Abstract
This protocol describes a method to determine both the average number and variance of proteins in the few to tens of copies in isolated cellular compartments, such as organelles and protein complexes. Other currently available protein quantification techniques either provide an average number but lack information on the variance or are not suitable for reliably counting proteins present in the few to tens of copies. This protocol entails labeling the cellular compartment with fluorescent primary-secondary antibody complexes, TIRF (total internal reflection fluorescence) microscopy imaging of the cellular compartment, digital image analysis, and deconvolution of the fluorescence intensity data. A minimum of 2.5 days is required to complete the labeling, imaging, and analysis of a set of samples. As an illustrative example, we describe in detail the procedure used to determine the copy number of proteins in synaptic vesicles. The same procedure can be applied to other organelles or signaling complexes.
Keywords: Protein Copy Number, Fluorescence, Protein Expression, Single-Molecule, Total Internal Reflection (TIRF) Microscopy
Introduction
Determining the copy number and stoichiometry of bio-molecules present in sub-cellular structures, such as organelles and complexes, is a pre-requisite to developing an accurate and predictive model of cellular function. Knowing how protein number varies within a class of sub-cellular structures can provide insight into the assembly, function and regulation of cellular structures. It also provides fundamental information with which to determine how changes in protein expression and targeting contribute to cellular dysfunction and disease states.
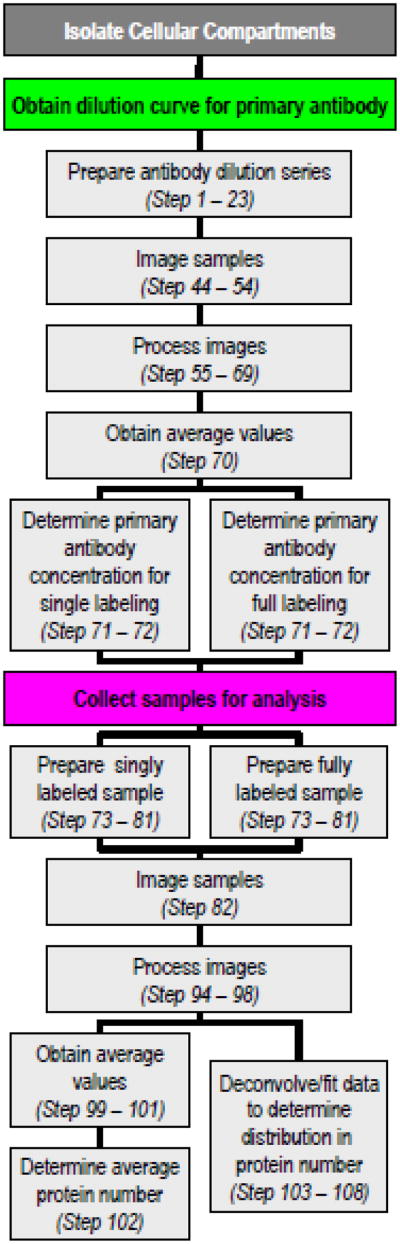
In this protocol we describe a technique which utilizes total internal reflection fluorescence (TIRF) microscopy to quantify low-copy number proteins. Proteins in isolated sub-cellular compartments are labeled with fluorescent antibodies by incubating the compartments with a primary antibody that targets the protein of interest and a fluorescent secondary antibody. Fluorescently labeled sub-cellular compartments deposited on a glass surface are then imaged with TIRF microscopy. Images are processed to obtain a fluorescence intensity distribution. Proteins are counted from the average intensity and the variation in protein number is found by deconvolving1 the intensity distributions2. Below, we provide a detailed discussion on how to find the average number of proteins in a cellular compartment and how to deconvolve the fluorescence intensities to determine the distribution of copy number around the average value. A summary of the steps is provided in Figure 1.
Figure 1. Flow chart outlining the analysis of isolated cellular compartments.
A dilution curve is first carried out to determine limits for obtaining fully and singly labeled cellular compartments (left side). Once the limits are determined, singly labeled and fully labeled samples are prepared and analyzed (right side).
Techniques currently available for quantifying protein number are limited to bulk approaches and single-molecule detection, which are either unable to provide information on the distribution around the average protein number or unable to count proteins present in the few to tens of copies. Bulk biochemical approaches, such as quantitative Western blot3, can only offer information about average values and not about the distribution of copy number. Fluorescence-based techniques that measure proteins present in high copy number (≥100) work by comparing the measured fluorescence intensity of biological samples to the average intensity estimated for a known number of proteins4,5. These techniques can be applied to determine the distribution of protein copy number but it has been difficult to extend these methods to low-copy-number bio-molecules, that is, those that range from a few to tens of copies. This is due to the fact that variability in the environment surrounding fluorophores can influence their measured intensity distribution, thus resulting in an observed intensity distribution that reflects more than just the distribution of copy number1. Currently, these fluorescence-based techniques have been applied to samples with large copy numbers inside cells, as is the case reported by Wu et al4 and Sugiyama et al5. In cases as these, the effect that the variability in the environment has on the observed intensity is averaged out and does not affect the results. However, for small copy numbers this is not necessarily true and the relationship between the width of the measured fluorescence intensity distribution and the width of the distribution of protein copy number is more complicated. If these techniques were to be applied to count smaller copy numbers without accounting for the fluorescence variability, the determined copy number distribution will be broader than the actual copy number distribution. The technique presented in this paper takes into consideration the environmental effect on the fluorescence intensity distribution during the fitting process providing a more precise determination of the copy number distribution.
The potential for environmental effects also makes it difficult to use sequential single-molecule photobleaching to reliably count molecules that are present in more than a few copies1. As the copy number increases, it becomes difficult to distinguish a step corresponding to bleaching of a single molecule because the potential for multiple molecules bleaching simultaneously increases. Though the technique discussed in this protocol can be applied to counting less than a few copies, it provides no advantage over sequential single-molecule photobleaching when less than a few copies of dyes are present. Also, application of this technique to large copy numbers (≥100) is unreasonable because an overwhelming number of data points would be needed to successfully apply the statistical analysis. This technique is most applicable to the counting of molecules in the few to tens of copies. Many proteins in signaling complexes or on organelles are present in the few to tens of copies and it is important to be able to quantify proteins in this number range.
Beyond merely counting proteins at low-copy numbers, this technique can also provide insight to the variability in the number of proteins2, which provides insight into the regulation of cellular processes. An additional benefit of this technique is the ability to count the proteins in situ but this leads to the greatest limitation as well. Often a protein is found in complexes with other proteins potentially altering that protein's conformation and possibly hindering antibody binding. In such cases as this, an absolute protein count may not be determined; however, observations relating to how distributions of protein number change can still be made. Changes to the apparent protein numbers caused by an environmental change can signify a change in the conformation/interaction state of that protein. Therefore, in such situations, this technique can still be a valuable tool for observing changes in the conformation of the protein complexes.
Currently this method has been applied to count protein copy number in a protein complex (avidin-biotin complexes1) and in an isolated sub-cellular organelle (synaptic vesicles2). However, this technique is suitable for counting proteins present in the few to tens of copies for most sub-cellular organelles or protein complexes. Procedures have been developed for the isolation and purification of many organelles and protein complexes6, including immuno-isolation7,8, sub-cellular fractionation9, and fluorescence-activated particle sorting10,11. This approach is most applicable for sub-cellular organelles and protein complexes that can be imaged within the TIRF layer (∼300nm), but alterations to the Procedure for use with epi-fluorescence would allow imaging of larger complexes. Additionally, instead of counting proteins via fluorescent antibodies, this method could be extended to counting endogenously labeled proteins in sub-cellular compartments. In such cases, photobleaching the endogenously labeled proteins to a single copy will provide the necessary intensity distribution for calibration.
Although it is possible to employ this technique to determine protein copy number in intact cells, we focus here instead on the quantification of proteins in isolated and purified sub-cellular compartments, specifically synaptic vesicles2. There are several advantages to using isolated sub-cellular compartments rather than intact cells: (1) The use of isolated compartments circumvents problems with auto-fluorescence that often plague imaging in situ. Thus it provides a high signal-to-noise ratio. (2) The ability to concentrate the cellular compartment of interest means that one can obtain large data sets with a limited number of images. (3) The use of isolated sub-cellular compartments facilitates antibody labeling, minimizing the potential for incomplete labeling. In addition, because we exogenously label protein, our procedure avoids the potential for altered protein trafficking, which can occur when fluorescent proteins are expressed in cells.
Experimental Design and Equipment Setup
Two-color labeling and selection of antibody
We developed a two-color labeling and imaging scheme where the cellular compartments are labeled with two different primary-secondary antibody pairs of two different colors (see Figure 2a), where the first antibody pair targets the protein of interest and the second antibody pair targets a protein that is abundant on the cellular compartment but different from the protein of interest. Images are collected in both colors and the analysis compares the images for two-color overlay, and only spots with both colors co localized are utilized in downstream statistical analysis. Although two-color labeling may not be necessary in all situations, its use increases the robustness of the technique by ensuring that only labeled components are studied.
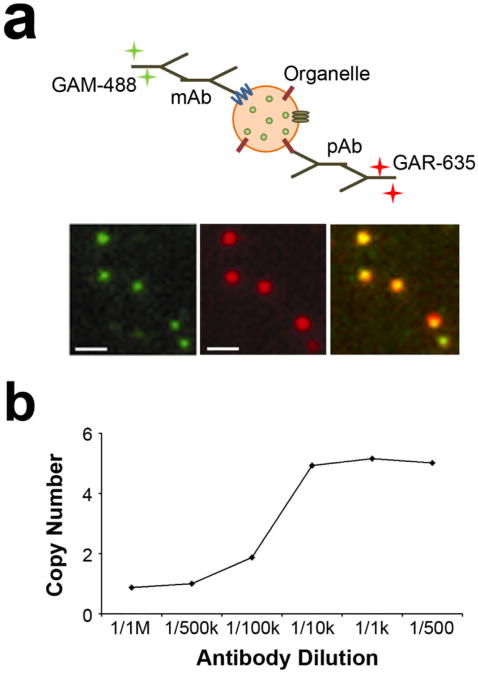
Figure 2. Antibody labeling of cellular compartments.
(a) Two-color labeling of cellular compartments, which in this case are synaptic vesicles. The mAb labels the protein of interest while the pAb labels a protein that is highly abundant on the cellular compartment and serves as verification that the fluorescent spot is a cellular compartment. Abbreviations: monoclonal antibody (mAb), Alexa-488 goat anti-mouse antibody (GAM-488), polyclonal antibody (pAb), Alexa-635 goat anti-rabbit antibody. (b) An antibody dilution curve showing the effects of antibody concentration on measured number of SV2 proteins per vesicle. Vesicles were labeled with mouse anti-SV2 antibody at different dilutions (concentration of stock solution was 1.3 mg/ml; 1/1M and 1/1k denote one million and one thousand fold dilutions, respectively). The average intensity per labeled vesicle was determined and the copy number with respect to the smallest antibody concentration was plotted (Mutch et al 2011).
The first primary antibody will be used to count the protein of interest and for this technique to be accurate, the first primary antibody must label the protein in a one-to-one fashion and must label all proteins present on the cellular compartment. Therefore, we suggest the use of monoclonal antibodies or peptide polyclonal antibodies that have been tested for specificity and which have a low dissociation constant (tight binding). To ensure tight binding, the labeled samples should be checked for degradation (e.g. by monitoring changes in the average fluorescence intensity) over the course of several hours or a couple of days. If the fluorescence intensity drastically decreases over the course of one day, then a different primary antibody should be used for quantification.
The second primary antibody should target a well known and highly abundant protein on the cellular compartment that is different from the protein of interest. The second primary antibody should be distinct from the first primary antibody in type or in subclass to ensure that each primary antibody may be labeled with a distinct fluorescent secondary antibody of different colors. This will provide a distinction between labeled cellular compartments (two-colors) and excess unbound antibody (one-color) and will ensure that only labeled compartments are studied. No quantification of the protein targeted by the second primary antibody is done; so, the second primary antibody does not need to label all of the targeted proteins on the cellular compartment.
Proper secondary antibodies to pair with each primary antibody need to be selected. Here, a highly cross-absorbed secondary antibody should be used to minimize non-specific labeling. Samples of the sub-cellular compartments should be labeled with the secondary antibodies in the absence of the primary antibodies to verify that non--specific binding does not occur. The two different color fluorophores on the two antibodies should have non-overlapping emission spectra and they should be photostable to avoid bleaching, especially for the fluorophore that labels the first primary antibody (for counting). A useful tool for determining spectral properties of different fluorophores can be found at the Invitrogen website (http://www.invitrogen.com/site/us/en/home/support/Research-Tools/Fluorescence-SpectraViewer.html). We found the Alexa dye series to have the needed photo-physical properties for this technique. Figure 2a shows a monoclonal mouse antibody paired with a goat-anti-mouse Alexa-488 antibody, and a rabbit polyclonal antibody paired with a goat-anti rabbit Alexa-635 antibody. This antibody and dye pairing was determined to have sufficiently non-overlapping emission spectra, and thus had little to no fluorescence bleed through from one color to another.
Determining antibody concentration and the average protein copy number
The first step of the Procedure is to carry out antibody dilution analyses in order to determine the concentration of antibodies that will label to saturation the protein of interest on the cellular compartment as well as the antibody concentration that corresponds to a situation where the labeled vesicles only contain one primary antibody (most vesicles will not be labeled). Saturated labeling is required to obtain information on the protein number on the cellular compartment, while minimally labeling vesicles correspond to the labeling of a single protein and this is essential for generating calibration distributions used for deconvolution.
The antibody dilution analyses are accomplished by serial dilutions of the amount of the first primary antibody added to the samples. In these experiments, the concentration of fluorescently labeled secondary antibody remains constant and any excess is removed later. The average intensity per labeled cellular compartment is obtained by averaging the intensities of the labeled spots in the TIRF microscopy images. The cellular compartments are deemed fully labeled when the average intensity per labeled cellular compartment reaches a stable high value (typically two consecutive high values), and singly labeled when the average intensity per labeled cellular compartment reaches a stable low value (see Figure 2b). The concentration of antibody needed to create an appropriate dilution series will have to be optimized for each system; however, the recommended dilutions in this protocol will serve as an initial starting point.
Once the antibody dilution analysis is finished, we use appropriate concentrations of antibody to prepare and analyze a set of three fully labeled and two singly labeled samples in each analysis. Preparing and analyzing the fully labeled samples in triplicate helps to quickly increase the number of data points for a particular protein; we determined that 5-6 samples are about the most samples that can be collected in a single day. The average number of proteins on cellular compartments is easily determined by dividing the average fluorescence intensity of the fully labeled sample with the average intensity of the singly labeled sample.
Design and fabrication of microfluidic chips
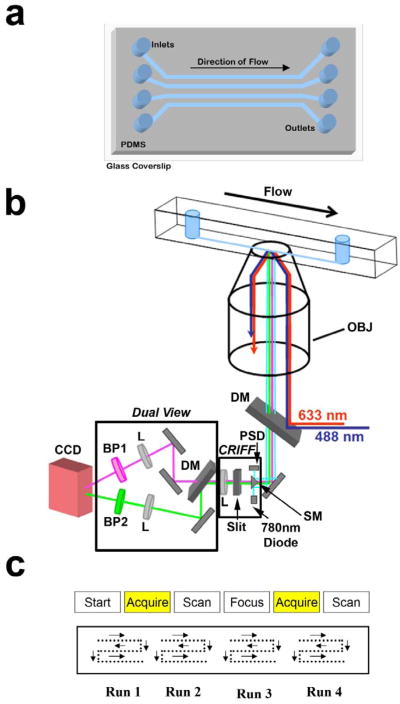
We use microchannels to deliver samples onto the surface of a coverslip for imaging using TIRF microscopy. We chose the microfluidic design depicted in Figure 3a because it minimized the channel-to-channel variability in the measurement. The microfluidic channels are fabricated in poly (dimethylsiloxane) (PDMS) with rapid prototyping12. Briefly, our high-resolution mask is generated from a computer-aided drawing file and imprinted with the channel design. A high quality print of the master design is commercially printed (http://www.finelineimaging.com/), or printed in house if a high-resolution printer is available. The mask is used in contact photolithography with SU-8 photoresist (MicroChem, Newton, MA) to create a positive relief pattern on a silicon wafer, which becomes our silicon master in subsequent steps of replica molding to form microchannels in PDMS. This silicon master is easily created using standard equipment for photolithography found at most university technology centers. From the master, PDMS channels are molded and then sealed irreversibly to a borosilicate glass coverslip by oxidizing the PDMS surface in oxygen plasma. We found that when the PDMS is sealed to a glass coverslip (described below) it causes a slight warping of the coverslip. Warping of the coverslip is minimized in the center of the chip, and therefore the imaging environment of the sample is more uniform there. Also, it is essential that each channel has enough surface area (800 μm × 200 μm × 2 cm, w × h × l) so that hundreds of images can be acquired in the same channel. Gravity driven flow is induced by placing 100 μL of ultrapure water into the inlet reservoir within minutes of sealing.
Figure 3. Imaging isolated cellular compartments.
(a) We use a microfluidic chip fabricated in PDMS and sealed to a glass coverslip with four large (800μm (w) × 200μm (h) × 2 cm (l)) channels to image samples. (b) Schematic showing the optical path for the TIRF microscope coupled with the CRIFF and dual view. Abbreviations: Lens (L), Dichroic Mirror (DM), Continuous Reflective-Interface Feedback Focus System (CFIFF), Charged Coupled Device (CCD), Position Sensitive Diode (PSD), Scanning Mirror (SM), Band Pass Filter (BP). (c) Diagram showing the scanning and acquisition pattern we used to collect images along the channel.
The use of microchannels to deliver samples for TIRF imaging is both convenient and important for our measurements. It is convenient because the samples non-specifically adsorb onto the surface of the coverslip as they flow through the channel, and thus the channel allows for facile solution exchange and a controllable way to deposit samples to an acceptable confluence on the surface. It is important because the channel offers an additional step of purification of the antibody labeled samples. We found that free antibodies only adhere to the glass surface in deionized water, but not in PBS buffer. As a result, only labeled samples are deposited onto the glass surface in the presence of buffer, which is important because we cannot have any free-antibody intensities present in the intensity distribution of our fully labeled samples. If a single antibody distribution is going to be collected, freshly collected ultrapure water should be used to allow free antibodies to adhere to the surface of the glass.
Microscope Setup
The data in this protocol is collected using a total-internal-reflection-fluorescence (TIRF) microscope. Figure 3b depicts a schematic of our optical setup, which is equipped with an objective-type TIRF, a continuous reflective-interface feedback focus system (CRIFF), a motorized stage, and a dual-view. The CRIFF automatically maintains the microscope focus as the sample is scanned and the dual view allows simultaneous two color imaging. Our statistical analysis requires a high signal-to-noise ratio, low background fluorescence, and a way to collect reproducibly hundreds of two-color fluorescence images. The use of TIRF greatly reduces background fluorescence by illuminating only ∼300 nm above the coverslip, thereby increasing the signal-to-noise ratio of the image13. The details of setting up a two-color TIRF microscope are beyond the scope of this protocol, and can be found elsewhere13,14. The two lasers chosen for the experiment must be wavelength matched with the fluorophores used, and they must be stable over the time-course of the experiment. The CRIFF combined with a motorized stage allows for the complete automation of focusing, scanning, and image acquiring along the channel. Set-ups with the above features built in are commercially available.
Before image acquisition, the intensities of the lasers and the settings of the CCD must be optimized to maximize the signal while maintaining the minimal amount of noise. Lasers should be set at the minimum intensity needed to visualize 1-2 fluorescent antibodies. If the intensity is set too high, saturation of the CCD may occur or the fluorescent antibody may photobleach during the exposure time resulting in the inability to successfully determine copy number. The optimum settings on the CCD will vary between CCDs. Exposure time should be long enough to optimize the signal-to-noise ratio but short enough that photobleaching of the fluorescent antibody does not occur. We found that the Alexa dye series antibodies provided optimum signal-to-noise over an exposure time of 300 msec even though there was minimal photobleaching over ∼900 msec2. Finally, the gain setting for the CCD should be set so that signal from a fully labeled sub-cellular compartment falls in approximately the middle of the CCD range. Comparison of images of fully labeled sub-cellular compartments with varying gain settings will help determine the optimal gain setting.
Image Acquisition
It is necessary to have hundreds or thousands of fluorescent spots for statistical analysis. The number of images needed is dependent on the concentration of spots in the imaging area. Using a motorized stage with a continuous focusing module to automate data collection greatly speeds up this process. As an alternative, the data can be collected manually by scanning and focusing each image. We found that this method works just as well as the automated method, though it is more time consuming. If a motorized stage is used a custom program, unique to each setup, is needed to drive the stage. Our program was implemented in Labview. The timing of the stage movement is synchronized with the focusing of the CRIFF and the image acquisition. The Procedure below assumes the use of an automated scanning and focusing device. Figure 3c shows a schematic of the method of data collection in order to optimally use the area of the microfluidic channel. Once an image is acquired the stage is translated a set distance and the CRIFF adjusts the focus before another image is acquired. It is pertinent during the stage translation that each new imaging area is well outside of the width of the previously illuminated area. This avoids any possibility of imaging samples that may have been partially photobleached during the previous acquisition. This cycle is repeated until the indicated number of images is collected.
Image Analysis
Analysis of images can be accomplished with several different methods. The most general method is to select regions of interest (ROIs) by hand. This method works well, but is slow and hampers further analysis (such as checking the spots for diffraction limit). This approach also has the potential to introduce scorer bias. A more automated method of image analysis can be done with any software that allows for the creation of binary image masks (Supplementary Discussion 1). Examples include open-source (ImageJ, NIH), and commercial (Metamorph, Universal Imaging) software. We used the image analysis tool box available in Matlab (Mathworks), which allows for nearly fully automated processing. Supplementary Method gives a detailed description of the steps we used for automated image analysis. If Matlab is not available, we found the Metamorph Image Morphology Analysis (IMA) tool to be useful for automating many of the steps described in the protocol. The steps to use Metamorph for analysis are described in detail in the Procedure.
Deconvolution of fluorescence intensity distributions
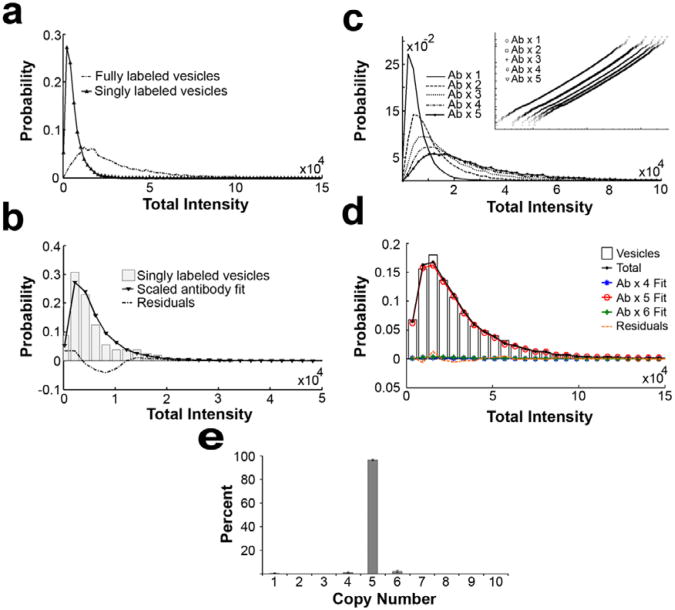
In principle, the intensity distribution of the fully labeled and singly labeled samples (Figure 4a), can be used for statistical analyses and deconvolution to determine the distribution of protein number. In practice, however, singly labeled samples typically provide < 1000 data points due to the extreme dilution needed to generate the sample. This number of data points is sufficient to determine an accurate average value, but not enough for our statistical analysis and deconvolution to obtain the distribution in copy number. A single primary-secondary antibody complex should be representative of a single labeled protein. We found that single primary-secondary antibody complexes provided thousands to tens of thousands of data points and the shape of the fluorescence intensity distribution is the same compared to the singly labeled sample; however, we found that the mean intensities differ between the two samples. The difference in mean intensity is likely caused by the fact that single primary-secondary antibody complexes are directly in contact with the surface of the coverslip while the antibody on singly labeled samples (e.g. synaptic vesicles) often are not. To address these issues, we developed a procedure where we use the mean intensity obtained from the singly labeled samples to scale the single primary-secondary antibody distribution (see Figure 4b). Note that this scaling is possible andjustified statistically only if the shapes of the two distributions are the same, which we found to be the case.
Figure 4. Deconvolution/fitting of fully labeled sample using single-antibody distribution to obtain protein copy number.
(a) Plot comparing the intensity distribution of fully labeled and singly labeled vesicles. (b) A histogram showing scaled single primary-secondary antibody complex distribution compared with singly labeled (i.e. with only one antibody per vesicle) vesicle distribution. Here, a measured single-line. (c) Plots showing the multiplication of the scaled single-antibody distribution to generate calibration curves for fitting. (d) A plot showing the fit for the protein SV2 on synaptic vesicles. The best fit was obtained with 95% of 5 proteins and 5% of 6 proteins. The small residuals of the fit (dotted line) indicate a good fit to the data. (e) Results of fitting six different SV2 data sets. (Mutch et al 2011)
Once we have obtained the scaled single-antibody calibration distribution, we can proceed with the deconvolution of the fluorescence intensity distributions. For fitting the intensity distributions, the scaled single-antibody intensities are first multiplied by integer values (1 through x) to create theoretical distributions (calibration curves) for x number of antibodies attached to the cellular compartments (Figure 4c). The best fit of a linear combination of the theoretical distributions to the measured intensity distribution of the fully labeled cellular compartments is obtained using least squares minimization. From the fit, we obtain both the average number and variation in the number of antibodies bound to the cellular compartments. Detailed information about best-fit analysis, and comparisons of minimization algorithms as it pertains to this procedure can be found elsewhere1,2 (also see Supplementary Discussion 2)
Materials
Reagents
Enriched isolated cellular compartments (e.g. synaptic vesicles, organelles, sub-cellular components)
Phosphate Buffered Saline (PBS, 1x, pH 7.2) liquid (Invitrogen, cat. no. 20012-027)
P10 size exclusion media (Bio-Gel P-10 Gel; Bio-Rad, cat. no. 150-4144)
Protein Assay Reagent (Bio-Rad, cat. no. 500-0205) CRITICAL Store at 4°C for up to a year.
Bovine serum albumin (BSA, Sigma-Aldrich, cat. no. A9418)
-
Primary antibodies specific to each sample (e.g. from Millipore, Abcam, Synaptic Systems). CRITICAL Primary antibodies should be stored frozen in small aliquots. A freshly thawed antibody should be used for each experiment.
CAUTION: We found that antibodies which have been stored at 4°C are more likely to aggregate.
Fluorescent secondary antibodies (Invitrogen). CRITICAL Secondary antibodies should be stored frozen in small aliquots. A freshly thawed antibody should be used for each experiment.
IgG beads tailored to each primary and secondary antibody (Sigma-Aldrich) CRITICAL Store at 4°C.
-
Silanes (tridecafluoro-1,1,2,2-tetrahydrooctyl trichlorosilane; Gelest, cat. no. SIT8174.0)
CAUTION: Silanes are toxic when inhaled, and reactive toward air.
PDMS elastomer kit (Dow Corning, Sylgard 184 silicone Elastomer Kit)
Ultrapure (MilliQ) water
30% hydrogen peroxide (JT Baker, cat. no. 2186-01)
Ammonium hydroxide (Sigma-Aldrich cat. no. 221228)
Equipment
Disposable column that can hold 3 mL (Econo-Pac Chromatography Columns; Bio-Rad, cat. no. 732-1010)
Sterile microcentrifuge tubes (200 μl, 1.5 ml; Axygen)
Pipette (Rainin or comparable)
Sterile filter tips (10 μl, 200 μl, 1000 μl; Rainin or comparable)
Vortex mixer (VWR, cat. no. 14005-824 or comparable)
Benchtop rotator (Labquake Tube Shaker/Rotators; Thermo Scientific or comparable)
Microcentrifuge (Centrifuge 5417c; Eppendorf or comparable)
Small glass vials
Micro-fabricated microfluidic channel master
2 Desiccators (One for silanizing microfluidic channel master and one for degassing PDMS; VWR)
Vacuum pump
Glass pipette
Pipette bulb
Disposable cups and plastic spoons for mixing PDMS
Petri Dishes (BD Falcon)
Oven
Large glass coverslips (Gold Seal Cover Glass)
Teflon holder for boiling glass coverslips
500ml beaker for cleaning glass
Watch glass for covering glass as it is boiling
Hotplate
Heat/Chemical resistant gloves
Laboratory hood
Scalpels for cutting PDMS
5 mm metal punch
N2 stream for drying coverslips
3″ × 3″ glass slide to hold PDMS and coverslip while plasma cleaning
Kimwipes (Betawipe; Texwipe, cat. no. TX2009 or comparable low particle wipe)
Small plastic tweezers
Plasma cleaner equipped with O2 (Harrick or comparable)
Digital timer
- Inverted microscope (Nikon or comparable) equipped with
- Two-color TIRF
- Scanning microscope stage
- Continuous Reflective-Interface Feedback Focus System (CRIFF), (ASI, Eugene, OR)
- Cooled CCD camera (such as Cascade, Photometrics, Tucson, AZ)
- Dual-view (Photometrics)
- Control software (Metamorph)
- Labview software (National Instruments)
High Speed Shutter (Uniblitz Electronic or comparable)
Image analysis software (Matlab, Metamorph, ImageJ)
Reagent Setup
Preparation of PBS buffer
Filter-sterilize the 1× PBS buffer using a 0.22 micron filter and store at 4°C for up to 2 weeks.
Preparation of “first primary antibody” dilution series
Prepare first primary antibody solutions with the following dilutions in PBS buffer (150 μl): 1:2 × 106, 1:106, 1:2 × 105, 1:2 × 104, 1:2 × 103, 1:103. Prepare antibody dilutions the same day the biological sample is prepared and store on ice until use.
Procedure
Labeling cellular compartments for antibody dilution analysis TIMING 1.5 days
Exchanging samples into fresh PBS buffer. Obtain 0.5 ml of an enriched sub-cellular compartment solution from a procedure suitable for the sub-cellular compartment of interest. A procedure for isolating and purifying synaptic vesicles can be found in Mutch et al 2. CRITICAL STEP: The cellular compartment solution must be fairly pure and free of excess debris. A highly purified sample will yield better data with lower background fluorescence. We store frozen aliquots of enriched cellular compartments at −80°C for up to 4 months. CAUTION: Storing samples for more than 4 months at −80°C may result in sample degradation, and incomplete labeling.
Equilibrate a 3 ml P10 column with ∼10 ml fresh PBS buffer. The P10 column is used to transfer the sub-cellular compartment into fresh PBS and remove any undesired material that may be present due to the procedure used to isolate and purify the compartment (such as sugars). CAUTION: We optimized this method for an organelle which is available in a highly enriched format. The use of a rarer organelle may require the adjustment of the stated volumes.
Load 0.5ml of the enriched cellular compartment onto column and collect 200 μl fractions.
Test each fraction for protein concentration using a standard Protein Assay Kit, following manufacturer's instructions.
Based on the protein concentrations found in Step 4, add a total of 50μg of protein (using the highest concentrated fraction first and combining fractions if needed) to a 1.5 ml microcentrifuge tube and dilute to 150μl with PBS buffer.
Repeat Step 5 for multiple samples.
Labeling the samples with primary antibody. Add the 150μl of first primary mouse antibody diluted to 1:2 × 106 (see Reagent Setup section) to the 150μl of sample (from Step 5) and briefly vortex (∼2 s). Final sample volume will be 300μl with a final antibody dilution of 1:106. CRITICAL STEP: The primary antibody used here will be the antibody that targets the protein of interest. The concentration of primary antibody needed to saturate a sample can vary widely and needs to be optimized to obtain accurate data.
Repeat Step 7 for the remaining first primary antibody dilutions resulting in 300μl samples with the following first primary antibody dilutions: 1:5 × 105, 1:105, 1:104, 1:103, 1:500.
Incubate samples with agitation (rotate or rock end-over-end) with ∼8 rpm at 4°C for 4-5 hours.
Add ∼20μl of anti-mouse IgG beads to each sample and incubate the samples with agitation at ∼8 rpm for 30 min at 4°C. CRITICAL STEP: The IgG beads serve to remove any excess unbound antibodies; so, use the appropriate anti-(host animal) beads if using an antibody other than a mouse primary antibody for counting. Also, the volume of IgG beads added to the samples need to be optimized according to the antibody concentrations used to ensure that the bulk of the excess antibody is removed. This can be accomplished by checking if an increased volume of beads added to fully labeled samples affects the average intensity of a sample. Optimize this step until a maximum intensity is reached.
Centrifuge samples at 200 × g for 1 min.
Transfer supernatant for each sample to a new tube and discard the beads.
Labeling the samples with fluorescent secondary antibody and second primary. To each sample (supernatant from Step 12), add 0.5μl of diluted fluorescently labeled goat secondary antibody (1:1 in PBS buffer). CRITICAL STEP: This antibody will be specific to the primary antibody added in Step 7 (see the Experimental Design for instructions on how to pick primary-secondary antibody pairs).
Incubate samples with agitation at ∼8 rpm overnight at 4°C.
Add 0.5μl second primary antibody to samples. We use a 1 to 1000 dilution of a 1.5 mg/ml rabbit pAb antibody for the second labeling. CRITICAL STEP: This antibody has a shorter incubation time than the first primary antibody because it is not used for quantification. See Experimental Design for instructions on how to pick primary-secondary antibody pairs.
Incubate samples with agitation at ∼8 rpm for 2 hrs at 4°C.
To each sample, add 0.5μl of diluted fluorescently labeled goat secondary antibody (1:1 in PBS buffer). CRITICAL STEP: This antibody will be specific to the primary antibody added in Step 15, and have a different non-overlapping fluorescence emission than the secondary antibody used in Step 13 (see Experimental Design for instructions on how to pick primary-secondary antibody pairs).
Incubate samples with agitation at ∼8 rpm for 1 hr at 4°C.
Add ∼ 20 μl anti-goat IgG beads. CRITICAL STEP: These beads are intended to remove both goat secondary antibodies used (Step 13 and Step 17). If using a non-goat secondary antibody in either step, use the appropriately matched anti-(host animal) beads. Repeat this step with appropriately matched anti-(host animal) beads if the secondary antibodies from Step 13 and 17 are from different same host animals. Before using non-goat secondary antibodies check that appropriately matched IgG beads are available.
Incubate samples with agitation at ∼8 rpm for 30 min at 4°C.
Centrifuge samples at 200 × g for 1 min.
Transfer supernatant for each sample to a new tube and discard beads.
Store samples (supernatant from Step 22) on ice until imaging is performed. CRITICAL STEP: Samples should be imaged within 8 hr for the best results. Degradation of the sample is unique to each specific antibody and can be checked by imaging the same sample over time. We found that most samples were stable for up to a day after labeling was complete if stored at 4°C.
Preparing microfluidic channels for use TIMING 29hr (these steps can be performed ahead of time)
24. Silinization of silicon master. Add 1-2 drops of silane to a small glass vial using a glass pipette, then place the vial beside a newly fabricated silicon master in a desiccator. The silicon master is a silicon wafer that is patterned with the SU-8 photoresist and is used for replica molding to form microchannels in PDMS. The fabrication of the silicon master has been described in detail elsewhere12. Leave under vacuum overnight. CRITICAL STEP: This step only needs to be done before the first use of a newly fabricated silicon master. CAUTION: Silanes are toxic when inhaled, and reactive toward air. PAUSE POINT: This step can be done ahead of time and silanized silicon masters can be stored for several months at room temperature.
25. Preparation of PDMS chip. Combine Sylgard 184 silicone elastomer base with curing agent at a 10:1 wt/wt ratio (final mass of ∼15 g) in a disposable plastic cup and stir with a disposable plastic spoon.
26. To degas the mixture (PDMS), place the PDMS in a dessicator and place under vacuum for ∼10-20 min until all visible bubbles are gone. CRITICAL STEP: PDMS will cure if left (> 2 hr) at room temperature.
27. Place silanized silicon master in petri dish.
28. Pour ∼3-4 mm thick layer of degassed PDMS from Step 26 onto silanized silicon master in the petri dish and cure for at least 2 hr at 60°C. CRITICAL STEP: The thickness of the PDMS can be approximated by eye, but if the PDMS is too thick, excessive warping of glass coverslip will occur when the two are sealed together. PAUSE POINT: This step can be done ahead of time and stored for several months at room temperature.
29. Boil glass coverslips for 2 hr in a 500 ml glass beaker with a solution of (3:2:1) H2O, H2O2, NH3OH, then immediately store in ultrapure (MilliQ) water for up to 5 days. CRITICAL STEP: Cover glass beaker with a watch glass as it is boiling to avoid the loss of cleaning reagent. CAUTION: Glass cleaning mixture is extremely corrosive and toxic. Boiling must be done in a laboratory fume hood.
30. Cut out PDMS chip from Step 28 with a sharp scalpel and peel chip off the master. CAUTION: Be careful not to cut over SU-8 photoresist because it will damage the silicon master. Also, use light pressure when cutting because the silicon is easily fractured.
31. Bore holes in PDMS channels with a 5 mm punch, rinse with ultrapure water and dry with a nitrogen stream. CRITICAL STEP: Cover PDMS chip with Kimwipe after cleaning to avoid dust from settling on cleaned surface. Excessive dust will inhibit the PDMS from sealing to the glass coverslip and may clog the microchannels.
32. Place PDMS chip face up on a 3″ × 3″ glass slide.
33. Remove glass coverslip from beaker with plastic tweezers, rinse with fresh ultrapure water and dry under nitrogen stream. CRITICAL STEP: Cover coverslip with Kimwipe after cleaning to avoid dust from settling on cleaned surface. Excessive dust will inhibit the PDMS from sealing to the glass coverslip and may clog the microchannels.
34. Place glass coverslip on 3″ × 3″ glass slide next to PDMS chip.
35. Remove Kimwipe and insert 3″ × 3″ slide with PDMS and glass coverslip into the plasma cleaner.
36. Close chamber door and pump down chamber to ∼150 mTorr.
37. Fill the chamber with ∼1000 mTorr of oxygen gas.
38. Pump down chamber to ∼200 mTorr.
39. Turn on current and watch for plasma to start. As soon as the blue-grey plasma is visible set timer to 1 minute and start it.
40. After 1 min of plasma treatment, turn off current and open chamber.
41. Flip PDMS chip onto glass coverslip and allow to seal. Very gentle pressure may be applied to aid in the sealing. CRITICAL STEP: Careful alignment of chip with glass is necessary before contact is made, so that the microwells are centered on the glass coverslip. TROUBLESHOOTING.
42. Fill the PDMS inlet wells with fresh ultrapure water within 1 min of sealing. Water should readily flow from the inlet well to the outlet well (see Figure 3a). Place freshly sealed chip in a clean covered petri dish to help prevent evaporation from wells. CRITICAL STEP: The PDMS wells must not dry out prior to experiment. Chips must be used within 10-12 hr of plasma sealing. TROUBLESHOOTING
43. Repeat Steps 25-42 if additional channels are needed. CRITICAL STEP: Channels are not reusable; therefore, enough chips must be prepared to provide at least 1 channel for each sample.
TIRF imaging of labeled cellular compartments TIMING 2-3hr per sample
44. Position the objective at the beginning of a channel.
45. Exchange the water in the fluid reservoir for pH 7.2 PBS buffer. Load ∼0.1μl of isolated compartment labeled with the lowest first primary antibody concentration (1:106) from Step 23 into the sample inlet well.
46. View the surface of the channel as the sample adsorbs onto the glass coverslip. TROUBLESHOOTING
47. Allow the solution to flow until desired confluence is obtained. CRITICAL STEP: If too much sample is loaded onto the surface of the glass (if the confluence is too high), the average intensity and the distribution of the intensity values will be too high. If the spots of interest are too close together multiple spots cannot be distinguished and may be grouped together by the imaging software as a single spot, and the resulting average intensity will be too high.
48. Rinse channel of excess sample by removing the PBS buffer containing the sample from the inlet and outlet well then fill the inlet well with clean PBS buffer.
49. Set the intensity of the lasers as low as possible to avoid photobleaching of the samples during acquisition. Optimize the camera (CCD) settings (gain, exposure). CRITICAL STEP: The same laser and camera settings must be used for all samples that are to be compared.
50. Position objective at one end of the channel
51. Focus and calibrate CRIFF as per manufacturer's instructions.
52. Begin image acquisition as described in Experimental Design and as illustrated in Figure 3c if using an automated system, or acquire images by hand. Collect 100-200 images of this channel. CRITICAL STEP: A shutter is used in-between acquisitions while CRIFF is focusing (∼10 s) to avoid photobleaching of the sample. The image acquisition software should be set to open and close the shutter in sync with image acquisition. A low vibration, quick acting shutter should be used (see Materials) so that images are not blurred when the shutter opens.
53. Save images as tiff files.
54. Repeat imaging (Steps 44-53) for each of the samples to be analyzed.
Image processing (described for use with Metamorph software) TIMING ∼5 min per image
55. Scan through images and remove out of focus images or test images for focus as described in the Supplementary Method.
56. Split dual-view image into two separate images representing the two fluorophores used for labeling using the Split-View function found under the Display drop down menu. When using this function choose Separate Images and then Apply.
57. Crop images to center region. CRITICAL STEP: The size of the center region will vary between laser setups. The center is defined to cover the most uniform laser excitation region (a uniform laser excitation region can be found by imaging concentrated fluorescent dye). When using a 100× objective on an in-house built TIRF microscope, the center region was found to be 210 × 210 pixels with the upper left corner located at pixel (25, 115). The exact same region should be cropped for each image.
58. Pseudo-color (green and red) the two images (from Step 56) representing the two different color fluorophores, and then overlay the images such that each overlaid green and red spot is represented by a yellow spot. This can be done in Metamorph using the Color Alignment function found in the Display drop down menu. Select one of the images from Step 56 to be green and the other image to be red then Apply. CRITICAL STEP: Adjust brightness and contrast for both the green and red images to ensure that every fluorescent spot can be seen.
59. Create regions of interest (ROIs) around spots that are yellow using the Create Region function found in the Region drop down menu. Before creating a region with this function select Ellipse and define the Width and Height of the region. The region Width and Height should be the minimum size needed to contain the entire fluorescent spot. Once this is set, input the approximate coordinate for the center of the fluorescent spot (x, y) and Create. Input coordinates and Create for all yellow spots in the image. CRITICAL STEP: All ROIs should be of the same size. This is to ensure accurate background subtraction in later processing.
60. Transfer ROIs back to the original non pseudo-colored image for counting using the Transfer Regions function in the Regions drop down menu. When using this function select the Source Image as the pseudo-colored image from Step 58 containing the regions defined in Step 59 and select the Destination Image as the image of the fluorescent antibodies labeling the protein of interest from Step 56. Verify that All Regions will transfer and select OK. This step selects the spots in the original image of the labeled proteins of interest for counting.
61. Measure total intensity (I) of each spot with the Region Measurements tool in the Measure drop down menu. Verify that I is being measured by selecting Integrated Intensity in the Configure tab. A list of all I for the selected regions can be viewed in the Measurement tab.
62. Log intensity data to a text file or spreadsheet.
63. Measure background of each image by creating 10-20 regions in areas where there are no ROIs. Create regions on the original image of the proteins of interest as described in Step 59 but input coordinates where no spots are present then repeat Step 61 to measure the intensity of the selected background. CRITICAL STEP: It is critical to accurately measure background where there are no ROIs, otherwise the background estimates will be too high, and the protein count will be too low.
64. Log background data.
65. Repeat Steps 55-64 for all images.
Data analysis of antibody dilution analysis TIMING 1hr
66. Calculate the average background (B̄) for each image by taking the average of the total intensities of the logged background data. CRITICAL STEP: Background values can drift slightly, so it is important to calculate and subtract the values individually for each image.
- 67. Subtract the average background (from Step 66) from the intensity measurements (I; from Step 61) to obtain total fluorescence intensity (F) for each spot.
68. Repeat Steps 66-67 for each image.
69. Compile a list of background-subtracted intensity values from all the images in a data set. There should be a list of intensity values for each first primary antibody concentration ({Fc}) where c denotes the concentration of the first primary antibody used for that cellular compartment.
70. Calculate the average background-subtracted intensity value (F̄c) for each {Fc}.
71. Graph all F̄c to determine proper first primary antibody concentrations for fully labeling and singly labeling cellular compartments (see Figure 2b for an example). TROUBLESHOOTING.
72. Determine the first primary antibody concentrations that corresponded to samples representing fully labeled and singly labeled compartments. A sample is considered fully labeled when a minimum of 2 consecutive antibody concentrations have the same high F̄c. Any of these corresponding concentrations can be used when generating fully labeled samples but the lowest of the potential concentrations was found to be satisfactory. Similarly, a sample is considered singly labeled when a minimum of 2 consecutive antibody concentrations have the same low F̄c and any of these suitable concentrations can be used for singly labeling samples.
Generating Fully Labeled and Singly Labeled Cellular Compartments TIMING 1.5 days
73. Fully labeling and singly labeling the samples with primary antibody. Make 3-150μl aliquots of first primary antibody in PBS buffer at the full labeling dilution found in Step 72 and 2-150μl aliquots of first primary antibody in PBS buffer at the single labeling dilution found in Step 72. CRITICAL STEP: The primary antibody used here will be the antibody that is counted in later steps.
74. Obtain 0.5 ml of an enriched sub-cellular compartment solution and repeat Steps 1-6.
75. Add one of the 150μl of first primary mouse antibody diluted to the full labeling concentration (from Step 73) to the 150μl sample (from Step 74) and briefly vortex (∼2 s). Final sample volume will be 300μl.
76. Repeat Step 75 for the remaining diluted antibody aliquots from Step 73.
77. Incubate samples with agitation at ∼8 rpm for 4-5 hours at 4°C.
78. Add ∼20μl of anti-mouse IgG beads to each sample and incubate the samples with agitation at ∼8 rpm for 30 min at 4°C. CRITICAL STEP: The IgG beads serve to remove any excess unbound antibodies; so use the appropriate anti-(host animal) beads if using an antibody other than a mouse primary antibody for counting.
79. Centrifuge samples at 200 × g for 1 min.
80. Transfer supernatant for each sample to a new tube and discard the beads.
81. Labeling the samples with fluorescent secondary antibody and second primary. Repeat Steps 13-23 for each sample.
Prepare microfluidic channels and image fully and singly labeled samples TIMING 5hr (channel preparation), 2-3hr per sample (imaging)
82. Repeat Steps 25-54 to prepare microfluidic channels and image samples.
Fluorescence imaging of single primary-secondary antibody complexes (optional steps used to determine the distribution in protein copy number) TIMING 2hr
83. Prepare an antibody solution by incubating a 0.5 μl of 1 μg/ml solution of primary antibody with 0.5 μl of 2 mg/ml fluorescent secondary antibody, then dilute solution to ∼200 μl and incubate for 30 min.
84. Place microfluidic chip on microscope stage and position objective in the center of the first channel.
85. Exchange the water in the channel with fresh ultrapure water.
86. Load 0.5 μl antibody solution into the sample well.
87. View the surface of the channel as the antibody adsorbs onto the glass.
88. Allow the solution to flow until desired confluence is reached. CRITICAL STEP: If antibody is not readily sticking to channel, the surface properties of channel have been compromised and a new channel must be tried. If too much sample is loaded onto the surface of the glass (if the confluence is too high) the average intensity and the distribution of the intensity values will be too high, because the imaging software may group these spots together as one spot.
89. Rinse channel of excess antibody by removing the water containing the antibody from the inlet and outlet well then fill the inlet well with fresh water.
90. Position objective at one end of the channel.
91. Focus and calibrate CRIFF.
92. Begin image collection described in Step 52 and as illustrated in Figure 3c. Collect ∼100 images. CRITICAL STEP: A shutter is used in-between acquisitions to prevent photobleaching of samples while CRIFF is focusing (∼ 10s) to avoid photobleaching of the sample.
93. Save images as tiff files
Image processing TIMING ∼5min per image
94. Repeat Steps 55-65 to process images.
Data analysis of fully and singly labeled samples TIMING 1hr
95. Calculate the average background (B̄) for each image by taking the average of the total intensities of the logged background data. CRITICAL STEP: Background values can drift slightly, so it is important to calculate and subtract the values individually for each image.
- 96. Subtract the average background (from Step 95) from the intensity measurements (I; from Step 94) to obtain total fluorescence intensity (F) for each spot.
97. Repeat Steps 95-96 for each image.
- 98. Compile a list of background-subtracted intensity values from all of the images in a data set. There should be the following data sets:
- A list of intensity values for fully labeled cellular compartments ({Ff}).
- A list of intensity values for singly labeled cellular compartments ({Fs}); there may be very few data points for this distribution.
-
A list of intensity values for single primary-secondary antibody complexes ({Fa}) (optional).CRITICAL STEP: The number of data points will determine the accuracy of the measurement. We found that several thousand data points are needed for fitting distributions and to obtain distributions in copy number. A few hundred data points, however, is enough to accurately determine the average value.
99. Calculate the average background-subtracted intensity value for fully labeled cellular compartments (F̄f).
100. Calculate the average background-subtracted intensity for singly labeled cellular compartments (F̄s).
101. Calculate the average background-subtracted intensity for single primary-secondary antibody complexes (F̄a) (optional).
- 102. The average number of labeled proteins (N̄) can be measured by dividing the average total (or peak) intensity of the fully labeled cellular compartments (F̄f) by the average total (or peak) intensity of the calibration sample, that is, the singly labeled cellular compartments (F̄s):
Using scaled antibody distribution to fit fully labeled compartment distribution TIMING Variable
103. Import intensity distributions ({Ff}, {Fs}, {Fa}) into Matlab or another appropriate curve fitting software (see Experimental Design).
- 104. Determine the scaling factor (S) for scaling the antibody distribution by calculating the ratio of the average intensity of the singly labeled cellular compartments (F̄s) to the average intensity of the primary-secondary antibody complex (F̄a).
-
105. Scale the antibody intensity distribution ({Fa}) to the same average intensity value as the singly labeled sample (F̄s) using the scaling factor (S) found in Step 104 to generate the scaled antibody distribution ({ }).
CAUTION: A scaled antibody intensity distribution ({ }) is used to fit the fully labeled compartment intensity distribution ({Ff}) because the singly labeled compartment intensity distribution ({Fs}) does not contain a sufficient number of data points. This scaling is only valid if the shape of the distributions for the singly labeled sample ({Fs}) and the antibody distribution ({Fa}) is the same.
- 106. Create calibration curves ({Cn}) by multiplying the scaled antibody distribution ({ }). by integer values (n = 1, 2, 3…, x) to represent x number of antibodies attached to the sub-cellular compartment.
107. Fit the fully labeled samples ({Ff}) using the antibody calibration curves from Step 106 by minimizing the parameters described in Supplementary Discussion 2. TROUBLESHOOTING
108. Confirm a reasonable fit15 by verifying that the reduced chi-squared of the fit is near one. Only data with a reasonable reduced chi-squared should be used.
Timing
Preparation of reagents: 20 min
Steps 1-23 (Labeling cellular compartment): 1.5 days
Step 24 (Silanization of silicon master): 24 hr
Steps 25-43 (Preparation of PDMS chips): 5 hr
Steps 44-54 (TIRF imaging of labeled cellular compartments): 2-3 hr per sample
Steps 55-65 (Image processing): variable, depending on method (∼5 min per image if done manually)
Steps 66-72 (Data analysis of antibody dilution analysis): 1 hr
Steps 73-81 (Generating fully labeled and singly labeled samples): 1.5 days
Step 82 (Channel preparation): 5 hr
Step 82 (Imaging fully and singly labeled samples): 2-3 hr per sample
Steps 83-93 (Fluorescence imaging of single primary-secondary antibody complexes): 2 hr
Step 94 (Image processing): variable, depending on method (∼5 min per image if done manually)
Steps 95-102 (Data analysis of fully and singly labeled samples): 1 hr
Steps 103-108 (Fitting): variable, depending on algorithm and program used
Troubleshooting
See Table 1 for troubleshooting of problems.
Table 1. Troubleshooting Table.
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 41 | PDMS is not sealing to glass | PDMS not properly plasma treated | Remake channels |
| Glass coverslip dirty | Boil new glass and remake channels | ||
| 42 | Channels will not fill with water after plasma sealing | Channels may be clogged with debris | Remake channels |
| Channels not oxidized thus not hydrophilic | Remake channels | ||
| Channels are leaking when filled with water | Channels not sealed with glass | Remake channels, check oxygen supply to plasma cleaner for leaks, pump down and re-fill oxygen line | |
| Surface of PDMS coated with contamination | Clean plasma cleaner and remake channels | ||
| 46 | Samples are not labeled or two-color labeled | Protein being labeled is not present or poor antibody | Try a different primary antibody |
| Secondary antibodies are not tagging primary | A longer incubation may be required, try a different secondary antibody, use a fresh aliquot of secondary antibody | ||
| Laser not exciting fluorophore | Try a different secondary with a different fluorophore, increase laser power, change camera settings, re-align setup or change filters | ||
| Primary antibody not-reactive or expired | Use a fresh aliquot of primary, try different primary | ||
| Individual regions of interest cannot be distinguished from background | The confluence of spots is too high | Re-load sample in channel with less sample | |
| Channels contaminated with fluorescence | Check buffer solutions for contamination | ||
| Laser power too high | Adjust laser power to lowest possible setting | ||
| Image not properly scaled | Adjust brightness and contrast in imaging software | ||
| Samples mistakenly loaded in water | Reload samples in buffer | ||
| 71 | Dilution curve will not flatten out | Sample aggregation | Prepare new samples with sub-cellular compartments that have first been filtered with a size exclusion column or other similar technique. |
| Excess free primary reacting with secondary antibody | Increase the amount of IgG beads added to sample, increase the amount of secondary antibody | ||
| 107 | Data is not reproducible | Samples not singly labeled or saturated | Re-check dilution curve |
| Laser drifting | Check laser power over time, replace laser if needed | ||
| Antibodies dissociating from sample | Image samples quickly after purification, try different antibody | ||
| Incomplete labeling | Try longer incubation, optimize secondary antibody concentration | ||
| Samples may be aggregating | Prepare new samples with sub-cellular compartments that have first been filtered with a size exclusion column or other similar technique. | ||
| Improper background selection | Make sure there are no active ROIs near background selection area | ||
| Camera or laser settings may have been changed between calibration and fully labeled samples | Use same settings |
Anticipated Results
The results obtained from using this protocol will vary widely depending on the biological sample used. We present here a set of data where we labeled enriched ratbrain synaptic vesicles, and counted the endogenous amount of SV2 protein on the vesicles2. The vesicles were enriched using a fractionation method9. Synaptic vesicles were labeled with mouse anti-SV2 monoclonal antibody (primary antibody) and Alexa-488 goat-anti-mouse secondary antibody (fluorescent secondary antibody), rabbit anti-synaptotagmin polyclonal antibody (second primary antibody) and Alexa-635 goat-anti-rabbit secondary antibody (second fluorescent secondary antibody). The reason we use two-color labeling and two different primary antibodies is so we are sure we only measure synaptic vesicles (where the two colors co-localize) and not free antibodies or fluorescent contamination (where only one color would be present).
Figure 2b shows the antibody dilution curve for SV2, which tells us the amount of SV2 primary antibody needed to both fully label and singly label the vesicles. This curve shows the samples were saturated at a 1:10,000 dilution and were singly labeled at a dilution of 1:500,000; the starting concentration of the SV2 primary antibody was 1.32 mg/ml. Figure 4a shows an example of intensity distributions for singly and fully labeled vesicles. The average number of proteins for a sample is easily determined by dividing the average fluorescence intensity of the fully labeled sample by the average fluorescence intensity of the singly labeled sample.
Figure 4b shows the scaled single-antibody distribution along with a singly labeled vesicle distribution. This scaled single-antibody distribution is used to create theoretical calibration curves (shown in Figure 4c), which were used to fit fully labeled synaptic vesicles. The calibration curves shown in Figure 4c represent theoretical intensity distributions for 1-5 proteins, which were created by multiplying each value in the scaled single-antibody distribution by an integer value. Figure 4d shows a sample fitting; the minimization program was given a set of 10 calibration curves to fit the labeled vesicles. The best fit for this data set used a sum of two calibration curves which were scaled by their representative amount of protein on the organelle (95% 5 proteins, and 5% 6 proteins). Figure 4e shows the compiled fitting data for 6 different SV2 fits. The data indicates that this protein is highly monodispersed in copy number and tightly regulated during the biogenesis and recycling of synaptic vesicles.
Supplementary Material
Discussion on semi-automated identification of regions of interest in images.
Discussion on the fitting procedure.
Acknowledgments
We gratefully acknowledge support of this work by NIH (NS052637 to D.T.C. and S.M.B.).
Footnotes
Author Contributions: S.A.M. designed and performed the experiments, worked on image and data analysis, and wrote the manuscript; J.C.G. performed all aspects of the experiments and helped prepare the manuscript; B.S.F. wrote the image processing and statistical fitting programs and helped with data analysis; P.K.H. prepared the samples used in the experiment; P.G.S. worked on the design and construction of the microscopy setup and helped with image acquisition; S.M.B. and D.T.C provided overall input into the project and helped to prepare the manuscript.
Competing Financial Interests: The authors declare that they have no competing financial interests.
Supplementary Method: Masking two-color images for identification of regions of interest.
References
- 1.Mutch SA, et al. Deconvolving single-molecule intensity distributions for quantitative microscopy measurements. Biophys J. 2007;92:2926–2943. doi: 10.1529/biophysj.106.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutch SA, et al. Protein quantification at the single vesicle level reveals that a subset of synaptic vesicle proteins are trafficked with high precision. J Neurosci. 2011;31:1461–1470. doi: 10.1523/JNEUROSCI.3805-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama Y, Kawabata I, Sobue K, Okabe S. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat Methods. 2005;2:677–684. doi: 10.1038/nmeth783. [DOI] [PubMed] [Google Scholar]
- 6.Pasquali C, Fialka I, Huber LA. Subcellular fractionation, electromigration analysis and mapping of organelles. J Chromatogr B. 1999;722:89–102. doi: 10.1016/s0378-4347(98)00314-4. [DOI] [PubMed] [Google Scholar]
- 7.Howell KE, Gruenberg J, Ito A, Palade GE. Immuno-isolation of subcellular components. Prog Clin Biol Res. 1988;270:77–90. [PubMed] [Google Scholar]
- 8.Burger PM, et al. Synaptic vesicles immunoisolated from rat cerebral cortex contain high levels of glutamate. Neuron. 1989;3:715–720. doi: 10.1016/0896-6273(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 9.Hell JW, Maycox PR, Stadler H, Jahn R. Uptake of GABA by rat brain synaptic vesicles isolated by a new procedure. Embo J. 1988;7:3023–3029. doi: 10.1002/j.1460-2075.1988.tb03166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock G, Steinlein P, Huber LA. Cell biologists sort things out: analysis and purification of intracellular organelles by flow cytometry. Tr Cell Biol. 1997;7:499–503. doi: 10.1016/S0962-8924(97)01160-4. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier DJ, Sobota JA, Ferraro F, Mains RE, Lazure C. Flow cytometry-assisted purification and proteomic analysis of the corticotropes dense-core secretory granules. Proteomics. 2008;8:3848–3861. doi: 10.1002/pmic.200700969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiorini GS, Chiu DT. Disposable microfluidic devices: fabrication, function, and application. Biotechniques. 2005;38:429–446. doi: 10.2144/05383RV02. [DOI] [PubMed] [Google Scholar]
- 13.Axelrod D. Total internal reflection fluorescence microscopy. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
- 14.Kuyper CL, Kuo JS, Mutch SA, Chiu DT. Proton permeation into single vesicles occurs via a sequential two-step mechanism and is heterogeneous. J Am Chem Soc. 2006;128:3233–3240. doi: 10.1021/ja057349c. [DOI] [PubMed] [Google Scholar]
- 15.Bevington PR. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; New York: 1969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Discussion on semi-automated identification of regions of interest in images.
Discussion on the fitting procedure.