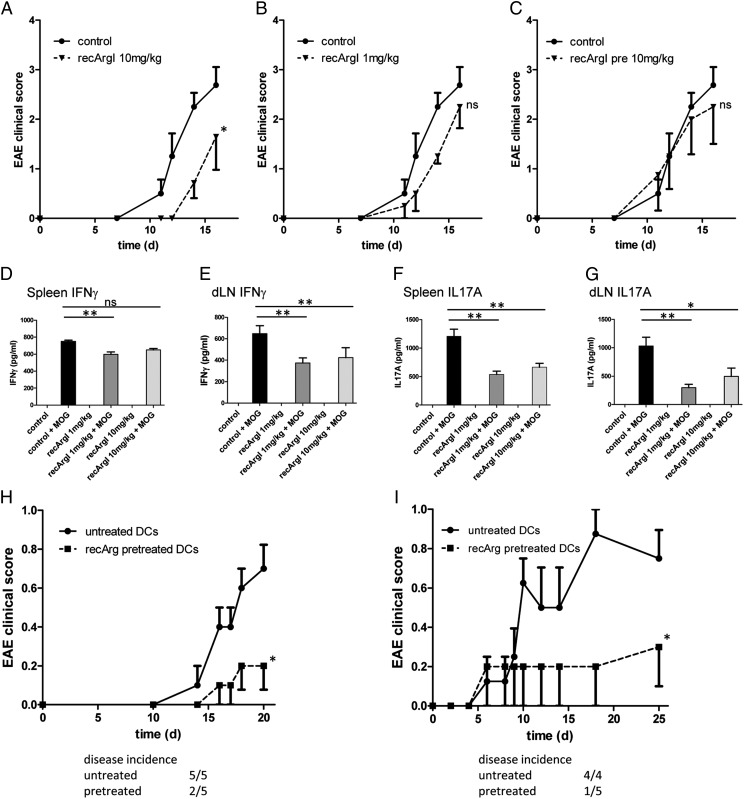
Autoimmune diseases are characterized by a dysregulated immune system that targets self-Ags. The murine model for multiple sclerosis, the EAE, reflects some of the features of the human autoimmune pathology and is a T cell–driven model upon immunization with the MOG35–55 peptide (32). EAE was induced in WT mice by immunization with MOG35–55 peptide in CFA, and pertussis toxin was administered at the indicated time points (Table I). We aimed to determine the potential efficacy of recArgI to ameliorate disease progression. Furthermore, we wanted to treat in a narrow time frame, although Ag presentation and T cell polarization were still ongoing, but leave out the later phases of the disease model. Therefore, we administered recArgI in two different concentrations before and shortly after immunization (Table I). The control group had signs of EAE visible after day 12, further increasing until day 17 as expected. Pretreatment with recArgI (10 mg/kg) did not have any effect on the course of disease (Fig. 5C). Pretreatment and posttreatment with recArgI (10 mg/kg), in contrast, showed a significant reduction in the onset, as well as the magnitude, of the disease (Fig. 5A). In the same experimental setup, we also tried a lower dose of recArgI (1 mg/kg). Treatment delayed the onset of disease, but disease progression and severity were not significantly inhibited by the treatment with the lower dose of recArgI (Fig. 5B).
FIGURE 5.
Treatment with recombinant Arginase I at early stages of disease development protects mice from development of EAE. (A–C) Disease development as indicated by clinical score in recArgI-treated versus control WT mice. (D–G) Cytokine secretion after ex vivo restimulation with MOG35–55 peptide of spleen cells and draining inguinal lymph node (dLN) cells from MOG35–55–immunized mice belonging to groups 1 and 2 (see Table I). (H) Clinical score of 2D2 TCR tg mice after transfer of recArgI pretreated or untreated BMDCs, which were pulsed with MOG35–55 peptide (30 μg/ml) and LPS (100 ng/ml). (I) Disease progression of 2D2 TCR tg mice receiving recArgI pretreated or untreated DCs pulsed with MOG35–55 peptide (50 μg/ml) and primed with LPS (100 ng/ml) and additional administration of CFA and pertussis toxin. *p < 0.05 in the EAE, (two-way ANOVA between the groups as indicated) *p < 0.05, **p < 0.01 for the evaluation of cytokine release. ns, not significant. Data are representative of experiments with four to eight mice per group (error bars represent SD).
To analyze the MOG-specific T cell response on a molecular level, we harvested spleens and the draining lymph node (inguinal), and restimulated the splenocytes and the lymph node cells, derived from recArgI-treated and control EAE mice, in vitro with the MOG35–55 peptide. Restimulation of cells without further activation by exogenous stimuli led to a marked increase in IFN-γ and IL-17A secretion into the SNs (see control versus control + MOG in Fig. 5D–G). Interestingly, we found highly significant differences for reduced IFN-γ (Fig. 5D, 5E) and IL-17A (Fig. 5F, 5G) levels in both pretreatment and posttreatment groups (1 and 10 mg/kg recArgI), in particular in the draining lymph node. To further elaborate on cell-specific effects of recArgI treatment in vivo, we established a DC transfer model into tg mice expressing a MOG35–55 specific TCR on the majority of CD4+ T cells (2D2 TCR tg). GM-CSF differentiated DCs were pretreated with or without recArgI (30 μg/ml) before loading with MOG35–55 peptide and priming with LPS. Then the DCs were transferred into 2D2 TCR tg recipient mice. We chose two different approaches with and without the administration of CFA and pertussis toxin, which is also used in the standard EAE model. The immunologic boost by CFA and pertussis promoted the onset of disease, day 6 versus day 14. In both experimental setups, we observed significant protection from the development of signs of the disease when DCs were pretreated ex vivo with recArgI (Fig. 5H, 5I).
These data suggest that recArgI treatment ex vivo on DCs and in vivo during the phase of Ag presentation at least at the higher concentration efficiently blunts EAE pathology in mice through a diminished capacity of CD4+ T cells to produce cytokines IFN-γ and IL-17A indispensable for the development of EAE.
Discussion
In this article, we show that sustained activation of the PI3K signaling pathway by deletion of PTEN upregulates Arginase I in cells of monocytic origin. Arginase I, in turn, acts as an immunomodulating protein on the intracellular and extracellular levels. We could previously show that deletion of PTEN in myeloid cells leads to reduced TNF-α but increased IL-10 expression in the lungs of mice infected with Streptococcus pneumoniae (9). Increased IL-10 secretion together with increased expression of Arginase I points toward an alternative activation of PTEN-deficient macrophages, which is supported by our observation of augmented levels of stabilin 1 in naive macrophages in vitro. Our findings thus indicate that PTEN modulates an M2-like state of macrophages. Paradoxically, prominent markers for alternative activation including chi3l3/YM1 and retnla/FIZZ were even downregulated upon PTEN deficiency, and the typical M1 marker iNOS is strongly upregulated in response to LPS in pten-deficient macrophages. Therefore, it is difficult to argue that unopposed PI3K activation leads to an alternatively activated phenotype of macrophages deleted for PTEN, but it may contribute to it to a certain extent, which has already been reported for the phosphatase SHIP1 (33).
Importantly, Arginase I was one of the genes in macrophages, where the most apparent differences in expression were observed, when PTEN is deficient. We propose that PI3K/PTEN-dependent Arginase expression is probably mediated by differential expression and increased transactivation properties of C/EBPβ, leading to the activation of the arginase I gene promoter. It has been shown that certain enhancer elements upstream of the transcriptional start site are indispensable for full promoter activation. This enhancer element contains consensus DNA binding sites for the TFs STAT6, PU1, and C/EBPβ (26). Because STAT6 was unaffected by PTEN deletion, we focused on C/EBPβ. One more argument in favor of the involvement of C/EBPβ is based on the fact that activation of CREB is dependent on PI3K activation and subsequent GSK3β inhibition (34, 35), and at the same time CREB transactivating properties are required for full C/EBPβ induction under inflammatory conditions (36). Our data showing that C/EBPβ, in particular the isoform LAP*, is not only upregulated but binds to the Arginase promoter consensus DNA sequences under normal conditions, are clearly supported by the previously published findings. In addition, we found activation of STAT3 under normal conditions in PTEN−/− macrophages, which goes in line with increased IL-10 cytokine release under nonstimulated conditions in tPMs (7). This observation is supported by recent findings suggesting that different STAT3-inducing cytokines, such as IL-6, IL-10, and G-CSF, can activate Arginase I expression in a paracrine or autocrine fashion (31). However, our data do not fully explain the Arginase I upregulation upon LPS induction in PTEN-deficient macrophages. Under these conditions, a multitude of STAT3-inducing cytokines is released and we could not see significant differences for C/EBPβ expression or STAT3 phosphorylation.
Unexpectedly, we found that Arginase I is secreted in high amounts by PTEN−/− macrophages, which cannot be attributed to apoptosis or necrosis. Besides the fact that dying apoptotic cells, in particular neutrophil granulocytes, passively release Arginase I (13), recent findings in human neutrophils indicate that Arginase I might have extracellular functions after its release together with azurophil granules (37). However, the finding that PTEN modulates this phenomenon to such an extent has not been described yet. Extracellular Arginase contributes to T cell dysfunction through Arginine depletion. We tested the hypothesis that extracellular Arginase I might have anti-inflammatory functions not only on T cells, but also specifically on APCs, namely, GM-CSF differentiated DCs. SNs derived from PTEN−/− macrophages with high amounts of Arginase I, as well as recombinant Arginase I, proved to limit the expression of T cell polarizing cytokines IL-6 and the common subunit for IL-12/23, although we speculate that other factors such as IL-10 contribute to the anti-inflammatory effects observed by the addition of SNs. This is in line with the fact that Arginase I is an attractive candidate to contribute to the observed anti-inflammatory phenotype. It is assumed that secondary metabolites of the urea cycle, so-called polyamines, such as Spermine, repress cytokine production in myeloid cells (28). Also, other arginine metabolites exhibit potent anti-inflammatory properties reducing, in particular, the expression of IL12 p40 in macrophages (38). In summary, our findings led us to the assumption that PTEN deficiency with concomitant sustained PI3K activation results in massive Arginase overexpression and release from macrophages, which might contribute to the anti-inflammatory phenotype observed in PTEN-deficient myeloid cells.
In vitro and in vivo experiments point at the immunomodulatory properties of extracellular Arginase I. Our findings suggest that DCs are an additional target for Arginase I. In vitro analyses, as well as in vivo EAE Ag-presenting cell transfer experiments, clearly show that treatment of DCs with recombinant Arginase I diminishes production of T cell polarizing cytokines and ameliorates disease progression in vivo. This is confirmed by experiments with recArgI-treated, OVA-presenting DCs using OTII T cells, which cannot polarize properly into Th1 or Th17 cells without significantly hampering their ability to proliferate.
Although there are numerous reports regarding Arginase I showing its contribution to inflammation-associated disease progression, recent findings indicate cell-type–specific and organ-specific immunologic effects of Arginase I. In murine lungs, myeloid cell–specific Arginase I deletion did not significantly alter the pathophysiology of several different clinically relevant pulmonary animal models (39). To test the effects of recombinant Arginase in in vivo settings involving Ag presentation, cytokine release, and T cell polarization, we have applied two different experimental approaches and found a robust beneficial effect in the murine EAE model: either treat DCs ex vivo with recArgI and adoptively transfer the cells or treat in vivo with recArgI at the time of Ag presentation and T cell polarization. Treatment with Arginase I reduces the capacity of Ag-specific CD4+ Th cells to produce pathogenic IFN-γ and IL-17A. Our results are in agreement with recently published findings supporting the role of Arginase I in myeloid-derived suppressor cells, which clearly modulates T cell functions in graft versus host disease (40). Although experiments using mouse models with Arginase deficiency advise the opposite, the beneficial effects we could observe in the EAE model, together with others in GVHD, suggest that under certain conditions where immunological imbalances occur, including autoimmune disease, Arginase I is an important modulator of the innate and adaptive immune responses. Therefore, the administration of clinically approved recArgI might be a promising future option in the treatment of autoimmune disorders.
Acknowledgments
We thank all colleagues, in particular Omar Sharif, for critically reviewing the manuscript. We thank Johannes Schmid and Alice Assinger for expert scientific and technical advice.
This work was supported by Austrian Science Fund (FWF) Grants P23730 (to S.B. and G.S.) and P24802 (to G.S.).
The online version of this article contains supplemental material.
- ABCD
- avidin–biotin complex DNA
- BCG
- bacillus Calmette–Guerin
- BMDC
- bone marrow–derived DC
- DC
- dendritic cell
- EAE
- experimental autoimmune encephalomyelitis
- iNOS
- inducible NO synthase
- LDH
- lactate dehydrogenase
- MOG
- myelin oligodendrocyte glycoprotein
- recArgI
- recombinant human pegylated Arginase I
- SN
- supernatant
- TF
- transcription factor
- tg
- transgenic
- tPM
- thioglycollate-elicited peritoneal macrophage
- WT
- wild type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Engelman J. A., Luo J., Cantley L. C. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7: 606–619 [DOI] [PubMed] [Google Scholar]
- 2.Ferguson G. J., Milne L., Kulkarni S., Sasaki T., Walker S., Andrews S., Crabbe T., Finan P., Jones G., Jackson S., et al. 2007. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat. Cell Biol. 9: 86–91 [DOI] [PubMed] [Google Scholar]
- 3.Heit B., Robbins S. M., Downey C. M., Guan Z., Colarusso P., Miller B. J., Jirik F. R., Kubes P. 2008. PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat. Immunol. 9: 743–752 [DOI] [PubMed] [Google Scholar]
- 4.Aksoy E., Taboubi S., Torres D., Delbauve S., Hachani A., Whitehead M. A., Pearce W. P., Berenjeno I. M., Nock G., Filloux A., et al. 2012. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. [Published erratum appears in 2013 Nat. Immunol. 14: 877.] Nat. Immunol. 13: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaurasia B., Mauer J., Koch L., Goldau J., Kock A. S., Brüning J. C. 2010. Phosphoinositide-dependent kinase 1 provides negative feedback inhibition to Toll-like receptor-mediated NF-kappaB activation in macrophages. Mol. Cell. Biol. 30: 4354–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukao T., Koyasu S. 2003. PI3K and negative regulation of TLR signaling. Trends Immunol. 24: 358–363 [DOI] [PubMed] [Google Scholar]
- 7.Günzl P., Bauer K., Hainzl E., Matt U., Dillinger B., Mahr B., Knapp S., Binder B. R., Schabbauer G. 2010. Anti-inflammatory properties of the PI3K pathway are mediated by IL-10/DUSP regulation. J. Leukoc. Biol. 88: 1259–1269 [DOI] [PubMed] [Google Scholar]
- 8.Luyendyk J. P., Schabbauer G. A., Tencati M., Holscher T., Pawlinski R., Mackman N. 2008. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J. Immunol. 180: 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schabbauer G., Matt U., Günzl P., Warszawska J., Furtner T., Hainzl E., Elbau I., Mesteri I., Doninger B., Binder B. R., Knapp S. 2010. Myeloid PTEN promotes inflammation but impairs bactericidal activities during murine pneumococcal pneumonia. J. Immunol. 185: 468–476 [DOI] [PubMed] [Google Scholar]
- 10.Munder M. 2009. Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol. 158: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peranzoni E., Marigo I., Dolcetti L., Ugel S., Sonda N., Taschin E., Mantelli B., Bronte V., Zanovello P. 2007. Role of arginine metabolism in immunity and immunopathology. Immunobiology 212: 795–812 [DOI] [PubMed] [Google Scholar]
- 12.Makarenkova V. P., Bansal V., Matta B. M., Perez L. A., Ochoa J. B. 2006. CD11b+/Gr-1+ myeloid suppressor cells cause T cell dysfunction after traumatic stress. J. Immunol. 176: 2085–2094 [DOI] [PubMed] [Google Scholar]
- 13.Munder M., Schneider H., Luckner C., Giese T., Langhans C. D., Fuentes J. M., Kropf P., Mueller I., Kolb A., Modolell M., Ho A. D. 2006. Suppression of T-cell functions by human granulocyte arginase. Blood 108: 1627–1634 [DOI] [PubMed] [Google Scholar]
- 14.Munder M., Eichmann K., Morán J. M., Centeno F., Soler G., Modolell M. 1999. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J. Immunol. 163: 3771–3777 [PubMed] [Google Scholar]
- 15.Rutschman R., Lang R., Hesse M., Ihle J. N., Wynn T. A., Murray P. J. 2001. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J. Immunol. 166: 2173–2177 [DOI] [PubMed] [Google Scholar]
- 16.El Kasmi K. C., Qualls J. E., Pesce J. T., Smith A. M., Thompson R. W., Henao-Tamayo M., Basaraba R. J., König T., Schleicher U., Koo M. S., et al. 2008. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 9: 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munder M., Mollinedo F., Calafat J., Canchado J., Gil-Lamaignere C., Fuentes J. M., Luckner C., Doschko G., Soler G., Eichmann K., et al. 2005. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 105: 2549–2556 [DOI] [PubMed] [Google Scholar]
- 18.Mattila J. T., Ojo O. O., Kepka-Lenhart D., Marino S., Kim J. H., Eum S. Y., Via L. E., Barry C. E., III, Klein E., Kirschner D. E., et al. 2013. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. 191: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswas S. K., Mantovani A. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11: 889–896 [DOI] [PubMed] [Google Scholar]
- 20.Gordon S., Martinez F. O. 2010. Alternative activation of macrophages: mechanism and functions. Immunity 32: 593–604 [DOI] [PubMed] [Google Scholar]
- 21.Cheng P. N., Lam T. L., Lam W. M., Tsui S. M., Cheng A. W., Lo W. H., Leung Y. C. 2007. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 67: 309–317 [DOI] [PubMed] [Google Scholar]
- 22.Domingues H. S., Mues M., Lassmann H., Wekerle H., Krishnamoorthy G. 2010. Functional and pathogenic differences of Th1 and Th17 cells in experimental autoimmune encephalomyelitis. PLoS ONE 5: e15531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki A., Yamaguchi M. T., Ohteki T., Sasaki T., Kaisho T., Kimura Y., Yoshida R., Wakeham A., Higuchi T., Fukumoto M., et al. 2001. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14: 523–534 [DOI] [PubMed] [Google Scholar]
- 24.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8: 265–277 [DOI] [PubMed] [Google Scholar]
- 25.Baumann S., Dostert A., Novac N., Bauer A., Schmid W., Fas S. C., Krueger A., Heinzel T., Kirchhoff S., Schütz G., Krammer P. H. 2005. Glucocorticoids inhibit activation-induced cell death (AICD) via direct DNA-dependent repression of the CD95 ligand gene by a glucocorticoid receptor dimer. Blood 106: 617–625 [DOI] [PubMed] [Google Scholar]
- 26.Pauleau A. L., Rutschman R., Lang R., Pernis A., Watowich S. S., Murray P. J. 2004. Enhancer-mediated control of macrophage-specific arginase I expression. J. Immunol. 172: 7565–7573 [DOI] [PubMed] [Google Scholar]
- 27.Pesce J. T., Ramalingam T. R., Mentink-Kane M. M., Wilson M. S., El Kasmi K. C., Smith A. M., Thompson R. W., Cheever A. W., Murray P. J., Wynn T. A. 2009. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 5: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M., Caragine T., Wang H., Cohen P. S., Botchkina G., Soda K., Bianchi M., Ulrich P., Cerami A., Sherry B., Tracey K. J. 1997. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J. Exp. Med. 185: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guha M., Mackman N. 2002. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J. Biol. Chem. 277: 32124–32132 [DOI] [PubMed] [Google Scholar]
- 30.Schabbauer G., Tencati M., Pedersen B., Pawlinski R., Mackman N. 2004. PI3K-Akt pathway suppresses coagulation and inflammation in endotoxemic mice. Arterioscler. Thromb. Vasc. Biol. 24: 1963–1969 [DOI] [PubMed] [Google Scholar]
- 31.Qualls J. E., Neale G., Smith A. M., Koo M. S., DeFreitas A. A., Zhang H., Kaplan G., Watowich S. S., Murray P. J. 2010. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Sci. Signal. 3: ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassmann H., van Horssen J. 2011. The molecular basis of neurodegeneration in multiple sclerosis. FEBS Lett. 585: 3715–3723 [DOI] [PubMed] [Google Scholar]
- 33.Rauh M. J., Ho V., Pereira C., Sham A., Sly L. M., Lam V., Huxham L., Minchinton A. I., Mui A., Krystal G. 2005. SHIP represses the generation of alternatively activated macrophages. Immunity 23: 361–374 [DOI] [PubMed] [Google Scholar]
- 34.Martin M., Rehani K., Jope R. S., Michalek S. M. 2005. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., Brown J., Gu Z., Garcia C. A., Liang R., Alard P., Beurel E., Jope R. S., Greenway T., Martin M. 2011. Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-β-signaling pathways regulates the innate inflammatory response. J. Immunol. 186: 5217–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R. G., Rosenthal N., Nerlov C. 2009. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 106: 17475–17480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotondo R., Bertolotto M., Barisione G., Astigiano S., Mandruzzato S., Ottonello L., Dallegri F., Bronte V., Ferrini S., Barbieri O. 2011. Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J. Leukoc. Biol. 89: 721–727 [DOI] [PubMed] [Google Scholar]
- 38.Haskó G., Kuhel D. G., Marton A., Nemeth Z. H., Deitch E. A., Szabó C. 2000. Spermine differentially regulates the production of interleukin-12 p40 and interleukin-10 and suppresses the release of the T helper 1 cytokine interferon-gamma. Shock 14: 144–149 [DOI] [PubMed] [Google Scholar]
- 39.Barron L., Smith A. M., El Kasmi K. C., Qualls J. E., Huang X., Cheever A., Borthwick L. A., Wilson M. S., Murray P. J., Wynn T. A. 2013. Role of arginase 1 from myeloid cells in th2-dominated lung inflammation. PLoS ONE 8: e61961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Highfill S. L., Rodriguez P. C., Zhou Q., Goetz C. A., Koehn B. H., Veenstra R., Taylor P. A., Panoskaltsis-Mortari A., Serody J. S., Munn D. H., et al. 2010. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood 116: 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]