Abstract
To define microRNA (miRNA) involvement during arbovirus infection of Aedes aegypti, we mined deep sequencing libraries of Dengue type 2 (DENV2) -exposed mosquitoes. Three biological replicates for each timepoint (2, 4, and 9 days post-exposure (dpe)) and treatment group allowed us to remove outliers associated with sample-to-sample variability. Using edgeR (R Bioconductor), designed for use with replicate deep sequencing data, we determined the log fold-change (logFC) of miRNA levels (18–23 nts). The number of significantly modulated miRNAs increased from 5 or fewer at 2 and 4 dpe to 23 unique miRNAs by 9 dpe. Putative miRNA targets were predicted by aligning miRNAs to the transcriptome, and the list was reduced to include the intersection of hits found using the Miranda, PITA, and TargetScan algorithms. To further reduce false positives, putative targets were validated by cross-checking them to mRNAs reported in recent DENV2 host response transcriptome reports; 4076 targets were identified. Of these, 464 gene targets have predicted miRNA binding sites in 3′UTRs. Context-specific target functional groups include proteins involved in transport, transcriptional regulation, mitochondrial function, chromatin modification and signal transduction processes known to be required for viral replication and dissemination. The miRNA response is placed in context with other vector host response studies by comparing the predicted targets to those of transcriptome studies. Together, these data are consistent with the hypothesis that profound and persistent changes to gene expression occur in DENV2-exposed mosquitoes.
Introduction
By definition, arbovirus transmission cycles require alternating replication in vector arthropods and vertebrate hosts. Mosquitoes mount a multi-pronged anti-viral response to arboviruses (Chauhan et al. 2012; Colpitts et al. 2011; Ocampo et al. 2013; Sanchez-Vargas et al. 2009; Xi et al. 2008). One avenue of the innate immune response relies on toll receptors, the jak-stat pathway, apoptosis and other factors (Bartholomay et al. 2010; Ramirez and Dimopoulos 2012; Rodriguez-Andres et al. 2012; Sanders et al. 2005; Souza-Neto et al. 2009; Xi et al. 2008). Another avenue uses a small RNA regulatory pathway (SRRP) specific for short interfering RNAs (siRNAs) to cleave viral targets via an Argonaute-2 (Ago2)/Dicer-2 dependent RNA interference (siRNAi) mechanism. A closely related RNAi pathway (miRNAi) uses miRNAs (~18–23nts) to modulate gene expression of house-keeping and developmental cellular processes in an Argonaute-1/Dicer-1 dependent manner. There are at least 88 unique miRNA genes in Ae. Aegypti (MirBas rg; MirBase release 19). Moreover, miRNAs control gene expression of at least 15% of the Drosophila genome (Grun et al. 2005).
A variety of studies indicate that vector competence is a complex multigenic trait (Behura et al. 2011; Campbell et al. 2008; Franz et al. 2006; Keene et al. 2004; Sanchez-Vargas et al. 2009). Quantitative genetic analysis has shown that about 40% of variation in vector competence is due to traits present at several loci (Bennett et al. 2005; Bosio et al. 2000; Gomez-Machorro et al. 2004). Included in these phenotypes are barriers that prevent the virus from infecting midguts or salivary glands, or, for example, escaping the midgut, as happens with the midgut escape barrier (MEB) (Bennett et al. 2005; Bosio et al. 2000). Importantly, Dicer-1 may be part of the MEB in wild mosquito populations (Bernhardt et al. 2012). Our earlier work showed evidence of the production of viRNAs in DENV2-infected Ae. aegypti at 2, 4 and 9 dpe (Hess et al. 2011). The recently identified association of Dicer-1 to the MEB in mosquitoes led us to investigate possible roles played by miRNAi in DENV2 infection. Importantly, the limits of anti-viral defense in vector mosquitoes and the complexity of these converging pathways are poorly understood, but may be critical to the biology of host-virus interactions.
Recent transcriptome studies of DENV2-infected Ae. aegypti provide a contextual framework for the study of miRNA pathway involvement in virus infection (Behura et al. 2011; Bonizzoni et al. 2012; Colpitts et al. 2011; Guo et al. 2010; Sim et al. 2012). Using these data to cross-validate predicted miRNA targets would reduce false-positives and allow us to move toward converging the existing SRRP data in virus-infected arthropods. However, there is limited concordance between differentially expressed mRNAs among recent transcriptome dataset publications (Figure S1). This variation may be due to differences in mosquito host or virus strains, inoculation routes, length of extrinsic incubation period or a natural heterogeneity in the overall mosquito response. By characterizing modulated miRNAs during DENV2 infection and placing them in the context of mosquito transcriptional responses to DENV infection, we hope to define common features of gene expression control that underpin host defense mechanisms. Moreover, miRNA target prediction allows us to identify coordinated miRNA responses that could work together to alter infection outcomes. A challenge to the use of deep sequencing data is the variability associated with biological replicates. Here, we used a method developed specifically for analysis of deep sequencing data with biological replicates, edgeR (R, Bioconductor). We follow up with miRNA target prediction and conclude with a discussion of the implications for the mosquito host response to DENV2 infection.
Results and Discussion
miRNA modulation
Eighteen sRNA libraries representing 3 biological replicates of pooled DENV2-exposed and unexposed control mosquitoes were mined to identify significantly modulated miRNAs (Hess et al. 2011). sRNAs were aligned to miRBase hairpin release 17 (Griffiths-Jones 2006; Kozomara and Griffiths-Jones 2011). Mapped reads in the miRNA size range (18–23 nts) showed a marked predominance of forward strand reads, whereas reads <18nts showed a more balanced representation of both forward and reverse strands (data not shown). This evidence supports the current understanding of miRNA biogenesis mechanisms, wherein the guide strand is retained and the complementary strand is degraded. In miRNA biogenesis, this process occurs via a two-step RISC-loading process, wherein the partial complementarity of the double-stranded precursor is sensed by the RISC, one strand is nicked by Ago2, and the guide strand is loaded into a second RISC, with concomitant loss of the passenger strand and subsequent cleavage of target mRNAs (Diederichs and Haber 2007; O’Toole et al. 2006; Preall and Sontheimer 2005), In contrast, siRNA biogenesis relies on a single cleavage-dependent RISC loading event of dsRNA precursors that presumably results in either strand serving as guide strand.
DENV2-exposed Ae. aegypti sRNA libraries showed modulation of miRNA profiles compared to un-exposed controls at 2, 4, and 9 dpe. Age-matched DENV2-fed and un-exposed controls were analyzed for each timepoint. Only those miRNAs homologous to previously reported mature -5p and -3p miRNAs, previously termed miRNAs and *miRNAs, respectively, were analyzed further (MirBase.org)(Griffiths-Jones 2006). Conserved miRNAs from 31 miRNA genes showed significant modulation (edgeR, p<0.05) (Figure 1, Table 1). Of these, 26 unique miRNAs have orthologs in insects, and 24 have been reported for Ae. Aegypti (Mirbase.org).
Figure 1.
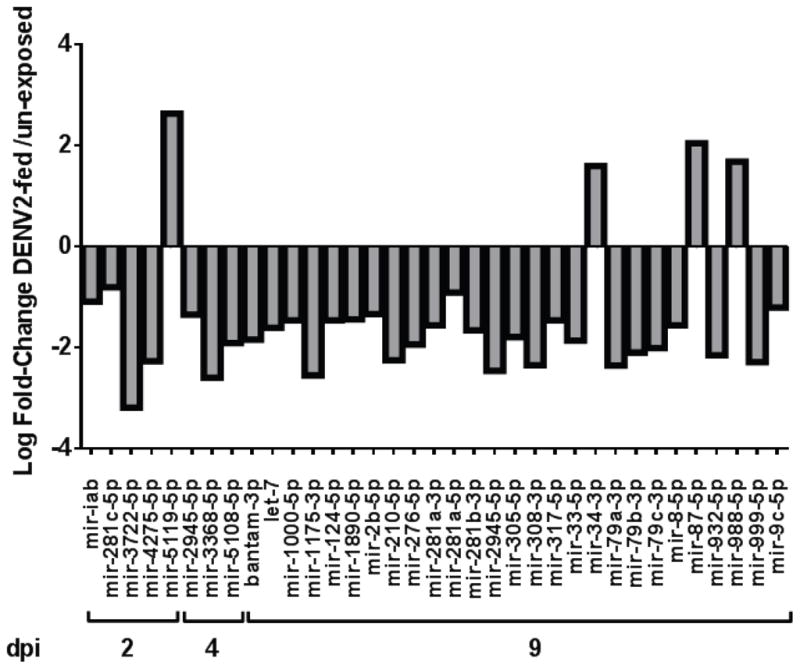
Significantly modulated miRNAs and indicated LogFC at each timepoint, relative to un-exposed controls (p< 0.05).
Table 1. miRNAs altered post- DENV2 exposure.
miRNA LogFC. Table lists miRNA sequence, length and edgeR output, log fold-change, and p value.
| miRNA | Sequence | nt # | Control* | DENV2* | Total count | logFC | p Value |
|---|---|---|---|---|---|---|---|
| mir-iab(2d) | ACGUAUACUGAAUGUAUCCUGAG | 23 | 7141 | 2435 | 9576 | −1.09 | 0.024 |
| bantam-3p(9d) | GAGAUCAUUUUGAAAGCUGAUUU | 23 | 52496 | 18229 | 70725 | −1.84 | 0.014 |
| let-7(9d) | UGAGGUAGUUGGUUGUAUAG | 20 | 10298 | 4193 | 14491 | −1.61 | 0.025 |
| mir-1000-5p(9d) | AUAUUGUCCUGUCACAGCAGU | 21 | 3132 | 1415 | 4547 | −1.46 | 0.015 |
| mir-1175-3p(9d) | GAGAUUCUACUUCUCCGACUUAA | 23 | 8961 | 1899 | 10860 | −2.55 | 0.001 |
| mir-124-5p(9d) | UAAGGCACGCGGUGAAUGCC | 20 | 6425 | 2912 | 9337 | −1.46 | 0.025 |
| mir-1890-5p(9d) | GAAAUCUUUGAUUAGGUCUGG | 21 | 904 | 414 | 1318 | −1.44 | 0.030 |
| mir-210-5p(9d) | UUGUGCGUGUGACAACGG | 18 | 1423 | 371 | 1794 | −2.25 | 0.006 |
| mir-276-5p(9d) | AGCGAGGUAUAGAGUUCCUAU | 21 | 7334 | 2374 | 9708 | −1.94 | 0.017 |
| mir-281a-3p(9d) | UGUCAUGGAAUUGCUCUCUUUAC | 23 | 11068 | 4666 | 15734 | −1.56 | 0.026 |
| mir-281a-5p(9d) | AAGAGAGCUAUCCGUCGACAGUA | 23 | 57847 | 38449 | 96296 | −0.91 | 0.038 |
| mir-281b-3p(9d) | UGUCAUGGAAUUGCUCUCUUGAU | 23 | 4926 | 1940 | 6866 | −1.66 | 0.012 |
| mir-281c-5p(2d) | AGAGAGCUAUCCGUCGACAGUAU | 23 | 15153 | 6250 | 21403 | −0.81 | 0.049 |
| mir-2945-5p(4d) | UGACUAGAGGCAGACUCG | 18 | 419 | 331 | 750 | −1.35 | 0.037 |
| mir-2945-5p(9d) | GACUAGAGGCAGACUCGUUUAGG | 23 | 9068 | 2046 | 11114 | −2.46 | 0.004 |
| mir-2b-5p(9d) | AUCACAGCCAGCUUUGUA | 18 | 1655 | 814 | 2469 | −1.34 | 0.046 |
| mir-305-5p(9d) | AUUGUACUUCAUCAGGUG | 18 | 2383 | 857 | 3240 | −1.79 | 0.015 |
| mir-308-3p(9d) | AUCACAGGAGUAUACUGUGA | 20 | 320 | 78 | 398 | −2.35 | 0.021 |
| mir-317-5p(9d) | GAACACAGCUGGUGGUAUCUUG | 22 | 10870 | 4919 | 15789 | −1.46 | 0.046 |
| mir-33-5p(9d) | GUGCAUUGUAGUUGCAUUGC | 20 | 1327 | 455 | 1782 | −1.86 | 0.015 |
| mir-3368-5p(4d) | UCAGUCUUUUAGAGAAGAAG | 20 | 233 | 64 | 297 | −2.6 | 0.001 |
| mir-34-3p(9d) | AACCACUAUCCGCCCUGCCGCC | 22 | 6838 | 25628 | 32466 | 1.59 | 0.027 |
| mir-3722-5p(2d) | CGAUUCGAGGUGgCGGUUUC | 20 | 15 | 1 | 16 | −3.19 | 0.047 |
| mir-4275-5p(2d) | CCAAUUACCACUucUUUUU | 19 | 488 | 73 | 561 | −2.27 | 0.020 |
| mir-5108-5p(4d) | GGUAGAGCACUGGAUGGUU | 19 | 269 | 82 | 351 | −1.91 | 0.031 |
| mir-5119-5p(2d) | CAUCUCAUCCUGGGGCUG | 18 | 10 | 46 | 56 | 2.632 | 0.008 |
| mir-79a-3p(9d) | AUAAAGCUAGAUUACCAAAGCAU | 23 | 2436 | 591 | 3027 | −2.36 | 0.001 |
| mir-79b-3p(9d) | UAAAGCUAGAUUACCAAAGCAUA | 23 | 573 | 166 | 739 | −2.1 | 0.010 |
| mir-79c-3p(9d) | UAAAGCUAGAUUACCAAAGCAUG | 23 | 159 | 49 | 208 | −2.01 | 0.006 |
| mir-8-5p(9d) | AAUACUGUCAGGUAAAGAUGUCU | 23 | 86028 | 36245 | 122273 | −1.56 | 0.023 |
| mir-87-5p(9d) | UGAGCAANNUUUCAGGUGUGCG | 22 | 426 | 2192 | 2618 | 2.047 | 0.002 |
| mir-932-5p(9d) | UCAAUUCCGUAGUGCAUUGCAG | 22 | 3179 | 892 | 4071 | −2.15 | 0.001 |
| mir-988-5p(9d) | CCCCUUGUUGCAAACCUCACGC | 22 | 78319 | 312139 | 390458 | 1.679 | 0.026 |
| mir-999-5p(9d) | GUUAACUGUAAGACUGUGUCUCG | 23 | 1015 | 258 | 1273 | −2.29 | 0.009 |
| mir-9c-5p(9d) | UCUUUGGUAUUCUAGCUGUAGA | 22 | 1505 | 812 | 2317 | −1.21 | 0.041 |
Counts are totaled across sequencing libraries
Over the course of infection, out of a total of 35 modulated miRNAs, the number of unique modulated miRNAs increases from 5 and 3 at 2 dpe and 4 dpe, respectively, to 23 by 9 dpe (Figure 1, Table 1). At all timepoints, DENV2 viral small RNAs (viRNAs) were detectable in DENV-exposed pools (Hess et al. 2011). Importantly, the libraries were constructed from whole mosquitoes that had received a virus-laden bloodmeal, and 50% of the mosquitoes sampled did not retain detectable virus by plaque titration at 9 dpe (Hess et al. 2011). The retention of significantly modulated miRNAs at 9 dpe is remarkable, considering that 50% of any given pool would have no detectable virus by plaque titration. In our previous work, we found that significant differences in overall host sRNA profiles were largely abrogated by 9 dpe, likely due to the mixed infection status. Persistent changes to the miRNA pathway have implications for life-long alteration of mosquito metabolism. Hypotheses with similar implications have been tested previously to assess behavioral changes in DENV-infected aedines, but this is the first evidence of a regulatory response at the global level (Platt et al. 1997; Sim et al. 2012).
Target Prediction
Although 3′UTRs were originally identified as being the key sites of miRNA-target interactions, recent target prediction studies set a precedent for probing full transcriptomes rather than merely 3′ UTRs (Fang and Rajewsky 2011; Hafner et al. 2010); this approach seems especially suitable for an organism whose genome is not extensively annotated, such as Ae. aegypti. Putative miRNA targets were identified from transcriptome release 1.3 (Vectorbase.org) using the Miranda, PITA and TargetScan prediction methods (Enright et al. 2003; Kertesz et al. 2007; Lewis et al. 2005). miRNA targets are commonly identified by comparing the base complementarity of a portion of miRNA to that of a given target sequence. In animals, miRNAs show partial rather than complete complementarity to targets (Brennecke et al. 2005). This partial complementarity is most important at the 5′ end of the miRNA and typically spans nt positions 2–8, thus defining the miRNA seed regions that are used in target prediction algorithms. The Targetscan prediction algorithm relies on base complementarity of seed region positions 2–8, whereas PITA also considers the free-energy of association between the miRNA and target along the length of the seed (Enright et al. 2003; Kertesz et al. 2007; Lewis et al. 2005). Miranda, one of the earliest prediction methods, considers free energy of association along the entire length of the microRNA-target region (Enright et al. 2003; Kertesz et al. 2007; Lewis et al. 2005). Putative targets were cross-validated with genes previously reported to be associated with DENV2 infection in Ae. aegypti (Behura et al. 2011; Bonizzoni et al. 2012; Colpitts et al. 2011; Guo et al. 2010; Sim et al. 2012). Only those targets contained within the intersection of the 3 target prediction methods and cross-validated by RNA-Seq reports were analyzed further.
The 35 modulated miRNAs share 4076 in silico validated transcriptome targets among all three prediction methods (Figure 2A). PITA and TargetScan identified many more sites per target than Miranda. The Miranda prediction algorithm with stringent cut-off criteria produced the most inclusive target set of all approaches used (see Materials and Methods).
Figure 2.
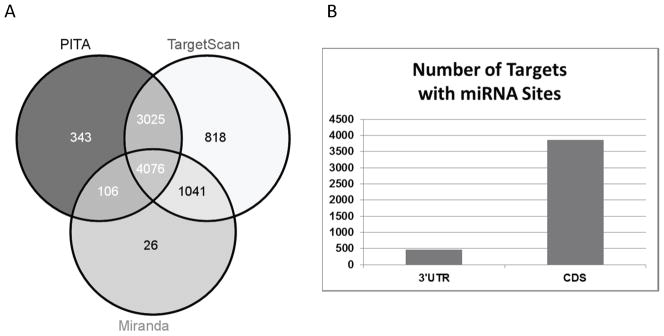
Context-specific canonical miRNAs common to the target prediction methods PITA, Miranda, and TargetScan. A. Context-specific targets for each prediction method. B. Context-specific targets identified in 3′ UTR and coding sequences (CDS) using the Miranda package.
Target sites in both the 3′UTR and the coding sequence (CDS) have been suggested to be synergistic for miRNA regulation (Fang and Rajewsky 2011). Upon interrogation of 3′ UTRs alone, we found 464 unique gene targets (Figure 2B). Of these, 365 targets contain miRNA binding sites in both the CDS and the 3′ UTR, further strengthening their predicted significance as targets.
To characterize miRNA target biological attributes over the course of infection, 4076 targets common to the 3 target prediction methods were graphed according to miRNA and functional group (Table S2). Targets were classified by processes known to be associated with viral replication (Le Breton et al. 2011; Perera et al. 2012). The functional groups chosen account for ≥ 85% of all targets (data not shown). The processes involving transport (TRP), transcription/translation (TT) and cytoskeletal/structural (CYT/STR) components are required for successful DENV2 replication and dissemination. Transport, signal transduction, cytoskeletal/structural, and metabolism make up the four most abundant target functional groups (Table S2). It is important to acknowledge the limitations inherent to studies of miRNA expression analysis. Any post-transcriptional or post-translational modifications of predicted targets would not be detected.
Implications for Viral Replication
The DENV2 miRNAi response could be a manifestation of several possible host responses. The following hypotheses are founded upon a variety of vector host response studies (Behura et al. 2011; Bonizzoni et al. 2012; Chauhan et al. 2012; Colpitts et al. 2011; Ocampo et al. 2013; Xi et al. 2008). For example, modulation of miRNA activity could regulate cell autonomous responses, such as those that occur within infected cells, or be indicative of signal transduction events that influence processes in distal tissues. Alternatively, modulation of miRNA levels could be evidence of viral exploitation of cellular processes. Some proteins, such as Loquacious, interact with proteins involved in both the miRNAi and siRNAi pathways (Fukunaga et al. 2012; Martinez and Gregory 2013).
Importantly, because of the mixed infection status by 9 dpe, another possibility is that the significantly changed miRNAs at this timepoint represent responses to repair mechanisms in un-infected mosquitoes. To explore the possibilities, we compared our results with other host response studies. We used data from two recent reports describing the comparison of gene expression responses in DENV2 resistant and susceptible Ae. aegypti strains (Chauhan et al. 2012; Ocampo et al. 2013). For example, Chauhan et al. reported that expressed genes representing glycolysis, gluconeogenesis, and the Wingless (Wnt) signaling functions are enriched in resistant strains, while transcripts with functions related to ER protein processing, nucleotide excision repair, the pentose phosphate pathway and the proteasome, are enriched in susceptible mosquitoes. In another study, Ocampo et al. described increased caspase expression in DENV2-infected tissues of resistant mosquitoes. To determine whether the predicted targets in the present work are supportive of resistant or susceptible phenotypes described by Ocampo and Chauhan, we looked at the number of targets listed in Table S1 in each of these categories, There are 43 targets associated with ER protein processing and 84 associated with proteasome function. There are 11 targets associated with glycolysis/gluconeogenesis and 2 for Wnt signaling genes. The caspases Caspase-16, Dronc, and Dredd were not present on the target list. Of the 35 miRNAs modulated, 4 are enriched at the remaining 31 are depleted. Upon miRNA depletion, given that the predicted target mRNAs would be expected to be retained rather than degraded, and therefore expected to subsequently produce more protein, we speculate that our results are consistent with a susceptible rather than a resistant phenotype.
Modulation of miRNAi at early timepoints in DENV2 infection suggests that upstream regulatory processes are altered early during infection and raises the important question of whether the miRNAi host response is a defense response or a result of viral exploitation. Alternative explanations for changes to miRNA levels could include the removal or repair of infected cells containing these non-coding RNAs. To explore possible implications of modulation to miRNA levels, targets orthologous to human flavivirus host response proteins were identified among the predicted targets. A subset of the targets is orthologous to a group of human proteins that physically interact with flavivirus non-structural proteins NS3 and NS5 (Le Breton et al. 2011). Human chromatin structural modification proteins also directly interact with flavivirus NS3 and S5 (Le Breton et al. 2011). In this study, we identified 141 putative targets in the chromatin structure and dynamics functional category. DENV2 NS3 and NS5 are essential for successful flavivirus replication in endoplasmic reticulum-associated replication vesicles (Luo et al. 2008; Welsch et al. 2009). Importantly, 50 predicted miRNA targets are identical to a subset of human proteins that physically interact with flavivirus NS3 and NS5 (Table S1) (Le Breton et al. 2011). Fifteen of these are targets of miRNAs modulated at 2 dpe. Moreover, we found 33 unique targets with innate immunity or defense descriptors (Table S1). As discussed above, because 31 of the 35 miRNAs are depleted rather than enriched, any such mRNA targets would be expected to be retained rather than depleted. These results suggest that suppression of the innate immune response in vectors, should it occur, may be mediated through other response mechanisms. Clearly, these preliminary indications require closer investigation to clarify possible gene for gene interactions between viral proteins and the host response.
Mitochondrial function is essential for cellular energy production and often associated with DENV2 infection (El-Bacha et al. 2007; Perera et al. 2012; Sessions et al. 2009). We found 235 mitochondrial protein targets within the common target set (Table S1, Table S2). The most likely explanation for these target effects is that mitochondrial function increases to accommodate increased demand for cellular energy needs. A major component of dipteran mitochondrial membranes, phosphatidylethanolamine, is significantly altered during DENV2 infection of mosquito cells (Chan 1970; Perera et al. 2012). It’s tempting to speculate that the predicted mosquito host response produces changes to mitochondrial membrane composition, as well, although this hypothesis remains to be validated experimentally.
The presence of modulated miRNAs in DENV2-exposed mosquitoes suggests that miRNA pathway activity is altered during infection, although the causal factors remain unknown. The characteristics of that involvement must rely on characterization of miRNA activity on a gene-by-gene level in future studies. The importance of this is illustrated in a recent work, wherein it was demonstrated that a bloodmeal-induced miRNA, miR-375, stimulates enrichment of one mRNA and depletion of another (Hussain et al. 2012). In our dataset, alteration of mIR-375 was depleted in DENV2-exposed mosquitoes at 9 dpe, but the change was not statistically significant (−1.6 LogFC, p=0.08, edgeR).
Conclusion
This work describes the characterization of miRNA levels for a mixed population of mosquitoes that showed a 50% infection rate by 9 dpe (Hess et al. 2011). This mixed population, tested as three biological replicates, showed a pattern of miRNA modulation that was most marked at 9 dpe. This result supports the hypothesis that persistent metabolic changes occur in DENV2-fed mosquitoes, regardless of resistant or susceptible infection status and begs the question of whether there may be some unknown fitness benefit for DENV2-exposed mosquitoes.
These data provide the foundation for future studies to determine whether modulation of the miRNA pathway during DENV2 infection is a host defense response attempting to clear the virus or evidence of viral hijacking of host cellular processes.
Experimental Procedures
Mosquito Infections and Library Preparation
Infection conditions were reported previously (Hess et al. 2011). Briefly, 3 independent cohorts of RexD colony mosquitoes (originally collected from Rexville, San Juan, Puerto Rico (Miller and Mitchell 1991)) were fed either meals containing mosquito cell culture-derived DENV2 virus, strain Jamaica 1409, or conditioned cell culture medium mixed 1:1 with defibrinated sheep’s blood. The DENV meal titers ranged from 6.7 to 7.8 log plaque-forming units per ml. Pools of 20 whole mosquitoes were harvested at 2, 4, or 9 dpe from both DENV2-fed and un-exposed controls. Infection rates, as determined by plaque assay, were 50% at 9 dpe. Following RNA extraction, small RNAs were isolated using the FLASHPAGE system (Applied Biosystems), and small RNA libraries were prepared using the SOLiD small RNA library preparation kit (Applied Biosystems). A total of 18 libraries, 3 each from un-exposed and exposed mosquitoes at each timepoint, were sequenced on a SOLiD 3 platform. Sequences are archived at the NCBI Sequence Read archive, with a study accession number of SRP026241. The exposure status of all pooled samples was confirmed by the detection of viral sRNAs (Hess et al. 2011).
Fold-Change Analysis
Small RNAs 18–23 nts in length were aligned to the miRBase hairpin database (Release 17) (mIRBase.org) using the NextGene suite (Softgenetics, LLC) and parameters reported previously (Hess et al. 2011). On a preliminary screen of the miRNA hairpins, differentially expressed miRNA candidates were identified among all libraries. Peak calls of miRNA read data were performed using the R statistical package (Gentleman et al. 2004). Peaks, representing mature miRNAs, were identified along each hairpin and classified in the following manner: conserved (or -5p, previously reported miRNA), *miRNA (or -3p, complementary to miRNA), or unclassified. Final cut-off values for confirmed mature miRNAs required ≥10 reads across all libraries. In compliance with changes to miRNA naming conventions, all complementary miRNAs identified in this study have been named according to the -5p/-3p nomenclature. miRNA naming conventions were confirmed with updates presented in miRBase Release 19 (http://www.mirbase.org/). miRNA read counts were analyzed by edgeR as previously described, using total mapped reads for each library for scaling (Hess et al. 2011). edgeR uses a negative binomial distribution and employs an empirical Bayes strategy to calculate changes to relative gene expression levels; in addition, it is designed specifically for biological replicate data (Robinson et al. 2010). Only those miRNAs showing a LogFC p value of < 0.05 are reported.
miRNA target prediction and analyses
Differentially expressed miRNAs were analyzed for complementarity to the Ae. aegypti transcriptome release (1.3) using three widely used algorithms; an intersection of targets identified by all three methods was used for analyses. Miranda was used with the following stringent parameters: Energy value ≤ −20 kcal/mol, Score ≥ 140 (Enright et al. 2003). PITA was implemented using high stringency parameters for seeds of 7–8, and excluded 6 seed matches (Enright et al. 2003; Kertesz et al. 2007) (http://genie.weizmann.ac.il/pubs/mir07/mir07_exe.html). Excluding 6nt seed matches in the PITA output has been shown to significantly reduce false positive target identification (Fang and Rajewsky 2011). Targetscan was used with default parameters (Lewis et al. 2005). Analysis of 3′UTR versus coding sequence miRNA binding sites relied on Miranda targets.
Predicted miRNA targets were annotated using Gene Ontology terms and SwissProt functional annotation data from AegyXcel (Ribeiro). (http://exon.niaid.nih.gov/transcriptome.html#aegyxcel). Data analysis scripts were implemented in Java and Python. Targets were validated in silico by cross-checking with transcriptome reports (Behura et al. 2011; Bonizzoni et al. 2012; Colpitts et al. 2011; Guo et al. 2010; Sim et al. 2012). Only those targets that were present in the aforementioned RNA-Seq reports were analyzed further. Venn diagrams were generated using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) (Oliveros 2007). Other graphs were prepared using Prism Graphpad, or Microsoft Excel.
Supplementary Material
Predicted miRNA targets.
Targets from the intersection of 3 target prediction methods were categorized by functional group. Functional categories: transport (TRP) (ie., receptor transport and secretory pathway); defense (DEF); lipid metabolism (LIP); signal transduction (SigT); apoptosis (APOP); mitochondrial function (MIT); chromatin structure and dynamics (CSD); DNA replication and repair; protease (PRO), including protein turnover; oxidation/reduction (REDOX); cytoskeletal or structural functions (CYT/STR); transcriptional activation or repression (TF/TR), transcription/translation (TT), unknown (UNK), metabolism (MET), or diverse functions (DIV).
Acknowledgments
We thank Mariangela Bonizzoni for providing RNA-Seq data. We also thank the reviewers for their insightful suggestions.
Footnotes
Author Contributions
Conceived, designed and performed experiments: CLC. Analyzed the data: CLC, TDH and AMH. Contributed reagents, materials, tools: TH, GDE, AMH and CLC. Wrote the paper: CLC and GDE. GDE and TH were supported by NIH-R01 AI067380.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–63. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, Waterhouse RM, Mayhew GF, Campbell CL, Michel K, Zou Z, Ramirez JL, Das S, Alvarez K, Arensburger P, Bryant B, Chapman SB, Dong Y, Erickson SM, Karunaratne SH, Kokoza V, Kodira CD, Pignatelli P, Shin SW, Vanlandingham DL, Atkinson PW, Birren B, Christophides GK, Clem RJ, Hemingway J, Higgs S, Megy K, Ranson H, Zdobnov EM, Raikhel AS, Christensen BM, Dimopoulos G, Muskavitch MA. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Gomez-Machorro C, Harker BW, deBruyn B, Lovin DD, Hemme RR, Mori A, Romero-Severson J, Severson DW. Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1385. doi: 10.1371/journal.pntd.0001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KE, Flick D, Fleming KH, Jochim R, Beaty BJ, Black WCt. Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti. Genetics. 2005;170:85–94. doi: 10.1534/genetics.104.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt SA, Simmons MP, Olson KE, Beaty BJ, Blair CD, Black WC. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PLoS One. 2012;7:e44198. doi: 10.1371/journal.pone.0044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. Complex Modulation of the Aedes aegypti Transcriptome in Response to Dengue Virus Infection. PLoS One. 2012;7:e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WCt. Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics. 2000;156:687–98. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK. Phospholipid composition in the mitochondria of the housefly, Musca domestica: a re-examination. J Insect Physiol. 1970;16:1575–7. doi: 10.1016/0022-1910(70)90256-8. [DOI] [PubMed] [Google Scholar]
- Chauhan C, Behura SK, Debruyn B, Lovin DD, Harker BW, Gomez-Machorro C, Mori A, Romero-Severson J, Severson DW. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. PLoS One. 2012;7:e47350. doi: 10.1371/journal.pone.0047350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, Wang P, Krishnan MN, Higgs S, Fikrig E. Alterations in the Aedesaegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011;7:e1002189. doi: 10.1371/journal.ppat.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- El-Bacha T, Midlej V, Pereira da Silva AP, Silva da Costa L, Benchimol M, Galina A, Da Poian AT. Mitochondrial and bioenergetic dysfunction in human hepatic cells infected with dengue 2 virus. Biochim Biophys Acta. 2007;1772:1158–66. doi: 10.1016/j.bbadis.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One. 2011;6:e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103:4198–203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–46. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Machorro C, Bennett K, del Lourdes Munoz M, Black Wt. Quantitative trait loci affecting dengue midgut infection barriers in an advanced intercross line of Aedes aegypti. Insect Mol Biol. 2004;13:637–648. doi: 10.1111/j.0962-1075.2004.00522.x. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Xu Y, Bian G, Pike AD, Xie Y, Xi Z. Response of the mosquito protein interaction network to dengue infection. BMC Genomics. 2010;11:380. doi: 10.1186/1471-2164-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C, Monighetti C, Campbell CL. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011;11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Walker T, O’Neill SL, Asgari S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem Mol Biol. 2012;43:146–52. doi: 10.1016/j.ibmb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:17240–5. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. Therole of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton M, Meyniel-Schicklin L, Deloire A, Coutard B, Canard B, de Lamballerie X, Andre P, Rabourdin-Combe C, Lotteau V, Davoust N. Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol. 2011;11:234. doi: 10.1186/1471-2180-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–19. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013 doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Mitchell CJ. Genetic selection of a flavivirus-refractory strain of the yellow fever mosquito Aedes aegypti. Am J Trop Med Hyg. 1991;45:399–407. doi: 10.4269/ajtmh.1991.45.399. [DOI] [PubMed] [Google Scholar]
- Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006;281:8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- O’Toole AS, Miller S, Haines N, Zink MC, Serra MJ. Comprehensive thermodynamic analysis of 3′ double-nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006;34:3338–44. doi: 10.1093/nar/gkl428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo CB, Caicedo PA, Jaramillo G, Ursic Bedoya R, Baron O, Serrato IM, Cooper DM, Lowenberger C. Differential expression of apoptosis related genes in selected strains of Aedes aegypti with different susceptibilities to dengue virus. PLoS One. 2013;8:e61187. doi: 10.1371/journal.pone.0061187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. Book Venny. 2007. Venny. City. [Google Scholar]
- Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt KB, Linthicum KJ, Myint KS, Innis BL, Lerdthusnee K, Vaughn DW. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am J Trop Med Hyg. 1997;57:119–25. doi: 10.4269/ajtmh.1997.57.119. [DOI] [PubMed] [Google Scholar]
- Preall JB, Sontheimer EJ. RNAi: RISC gets loaded. Cell. 2005;123:543–5. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2012;34:625–9. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM. Book AegyXcel. AegyXcel. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Andres J, Rani S, Varjak M, Chase-Topping ME, Beck MH, Ferguson MC, Schnettler E, Fragkoudis R, Barry G, Merits A, Fazakerley JK, Strand MR, Kohl A. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 2012;8:e1002977. doi: 10.1371/journal.ppat.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, Gill SS. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem Mol Biol. 2005;35:1293–307. doi: 10.1016/j.ibmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–50. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl AcadSci U S A. 2009;106:17841–6. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assemblysites. Cell Host Microbe. 2009;5:365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted miRNA targets.
Targets from the intersection of 3 target prediction methods were categorized by functional group. Functional categories: transport (TRP) (ie., receptor transport and secretory pathway); defense (DEF); lipid metabolism (LIP); signal transduction (SigT); apoptosis (APOP); mitochondrial function (MIT); chromatin structure and dynamics (CSD); DNA replication and repair; protease (PRO), including protein turnover; oxidation/reduction (REDOX); cytoskeletal or structural functions (CYT/STR); transcriptional activation or repression (TF/TR), transcription/translation (TT), unknown (UNK), metabolism (MET), or diverse functions (DIV).


