Summary
Recurrent mutations in H3F3A at K27 and G34 are frequent in pediatric GBM, but it is unclear how these mutations promote tumorigenesis. In this issue of Cancer Discovery, Jones and colleagues demonstrate that mutation at G34 in H3F3A result in elevated expression of MYCN as a potential mechanism in gliomagenesis.
Brain tumors represent the most common malignancy of childhood. Of these, glioblastoma (GBM) is among the most lethal, with long-term survival rates of 10-15%. While multiple driver mutations have been defined in adult GBM, including genes activating RTK/RAS/PI3K, p53, IDH1/2, and RB signaling, comparatively little is known about pathways that drive pediatric GBM. Within the last year, however, much attention has been brought to the role that epigenetics plays in pediatric GBM. Whole-genome and whole-exome sequencing studies independently identified recurrent mutations in the gene encoding histone 3 variant H3.3 (H3F3A) (1,2).
Nuclear DNA is condensed into chromatin by winding around nucleosomes. Nine histone subunits comprise each nucleosome: two of each core histone (H2A, H2B, H3, and H4) and one histone H1, which together are wrapped by 2 turns of DNA. Histones contribute to epigenetic regulation of the genome through myriad posttranslational modifications and substitution of different histone isoforms. Histone H3 can localize to either centromeric or chromosomal arm locations, and is encoded by at least 15 histone H3 genes. Of these, the histone H3.3 isoform mainly localizes to chromosomal arms, is not associated with DNA replication, and contributes to transcriptional regulation (3). Mutations in histone H3.3 in pediatric glioma occur at two distinct residues, either K27 or G34 (1,2). Anatomically, the K27-mutant tumors arise in pontine and more rostral midline brain structures, whereas the G34-mutant tumors are typically hemispheric. Both mutations localize to the histone tail at or near K27/K36 sites accessible to posttranslational methylation and acetylation, normally associated with gene silencing and expression, respectively. In addition to H3F3A mutations, pediatric GBM shows frequent mutations in the chromatin remodeling protein ATRX (31%) and tumor suppressor p53 (54%). Interestingly, all patients with mutations in H3F3A G34 also had ATRX mutations, suggesting cooperation between these proteins in the pathogenesis of pediatric GBM (1).
A genome-wide DNA methylation analysis of both adult and pediatric GBM observed that TP53 mutations were often found with H3F3A mutations in pediatric GBM, reminiscent of the association between TP53 and the epignetic modulator IDH1 in adult GBM (4). Histone H3.3 K27 and G34 mutations may therefore affect genome stability and global gene expression. Sturm and coworkers (4) revealed that mutations in IDH1 and H3F3A were mutually exclusive, as were the K27 and G34 mutations within H3F3A. In fact, a clear age distribution exists among patients with H3F3A or IDH1 mutant tumors, with H3F3A K27 occurring in children, H3F3A G34 in adolescents, and IDH1 in young adults. The developmental gene signatures for mutations in K27 and G34 appeared distinct as well. K27 was linked with mid to late fetal stages of the striatum and thalamic development while G34 was associated with early to mid-fetal stages in striatum and neocortex development (3,4). Together with the anatomical bias of the mutations, these distinct spatiotemporal expression signatures further suggest that K27 and G34 mutations arise in different cells of origin and are distinct diseases.
K27 is commonly methylated, raising the possibility that mutation alters regulation of a critical methylation site. The large number of distinct histone H3 genes and proteins would suggest that a point mutation in a single H3 gene would be unlikely to affect methylation of all histone H3 proteins. Surprisingly in this regard, Judkins and colleagues recently described a significant decrease in H3K27me3 in GBM with H3F3A K27M, as compared to tumors wild type for H3F3A (1,2,5). The most compelling explanation for this widespread effect on H3K27me3 methylation is that the H3K27M mutation acts as a dominant inhibitor of the K27 methylase, EZH2. Indeed, there is evidence that the H3K27M peptide can inhibit EZH2 methylase activity at nanomolar concentrations (6). Though it is unclear how decreased trimethylation of H3.3 K27 would achieve gliomagenesis, it has been proposed to affect differentiation pathways, as expression of the neural developmental gene FOXG1 is suppressed in the K27M tumors (1,4).
While G34 itself is not methylated, it is located near K36, a frequently methylated position. Sturm et al found that mutations at G34 also resulted in global hypomethylation of DNA particularly prominent at the ends of chromosomes. Thus, loss of DNA methylation at chromosome ends may provide insights into the association between G34 mutations and alternative lengthening of telomeres (1,4) Similar to K27 mutations, and despite the fact that DNA hypomethylation is typically associated with increased gene expression, G34 mutants down-regulate the expression of differentiation genes such as OLIG1 and OLIG2 (4). The mechanism by which G34 mutations promote tumorigenesis, however, has yet to be elucidated.
In this issue of Cancer Discovery, Jones and colleagues investigated the effect of G34 mutations in GBM and identified a potential therapeutic opportunity. Importantly, analysis of two independent data sets in this study validated several distinctions between K27 and G34 mutant tumors. First, G34 and K27 mutations exhibited differential gene expression patterns. Second, patients with K27 mutations had worse overall survival than G34 or H3F3A wild-type patients. Third, K27 mutations were observed in younger patients compared with G34 mutations. To analyze signaling pathways activated in G34 tumors, the authors utilized the KNS42 pediatric GBM cell line, which harbors a G34V mutation. Consistent with the previous finding that ATRX mutations coincided with G34 mutants, the KNS42 line had a Q891E mutation in the ATRX gene.
Because of the close proximity of G34 with the K36 methylation site, the authors initially evaluated K36 methylation status in KNS42 cells, observing no differences as compared to pediatric GBM lines wild-type for H3F3A. This observation stands in contrast to a report that H3K36 was hypermethylated in a different pediatric GBM line harboring a G34V mutation (1). Nevertheless, Bjerke et al hypothesized that mutation at G34 influenced the H3K36me3 binding profile throughout the genome. Indeed, an unbiased ChIP-Seq screen for H3K36me3 revealed differential binding at 5130 regions representing 156 genes. Differential binding to H3K36me3 correlated with differential binding to RNA polymerase II, suggesting that expression of these 156 genes was altered. This gene set was enriched for regulators of forebrain and cortical development (e.g. DLX6, FOXA1, SOX2), up-regulated from embryonic and early fetal time points, and down-regulated by mid-late fetal development. In contrast, K27 mutant tumors exhibited a gene signature correlating with midline structures including embryonic upper rhombic lip, early-mid fetal thalamic and cerebellar structures; and showed maximal expression during the mid-late fetal period. These expression details further validate spatiotemporal differences between K27 and G34 mutations, and provide insights into the distinct anatomical localization of G34 and K27 mutant tumors.
The most significant differentially bound gene encoded the proto-oncogene MYCN (~33 fold change), a gene that may alternatively be amplified in pediatric GBM, and that predicts poor overall survival in other pediatric tumors including neuroblastoma and medulloblastoma. A general challenge in this area, is the relative scarcity of GBM cell lines mutated at IDH1/2 or H3F3A, and the inability, to date, to model these mutant cancers in genetically engineered mouse models. To circumvent this issue, the authors mis-expressed mutant G34 in normal human astrocytes and transformed human fetal glial cells, observing a 2-3 fold induction of MYCN levels (Figure 1A, B). This study implicates activation of MYCN as a mechanism of G34 mutation-induced tumorigenesis and should be validated in additional primary tumors and tumor-derived cell lines.
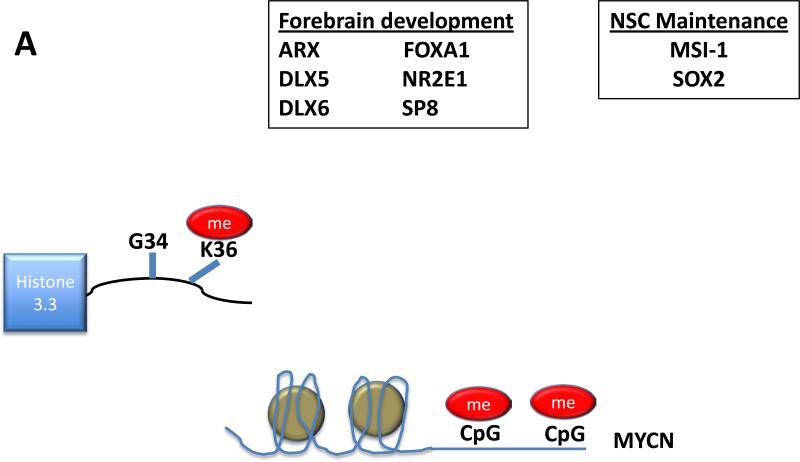
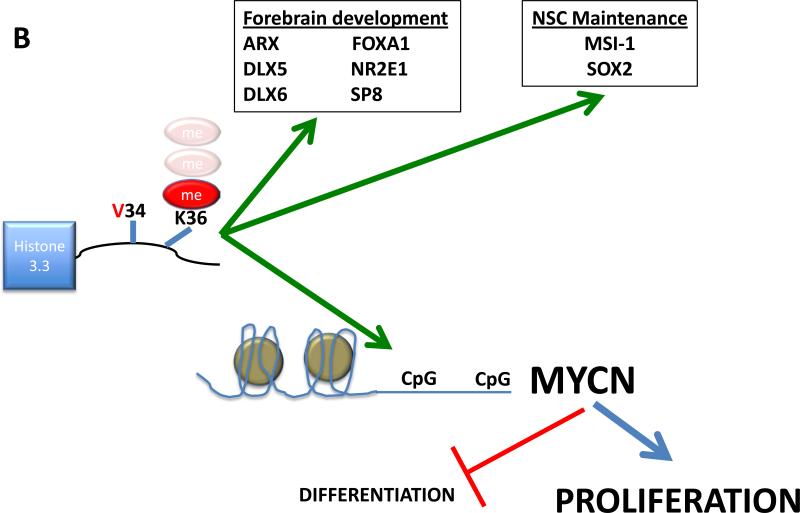
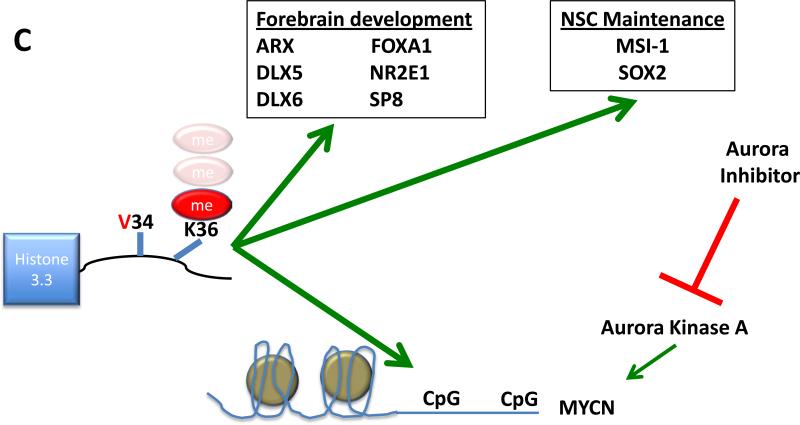
Figure 1.
(A) In pediatric GBM wildtype for H3F3A, upstream CpG islands are methylated, silencing expression of MYCN.
(B) Mutation at G34 leads to hypomethylation and differential binding of H3K36 at genes implicated in forebrain development and neural stem cell maintenance. Increased expression of MYCN associated with this mutation drives proliferation and suppresses differentiation. K36 methylation status remains in question.
(C) VX-689, a small molecule inhibitor targeting Aurora Kinase A, blocks MYCN, representing a candidate therapy for this disease.
MYCN is considered undruggable, as its structure is composed almost entirely of alpha helices with no surfaces for ligand binding. Therefore, Bjerke et al performed a synthetic lethal screen to identify kinases that could represent a therapeutic target. Out of 714 kinases tested, knockdown of Checkpoint Kinase 1 (CHK1) or Aurora Kinase A (AURKA), proteins that promote MYCN stability, reduced viability in KNS42 cells. In a dose dependent manner, the Aurora Kinase A inhibitor VX-689 decreased levels of MYCN protein and cell viability (Figure 1C). Interestingly, the bromodomain inhibitor, JQ1 was demonstrated recently to target MYCN amplified tumors by blocking MYCN-dependent transcription (7), suggesting BET-bromodomain inhibitors as an alternative or combinatorial approach in this tumor type.
The recent identification of H3F3A mutations in pediatric GBM was based on analysis of primary patient samples. How these mutations affect DNA and histone methylation, and whether these effects drive tumor formation, remains unclear. This paper provides initial mechanistic clues into how G34 mutations influence gene expression, namely via differential binding of H3K36me3 and increased expression of MYCN (Figure 1). While the precise mechanism for how G34 mutations selectively upregulate MYCN expression remain to be elucidated, the elevated levels of MYCN in this human pediatric GBM line parallel the finding that mis-expression of a stabilized allele of MYCN in P0 mouse forebrain neural stem and progenitor cells can generate glioma (8). Moreover, MYCN classically suppresses differentiation pathways, providing a potential insight into how G34 mutant tumors downregulate the expression of differentiation genes including OLIG1 and OLIG2 (4). In addition to validating previous distinctions between K27 and G34 mutant tumors, and describing a molecular explanation for mutant G34-induced tumorigenesis, Bjerke et al also show data that, if substantiated in additional pediatric GBM cell lines and xenografts, suggest Aurora Kinase A as a relevant target for children with G34 mutant GBM.
Acknowledgements and grant support
We thank Erin Simonds for critical review and comments. M. Huang is supported by the Pediatric Brain Tumor Foundation and by NIH grant F32CA174259. The Weiss lab is supported by NIH grants CA133091, CA102321, CA128583, CA148699, CA159859, CA163155, CA081403, Katie Dougherty, Pediatric Brain Tumor and Samuel G. Waxman Foundations.
Footnotes
Conflict of Interest
The authors declare no conflict of interest
References
- 1.Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012 Feb.482(7384):226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 2.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012 Mar.44(3):251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maze I, Noh K-M, Allis CD. Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacology. 2013 Jan.38(1):3–22. doi: 10.1038/npp.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012 Oct.22(4):425–37. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, et al. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013 Feb. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simard JR, Plant M, Emkey R, Yu V. Development and Implementation of a High-Throughput AlphaLISA Assay for Identifying Inhibitors of EZH2 Methyltransferase. Assay Drug Dev Technol. 2013 Feb. doi: 10.1089/adt.2012.481. [DOI] [PubMed] [Google Scholar]
- 7.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in Neuroblastoma by BET Bromodomain Inhibition. Cancer Discov. 2013 Mar.3(3):308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012 May 15;21(5):601–13. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]