Abstract
The goal of our research is to identify genes and mutations causing auto-somal dominant retinitis pigmentosa (adRP). For this purpose we established a cohort of more than 250 independently ascertained families with adRP in the Houston Laboratory for Molecular Diagnosis of Inherited Eye Diseases. Affected members of each family were screened for disease-causing mutations in genes and gene regions that are commonly associated with adRP. By this approach, we detected mutations in 65 % of the families, leaving 85 families that are likely to harbor mutations outside of the “common” regions or in novel genes. Of these, 32 families were tested by several types of next-generation sequencing (NGS), including (a) targeted polymerase chain reaction (PCR) NGS, (b) whole exome NGS, and (c) targeted retinal-capture NGS. We detected mutations in 11 of these families (31 %) bringing the total detected in the adRP cohort to 70 %. Several large families have also been tested for linkage using Afymetrix single nucleotide polymorphism (SNP) arrays.
Keywords: Retinitis pigmentosa, Next-generation sequencing, Linkage mapping, Mutation prevalence, Retinal gene capture, Whole-exome sequencing
16.1 Introduction
Retinitis pigmentosa (RP) is a highly heterogeneous set of inherited retinopathies with many causative genes, thousands of reported mutations, and complicated relationships between genotypes and phenotypes [1, 2]. For example, mutations in more than 23 genes are known to cause autosomal dominant RP (adRP), and mutations in 36 genes may cause autosomal recessive RP [3]. Our research focuses on adRP and dominant-acting mutations in X-linked RP genes. Over the past two decades, we have enrolled more than 600 unrelated families with a provisional diagnosis of adRP in our studies. Of these, 256 currently meet or exceed criteria for inclusion in our adRP cohort. To establish prevalences for genes and mutations causing RP, we have applied a staged set of genetic tests to affected members of the cohort. (Newly detected genes are then confirmed in the extended collection of families.) Initial screening includes Sanger sequencing of 12 adRP genes or gene regions known to cause approximately 50 % of adRP cases among Americans of European origin and Europeans [4–6], followed by screening for deletions not detected by conventional sequencing [7], and sequencing of the X-linked RP genes RPGR and RP2 to detect mutations which may cause clinically significant retinal disease in carrier females [8].
Disease-causing mutations were found in 65 % of the adRP cohort families based on these sequential steps. The remaining families then became candidates for different sequencing approaches based on high-throughput, next-generation sequencing (NGS). Methods included targeted polymerase chain reaction (PCR) of 46 genes, linkage mapping using Affymetrix 6.0 single nucleotide variation (SNV) arrays, whole-exome sequencing, and targeted retinal-capture NGS.
16.2 Methods
16.2.1 AdRP Cohort
Families in the adRP cohort were ascertained by clinical collaborators including Dr. David Birch, Retina Foundation of the Southwest, Dallas and Dr. John Hecken-lively, Kellogg Eye Center, Univ. of Michigan, Ann Arbor. Clinical examinations included visual acuity, visual fields and dark adaptation, and, in most cases, optical coherence tomography and electroretinography [9]. Criteria for enrollment in the adRP cohort were three affected generations with affected females, or two affected generations with male-to-male transmission (to minimize X-linked RP). DNAs were first tested by Sanger sequencing in our CLIA-certified diagnostic laboratory and then, if no mutation was found, entered into our NGS research protocols. See Fig. 16.1 for sample staging.
Fig. 16.1.

Flow chart of testing stages for families in the adRP cohort of the Houston Laboratory for Molecular Diagnosis of Inherited Eye Diseases (LMDIED)
The study was performed in accordance with the Declaration of Helsinki, and was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston and by the respective human subjects’ review boards at each participating institution.
16.2.2 Targeted PCR NGS
Twenty-one families were tested by targeted PCR amplification of 46 known RP genes, equivalent to 1,000 PCR amplimer products, followed by NGS using 454GS FLX Titanium GAIIx and Illumina/Solexa platforms to an average sequence depth of 70X and 150X, respectively [10, 11].
16.2.3 Linkage Mapping
Whole-exome linkage testing was conducted using Affymetrix 6.0 single nucleotide polymorphism (SNP)/copy number variation (CNV) arrays and the data were analyzed using the Affymetrix Genotyping Console™ and PLINK [12]. Linkage testing was done in collaboration with Dr. Susan Blanton, Univ. of Miami.
16.2.4 Whole-Exome NGS
Whole-exome NGS was done at the Genome Institute, Washington Univ., St. Louis and in collaboration with Dr. Eric Pierce, Massachusetts Eye and Ear Infirmary, Boston. A variety of platforms, capture reagents and sequencing platforms were used, including Agilent and NimbleGen arrays, Illumina HiSeq and Roche 454 FLX sequencing, and custom resequencing and validation arrays [11, 13]. At least four affected family members and an unaffected parent were tested in each family.
16.2.5 Targeted Retinal-Capture NGS
Targeted retinal-capture NGS was done in collaboration with Dr. Rui Chen, Baylor College of Medicine, Houston. All coding exons, including retina-specific exons, of 172 genes known to cause inherited retinal diseases were captured using Agilent probes and sequenced using the Illumina platform.
16.3 Results
Starting with 253 families in the adRP cohort, we screened for disease-causing mutations by a variety of approaches (Fig. 16.1) Based on conventional Sanger sequencing, linkage mapping, and multiplex ligation-dependent probe amplification (MLPA) to detect large deletions [7], we determined the disease-causing gene and mutation in 164 families. Of the remaining families, 32 were tested by NGS using three approaches, targeted PCR NGS, whole-exome NGS and targeted retinal-capture NGS. We also mapped the disease locus in several large adRP families by whole-exome linkage mapping. Results, in summary, were as follows.
We applied targeted PCR NGS to 21 families. By this approach we identified disease-causing mutations in 5 families (24 %) [10] (see Table 16.1).
Linkage mapping and whole-exome NGS identified a dominant-acting mutation in RPE65 on chromosome 1p31 in a large Irish family. Novel mutations in known genes were identified in two additional families (Fig. 16.2). No other variants were detected; in vitro studies indicated protein instability; and two additional adRP families with this mutation were observed [13]. Mutations in RPE65 are usually associated with recessive Leber congenital amaurosis. This illustrates that mutations in retinal disease genes may be recessive acting in some cases but different mutations in the same gene may be dominant acting in other cases.
Further linkage mapping in other families identified potentially-novel adRP loci on 2q24, 19q13, and 20q13 in unrelated families (Table 16.2). These families are currently the subject of whole-exome sequencing, focusing on the minimal linkage region in each case.
Targeted retinal-capture NGS involves liquid capture of exons of 172 retinal disease genes (the “RetNet set”) using a NimbleGen capture panel, followed by Illumina NGS. In early trials of targeted retinal-capture NGS we detected novel disease-causing mutations in 3 of 13 adRP families tested (23 %). This is potentially a rapid, reliable, and highly effective method to detect disease-causing mutations in adRP patients.
Table 16.1.
Pathogenic variants detected in the 1,000 amplimer project
| Family | Gene | Chromosome | Functional class | Coding | Protein | Frequency in controls (%) |
|---|---|---|---|---|---|---|
| VCH010 | KLHL7 | 7p15.3 | missense | c.458C > T | p.Ala153Val | 0.00 |
| VCH012 | GUCY2D | 17p13.1 | missense | c.2512C >T | p.Arg838Cys | 0.00 |
| VCH017 | RPGR | Xp11.4 | nonsense | c.2212C >A | p.Gly738* | 0.00 |
| VCH018 | RPGR | Xp11.4 | missense | c.194C >T | p.Gly65asp | 0.00 |
| VCH020 | PRPF31 | 19q13.42 | splice-site | c.946-1 | unknown | 0.00 |
Fig. 16.2.

Pedigrees of Irish and Canadian adRP families with a dominant-acting mutation in RPE65, detected using whole-exome NGS
Table 16.2.
Families in the adRP cohort with mapped loci
| Family | People for linkage | Chromosome | Mb (genes) |
|---|---|---|---|
| RFS132 | 25 | 19p13.3 | 1.1 (15) |
| UTAD055 | 20 | 19q13.2–q13.42 | 15.7 (400 +) |
| UTAD562 | 15 | 20q13.33-qter | 4.1 (150) |
| UTAD569 | 32 | 1p31.3–p31.1 | 8.8 (50) |
| UTAD598 | 16 | 2q24.1–q31.1 | 14.0 (81) |
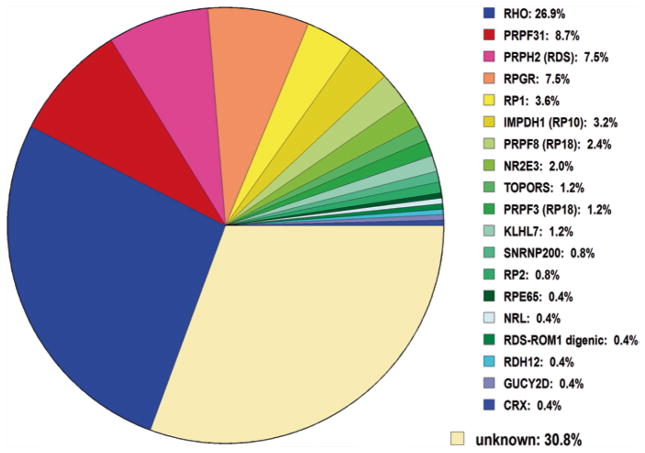
Altogether, using several NGS approaches and whole-exome linkage mapping, we detected the disease-causing mutation in 11 of 32 families tested (34 %) or 5 % of the total adRP cohort (Fig. 16.3 and Table 16.3).
Fig. 16.3.
Mutations detected to date in the Houston LMDIED adRP cohort (including dominant X-linked RP mutations)
Table 16.3.
Summary of NGS findings in the adRP cohort
| ID | WU-GSC 42 genes | WU-GSC exome | Baylor | Final result |
|---|---|---|---|---|
| RFS021 | – | RP2 | – | RP2 del EX04-flanking |
| RFS038 | KLHL7 | – | – | KLHL7 Ala153Val |
| RFS048 | – | – | SNRNP200 | SNRNP200 Ala542Val benign? |
| RFS066 | GUCY2D | – | – | GUCY2D Arg838Cys |
| RFS191 | RPGR | – | – | RPGR Gly738* |
| RFS296 | RPGR | – | – | RPGR Gly65Asp |
| RFS397 | – | PROM1 | – | PROM1 Arg373Cys |
| UTAD037 | PRPF31 | – | – | PRPF31 946-1 G > C |
| UTAD198 | – | NRL | NRL | NRL Pro51Ala |
| UTAD388 | – | – | PRPF31 | PRPF31 Gly272Val |
| UTAD565 | – | SNRNP200 | SNRNP200 | SNRNP200 Arg681Lys |
| UTAD569 | – | RPE65 | – | RPE65 Asp477Gly |
16.4 Discussion
NGS is a highly versatile and effective approach to detection of novel disease-causing genes and mutations in families with autosomal dominant forms of retinopathy. Linkage mapping is a critical adjunct to NGS, significantly reducing the number of genes to consider and helping to distinguish pathogenic variants from the many rare, potentially deleterious variants in the human genome.
Acknowledgments
This work was supported by NIH grant EY007142 and the Foundation Fighting Blindness. We thank Dr. David Birch, Dr John Heckenlively, Dr. Richard Lewis, Dr. Dianna Wheaton, Ms. Kari Branham, and Ms. Elizabeth Cadena for clinical assistance; and Ms. Cheryl Avery, Ms. Aimee Buhr, and Ms. Elizabeth Quimby for technical assistance.
Contributor Information
Stephen P. Daiger, Email: Stephen.P.Daiger@uth.tmc.edusss, Human Genetics Center, School of Public Health, Univ. of Texas HSC, 1200 Herman Pressler Dr., 77030 Houston, TX, USA
Sara J. Bowne, Human Genetics Center, School of Public Health, Univ. of Texas HSC, 1200 Herman Pressler Dr., 77030 Houston, TX, USA
Lori S. Sullivan, Human Genetics Center, School of Public Health, Univ. of Texas HSC, 1200 Herman Pressler Dr., 77030 Houston, TX, USA
Susan H. Blanton, Miami Institute for Human Genomics, Univ. of Miami, Miami, FL, USA
George M. Weinstock, Genome Institute, Washington Univ. School of Med., StsLouis, MO, USA
Daniel C. Koboldt, Genome Institute, Washington Univ. School of Med., St. Louis, MO, USA
Robert S. Fulton, Genome Institute, Washington Univ. School of Med., St. Louis, MO, USA
David Larsen, Genome Institute, Washington Univ. School of Med., St. Louis, MO, USA.
Peter Humphries, Dept. of Genetics, Trinity College, Dublin, Ireland.
Marian M. Humphries, Dept. of Genetics, Trinity College, Dublin, Ireland
Eric A. Pierce, Ocular Genomics Institute, Massachusetts Eye and Ear Infirmary, Boston, MA, USA
Rui Chen, Dept. of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Yumei Li, Dept. of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
References
- 1.Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–75. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125:151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daiger Stephen P., PhD, Administrator RetNet. The Retinal Information Network. The University of Texas Health Science Center; Houston: 2013. http://www.sph.uth.tmc.edu/RetNet/ [Google Scholar]
- 4.Daiger SP, Sullivan LS, Gire AI, Birch DG, Heckenlively JR, Bowne SJ. Mutations in known genes account for 58 % of autosomal dominant retinitis pigmentosa (adRP) Adv Exp Med Biol. 2008;613:203–209. doi: 10.1007/978-0-387-74904-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa (adRP): a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan LS, Bowne SJ, Seaman CR, Blanton SH, Lewis RA, Heckenlively JR, et al. Genomic rearrangements of the PRPF31 gene account for 2.5 % of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:4579–4588. doi: 10.1167/iovs.06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchill JD, Bowne SJ, Sullivan LS, Lewis RA, Wheaton DK, Birch DG, et al. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5 % of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013;54:1411–1416. doi: 10.1167/iovs.12-11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Y, Locke KG, Klein M, Bowne SJ, Sullivan LS, Ray JW, et al. Phenotypic characterization of 3 families with autosomal dominant retinitis pigmentosa due to mutations in KLHL7. Arch Ophthalmol. 2011;129:1475–1482. doi: 10.1001/archophthalmol.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowne SJ, Sullivan LS, Koboldt DC, Ding L, Fulton R, Abbott RM, et al. Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci. 2010;52:494–503. doi: 10.1167/iovs.10-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daiger SP, Sullivan LS, Bowne SJ, Birch DG, Heckenlively JR, Pierce EA, et al. Targeted high-throughput DNA sequencing for gene discovery in retinitis pigmentosa. Adv Exp Med Biol. 2010;664:325–331. doi: 10.1007/978-1-4419-1399-9_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowne SJ, Humphries MM, Sullivan LS, Kenna PF, Tam LCS, Kiang AS, et al. A dominant-acting mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Euro J Hum Genet. 2011;10:1074–1081. doi: 10.1038/ejhg.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]